Abstract
To design a vaccine that will remain potent against HIV-1, the immunogenic regions in the viral envelope that tend to change as well as those that remain constant over time must be identified. To determine the neutralization profiles of sequential viruses over time and study whether neutralization patterns correlate with sequence evolution, 12 broadly neutralizing plasmas from HIV-1 subtype B-infected individuals were tested for their ability to neutralize sequential primary HIV-1 subtype B viruses from four individuals. Three patterns of neutralization were observed, including a loss of neutralization sensitivity by viruses over time, an increase in neutralization sensitivity by sequential viruses, or a similarity in the sensitivity of sequential viruses to neutralization. Seven to 11 gp160 clones from each sequential virus sample were sequenced and analyzed to identify mutational patterns. Analysis of the envelope sequences of the sequential viruses revealed changes characteristic of the neutralization patterns. Viruses that evolved to become resistant to neutralizing antibodies also evolved with diverse sequences, with most of the changes being due to nonsynonymous mutations occurring in the V1/V2, as well as in the constant regions (C2, C3, C4), the most changes occurring in the C3. Viruses from the patient that evolved to become more sensitive to neutralization exhibited less sequence diversity with fewer nonsynonymous changes that occurred mainly in the V1/V2 region. The V3 region remained constant over time for all the viruses tested. This study demonstrates that as viruses evolve in their host, they either become sensitive or resistant to neutralization by antibodies in heterologous plasma and mutations in different envelope regions account for these changes in their neutralization profiles.
Introduction
A polyvalent vaccine designed to induce a humoral immune response to prevent infection by the human immunodeficiency virus (HIV)–type 1 would need to include immunogens from both variable and constant viral envelope regions, including those that are shared by/or unique to each strain. Data from longitudinal studies with sequential viruses and autologous plasma reveal that sequence changes in neutralization-sensitive epitopes within or in adjacent envelope regions will evolve over time either through point mutations, insertions and/or deletions, and changes in N-linked glycosylation patterns to escape neutralization.1–6 Thus, during acute HIV-1 infection, viruses undergo immune escape losing their neutralization-sensitive epitopes to autologous antibodies over time.4,6 Conversely, the host immune response also matures over time resulting in increasing titers of neutralizing antibodies in the host that are capable of potently neutralizing virus collected early in the infection.4,6 While mutations occur over time on viral envelopes in response to autologous antibodies generated by their hosts, it is not known how these mutations also affect epitopes that are sensitive to neutralization by heterologous antibodies.
Heterologous plasma (or sera) from HIV-1-infected individuals tested in cross-sectional neutralization experiments exhibit differential patterns of neutralization to primary HIV-1 isolates within and between clades.7–12 Several scenarios of neutralization of heterologous plasma with primary viruses do exist, including (1) plasmas that neutralize mainly their autologous viruses, suggesting recognition of strain-specific epitopes; (2) those that neutralize predominantly viruses from within the same clade, suggesting that there are clade-specific epitopes; and (3) those that neutralize viruses from many different clades, suggesting the presence of shared neutralization-sensitive epitopes across clades.7–12 While neutralization of primary isolates by heterologous plasma in cross-sectional studies represents a snapshot of an evolving virus, it is not known if in longitudinal studies these patterns of neutralization by these same heterologous plasma to viruses obtained sequentially will persist. However, what is certain is that epitopes recognized by autologous antibodies in plasma always evolve resulting in neutralization resistance.4,6 A successful vaccine candidate that would prevent infection by different HIV-1 strains and clades must be composed of immunogens that induce antibodies to epitopes that will remain stable over time and will be recognized by such antibodies. In the present study, we examined the neutralization profiles of sequential HIV-1 subtype B viruses by heterologous plasma samples from HIV-1 subtype B-infected individuals and determined the evolutionary sequence patterns of various regions of the envelope proteins of these sequential viruses in order to attempt to correlate changes in neutralization profiles over time with sequence evolution of the viral envelope.
Materials and Methods
Study subjects and virus isolation
Blood specimens were obtained sequentially at approximately 1 year intervals over a 3- to 4-year period from four HIV-1 subtype B-infected subjects attending the AIDS Reference Center at the Institute of Tropical Medicine, Antwerp, Belgium. The blood samples were used to obtain peripheral blood mononuclear cells (PBMCs) for virus isolation. A portion of the whole blood obtained at each time point was also used for CD4 determination.
PBMCs were obtained from each subject at each time point by Ficoll-Hypaque density gradient centrifugation; they were kept in liquid nitrogen in Antwerp and shipped in dry ice to New York for the studies described here. The thawed PBMCs were used to isolate viruses by cocultivation with donor PBMCs over a 2- to 3-week period. Virus growth in cultures was monitored twice weekly by determining p24 concentration in culture supernatant using a noncommercial p24 ELISA.13 When a p24 concentration of 100 ng/ml was attained, the virus culture supernatant was aliquoted in 1-ml tubes and stored in liquid nitrogen until used to prepare virus stocks. Stocks of viruses were prepared in HIV-negative donor PBMCs over a 2-week period as previously described,7,13 titrated in GHOST cells, and tested in the GHOST cell neutralization assays as described below and in previous studies.14,15
Plasma
A panel of 10 heterologous plasma and two sera (FDA-2 and UCLA) obtained from HIV-1 subtype B-infected subjects was used in the neutralization assays to test escape from neutralization by the sequential viruses from the four HIV-1 subtype B-infected subjects. The 10 plasma samples were obtained from HIV-1 treatment-naive, asymptomatic, chronically infected individuals attending the Veterans Affairs New York Harbor Healthcare Systems, New York. The two sera, FDA-2 and UCLA, were donated by Dr. L.K. Vujcic and Dr. S. Miles, respectively.
These plasma samples were selected based on studies in our laboratory showing a broad neutralizing capacity to many subtype B and non-B primary isolates as well as laboratory-adapted strains (unpublished data). The plasma samples were all from chronically HIV-1-infected drug-naive asymptomatic individuals. The plasma and serum samples from all 12 HIV-1 subtype B-infected individuals were tested for their ability to neutralize the sequential viruses from the study subjects.
Ghost cell neutralization assay
Neutralization assays were performed in GHOST cells as previously described.14,15 Briefly, equal volumes of patient-derived virus, at a dilution predetermined in earlier experiments to yield 200–1000 infected cells per 15,000–20,000 cells, and heat-inactivated patient's plasma or sera, at a final dilution of 1:40, were incubated for 1 h at 37°C. (We choose to use plasma at dilutions of 1/40 for all our experiments to avoid nonspecific neutralization and background noise at much lower dilutions and based on our prior experiments, the neutralization assays were best reproducible even with neutralizations <50%.) An HIV-negative human plasma pool was used as negative control. Prior experiments of coreceptor determination with the GHOST cell assay indicated that all the sequential viruses utilized the CCR5 coreceptor; thus, each virus/plasma mixture was added to GHOST-CD4-R5 cells that had been seeded in 24-well plates. The cells were maintained in 1 ml Dulbecco's modified essential medium (DMEM) containing 10% heat-inactivated fetal calf serum (FCS), 2% penicillin-streptomycin, 1% L-glutamine, 500 μg/ml geneticin, 50 μg/ml hygromycin, 1 μg/ml puromycin, and 25 μg/ml DEAE-Dextran and incubated at 37°C in a 7% CO2 incubator overnight. The next day the virus/plasma mixture was removed, the cells were washed once with 1 ml of phosphate-buffered saline (PBS) followed by the addition of 1 ml DMEM media and incubated at 37°C in 7% CO2 for 3–4 days. The cells were harvested by resuspension in 1 mM EDTA, fixed in 6% formaldehyde, and analyzed by acquisition of 15,000–20,000 cells with a FACScan flow cytometer as previously described.14,15 Tests of each plasma/virus combination in neutralization experiments were repeated two or three times. Percent neutralization was calculated by dividing the difference in the number of fluorescent cells of the negative control and the test well containing the patient plasma by the number of fluorescent cells in the negative control. The averages of the repeat experiments were computed and used to determine the neutralization sensitivity. Neutralization ≥50% was considered potent.
RNA extraction, reverse transcriptase-polymerase chain reaction (RT-PCR), cloning, sequencing, and phylogenetic analysis
RNA extraction and RT-PCR
Viral RNA was extracted from virus culture supernatant (the same aliquote used for neutralization) as described by Boom et al.16 followed by reverse transcription using oligo(dT) primer (Promega Corporation, Madison, WI) and AMV reverse transcriptase (Access RT-PCR system, Promega Corporation, Madison, WI). Viral RNA was heated at 70°C for 10 min with the oligo(dT) primer and immediately chilled on ice prior to the addition of the enzyme mixture, followed by one cycle of 48°C for 1 h. The Expand Long Template PCR System (Roche, Mannheim, Germany) was used for the first-round and second-round PCR amplification of the envelope gp160 gene according to the instructions given by the manufacturer. Amplification was performed with outer primers R8005B (forward) 5′ GCCCTGGAAGCATCCAGGAAGTCAGCCT 3′ (HXB2 location 5857–5884) and ENV N (reverse) 5′ CTGCCAATCAGGGAAGTAGCCTTGTGT 3′ (HXB2 location 9171–9145), and with inner primers R8003B (forward) 5′ AAAAGGCTTAGGCATCTCTATGGCAGGAAGAAGCG 3′ (HXB2 location 5950–5985) and R8004B (reverse) 5′ ATATGTCGACCTCGAGATACTGCTCCCACCCCTTCTGCTACTG 3′ (HXB2 location 8912–8870). The amplification protocol for the first-round PCR was one cycle of 94°C for 2 min, followed by 30 cycles of 94°C for 10 s, 55°C for 30 s, and 68°C for 8 min, ending with a single extension cycle at 68°C for 10 min. For the second-round PCR, the protocol started with one cycle of 94°C for 2 min, followed by 10 cycles of 94°C for 10 s, 60°C for 30 s, and 68°C for 8 min, continuing with an additional 20 cycles of 94°C for 15 s, 62°C for 35 s, and 68°C for 8 min, and ending with a single extension cycle at 68°C for 10 min.
Cloning and sequencing
PCR products were cloned into the Topo TA cloning vector and plasmids were transformed into chemically competent Escherichia coli cells according to the manufacturer's recommendations (Invitrogen, Carlsbad, CA). Ten colonies selected by Kanamycin (Kan) resistance and blue-white screening were grown up in 3 ml LB + Kan broth overnight. Plasmids were purified using the QIAprep Spin Miniprep kit (Qiagen Inc, Valencia, CA) and analyzed by PCR to determine insert size. Positive clones were sequenced at the 5′ and 3′ ends with the universal T3 and T7 primers. Primer walking was performed to obtain the full sequence. Sequences were assembled using the Pregap and Gap programs from the Staden software package.17 Sequences were assessed for nonsense mutations using the Sequence Locator program on the Los Alamos HIV Sequence database website (http://hiv-web.lanl.gov/content/hiv-db/LOCATE/locate.html).
Phylogenetic analysis
Sequences were automatically aligned with reference sequences from the Los Alamos HIV sequence database (http://hiv-web.lanl.gov/content/hiv-db/SUBTYPE_REF/align.html) of all known HIV-1 group M (sub)subtypes and CRFs (A1, A2, A3, B,C, D, F1, F2, G, H, J, K, and CRF01_AE–CRF34_01B) using CLUSTAL X with minor manual adjustments.18 Phylogenetic analyses were performed using the MEGA version 3.1 software package.19 Pairwise evolutionary distances were estimated using Kimura's two-parameter method, and phylogenetic trees were constructed by the neighbor-joining method of the MEGA 2.1 software package.20,21 The reliability of tree topologies was estimated by bootstrap analysis (1000 replicates).22 Bootstrap values ≥70% were considered significant for subtype determination.
Phylogenetic tree distance analysis
A phylogenetic tree of patient's first time point consensus sequence (Consensus Maker Tool, www.hiv.lanl.gov) and the cloned sequences from later time points for the selected conserved (C2, C3, and C4) and variable regions (V1/V2, V3, and V4) were individually constructed by the neighbor-joining method of the Mega 3.1 software using 1000 bootstrap cycles. Pairwise evolutionary distances were estimated using Kimura's two-parameter method and each tree was rooted using the HXB2 sequence of the given region analyzed. The branch length from the node of the individual's first time point consensus sequence to the node of each of the later time point clones was measured and the mean distance for each time point of each region was calculated.
Synonymous nonsynonymous analysis program (SNAP)
The proportion of synonymous versus nonsynonymous base substitutions as described in Nei and Gojobori23 was determined by comparing codon aligned sequences of the first time point viral clones pairwise with all clones from each of the subsequent time points using the synonymous nonsynonymous analysis program (SNAP) (www.hiv.lanl.gov).24 This analysis was performed individually for each of the variable (V1/V2, V3, V4) and constant (C2, C3, C4) regions of gp120.
Results
CD4 T cell profiles of study subjects
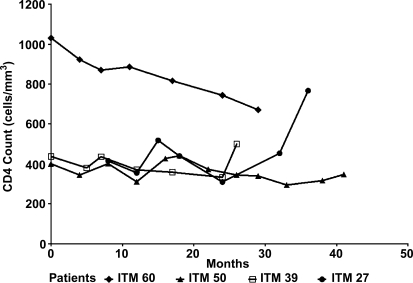
Sequential blood samples were collected at approximately 1 year intervals from the four HIV-1-infected subjects over a 3- to 4-year study period and used to determine the CD4 cell counts by FACScan. These subjects studied were asymptomatic during the study period and were naive to antiretroviral drugs. At the start of the study, the CD4 counts of three study subjects (ITM27, ITM39, and ITM50) ranged between 400 and 468 cells/mm3, while that of one subject (ITM60) was at 1031 cells/mm3. The CD4 T cell profiles of these subjects are shown in Fig. 1 and reveal that over the 3- to 4-year study period, the CD4 counts of three subjects (ITM27, ITM39, and ITM50) stayed relatively stable over time while that of ITM60 declined to 671 cells/mm3, though remaining higher than those of the three study subjects. This subject (ITM60), however, remained asymptomatic, as did all the three subjects with lower but stable CD4 counts. It was noted that in the last 6 months of analysis, the CD4 T cell count of subject ITM27 increased from 415 to 767 cells/mm3. Viruses isolated from a portion of the blood sample that was used for CD4 count determination were used for the neutralization studies described below.
FIG. 1.
CD4 profile of HIV-1 subtype B-infected, treatment-naive subjects. All the study subjects were asymptomatic. CD4 counts were determined by FACScan.
Neutralization profiles of sequential viruses from study subjects
Neutralization of virus obtained at the first visit
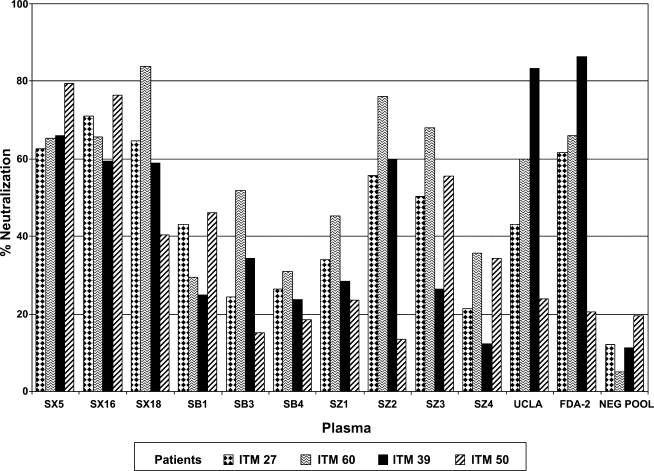
Neutralization was categorized as potent when plasma neutralized viruses with a neutralization capacity of ≥50%. Based on this cutoff, the plasmas were grouped into two categories of neutralization patterns: those that neutralized two or more of the four viruses and those that neutralized none or only one of the four viruses. As shown in Fig. 2, seven of the 12 plasma samples including SX5, SX16, SX18, SZ2, SZ3, UCLA, and FDA-2 neutralized the first visit viruses from two to four patients with a neutralization capacity ≥50%. The remaining five plasma including SB1, SB3, SB4, SZ1, and SZ4 showed weak neutralization capacities to the first visit viruses of the four patients, except for plasma SB3 that neutralized the first visit virus from patient ITM60.
FIG. 2.
Neutralization patterns of the heterologous plasma samples from HIV-1 subtype B-infected subjects against the HIV-1 subtype B viruses obtained from patients at the first visit. The plasma samples were tested at a final dilution of 1:40 with the virus in the GHOST cell neutralization assay.
Of note was that two of the seven potently neutralizing plasma samples (SX5 and SX16) were able to neutralize viruses from the four study subjects obtained at the first visit with similar potency (Fig. 2). For example, plasma SX5 neutralized virus from patients ITM27, ITM39, ITM50, and ITM60 with capacities ranging between 63% and 79% (Fig. 2). These data suggest that the antibodies in these plasma samples could be targeting antigenic regions that are shared among the different viruses.
Neutralization patterns of the sequential viruses
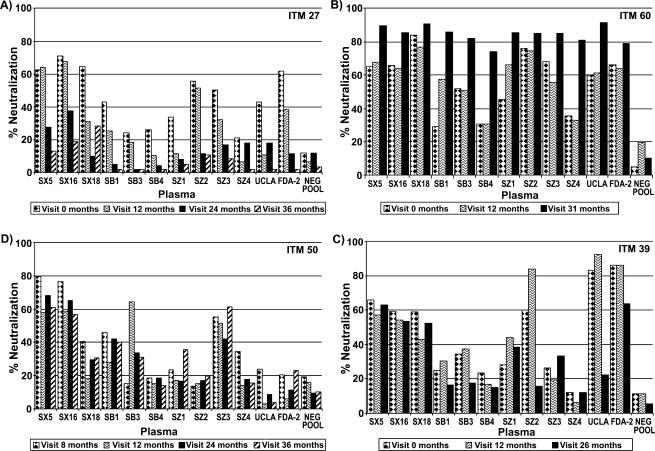
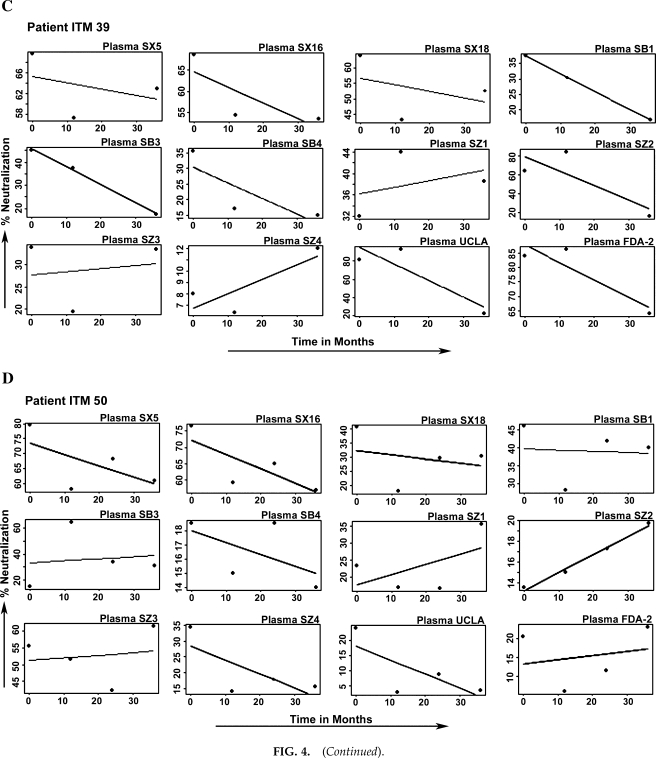
Three patterns of neutralization were noted among the sequential viruses from the four study subjects (Fig. 3). The first patterns included that seen with patient ITM27, in which over time there was loss of neutralization capacity by all 12 plasmas tested with the sequential viruses (Fig. 3A). The second pattern was that seen with patient ITM60, in which there was an increase in the neutralizing capacity of the 12 plasmas against the sequential viruses (Fig. 3B). A third pattern was seen in ITM 39 and ITM50, in which some plasma samples neutralized the sequential viruses from these patients with potency similar to the case with plasmas SX5, SX16, and SX18 (Fig. 3C and D).
FIG. 3.
Neutralization profile of the sequential viruses from HIV-1 subtype B-infected subjects (A–D). The sequential viruses were obtained from the four HIV-1 subtype B-infected subjects and tested in a GHOST cell neutralization assay with heterologous plasma samples from HIV-1 subtype B-infected individuals at a final dilution of 1:40. Virus obtained from the patient at the first visit (month 0) does not indicate virus taken after seroconversion. The subsequent visit months noted (e.g., visit 12 months) represent time points at which viruses were obtained after the first visit.
As noted with viruses from patient ITM27, loss of neutralization sensitivity occurred for viruses collected from later visits. Among the sequential viruses obtained from this patient, loss of neutralization by viruses collected from the patient at the month 36 visit was noted for six plasma samples (SX5, SX16, SX18, SZ2, SZ3, and FDA-2) that neutralized patient virus obtained at the first visit (month 0) (Fig. 3A). This pattern of loss of neutralization was also noted for the second category of plasma samples, which showed a weak neutralizing capacity to the initial virus collected at the first visit. For example, four plasma, SB1, SB3, SB4, and SZ1, were found to have lost their ability to even weakly neutralize the sequential viruses from patient ITM27 (Fig. 3A).
Unlike the pattern of neutralization observed for patient ITM27 described above, an entirely different pattern of neutralization was observed for the sequential viruses from patient ITM60 with eight potently and four weakly neutralizing plasma (Fig. 3B). Notably, for patient ITM60, virus collected at the last visit was more sensitive to neutralization than the virus obtained from the first visit. As well, the eight plasma samples (SX5, SX16, SX18, SB3, SZ2, UCLA, SZ3, and FDA-2) that potently neutralized (>50%) viruses from the first visit also potently neutralized viruses from the last visit, obtained 31 months later (Fig. 3B). Of particular interest was the pattern revealed by the remaining plasma (SB1, SB4, SZ1, and SZ4), which weakly neutralized viruses from the first visit but potently neutralized (>74–88%) viruses obtained 31 months after.
The third pattern of neutralization was exhibited by the sequential viruses of patients ITM39 and ITM50, in which their sequential viruses were either neutralized by some plasma with similar potency, as seen for plasma SX5 and SX16 against the viruses of patient ITM39, or their sequential viruses became more resistant to neutralization by the plasma, as seen for plasma SZ2 and UCLA for patient ITM39 and plasma SB3 for patient ITM50 (Fig. 3C and D).
Analyses that define the three neutralization profiles: increasing, decreasing, and unchanging
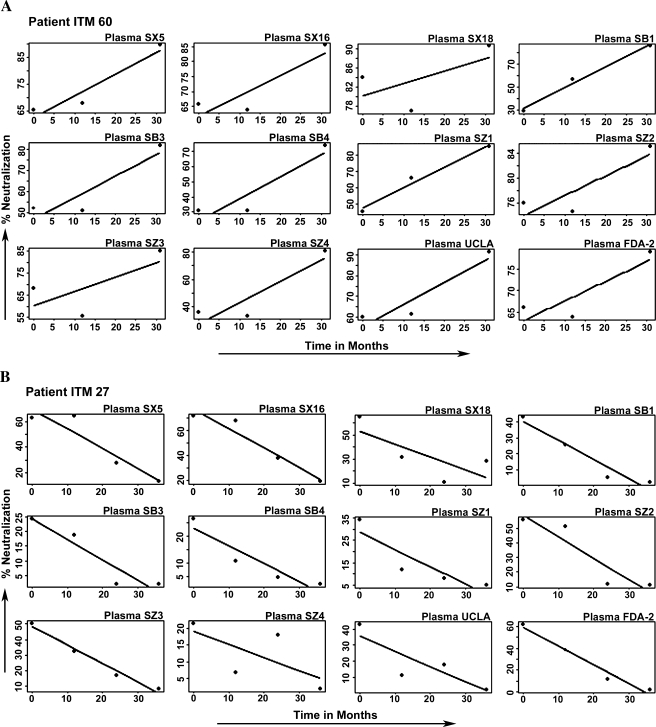
The percent neutralization of each of the 12 plasma samples for each patient's sequential viruses over time was plotted graphically and a linear regression line was then fitted as shown in Fig. 4. A positive, negative, or zero slope indicates the neutralization profile and can be interpreted as follows: a positive slope represents an increasing neutralization sensitivity, a negative slope represents a decreasing neutralization sensitivity (increasing resistance), and a zero slope represents an unchanging neutralization sensitivity over time. From the regression lines shown in Fig. 4 for each patient's sequential viruses, a positive slope can clearly be noted for the sequential viruses from patient ITM60 (Fig. 4A) with all the 12 plasma samples; conversely, a negative slope is noted for the sequential viruses from patient ITM27 with the same 12 plasma samples (Fig. 4B). No uniform slope was seen for the sequential viruses of patients ITM39 and ITM50 (Fig. 4C and D); thus it can be characterized as unchanging.
FIG. 4.
Analysis that defines HIV-1 neutralization profiles (A–D) with heterologous plasma: (a) increased, (b) decreased, and (c) unchanging neutralization sensitivity. The percent neutralization data were plotted as dots and a linear regression line was fitted to determine the neutralization profile. Positive and negative slopes represent increased and decreased neutralization sensitivity, respectively. The sequential viruses of patients ITM39 and ITM50 did not exhibit the same pattern with all the 12 plasma samples tested; thus, they are classified as unchanging neutralization sensitivity.
Because the fitted slopes from the linear models may represent unreliable measurements for predicting percent neutralization as a function of time, we tested the null hypothesis that N = 12 slopes, corresponding to 12 plasmas, were a sample from a distribution of plasma that has probability p = 1/2 for a positive slope and 1 − p = 1/2 for a negative slope. The alternative hypothesis is that p is not 1/2. We associate a true p > 1/2, p < 1/2, and p = 1/2 with the three patterns of neutralization. The number of positive slopes has a binomial distribution with parameter (N = 12, p = 1/2) and, under the null hypothesis, we expect six negative slopes. A test of the null hypothesis against the alternative that p > 1/2 rejects the null hypothesis if the number of positive slopes is too large. It follows for independent samples that the null probability of having as many as 12 slopes positive (as for patient ITM60) is 1/2 to the power 12, a vanishingly small p-value. It also follows similarly for the data of patient ITM27 where all the slopes are negative. Thus, for these two patients, the null hypothesis is resoundingly rejected, but not so for the remaining two patients; in this way, our data exhibit one or more examples of each pattern of neutralization.
Sequence analysis of the sequential viruses
Phylogenetic analysis of the sequential viruses from the study subjects
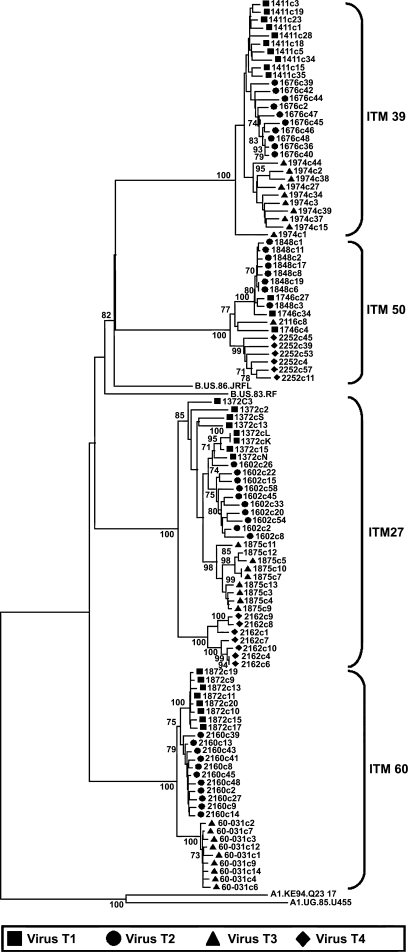
The envelope (gp160) of the sequential viruses infecting the study subjects were cloned, sequenced, and phylogenetically analyzed to confirm that all the viruses tested in neutralization assays were of the subtype B lineage and that these subjects were not dually infected with different virus subtypes. A total of 1–11 clones from each time point per subject were sequenced. The phylogenetic tree was rooted with subtype A1 sequences from the Los Alamos database. As shown in the phylogenetic tree, all the clones analyzed were of the subtype B lineage (Fig. 5). With the exception of the first two time point sequences intermixing (patient ITM27), sequences of all clones from each time point clustered together. These data reveal that at each time point (1 year interval) at which the samples were analyzed, the viral quasispecies had changed and did not contain viruses present at previous time points.
FIG. 5.
Phylogenetic analysis of the sequential HIV-1 sequences obtained from the HIV-1 cohort. Phylogenetic trees were initially constructed with all subtype references available at the Los Alamos Sequence DataBase. All the clones sequenced clustered with subtype B references. Subsequently all the sequences from the patients (ITM27, ITM60, ITM50, and ITM39) were then phylogenetically analyzed with subtype B references and the tree rooted with two subtype A references. The bootstrap values are shown at the internodes. Sequences obtained at sequential visits are shown in sequential order as virus T1, virus T2, virus T3, and virus T4 and represent the visits from which virus was obtained and used in the neutralization studies. Between 1 and 11 clones at each visit were sequenced and analyzed.
Genetic distance analysis and mutational patterns that reflect differences in neutralization sensitivity
Genetic distance and neutralization sensitivity
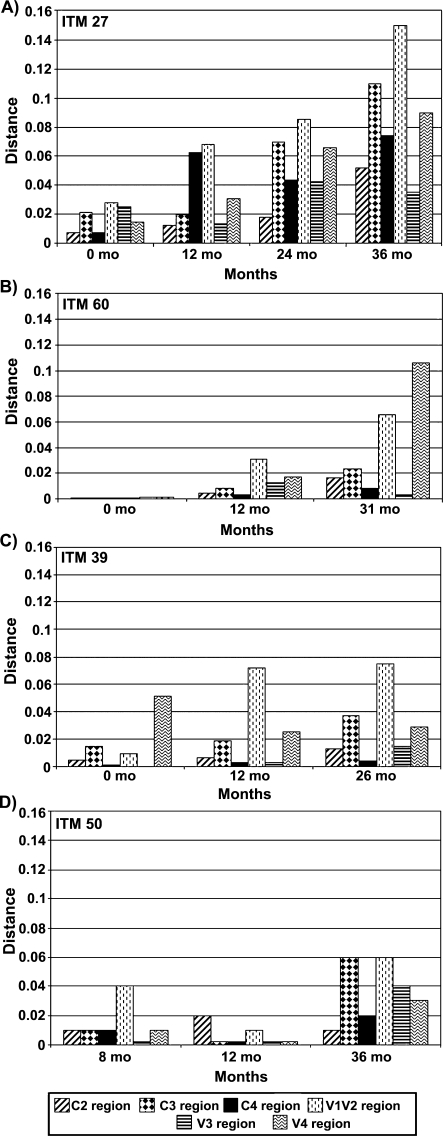
We examined the evolution of the various envelope regions of the viruses infecting these individuals by determining the phylogenetic tree distances of the sequential viral sequences (Fig. 6). In examining the genetic distances for patient ITM27 at months 0, 12, 24, and 36, we observed a continuous increase in the genetic distance over time (Fig. 6A). This increase in the genetic distance reflects each envelope region's contribution to the divergence and evolution of the virus as a whole. In particular, we noted the greatest divergence in the V1/V2, C3, and V4 regions of the viral envelope. These data reveal how different viral envelope regions evolve over time, consequently resulting in neutralization escape, as demonstrated by the neutralization studies presented in Fig. 3A. It was also noted that the V3 region exhibited the least change over time.
FIG. 6.
Phylogenetic tree distances of the sequential envelope gp120 regions from the subtype B-infected subjects. The distances were obtained through phylogenetic analysis from which the branch length from the node of the viral consensus sequence obtained from each subject's first visit to the node of each of the sequence clones from later visits was measured and the mean distance for sequences from each visit time point of each region was calculated. The consensus sequence for each subject's first visit was obtained from 8 clones each from patient ITM27 (A) and ITM60 (B), 10 clones from patient ITM39 (C), and 3 clones from patient ITM50 (D). The gp120 envelope regions analyzed include C2, C3, C4, V1V2, V3, and V4.
For patient ITM60, an increase in genetic distance was noted for the V1/V2 and V4 regions between months 0 and 12 and between months 12 and 31 (Fig. 6B). No significant changes were noted in the C2, C3, C4, or V3 regions. For patient ITM39, an increase in the genetic distance was noted in the V1/V2 and V4, as well as in the C3 regions (Fig. 6C). No significant difference was noted between months 12 and 26.
Nonsynonymous mutations and the differences in the neutralization sensitivities exhibited by the sequential viruses of patient ITM27 and ITM60
Sequence changes that contribute to virus evolution over time include insertions and deletions, synonymous and nonsynonymous changes, and changes in glycosylation patterns. Such changes have been documented to occur both in the constant and variable regions of gp160, and can result in escape from the neutralizing effect of antibodies.1–6 To analyze the nonsynonymous changes that occur among the viral quasispecies over time, the gp120 sequences obtained at the first visit for patients ITM27, ITM60, and ITM39 were compared with those of subsequent visits for each patient using the SNAP analysis as described in Nei and Gojobori23 and available at the Los Alamos website. Both variable and constant regions of gp120 were analyzed. The variable regions analyzed included V1/V2, V3, and V4. In analyzing the V1/V2 sequences from samples collected from the first and last visits, we identified eight and three mutations due to nonsynonymous changes for patients ITM27 and ITM60, respectively. In the V4 region, three and two changes were noted for ITM27 and ITM60, respectively. In the V3 region, two changes and no change were noted for ITM27 and ITM60, respectively. In analyzing the constant regions (C2, C3, C4), more nonsynonymous changes were observed for ITM27 than for ITM60. In particular, in the C2 region, four and one change, in the C3 region, six and one change, and in the C4, five and no changes were noted for ITM27 and ITM60, respectively. Though the number of changes between the sequential viruses of patients ITM27 and ITM60 identified in the various envelope regions appears small, we further analyzed the data to determine whether there is a statistical difference. Thus, we organized the number of changes in the variable and constant regions for patient ITM27 as n = 8, 3, 2, 4, 6, 5 and those for ITM60 as n = 3, 2, 0, 1, 1, 0. We then tested the null hypothesis that the mean difference between ITM27 and ITM60 is zero. Applying a one-sample t-test using the differences in the six regions as data results yields a p-value = 0.005, thus rejecting the null hypothesis and thereby indicating that the differences seen between ITM27 and ITM60 are statistically significant. We also applied the Wilcoxon rank sum test with continuity correction and obtained a p-value = 0.01.
The sequences were also examined for insertions and deletions. While one amino acid insertion was noted in the V3 and C3 regions of ITM27, no insertions were noted for ITM60. Five and four amino acid deletions were noted in the V1/V2 region of ITM27 and ITM60, respectively. There was no pattern of changes in the N-linked glycans in the sequences of both patients (data not shown).
Discussion
The fact that the heterologous plasma samples obtained from the HIV-1 subtype B-infected subjects neutralized early viruses from these study subjects suggests that these early viruses share common neutralization epitopes recognized by antibodies present in the plasma samples. Without relevant sequence changes in the envelope, the sequential viruses from these study subjects would be expected to remain sensitive to neutralization over time; however, this was not the case for many of the viruses described in this study. Our studies with heterologous plasma samples identified three patterns of neutralization against the sequential viruses from the four study subjects including (1) a loss of neutralization sensitivity of viruses over time, as exhibited by viruses from patient ITM27; (2) an increase in neutralization sensitivity of sequential viruses, as seen with those from patient ITM60; and (3) similar sensitivities of sequential viruses to neutralization, as seen with some plasma with viruses from patients ITM39 and ITM50 (Figs. 3 and 4). These patterns of neutralization identified may have several implications for vaccine design: (1) the loss of neutralization by heterologous plasma of sequential viruses over time suggests that some viruses could evolve to escape the effect of a potent vaccine. Thus, vaccines must target envelope regions that are less likely to evolve to escape recognition by antibodies induced by such a vaccine. (2) Viruses that evolve over time to become more sensitive to neutralization suggest that as some viruses evolve in their host, some antigenic epitopes become exposed and accessible to antibodies. If such viruses are transmitted, they may represent easy targets for vaccine-induced antibodies. The study presented here indicates the need to test the neutralization specificities of these sequential viruses using reagents such as anti-HIV-1 monoclonal antibodies directed at specific epitopes and techniques of epitope mapping to specifically identify the changes that account for the patterns of increase or decrease in neutralization sensitivity over time.
SNAP and phylogenetic distance analyses of the sequential viruses revealed changes that were characteristic of each neutralization profile. Of interest was the pattern seen with viruses from patient ITM27, whereby the sequential viruses became more resistant to neutralization by the heterologous plasma samples. This pattern of neutralization could, in part, be explained by the extensive mutations observed in the different envelope regions, as demonstrated by SNAP and phylogenetic tree distance analyses (Fig. 6). The pattern of neutralization escape exhibited by the sequential viruses from patient ITM27 has been documented in both acutely and chronically HIV-1-infected patients using sequential homologous plasma, whereby virus obtained at a later date is increasingly resistant to neutralization.4–6,15,25 Furthermore, neutralization studies of sequential viruses from a subtype B-infected individual with purified HIVIG, mAbs, and soluble CD4 have demonstrated neutralization escape26 similar to that seen with patient ITM27 (Fig. 3).
Of particular interest was patient ITM60, whose sequential viruses became more sensitive to neutralization by all the heterologous plasma tested. The increase in sensitivity of the viruses from this patient could, in part, be related to the higher CD4 counts in this patient compared to the other patients (Fig. 1), resulting in a better immune control of the viruses and limiting viral diversity. Indeed, our sequence analysis suggests that changes in certain envelope regions can render the virus more susceptible to neutralization. In comparing the changes that occurred in ITM60 and ITM27 as shown in the phylogenetic distance tree analysis, fewer changes occurred for ITM60 than for ITM27. The V3 sequences and sequences of the constant regions remained relatively unchanged, a reflection in their genetic distances (Fig. 6). Further studies are underway to characterize the amino acid changes that result in increased viral sensitivity to neutralizing antibodies. Such epitopes should be relevant when designing a potent HIV-1 vaccine.
While the sequential viruses of study subjects ITM39 and ITM50 exhibited a similar sensitivity to neutralization by many plasmas, it was noted that over time, the sequential viruses also became resistant to a few plasmas, as seen with plasma samples SZ2 and UCLA against the sequential viruses of ITM39 (Fig. 3A). This observation suggests the early emergence of escape mutants and the possibility that subsequent viruses from these individuals would be resistant to such plasma samples, as seen with patient ITM27.
Previous studies have identified immunogenic epitopes in the viral envelope gp120 to include the V1/V2, V3, the CD4 binding domain (CD4bd), and epitopes in the gp41 region.27–35 Little is known regarding whether other constant envelope regions such as C2, C3, and C4 are immunogenic, nor have antibodies been derived from HIV-1-infected individuals that are directed at these envelope regions; yet, we observed several nonsynonymous mutations that occurred in these regions over time. Mutations in the C3 have also been documented in analysis of HIV-136 and simian immunodeficiency virus (SIV) sequences.37 The question that remains is whether the mutations observed in these regions are due to immune pressure by neutralizing antibodies. If so, then these studies suggest that these regions may represent other important epitopes involved in viral escape from the host immune response. Alternatively, mutations that occur in these regions might cause a conformational change in other envelope regions that are more immunogenic, thereby obscuring them from the effect of antibodies. The changes identified, however, could also be relevant in the virus's escape from the effect of the host's cell-mediated immune response. Furthermore, though not studied here, other viral genomic regions such as gag and nef may also play a role in immune escape. Thus, extensive analysis that would examine the full genomic spectrum of HIV-1 and viral escape from the humoral and cell-mediated immune responses is critical to fully understand how to design an effective HIV-1 vaccine.
Despite the fact that several antibodies that have been derived from HIV-1-infected individuals are directed at epitopes in the V3 loop,27 indicating that this region of the virus is under immense immune pressure, it was surprising that the V3 region of the sequential viruses from all four subjects studied exhibited few sequence changes over time, irrespective of their neutralization profiles. In particular, few V3 sequence changes were noted for patient ITM27, whose initial viruses were sensitive to neutralization but whose later viruses were resistant. Similarly, the V3 sequence remained conserved in the sequential viruses from patient ITM60, whose later time point viruses became more sensitive to neutralization by the heterologous plasma. This was noted both in the SNAP (data not shown) and phylogenetic tree distance analyses (Fig. 6). Because the V3 loop plays a critical role in virus infectivity,38,39 significant changes in this envelope region may be detrimental to the virus.40,41 Therefore, the virus might escape from anti-V3 antibodies through different mechanisms not yet fully understood. For example, the virus may escape the immune response through few and specific point mutations occurring within or outside the V3 loop. Furthermore, studies also suggest that the V1/V2 region masks the V3 loop, thereby resulting in its inaccessibility to anti-V3 antibodies to neutralize the virus.31,42,43 This may also help explain why the V3 loop undergoes very little sequence change. Antibodies directed at the V3 loop are known to be both intraclade and interclade cross-reactive. Thus, the conformation as well as specific amino acids sequence contributes to V3-coreceptor binding, antibody binding, cross-reactivity, and cross-neutralization.30,43–51 That the V3 sequence remains constant over time and is very immunogenic is an indication that this region must be seriously considered as an important immunogenic epitope in a vaccine.
In conclusion, the data presented demonstrates that (1) viruses sensitive to neutralizing antibodies in a heterologous plasma sample may evolve over time to escape neutralization by the same plasma sample; and (2) viruses that are resistant or less sensitive to neutralization may evolve to become more neutralization sensitive. Because this study demonstrates that HIV-1 viruses can evolve over time in the V1/V2, V4, C2, and C3 regions to become resistant or sensitive to the effect of antibodies in plasma, changes that occur over time in these regions should be considered carefully when attempting to design a potent vaccine against HIV-1. Studies of epitope mapping using anti-HIV-1 monoclonal antibodies to different envelope regions should help identify changes that account for the increasing or decreasing neutralization sensitivities and should help improve our ability to design better HIV-1 vaccines.
Acknowledgments
We thank the individuals who have donated their blood for these studies. This work was supported with funds from the Department of Veterans Affairs (Merit Review Award and the Research Enhancement Program) and by Grant AI027742-17 from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) through the NYU Center for AIDS Research.
Disclosure Statement
No competing financial interests exist.
References
- 1.Back NK. Smit L. De Jong JJ, et al. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology. 1994;199(2):431–438. doi: 10.1006/viro.1994.1141. [DOI] [PubMed] [Google Scholar]
- 2.Chackerian B. Rudensey LM. Overbaugh J. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J Virol. 1997;71(10):7719–7727. doi: 10.1128/jvi.71.10.7719-7727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gram GJ. Hemming A. Bolmstedt A, et al. Identification of an N-linked glycan in the V1-loop of HIV-1 gp120 influencing neutralization by anti-V3 antibodies and soluble CD4. Arch Virol. 1994;139(3–4):253–261. doi: 10.1007/BF01310789. [DOI] [PubMed] [Google Scholar]
- 4.Richman DD. Wrin T. Little SJ, et al. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100(7):4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsang ML. Evans LA. McQueen P, et al. Neutralizing antibodies against sequential autologous human immunodeficiency virus type 1 isolates after seroconversion. J Infect Dis. 1994;170(5):1141–1147. doi: 10.1093/infdis/170.5.1141. [DOI] [PubMed] [Google Scholar]
- 6.Wei X. Decker JM. Wang S, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 7.Nyambi PN. Nkengasong J. Peeters M, et al. Reduced capacity of antibodies from patients infected with human immunodeficiency virus type 1 (HIV-1) group O to neutralize primary isolates of HIV-1 group M viruses. J Infect Dis. 1995;172(5):1228–1237. doi: 10.1093/infdis/172.5.1228. [DOI] [PubMed] [Google Scholar]
- 8.Nyambi PN. Nkengasong J. Lewi P, et al. Multivariate analysis of human immunodeficiency virus type 1 neutralization data. J Virol. 1996;70(9):6235–6243. doi: 10.1128/jvi.70.9.6235-6243.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nyambi PN. Willems B. Janssens W, et al. The neutralization relationship of HIV type 1, HIV type 2, and SIVcpz is reflected in the genetic diversity that distinguishes them. AIDS Res Hum Retroviruses. 1997;13(1):7–17. doi: 10.1089/aid.1997.13.7. [DOI] [PubMed] [Google Scholar]
- 10.van der Groen G. Nyambi PN. Beirnaert E, et al. Genetic variation of HIV type 1: Relevance of interclade variation to vaccine development. AIDS Res Hum Retroviruses. 1998;14(Suppl. 3):S211–221. [PubMed] [Google Scholar]
- 11.Weber J. Fenyo EM. Beddows S, et al. Neutralization serotypes of human immunodeficiency virus type 1 field isolates are not predicted by genetic subtype. The WHO Network for HIV Isolation and Characterization. J Virol. 1996;70(11):7827–7832. doi: 10.1128/jvi.70.11.7827-7832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostrikis LG. Cao Y. Ngai H, et al. Quantitative analysis of serum neutralization of human immunodeficiency virus type 1 from subtypes A, B, C, D, E, F, and I: Lack of direct correlation between neutralization serotypes and genetic subtypes and evidence for prevalent serum-dependent infectivity enhancement. J Virol. 1996;70(1):445–458. doi: 10.1128/jvi.70.1.445-458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nyambi PN. Burda S. Bastiani L, et al. A virus binding assay for studying the antigenic landscape on intact, native, primary human immunodeficiency virus-type 1. J Immunol Methods. 2001;253(1–2):253–262. doi: 10.1016/s0022-1759(01)00384-2. [DOI] [PubMed] [Google Scholar]
- 14.Cecilia D. KewalRamani VN. O'Leary J, et al. Neutralization profiles of primary human immunodeficiency virus type 1 isolates in the context of coreceptor usage. J Virol. 1998;72(9):6988–6996. doi: 10.1128/jvi.72.9.6988-6996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly HR. Urbanski M. Burda S, et al. Neutralizing antibody patterns and viral escape in HIV-1 non-B subtype chronically infected treatment-naive individuals. Hum Antibodies. 2005;14(3–4):89–99. [PubMed] [Google Scholar]
- 16.Boom R. Sol CJ. Salimans MM, et al. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28(3):495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Staden R. Beal KF. Bonfield JK. The Staden package, 1998. Methods Mol Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 18.Thompson JD. Gibson TJ. Plewniak F, et al. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S. Tamura K. Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5(2):150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 20.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 21.Saitou N. Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. Confidence limits on Phylogenies: An approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 23.Nei M. Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 24.Korber B. HIV Signature and sequence variation analysis. In: Rodrigo AG, editor; Learn GH, editor. Computational Analysis of HIV Molecular Sequences. Kluwer Academic Publishers; Dordrecht, Netherlands: 2000. pp. 55–72. [Google Scholar]
- 25.Albert J. Abrahamsson B. Nagy K, et al. Rapid development of isolate-specific neutralizing antibodies after primary HIV-1 infection and consequent emergence of virus variants which resist neutralization by autologous sera. AIDS. 1990;4(2):107–112. doi: 10.1097/00002030-199002000-00002. [DOI] [PubMed] [Google Scholar]
- 26.Beaumont T. van Nuenen A. Broersen S, et al. Reversal of human immunodeficiency virus type 1 IIIB to a neutralization-resistant phenotype in an accidentally infected laboratory worker with a progressive clinical course. J Virol. 2001;75(5):2246–2252. doi: 10.1128/JVI.75.5.2246-2252.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorny MK. Zolla-Pazner S. Human monoclonal antibodies that neutralize HIV-1. In: Korber B, editor; Brander C, editor; Haynes B, et al., editors. HIV Immunology and HIV/SIV Vaccine Database 2003. Theoretical Biology and Biophysics Group; Los Alamos: Los Alamos National Laboratory, Los Alamos, NM: p. 2003. [Google Scholar]
- 28.He Y. Honnen WJ. Krachmarov CP, et al. Efficient isolation of novel human monoclonal antibodies with neutralizing activity against HIV-1 from transgenic mice expressing human Ig loci. J Immunol. 2002;169(1):595–605. doi: 10.4049/jimmunol.169.1.595. [DOI] [PubMed] [Google Scholar]
- 29.Honnen WJ. Krachmarov C. Kayman SC, et al. Type-specific epitopes targeted by monoclonal antibodies with exceptionally potent neutralizing activities for selected strains of human immunodeficiency virus type 1 map to a common region of the V2 domain of gp120 and differ only at single positions from the clade B consensus sequence. J Virol. 2007;81(3):1424–1432. doi: 10.1128/JVI.02054-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zolla-Pazner S. Zhong P. Revesz K, et al. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Res Hum Retroviruses. 2004;20(11):1254–1258. doi: 10.1089/aid.2004.20.1254. [DOI] [PubMed] [Google Scholar]
- 31.Pinter A. Honnen WJ. He Y, et al. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J Virol. 2004;78(10):5205–5215. doi: 10.1128/JVI.78.10.5205-5215.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorny MK. Williams C. Volsky B, et al. Human monoclonal antibodies specific for conformation-sensitive epitopes of V3 neutralize human immunodeficiency virus type 1 primary isolates from various clades. J Virol. 2002;76(18):9035–9045. doi: 10.1128/JVI.76.18.9035-9045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorny MK. Zolla-Pazner S. Recognition by human monoclonal antibodies of free and complexed peptides representing the prefusogenic and fusogenic forms of human immunodeficiency virus type 1 gp41. J Virol. 2000;74(13):6186–6192. doi: 10.1128/jvi.74.13.6186-6192.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanchez-Martinez S. Lorizate M. Katinger H, et al. Membrane association and epitope recognition by HIV-1 neutralizing anti-gp41 2F5 and 4E10 antibodies. AIDS Res Hum Retroviruses. 2006;22(10):998–1006. doi: 10.1089/aid.2006.22.998. [DOI] [PubMed] [Google Scholar]
- 35.Parker CE. Deterding LJ. Hager-Braun C, et al. Fine definition of the epitope on the gp41 glycoprotein of human immunodeficiency virus type 1 for the neutralizing monoclonal antibody 2F5. J Virol. 2001;75(22):10906–10911. doi: 10.1128/JVI.75.22.10906-10911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gnanakaran S. Lang D. Daniels M, et al. Clade-specific differences between human immunodeficiency virus type 1 clades B and C: Diversity and correlations in C3–V4 regions of gp120. J Virol. 2007;81(9):4886–4891. doi: 10.1128/JVI.01954-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nyambi PN. Lewi P. Peeters M, et al. Study of the dynamics of neutralization escape mutants in a chimpanzee naturally infected with the simian immunodeficiency virus SIVcpz-ant. J Virol. 1997;71(3):2320–2330. doi: 10.1128/jvi.71.3.2320-2330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baritaki S. Dittmar MT. Spandidos DA, et al. In vitro inhibition of R5 HIV-1 infectivity by X4 V3-derived synthetic peptides. Int J Mol Med. 2005;16(2):333–336. [PubMed] [Google Scholar]
- 39.Rizos AK. Baritaki S. Tsikalas I, et al. Biophysical characterization of V3-lipopeptide liposomes influencing HIV-1 infectivity. Biochem Biophys Res Commun. 2007;355(4):963–969. doi: 10.1016/j.bbrc.2007.02.052. [DOI] [PubMed] [Google Scholar]
- 40.Pohlmann S. Munch J. Aziz S, et al. A simian immunodeficiency virus V3 loop mutant that does not efficiently use CCR5 or common alternative coreceptors is moderately attenuated in vivo. Virology. 2007;360(2):275–285. doi: 10.1016/j.virol.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 41.Yang ZY. Chakrabarti BK. Xu L, et al. Selective modification of variable loops alters tropism and enhances immunogenicity of human immunodeficiency virus type 1 envelope. J Virol. 2004;78(8):4029–4036. doi: 10.1128/JVI.78.8.4029-4036.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu CB. Zhu L. Holz-Smith S, et al. The role of the third beta strand in gp120 conformation and neutralization sensitivity of the HIV-1 primary isolate DH012. Proc Natl Acad Sci USA. 2001;98(26):15227–15232. doi: 10.1073/pnas.261359098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krachmarov CP. Honnen WJ. Kayman SC, et al. Factors determining the breadth and potency of neutralization by V3-specific human monoclonal antibodies derived from subjects infected with clade A or clade B strains of human immunodeficiency virus type 1. J Virol. 2006;80(14):7127–7135. doi: 10.1128/JVI.02619-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cardozo T. Kimura T. Philpott S, et al. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res Hum Retroviruses. 2007;23(3):415–426. doi: 10.1089/aid.2006.0130. [DOI] [PubMed] [Google Scholar]
- 45.Gorny MK. Williams C. Volsky B, et al. Cross-clade neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B clades of HIV-1. J Virol. 2006;80(14):6865–6872. doi: 10.1128/JVI.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stanfield RL. Gorny MK. Zolla-Pazner S, et al. Crystal structures of human immunodeficiency virus type 1 (HIV-1) neutralizing antibody 2219 in complex with three different V3 peptides reveal a new binding mode for HIV-1 cross-reactivity. J Virol. 2006;80(12):6093–6105. doi: 10.1128/JVI.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Parseval A. Bobardt MD. Chatterji A, et al. A highly conserved arginine in gp120 governs HIV-1 binding to both syndecans and CCR5 via sulfated motifs. J Biol Chem. 2005;280(47):39493–39504. doi: 10.1074/jbc.M504233200. [DOI] [PubMed] [Google Scholar]
- 48.Rosen O. Chill J. Sharon M, et al. Induced fit in HIV-neutralizing antibody complexes: Evidence for alternative conformations of the gp120 V3 loop and the molecular basis for broad neutralization. Biochemistry. 2005;44(19):7250–7258. doi: 10.1021/bi047387t. [DOI] [PubMed] [Google Scholar]
- 49.Stanfield RL. Gorny MK. Williams C, et al. Structural rationale for the broad neutralization of HIV-1 by human monoclonal antibody 447-52D. Structure. 2004;12(2):193–204. doi: 10.1016/j.str.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 50.Nyambi PN. Mbah HA. Burda S, et al. Conserved and exposed epitopes on intact, native, primary human immunodeficiency virus type 1 virions of group M. J Virol. 2000;74(15):7096–7107. doi: 10.1128/jvi.74.15.7096-7107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nyambi PN. Nadas A. Mbah HA, et al. Immunoreactivity of intact virions of human immunodeficiency virus type 1 (HIV-1) reveals the existence of fewer HIV-1 immunotypes than genotypes. J Virol. 2000;74(22):10670–10680. doi: 10.1128/jvi.74.22.10670-10680.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]