Abstract
Pan-American mitochondrial DNA (mtDNA) haplogroup C1 has been recently subdivided into three branches, two of which (C1b and C1c) are characterized by ages and geographical distributions that are indicative of an early arrival from Beringia with Paleo-Indians. In contrast, the estimated ages of C1d—the third subset of C1—looked too young to fit the above scenario. To define the origin of this enigmatic C1 branch, we completely sequenced 63 C1d mitochondrial genomes from a wide range of geographically diverse, mixed, and indigenous American populations. The revised phylogeny not only brings the age of C1d within the range of that of its two sister clades, but reveals that there were two C1d founder genomes for Paleo-Indians. Thus, the recognized maternal founding lineages of Native Americans are at least 15, indicating that the overall number of Beringian or Asian founder mitochondrial genomes will probably increase extensively when all Native American haplogroups reach the same level of phylogenetic and genomic resolution as obtained here for C1d.
While debate is still ongoing among scientists from several disciplines regarding the number of migratory events, their timing, and entry routes into the Americas (Wallace and Torroni 1992; Torroni et al. 1993; Forster et al. 1996; Kaufman and Golla 2000; Goebel et al. 2003, 2008; Schurr and Sherry 2004; Wang et al. 2007; Waters and Stafford 2007; Dillehay et al. 2008; Gilbert et al. 2008a; O'Rourke and Raff 2010), the general consensus is that modern Native American populations ultimately trace their gene pool to Asian groups who colonized northeast Siberia, including parts of Beringia, prior to the last glacial period. These ancestral population(s) probably retreated into refugial areas during the Last Glacial Maximum (LGM), where their genetic variation was reshaped by drift. Thus, pre-LGM haplotypes of Asian ancestry were differently preserved and lost in Beringian enclaves, but at the same time, novel haplotypes and alleles arose in situ due to new mutations, often becoming predominant because of major founder events (Tamm et al. 2007; Achilli et al. 2008; Bourgeois et al. 2009; Perego et al. 2009; Schroeder et al. 2009). The scenario of a temporally important differentiation stage in Beringia explains the predominance in Native Americans of private alleles and haplogroups such as the autosomal 9-repeat at microsatellite locus D9S1120 (Phillips et al. 2008; Schroeder et al. 2009), the Y chromosome haplogroup Q1a3a-M3 (Bortolini et al. 2003; Karafet et al. 2008; Rasmussen et al. 2010), and the pan-American mtDNA haplogroups A2, B2, C1b, C1c, C1d, D1, and D4h3a (Tamm et al. 2007; Achilli et al. 2008; Fagundes et al. 2008; Perego et al. 2009).
In the millennia after the initial Paleo-Indian migrations, other groups from Beringia or eastern Siberia expanded into North America. If the gene pool of the source population(s) had in the meantime partially changed, not only because of drift, but also due to the admixture with population groups newly arrived from regions located west of Beringia, this would have resulted in the entry of additional Asian lineages into North America. This scenario, sometimes invoked to explain the presence of certain mtDNA haplogroups such as A2a, A2b, D2a, D3, and X2a only in populations of northern North America (Torroni et al. 1992; Brown et al. 1998; Schurr and Sherry 2004; Helgason et al. 2006; Achilli et al. 2008; Gilbert et al. 2008b; Perego et al. 2009), has recently received support from nuclear and morphometric data showing that some native groups from northern North America harbor stronger genetic similarities with some eastern Siberian groups than with Native American groups located more in the South (González-José et al. 2008; Bourgeois et al. 2009; Wang et al. 2009; Rasmussen et al. 2010).
As for the pan-American mtDNA haplogroups, when analyzed at the highest level of molecular resolution (Bandelt et al. 2003; Tamm et al. 2007; Fagundes et al. 2008; Perego et al. 2009), they all reveal, with the exception of C1d, entry times of 15–18 thousand years ago (kya), which are suggestive of a (quasi) concomitant post-LGM arrival from Beringia with early Paleo-Indians. A similar entry time is also shown for haplogroup X2a, whose restricted geographical distribution in northern North America appears to be due not to a later arrival, but to its entry route through the ice-free corridor (Perego et al. 2009). Despite its continent-wide distribution, C1d was hitherto characterized by an expansion time of only 7.6–9.7 ky (Perego et al. 2009). This major discrepancy has been attributed to a poor and possibly biased representation of complete C1d mtDNA sequences (only 10) in the available data sets (Achilli et al. 2008; Malhi et al. 2010). To clarify the issue of the age of haplogroup C1d and its role as a founding Paleo-Indian lineage, we sequenced and analyzed 63 C1d mtDNAs from populations distributed over the entire geographical range of the haplogroup.
Results
The phylogeny of haplogroup C1d
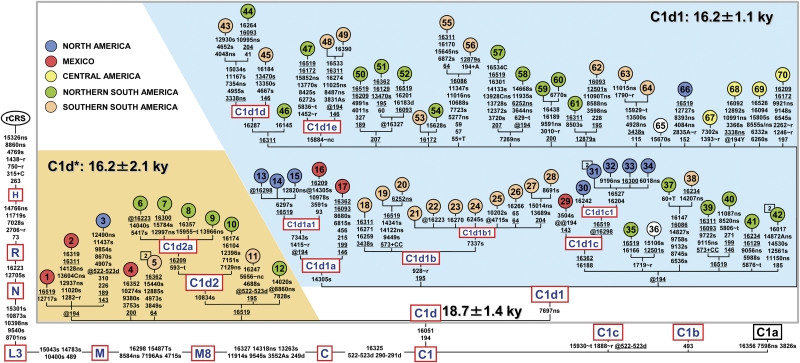
The phylogeny encompassing the novel 63 C1d sequences (Fig. 1), plus 10 previously published mtDNA genomes (Achilli et al. 2008; Malhi et al. 2010), revealed that not all C1d sequences are defined by the mutational motif 7697-16051, as previously suggested (Achilli et al. 2008; Malhi et al. 2010). About 18% of the C1d mtDNAs, with representatives in both North and South America, formed a paragroup (C1d*) lacking the coding-region transition at nucleotide position (np) 7697. This finding suggests that only the control-region mutation at np 16051 is ancestral to the entire haplogroup, and the mutational event at np 7697 occurred later, marking one (major) C1d branch, here termed C1d1, which is also represented all over the double continent. Moreover, the control region mutation at np 194 was observed in mtDNAs belonging to both C1d* and C1d1 and in ∼60% of the C1d samples in public databases, thus indicating that, alongside 16051, it is most likely a basal mutation for the entire C1d haplogroup, but somewhat prone to back mutation as also testified to by one heteroplasmic instance in Figure 1 and its mutation rate as scored (12) in Soares et al. (2009).
Figure 1.
Detailed tree of C1d in the context of haplogroup C1. All 73 C1d mtDNA sequences (63 novel and 10 published) are complete except for samples 36 and 65, for which only coding-region data are available. The basal motifs for Native American haplogroups C1b and C1c are also included together with the motif of the Asian-specific branch C1a. The position of the revised Cambridge reference sequence (rCRS) (Andrews et al. 1999) is indicated for reading off-sequence motifs. Mutations are shown on the branches; they are transitions unless a base is explicitly indicated. The prefix @ designates reversions, while suffixes indicate transversions (to A, G, C, or T), indels (+, d), gene locus (∼t, tRNA; ∼r, rRNA; ∼nc, noncoding region outside of the control-region), synonymous or nonsynonymous changes (s or ns), and T/C heteroplasmy (Y). Recurrent mutations within the phylogeny are underlined. We have followed the guidelines for standardization of the alignment in long C stretches (Bandelt and Parson 2008), but disregarded any length variation in the C-stretch between nucleotides 303 and 315, with the exception of the well-known 315+C insertion. Additional information regarding each mtDNA is available in Supplemental Table S1. Coalescence times shown for C1d, C1d*, and C1d1 are maximum-likelihood (ML) estimates, while the corresponding averaged distance (ρ) accompanied by a heuristic estimate of SE (σ) are shown in Table 1. As for the geographic affiliation (top left corner), North America refers to USA and Canada; northern South America refers to Colombia, Venezuela, Ecuador, Peru, and Brazil; southern South America corresponds to Chile, Argentina, Uruguay, and Paraguay.
Age estimates of haplogroup C1d
The maximum-likelihood (ML) divergence based on the complete mtDNA sequence for the entire C1d haplogroup of 0.0074 ± 0.00019 substitutions per site corresponds to a divergence time of 18.7 ± 1.4 ky according to the mutation rate calibrated by Soares et al. (2009). The ML divergences for C1d* and C1d1 are not much lower than that of the entire C1d and virtually identical to each other with values of 0.0061 ± 0.00019 and 0.0068 ± 0.00015 substitutions per site, corresponding to divergence times of 16.2 ± 2.1 ky and 16.2 ± 1.1 ky, respectively (Fig. 1). These divergence ages are confirmed when the average distance of the haplotypes from the root of C1d, C1d*, and C1d1 (ρ-statistics) are computed (Table 1). In this case, the time to the most recent common ancestor for C1d is 18.8 ± 2.8 ky when using the sequence variation of the entire genome (Soares et al. 2009), and 14.9 ± 1.9/15.1 ± 1.8 ky when only synonymous mutations are considered (Loogväli et al. 2009; Soares et al. 2009). As for C1d* and C1d1, rho age estimates are ∼14–18 ky and 14–17 ky, respectively.
Table 1.
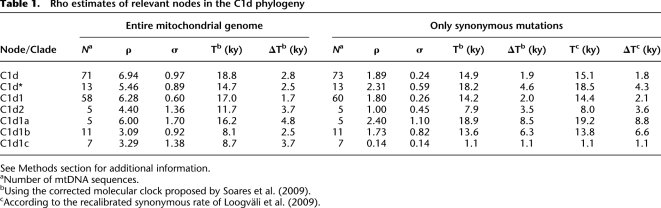
Rho estimates of relevant nodes in the C1d phylogeny
See Methods section for additional information.
aNumber of mtDNA sequences.
bUsing the corrected molecular clock proposed by Soares et al. (2009).
cAccording to the recalibrated synonymous rate of Loogväli et al. (2009).
Discussion
Overall, the new data confirm that the coalescence time previously reported for C1d was indeed heavily underestimated and indicate that C1d as a whole is ancient enough to be among the founding Paleo-Indian mtDNA lineages. The Americas present a particular difficulty for the identification of founder mitochondrial genomes. In other geographical contexts, founders can be identified as sequence matches between the putative source and settled regions. In our case, the source population does not exist anymore, so that the criterion of matching cannot be used. Thus, the identification of founder Paleo-Indian mtDNA sequences is based on the evaluation of two remaining parameters: the coalescence time and the geographical distribution of the derived haplogroup/subhaplogroup from the postulated founder. Coalescence times of C1d* and C1d1 are very similar to those reported for haplogroups A2, B2, C1b, C1c, D1, D4h3a, and X2a (Perego et al. 2009). Moreover, both C1d* and C1d1 mtDNAs are found in North, Central, and South America. Therefore, it is most likely that the founding Paleo-Indian population(s), who entered the Americas about 15–17 kya, harbored not only one, but two founding C1d sequences—one corresponding to the C1d node and one already characterized by the mutation at np 7697 corresponding to the C1d1 node (Fig. 1). As for the other newly defined sub-branches within C1d* and C1d1, both age estimates (Table 1) and geographical distributions (Fig. 1) are most compatible with an origin, either in North America (C1d1a, C1d1c) or South America (C1d1b, C1d2) at intermediate stages of the in situ differentiation of local Native American groups.
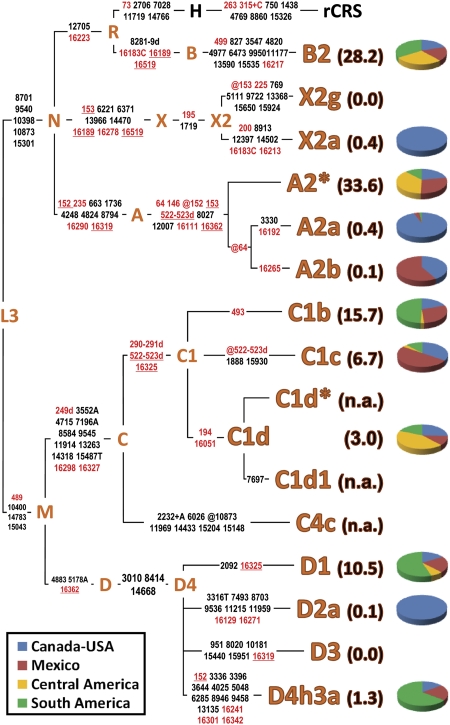
Also, in the Americas, similar to other continents (Kayser 2010; O'Rourke and Raff 2010; Renfrew 2010; Soares et al. 2010; Stoneking and Delfin 2010), a systematic survey of mtDNA variation based on whole-genome sequencing makes it possible to dissect haplogroups into branches and sub-branches (and so on) often distinguished, as in the case of C1d, C1d1, and C1d1a, by a single mutation. Once this (maximum) level of phylogenetic and genomic resolution is reached, it becomes possible to identify all different mtDNA sequences that might have participated in a colonization or migratory event. As for Native Americans, within the last few years the overall number of recognized maternal founding lineages has gone from just five (A2, B2, C1, D1, and X2a) to a current count of 15 (Fig. 2). Most likely, the number of Beringian or Asian founder mitochondrial genomes will further increase when Native American haplogroups reach the same level of resolution as obtained here for C1d, and as previously reported for D4h3a and X2a (Perego et al. 2009). This can be achieved, as demonstrated by the frequency patterns shown in Figure 3, through the analysis of not only Native American tribes or communities, but also the general mixed population of national states. Indeed, the substantial overlap of C1d distributions indicates that, despite the extensive genetic input from Old World populations (mainly from Europe and Africa), general populations of the double continent retain a substantial fraction of the local Native American mtDNA pool. If applied to the northern American haplogroups A2a, A2b, D2a, and D3, such a level of phylogenetic resolution will also allow an accurate evaluation of more recent (post-Paleo-Indian) events of gene flow from Beringia or Eastern Siberia, such as that recently identified by sequencing the genome of an ancient Palaeo-Eskimo (Rasmussen et al. 2010).
Figure 2.
MtDNA tree encompassing the roots of all known Native American haplogroups. The distinguishing mutational motifs for the 15 known Native American haplogroups are reported on the branches. The position of the revised Cambridge reference sequence (rCRS) (Andrews et al. 1999) is indicated for reading off-sequence motifs. Mutations in the control region are in red, while mutations in the coding region are listed in black; they are transitions unless a base is explicitly indicated. The prefix @ designates reversions, while suffixes indicate transversions (to A, G, C, or T), indels (+, d). Recurrent mutations within the tree are underlined. The percent frequency of each Native American haplogroup in the entire double continent is reported in parentheses and has been obtained from the Sorenson Molecular Genealogy Foundation Mitochondrial (SMGF) mtDNA database (http://www.smgf.org) (entire control region) excluding all non-Native American mtDNAs. For each haplogroup, the relative frequencies in northern America (Canada and USA), Mexico, Central America, and South America are reported in different colors in the corresponding pie chart. Some haplogroups are completely absent in the SMGF mtDNA database because either they are extremely rare (X2g) or harbor a restricted distribution range (D3 in the Eskimos and Aleuts). For C4c, C1d*, and C1d1 frequency values are not available (n.a.) due to the lack of distinguishing control region mutations, but the overall C1d incidence (C1d* plus C1d1) is reported.
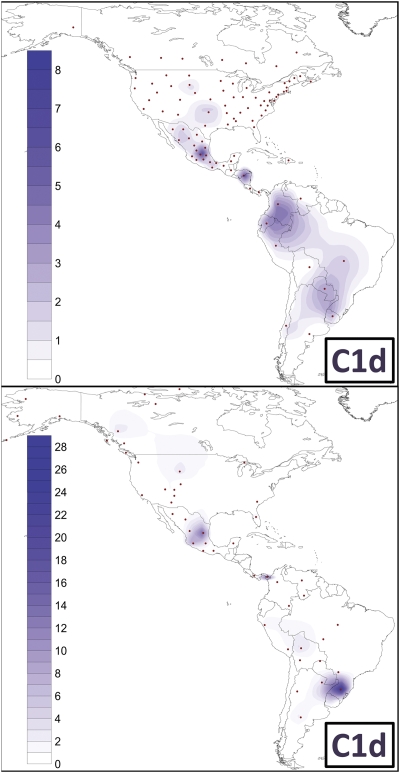
Figure 3.
Spatial frequency distribution of haplogroup C1d. The top map shows the frequency distribution of haplogroup C1d in general mixed populations of national states, while the bottom map illustrates the distribution in Native American tribes or communities. Note that the frequency scales (%) used in the two maps are different. The dots indicate the geographic location of the population samples included in each survey (Supplemental Tables S2, S3). Frequency maps were obtained as in Pala et al. (2009).
Methods
Analysis of mtDNA sequence variation
Candidate C1d mtDNAs were identified and selected based on the presence of the C1 control region motif (73, 249d, 263, 290–291d, 315+C, 489, 522–523d, 16223, 16298, 16325, 16327), plus the C1d diagnostic transition at np 16,051 (Achilli et al. 2008). For all subjects, an appropriate informed consent was obtained and institutional review boards at the various organizations involved with the current study approved all procedures. Sequencing of entire mtDNAs and phylogeny construction were performed as previously described (Torroni et al. 2001; Achilli et al. 2005).
Maximum likelihood analysis
We used PAML 3.13 (Yang 1997), assuming the HKY85 mutation model (with indels ignored, as usual) with gamma-distributed rates (approximated by a discrete distribution with 32 categories) and three partitions: HVS-I (positions 16051–16400), HVS-II (positions 68–263), and the remainder. We performed the analysis in two ways: (1) using the entire data set reported in Figure 1; and (2) using only the C1d* sequences in order to calculate the divergence of this paragroup. The age estimates were extrapolated using the corrected mutation rate of Soares et al. (2009).
Rho statistics
We compared the ML estimates with those directly obtained from converting the averaged distance (ρ) of the haplotypes of a clade to the respective root haplotype, accompanied by a heuristic estimate of the standard error (σ) calculated from an estimate of the genealogy (Saillard et al. 2000); see Table 1. This calculation was performed on the entire mtDNA haplotypes (excluding the mutations 16182C, 16183C, 16194C, and 16519) and repeated considering only synonymous mutations. Mutational distances were converted into years using the corrected molecular clock proposed by Soares et al. (2009) and the recalibrated synonymous rate of Loogväli et al. (2009). The differences between the ML and ρ estimators of the coalescence ages based on the entire mtDNA sequence are very minor (<1.5%) for the three major clades (C1d, C1d1, and C1d*).
Acknowledgments
This research received support from the Sorenson Molecular Genealogy Foundation (U.A.P. and S.R.W), Ministerio de Ciencia e Innovación-SAF2008-02971 (A.S.), Fundación de Investigación Médica Mutua Madrileña-2008/CL444 (A.S.), the FWF Austrian Science Fund grant TR397 (W.P.), Progetti Ricerca Interesse Nazionale 2007 (Italian Ministry of the University) (O.S. and A.T.), Fondazione Alma Mater Ticinensis (O.S. and A.T.). We thank all of the donors for providing biological specimens, Juan Carlos Jaime and José Edgar Gomez-Palmieri for their help in collecting the samples, Hans-Jürgen Bandelt for valuable comments and suggestions on this work, Diahan Southard for assistance in compiling data from the published literature, and everyone at the Sorenson Molecular Genealogy Foundation for their work on the preliminary data.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/genbank) under accession nos. HM107306–HM107368.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.109231.110.
References
- Achilli A, Rengo C, Battaglia V, Pala M, Olivieri A, Fornarino S, Magri C, Scozzari R, Babudri N, Santachiara-Benerecetti AS, et al. 2005. Saami and Berbers—an unexpected mitochondrial DNA link. Am J Hum Genet 76: 883–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achilli A, Perego UA, Bravi CM, Coble MD, Kong QP, Woodward SR, Salas A, Torroni A, Bandelt H-J 2008. The phylogeny of the four pan-American mtDNA haplogroups: Implications for evolutionary and disease studies. PLoS ONE 3: e1764 doi: 10.1371/journal.pone.0001764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N 1999. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23: 147 doi: 10.1038/13779 [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Parson W 2008. Consistent treatment of length variants in the human mtDNA control region: A reappraisal. Int J Legal Med 122: 11–21 [DOI] [PubMed] [Google Scholar]
- Bandelt H-J, Herrnstadt C, Yao Y-G, Kong Q-P, Kivisild T, Rengo C, Scozzari R, Richards M, Villems R, Macaulay V, et al. 2003. Identification of Native American founder mtDNAs through the analysis of complete mtDNA sequences: Some caveats. Ann Hum Genet 67: 512–524 [DOI] [PubMed] [Google Scholar]
- Bortolini MC, Salzano FM, Thomas MG, Stuart S, Nasanen SP, Bau CH, Hutz MH, Layrisse Z, Petzl-Erler ML, Tsuneto LT, et al. 2003. Y-chromosome evidence for differing ancient demographic histories in the Americas. Am J Hum Genet 73: 524–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeois S, Yotova V, Wang S, Bourtoumieu S, Moreau C, Michalski R, Moisan JP, Hill K, Hurtado AM, Ruiz-Linares A, et al. 2009. X-chromosome lineages and the settlement of the Americas. Am J Phys Anthropol 140: 417–428 [DOI] [PubMed] [Google Scholar]
- Brown MD, Hosseini SH, Torroni A, Bandelt H-J, Allen JC, Schurr TG, Scozzari R, Cruciani F, Wallace DC 1998. MtDNA haplogroup X: An ancient link between Europe/Western Asia and North America? Am J Hum Genet 63: 1852–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillehay TD, Ramirez C, Pino M, Collins MB, Rossen J, Pino-Navarro JD 2008. Monte Verde: Seaweed, food, medicine, and the peopling of South America. Science 320: 784–786 [DOI] [PubMed] [Google Scholar]
- Fagundes NJ, Kanitz R, Eckert R, Valls AC, Bogo MR, Salzano FM, Smith DG, Silva WA Jr, Zago MA, Ribeiro-dos-Santos AK, et al. 2008. Mitochondrial population genomics supports a single pre-Clovis origin with a coastal route for the peopling of the Americas. Am J Hum Genet 82: 583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P, Harding R, Torroni A, Bandelt H-J 1996. Origin and evolution of Native American mtDNA variation: A reappraisal. Am J Hum Genet 59: 935–945 [PMC free article] [PubMed] [Google Scholar]
- Gilbert MTP, Jenkins DL, Götherström A, Naveran N, Sanchez JJ, Hofreiter M, Thomsen PF, Binladen J, Higham TFG, Yohe RM II, et al. 2008a. DNA from pre-Clovis human coprolites in Oregon, North America. Science 320: 786–789 [DOI] [PubMed] [Google Scholar]
- Gilbert MTP, Kivisild T, Grønnow B, Andersen PK, Metspalu E, Reidla M, Tamm E, Axelsson E, Götherström A, Campos PF, et al. 2008b. Paleo-Eskimo mtDNA genome reveals matrilineal discontinuity in Greenland. Science 320: 1787–1789 [DOI] [PubMed] [Google Scholar]
- Goebel T, Waters MR, Dikova M 2003. The archaeology of Ushki Lake, Kamchatka, and the Pleistocene peopling of the Americas. Science 301: 501–505 [DOI] [PubMed] [Google Scholar]
- Goebel T, Waters MR, O'Rourke DH 2008. The late Pleistocene dispersal of modern humans in the Americas. Science 319: 1497–1502 [DOI] [PubMed] [Google Scholar]
- González-José R, Bortolini MC, Santos FR, Bonatto SL 2008. The peopling of America: Craniofacial shape variation on a continental scale and its interpretation from an interdisciplinary view. Am J Phys Anthropol 137: 175–187 [DOI] [PubMed] [Google Scholar]
- Helgason A, Pálsson G, Pedersen HS, Angulalik E, Gunnarsdóttir ED, Yngvadóttir B, Stefánsson K 2006. mtDNA variation in Inuit populations of Greenland and Canada: Migration history and population structure. Am J Phys Anthropol 130: 123–134 [DOI] [PubMed] [Google Scholar]
- Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, Hammer MF 2008. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res 18: 830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman T, Golla V 2000. Language groupings in the New World: Their reliability and usability in cross-disciplinary studies. In America past, America present: Genes and language in the Americas and beyond (ed. Renfrew C), pp. 47–57 McDonald Institute for Archaeological Research, Cambridge, UK [Google Scholar]
- Kayser M 2010. The human genetic history of Oceania: Near and remote views of dispersal. Curr Biol 20: R194–R201 [DOI] [PubMed] [Google Scholar]
- Loogväli EL, Kivisild T, Margus T, Villems R 2009. Explaining the imperfection of the molecular clock of hominid mitochondria. PLoS ONE 4: e8260 doi: 10.1371/journal.pone.0008260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi RS, Cybulski JS, Tito RY, Johnson J, Harry H, Dan C 2010. Brief communication: Mitochondrial haplotype C4c confirmed as a founding genome in the Americas. Am J Phys Anthropol 141: 494–497 [DOI] [PubMed] [Google Scholar]
- O'Rourke DH, Raff JA 2010. The human genetic history of the Americas: The final frontier. Curr Biol 20: R202–R207 [DOI] [PubMed] [Google Scholar]
- Pala M, Achilli A, Olivieri A, Hooshiar Kashani B, Perego UA, Sanna D, Metspalu E, Tambets K, Tamm E, Accetturo M, et al. 2009. Mitochondrial haplogroup U5b3: A distant echo of the Epipaleolithic in Italy and the legacy of the early Sardinians. Am J Hum Genet 84: 814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego UA, Achilli A, Angerhofer N, Accetturo M, Pala M, Olivieri A, Hooshiar Kashani B, Ritchie KH, Scozzari R, Kong QP, et al. 2009. Distinctive Paleo-Indian migration routes from Beringia marked by two rare mtDNA haplogroups. Curr Biol 19: 1–8 [DOI] [PubMed] [Google Scholar]
- Phillips C, Rodriguez A, Mosquera-Miguel A, Fondevila M, Porras-Hurtado L, Rondon F, Salas A, Carracedo A, Lareu MV 2008. D9S1120, a simple STR with a common Native American-specific allele: Forensic optimization, locus characterization and allele frequency studies. Forensic Sci Int Genet 3: 7–13 [DOI] [PubMed] [Google Scholar]
- Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, Moltke I, Metspalu M, Metspalu E, Kivisild T, Gupta R, et al. 2010. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature 463: 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfrew C 2010. Archaeogenetics—towards a ‘new synthesis’? Curr Biol 20: R162–R165 [DOI] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt H-J, Nørby S 2000. MtDNA variation among Greenland Eskimos: The edge of the Beringian expansion. Am J Hum Genet 67: 718–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KB, Jakobsson M, Crawford MH, Schurr TG, Boca SM, Conrad DF, Tito RY, Osipova LP, Tarskaia LA, Zhadanov SI, et al. 2009. Haplotypic background of a private allele at high frequency in the Americas. Mol Biol Evol 26: 995–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurr TG, Sherry ST 2004. Mitochondrial DNA and Y chromosome diversity and the peopling of the Americas: Evolutionary and demographic evidence. Am J Hum Biol 16: 420–439 [DOI] [PubMed] [Google Scholar]
- Soares P, Ermini L, Thomson N, Mormina M, Rito T, Rohl A, Salas A, Oppenheimer S, Macaulay V, Richards MB 2009. Correcting for purifying selection: An improved human mitochondrial molecular clock. Am J Hum Genet 84: 740–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares P, Achilli A, Semino O, Davies W, Macaulay V, Bandelt HJ, Torroni A, Richards MB 2010. The archaeogenetics of Europe. Curr Biol 20: R174–R183 [DOI] [PubMed] [Google Scholar]
- Stoneking M, Delfin F 2010. The human genetic history of East Asia: Weaving a complex tapestry. Curr Biol 20: R188–R193 [DOI] [PubMed] [Google Scholar]
- Tamm E, Kivisild T, Reidla M, Metspalu M, Smith DG, Mulligan CJ, Bravi CM, Rickards O, Martínez-Labarga C, Khusnutdinova EK, et al. 2007. Beringian standstill and spread of Native American founders. PLoS ONE 2: e829 doi: 10.1371/journal.pone.0000829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Yang CC, Szathmary EJE, Williams RC, Schanfield MS, Troup GA, Knowler WC, Lawrence DN, Weiss KM, et al. 1992. Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics 130: 153–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Schurr TG, Cabell MF, Brown MD, Neel JV, Larsen M, Smith DG, Vullo CM, Wallace DC 1993. Asian affinities and continental radiation of the four founding Native American mtDNAs. Am J Hum Genet 53: 563–590 [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Rengo C, Guida V, Cruciani F, Sellitto D, Coppa A, Calderon FL, Simionati B, Valle G, Richards M, et al. 2001. Do the four clades of the mtDNA haplogroup L2 evolve at different rates? Am J Hum Genet 69: 1348–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC, Torroni A 1992. American Indian prehistory as written in the mitochondrial DNA: A review. Hum Biol 64: 403–416 [PubMed] [Google Scholar]
- Wang S, Lewis CM, Jakobsson M, Ramachandran S, Ray N, Bedoya G, Rojas W, Parra MV, Molina JA, Gallo C, et al. 2007. Genetic variation and population structure in Native Americans. PLoS Genet 3: e185 doi: 10.1371/journal.pgen.0030185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Bedoya G, Labuda D, Ruiz-Linares A 2009. Brief communication: Patterns of linkage disequilibrium and haplotype diversity at Xq13 in six Native American populations. Am J Phys Anthropol 142: 476–480 [DOI] [PubMed] [Google Scholar]
- Waters MR, Stafford TWJ 2007. Redefining the age of Clovis: Implications for the peopling of the Americas. Science 315: 1122–1126 [DOI] [PubMed] [Google Scholar]
- Yang Z 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13: 555–556 [DOI] [PubMed] [Google Scholar]