Abstract
Chronic neuropathic pain is affected by specifics of the precipitating neural pathology, psychosocial factors, and by genetic predisposition. Little is known about the identity of predisposing genes. Using an integrative approach, we discovered that CACNG2 significantly affects susceptibility to chronic pain following nerve injury. CACNG2 encodes for stargazin, a protein intimately involved in the trafficking of glutamatergic AMPA receptors. The protein might also be a Ca2+ channel subunit. CACNG2 has previously been implicated in epilepsy. Initially, using two fine-mapping strategies in a mouse model (recombinant progeny testing [RPT] and recombinant inbred segregation test [RIST]), we mapped a pain-related quantitative trait locus (QTL) (Pain1) into a 4.2-Mb interval on chromosome 15. This interval includes 155 genes. Subsequently, bioinformatics and whole-genome microarray expression analysis were used to narrow the list of candidates and ultimately to pinpoint Cacng2 as a likely candidate. Analysis of stargazer mice, a Cacng2 hypomorphic mutant, provided electrophysiological and behavioral evidence for the gene's functional role in pain processing. Finally, we showed that human CACNG2 polymorphisms are associated with chronic pain in a cohort of cancer patients who underwent breast surgery. Our findings provide novel information on the genetic basis of neuropathic pain and new insights into pain physiology that may ultimately enable better treatments.
Chronic pain is a healthcare problem of enormous proportions, directly affecting nearly 20% of adults and associated with massive financial costs (Breivik et al. 2006). At least 25% of this burden is attributable to “neuropathic pain,” pain that follows nerve damage (Bouhassira et al. 2008). In patients with neuropathy spontaneous pain is typically the most prominent cause of suffering, rather than stimulus-provoked pain. A striking example is phantom pain. Virtually all limb amputees report feeling a phantom limb, and most report spontaneous burning, stabbing, or electric shock–like pain, at least occasionally (Sherman et al. 1996; Nikolajsen and Jensen 2001). Phantom pain is also common after breast removal (Tytherleigh et al. 1998; Rothemund et al. 2004; Vadivelu et al. 2008) and in body parts that have been denervated but are still present (“anesthesia dolorosa”) (Wynn Parry 1980).
Neuropathic pain, including phantom pain, is a complex trait affected by both the nature of the neural injury and by psychosocial factors. Its notorious variability among individuals, even when the underlying nerve pathology is identical, has prompted awareness of a significant genetic contribution to the amount of pain felt (Diatchenko et al. 2007; Lacroix-Fralish and Mogil 2008; LaCroix-Fralish et al. 2009). At present, the biological process linking nerve injury to chronic pain is incompletely understood (Devor 2006a), and treatments are inadequate. Identification of genes affecting predisposition to pain may shed light on the underlying biology and hence facilitate the development of more effective treatments (Mogil et al. 2005; Tegeder et al. 2006).
There are several animal models of neuropathic pain. We used one that emulates the spontaneous pain of phantom limb and anesthesia dolorosa, the Neuroma model (Wall et al. 1979). In this model, one hindpaw is completely denervated, and pain is monitored by scoring “autotomy.” This is a behavior that comprises scratching and biting of the numb paw, apparently in response to unpleasant phantom limb sensations (Devor 2007). Autotomy behavior is highly variable among individuals and across inbred strains. In previous studies using the Neuroma model we identified a quantitative trait locus (QTL), Pain1, that affects predisposition to this pain phenotype and mapped it to an ∼25-cM interval on chromosome 15 (Seltzer et al. 2001; Devor et al. 2005). No other significant QTLs were found in a whole-genome scan (Devor et al. 2007). Mapping QTLs to specific chromosomal regions is only the first step toward the ultimate goal of identifying the underlying gene(s), the aim of the present study. We accomplished this aim by implementing a broad, integrative approach that included fine-mapping experiments followed by application of several strategies for prioritizing candidate genes. The gene identified in this way is CACNG2. CACNG2 encodes for the gamma-2 transmembrane AMPA receptor protein (TARP) stargazin, known to be intimately involved in the trafficking of glutamatergic AMPA receptors and the modulation of their ion channel function (Priel et al. 2005; Bats et al. 2007; Cokic and Stein 2008; Milstein and Nicoll 2009). It also modulates neuronal Cav2 Ca2+ channels (Kang et al. 2001; Sandoval et al. 2007; Tselniker et al. 2010), although this role remains controversial. CACNG2 is known to play a role in cerebellar function and in epilepsy, but not neuropathic pain. We have now established that it also plays a functional role, in both mice and humans, in the heritable predisposition to neuropathic pain.
Results
Fine mapping of Pain1
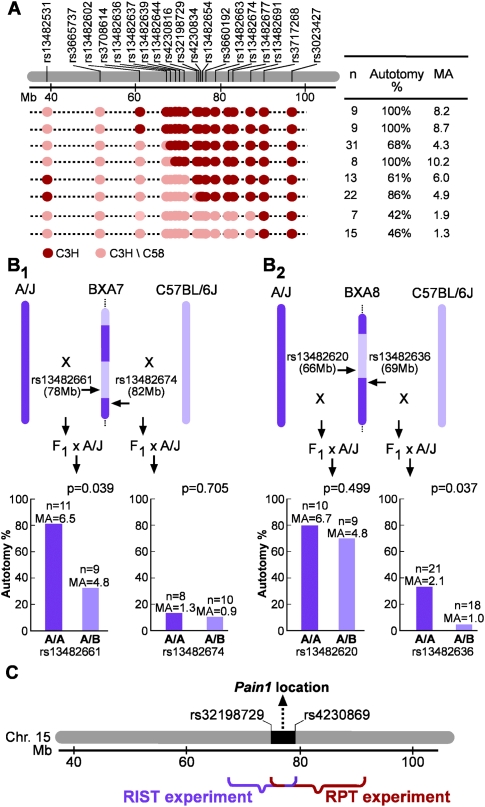
Identification of Cacng2 as the gene underlying Pain1 began with the implementation of two independent fine-mapping strategies: recombinant progeny testing (RPT) and recombinant inbred segregation testing (RIST) (Darvasi 1998). For RPT, 75 male backcross mice (BC; [C58/J × C3H/HeN] × C3H/HeN) were genotyped with 17 single nucleotide polymorphisms (SNPs) to identify recombinants. Eight of these mice were selected for further study because they had a recombination across the Pain1 interval (Fig. 1A). Male #6 was genotyped with two additional SNPs, rs32198729 and rs4230834 (between rs4230816 and rs13482654), to localize more precisely the recombination point. The eight males were mated with C3H/HeN females. Female offspring (seven to 31 per male) that carried their fathers' recombinant haplotype were phenotyped for autotomy. Only females were phenotyped since in this cross the effect of Pain1 is obscured in males by environment-related trait variation that is male-specific (Devor et al. 2007). Since the C3H/HeN strain is recessive for the trait (Devor et al. 2005), results of this phenotyping can establish the genotypic state of Pain1 for each recombinant progenitor male; little versus considerable autotomy in the offspring differentiates the heterozygous from the homozygous state at the QTL.
Figure 1.
Fine mapping of Pain1 using RPT and RIST. (A) The RPT experiment. The region of chromosome 15 containing Pain1 and the genotype of 17 SNPs for the eight recombinant progenitor males (and two additional SNPs for progenitor #6) are shown. Here, and in B, “Autotomy %” is the percentage of animals with an autotomy score >1; n = the number of female mice phenotyped, and MA is the mean autotomy score. (B) The RIST experiment. For each of the four BC populations—(B1) two BXA7 BCs; (B2) two BXA8 BCs—a comparison of A/A (homozygous A/J) versus A/B (heterozygous A/J-C57BL/6J) at the relevant SNP is shown. (C) RIST and RPT map integration.
Figure 1A presents the mean autotomy score and percentage of animals exhibiting autotomy (defined as an autotomy score > 1), for the progeny of each of the recombinant progenitor mice. The top six recombinant males in this figure seem to carry the homozygous genotype (considerable autotomy), whereas the bottom two seem to carry the heterozygous genotype (little autotomy). Mean autotomy scores in offspring of the top six (n = 92) and the bottom two (n = 22) differed significantly (P = 6.98 × 10−7, two-tail t-test), as did the proportion of offspring with and without autotomy (P = 2.2 × 10−5, Fisher's exact test). This locates Pain1 to the interval defined by the genotype of the recombinant progenitor males, between SNPs rs32198729 and rs3717268 (75.27–90.7 Mb on chromosome 15). Since throughout this study similar outcomes were obtained in statistical tests of score means and proportions and since the Fisher's exact test is the more robust, the more appropriate given the sample sizes used, and in some instances the more powerful, hereinafter we report results based on this test only.
In parallel with RPT, RIST experiments were carried out using BXA7 and BXA8, two recombinant inbred (RI) strains known to have recombinations in the region of Pain1. In RIST, the genotypic state of the QTL in each recombinant strain is assessed, localizing the QTL to a defined chromosomal interval (Darvasi 1998). To do this, two independent BC populations were generated for each RI (a total of four independent BC populations). Initially, for each RI strain, we crossed RI individuals to A/J (one of the progenitor strains used to create the RI strains) and also to C57BL/6J (the other progenitor). In both cases, the F1 offspring were backcrossed to the recessive A/J strain. For each of the resulting four BC populations, female mice were phenotyped for autotomy and then genotyped at a single SNP selected to be both polymorphic in the BC mice used and relatively close to the recombination breakpoint in the relevant RI strain (Fig. 1B). As appropriate, a different SNP was used for each of the four BC populations. The effect of the SNPs on autotomy behavior was assessed by comparing autotomy of A/A mice (homozygous for the A/J allele) and A/B mice (heterozygous, with an A/J-C57BL/6J genotype).
The results are summarized in Figure 1B. For BXA7 × C57BL/6J (F1) mice backcrossed to A/J, the rs13482674 SNP did not associate with the pain phenotype (P = 0.705). Therefore, in BXA7 mice, Pain1 appears not to be in a region originating from A/J. Knowing this, association is expected in the reciprocal BC population, BXA7 × A/J (F1) mice backcrossed to A/J. Indeed, a significant association was found in this population (P = 0.039 for rs13482661). Thus, in BXA7 mice, Pain1 is located in a region of chromosome 15 originating from C57BL/6J (Fig. 1B 1). The same logic holds for the BXA8 RIST experiment, with the results locating Pain1 to a chromosome 15 region originating from A/J (Fig. 1B 2). The overlapping localization of Pain1 in BXA7 and in BXA8 mice places the gene in an interval between 67.7 Mb and 79.5 Mb. Combining the information from the RPT and the RIST experiments places Pain1 within a 4.2-Mb interval on chromosome 15, between rs32198729 and rs4230869 (i.e., between 75.27 and 79.5 Mb) (Fig. 1C).
Bioinformatics
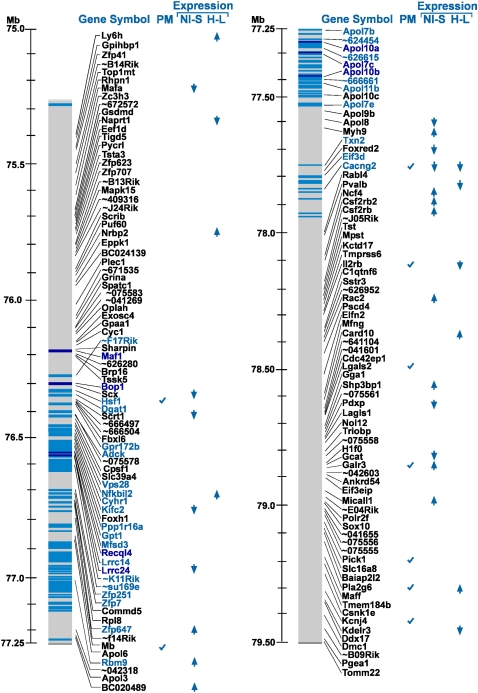
For seven inbred strains (129/J, AKR/J, A/J, BALB/cJ, C57BL/6J, C3H/HeN, and DBA/2J) with an established pain phenotype in the Neuroma model (Devor et al. 2007), sequence information is available for the same or a closely related strain (129S1/SvImJ, AKR/J, A/J, BALB/cByJ, C57BL/6J, C3H/HeJ, and DBA/2J, respectively). In total, 14,606 SNPs and 155 genes have been catalogued in the 4.2-Mb interval containing Pain1 (Mouse Phenome Database, http://phenome.jax.org, build 37.1). Three of the seven strains show high autotomy levels (A/J, C3H/HeN, BALB/cJ), and four show low autotomy (129/J, AKR/J, C57BL/6J, DBA/2J). Within the 4.2-Mb Pain1 interval, 393 SNPs showed complete cosegregation of allele and autotomy phenotype (marked by a blue bar in Fig. 2). That is, for all seven strains, these SNPs exhibited one allele in the three phenotypically high strains and a different allele in the four phenotypically low strains. These 393 cosegregating SNPs were located in 35 of the 155 genes, and all lay in the central region of the 4.2-Mb interval (Fig. 2). This suggests that Pain1 is probably located in the central region of the 4.2-Mb interval, most likely among these 35 genes. A small subset of the 393 SNPs are SNPs that alter protein sequence; eight such SNPs were found in seven genes (dark blue bars and dark blue gene names in Fig. 2). We also carried out a PubMed search on the 155 genes using “〈gene name〉 and pain” as keywords. Only nine genes showed potential relevance to pain based on current knowledge: Mb (Brown et al. 2007), Cacng2 (Tao et al. 2006), Il2rb (Yao et al. 2002), Lgals1 (McGraw et al. 2005), Galr3 (Mitsukawa et al. 2008), Hsf1 (Saban et al. 2007), Pick1 (Thorsen et al. 2010), Pla2g6 (Ma et al. 2010), and Kcnj4 (Dai et al. 2008).
Figure 2.
Integrative analysis of the 155 genes within the Pain1-containing interval. The gene names and their location on mouse chromosome 15 are shown on the left gray bar. (Blue bars) SNPs with a complete cosegregation among the seven mouse strains (see text); (dark blue bars) coding nonsynonymous SNPs; (light blue) other SNPs. Predicted/annotated genes and hypothetical protein names (long names) are given as “∼xxxxxx” (where xxxxxx is the six last characters of the gene name). For the full name, see Supplemental Table S3. The column “PM” indicates genes found to be related to pain physiology in a PubMed search. The columns “NI-S” (NI, nerve-injured; S, sham) and “H-L” (H, high autotomy; L, low autotomy) highlight genes with significant expression-fold change (the direction of the arrowheads indicates up-regulation or down-regulation). The “NI-S” column presents the results for the nerve-injured versus sham comparison, and the “H-L” column presents the results for the high-autotomy versus the low-autotomy strains.
Gene expression analysis and data integration
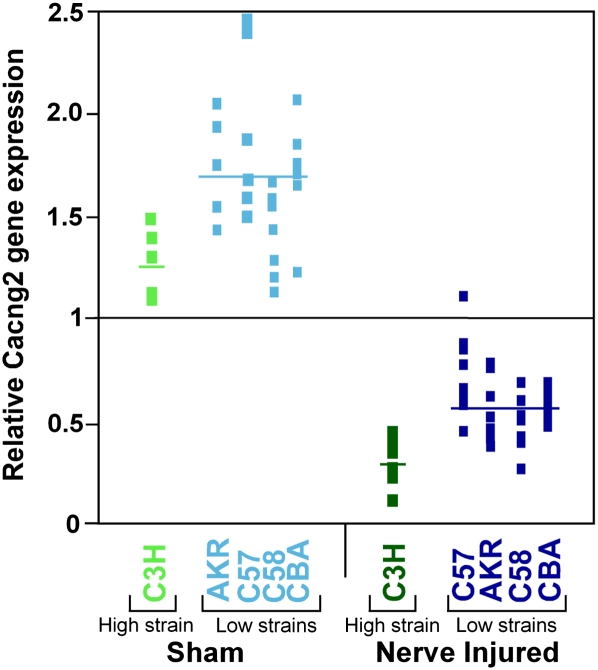
Gene expression analysis provided an additional approach for screening contenders among the 155 candidates for Pain1. Whole-genome expression analyses were obtained for the L5 dorsal root ganglion (L5DRG) of 73 individual mice of five different mouse strains: AKR/J, C3H/HeJ, CBA/J, C57BL/6J, and C58/J. In 41 of the mice the L5 spinal nerve was transected 3 d previously (“nerve-injured,” seven to nine mice per strain), and 32 underwent sham surgery (five to eight mice per strain). The expression microarray used included 244 transcripts in the 4.2-Mb interval containing Pain1, corresponding to 128 out of the 155 genes present in this interval (the gene expression raw data are presented in Supplemental Table S2). Among these 128 genes, 108 (84%) had at least one transcript that was expressed above noise level. Using these array data, we first compared gene expression levels of nerve-injured versus sham-operated mice. The average expression in all nerve-injured mice divided by the average in all sham operated mice represents the fold change in gene expression level following nerve injury for each given transcript. We also compared gene expression following nerve injury in C3H/HeJ mice (a high autotomy strain) against the average expression of AKR/J, CBA/J, C57BL/6J, and C58/J mice (all low autotomy strains). To obtain fold difference, the averages of the two groups were divided. In both analyses, one transcript per gene (the most significant one) is presented in Supplemental Table S3. For both the nerve-injured versus sham and the high versus low autotomy analyses, in Figure 2 we highlighted (with arrowheads indicating the direction of the change) genes in which expression differed by >1.5-fold (or <1/1.5) and had an associated nominal statistical significance of P < 0.001 (corresponding to P ∼ 0.05 after correcting for multiple comparisons). When comparing nerve-injured versus sham operated mice, the expression of 21 genes was highlighted as being significantly affected, and 10 genes were highlighted in the comparison of the phenotypically high versus low mouse strains. Only one gene, Cacng2, was highlighted in both analyses. For Cacng2 there was a 3.78-fold down-regulation following nerve injury (P = 3 × 10−31) and a 2.00-fold lower expression level in C3H/HeJ mice compared to the mean of the four low autotomy strains (P = 3 × 10−4). Standardized Cacng2 gene expression levels for all of the mice studied are presented in Figure 3.
Figure 3.
Cacng2 gene expression. For each mouse, the relative gene expression value at Cacng2 is shown (transcript 1440210_at/#BB342913). This is the expression value in a given mouse divided by the mean expression value across all mice. Results for the high autotomy strain C3H are shown for sham operated mice (light green) and nerve-injured mice (dark green). Results for the low autotomy strains C58, C57, CBA, and AKR are shown for sham-operated mice (light blue) and nerve-injured mice (dark blue). The Cacng2 expression average for each group is represented by a horizontal line. For reference, the horizontal line drawn at 1 represents the mean Cacng2 expression level of all mice.
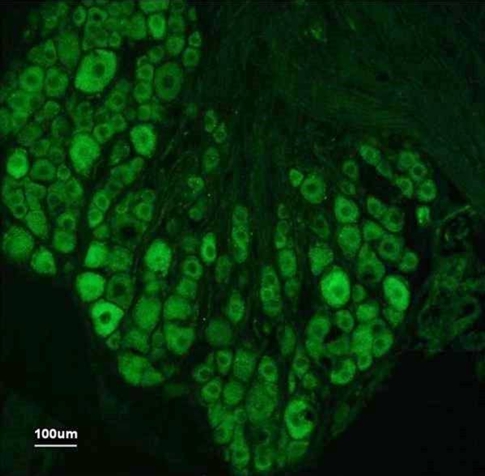
Considering all the results highlighted in Figure 2, Cacng2 is by far the most likely candidate for the Pain1 QTL, as it is the only gene within the fine-mapped interval to meet all four of our screening criteria (Fig. 2): (1) complete cosegregation of SNPs between genotype and phenotype; (2) functional relevance to pain by PubMed search; (3) significant regulation following nerve injury; and (4) differential expression in high versus low autotomy strains. No other gene met any three of the criteria, and only nine met two criteria: Galr3 (criteria 2 and 3), Hsf1 (criteria 1 and 2), Kifc2 (criteria 1 and 3), Il2rb (criteria 2 and 4), Lrrc24 (criteria 1 and 3), Nfkbil2 (criteria 1 and 4), Pla2g6 (criteria 2 and 4), Rbm9 (criteria 1 and 3), and Zfp647 (criteria 1 and 3). Immunolabeling on tissue sections confirmed that Cacng2 is, indeed, prominently expressed in the L5DRG and that it is localized to neurons, rather than glia or other cells. Neurons of all sizes were labeled, but a marked preference for large diameter somata was noted (Fig. 4). In situ hybridization data for Cacng2 in the mouse are available from the Allen Institute brain library (http://mouse.brain-map.org/welcome.do). Cacng2 mRNA is quite widely distributed in the brain, including in pain signaling pathways. A relatively intense signal is present in the DRG, all laminae of the spinal cord dorsal horn including the substantia gelatinosa, the rostroventromedial medulla (RVM), the parabrachial area, the periaqueductal gray (PAG), the ventrobasal thalamus, the amygdaloid complex, and much of the cerebral cortex.
Figure 4.
Cacng2 immunofluorescence in the mouse DRG. Fluorescence micrograph from a 12-μm cryosection of a naïve CBA/J mouse L5DRG incubated with antibodies against Stargazin (Cacng2 gene product). Cytoplasmic immunolabeling of varying intensities is seen in neurons of all sizes, but most prominently in neurons of larger sizes.
No SNP was found that alters protein sequence in Cacng2 and that also segregates between low and high autotomy strains. We suspect, therefore, that the relevant polymorphism is located in the noncoding region of the gene and probably affects gene expression rather than the structure of the gene product.
Cacng2 stargazer mice
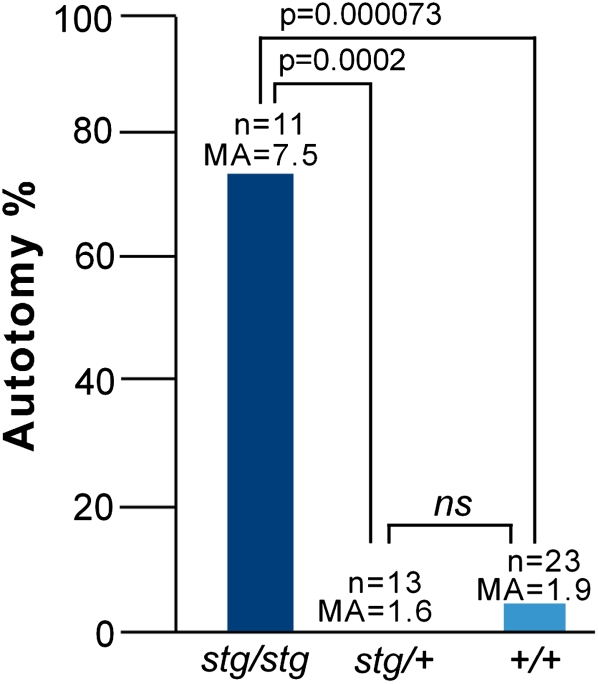
To directly test if Cacng2 has a functional role as the gene affecting pain susceptibility, we exploited the naturally occurring hypomorphic mutant “stargazer” (Letts et al. 1998). Heterozygous stargazer mice (stg/+) were intercrossed to produce an F2 population. An initial population of 70 F2 mice was phenotyped for pain behavior. Both sexes were tested since unlike RPT and RIST we had no prior information on the effect of sex on phenotype in this mutant. Overall, autotomy levels in this F2 population were quite low. Mean autotomy scores for stg/stg (n = 8), stg/+ (n = 42), and +/+ (n = 20) were 2.8, 0.8, and 2.0, respectively (on the 0–11 scale). The stg/stg showed the highest level of autotomy. However, the sample size of stg/stg mice was not large enough to achieve a statistically significant difference compared to the other genotypes. Nonetheless, the modest difference between stg/+ and +/+ was statistically significant (P = 0.0035, Fisher's exact test).
With such overall low autotomy, we suspected that genetic effects present might be difficult to detect using this experimental paradigm. With this in mind, we ran a second experiment in which background autotomy levels were enhanced. Prior studies have shown that the tendency to autotomize is enhanced by social interactions in a group caging paradigm (Devor and Raber 1990; Devor et al. 2007). Therefore, in a second, independent F2 population (47 mice), phenotyping was done in cages containing one or two F2 animals and one same-sex mouse of a high autotomy strain (C3H/Hen) that was nerve-injured and performed autotomy. As anticipated, group caging enhanced the overall incidence of autotomy, and this uncovered a marked effect of the genotype. Mean autotomy scores for stg/stg (n = 11), stg/+ (n = 13), and +/+ (n = 23) were now 7.5, 1.6, and 1.9, respectively. The difference between stg/stg and both stg/+ and +/+ was highly significant (P = 0.00022 and P = 0.000073) (Fig. 5). Since the autotomy background was artificially elevated, we applied a conservative definition of autotomy in this experiment, an autotomy score ≥ 9. This criterion for “high autotomy” has also been used in previous studies (Devor et al. 2005; Nissenbaum et al. 2008). For completeness, we also performed a combined analysis using all phenotyped mice from both F2 populations, with their respective autotomy scores as the dependent variable in the model. For this analysis we did not employ any autotomy threshold, but used caging (i.e., the first vs. the second F2 population), sex, and genotype at Cacng2 as independent variables. Multivariate ANOVA revealed that genotype (at Cacng2) was the most significant variable accounting for the autotomy score (P = 2.8 × 10−8). Group caging as a single factor also significantly increased autotomy (P = 4.1 × 10−4). Sex on its own did not have a significant effect (P = 0.105), but the sex–genotype interaction was highly significant (P = 3.7 × 10−8). Male stargazer mice had higher scores than females, while the opposite was true for wild-type mice. The genotype–caging interaction was also significant (P = 10−3) due to elevated autotomy in group-caged stargazer mice.
Figure 5.
Cacng2 stargazer analysis. Autotomy behavior, comparing alternative stargazer genotypes; stargazer hypomorph (stg/stg), heterozygote (stg/+), and wild-type (+/+). These F2 mice were phenotyped in the presence of a single C3H/HeN mouse, a device that enhances the tendency to autotomize. (Autotomy %) The percentage of animals with an autotomy score ≥ 9. n, the number of mice phenotyped in each group; MA, mean autotomy score; P, statistical significance for the indicated comparisons; ns, not significant.
A second functional test applied to Cacng2 stargazer mice was electrophysiological analysis of spontaneous ectopic discharge generated in afferent neurons axotomized by nerve injury. A considerable body of evidence in animal models and in man establishes spontaneous afferent discharge as an important substrate of spontaneous dysesthesias and pain in a variety of neuropathic pain conditions (Nordin et al. 1984; Ochoa et al. 2005; Devor 2006a, 2007). Teased fiber recordings were made from dorsal roots L4 and L5 of animals in which the corresponding peripheral (spinal) nerves had been cut 2–3 d previously. We compared the incidence of spontaneous firing (measured as the percent of afferent axons that exhibited spontaneous firing out of the total number of afferent axons sampled) in seven homozygous stargazer animals (stg/stg, approximately 1360 afferent axons sampled) and eight controls (six heterozygotes [stg/+] and two wild-type mice [+/+], approximately 1870 afferent axons sampled). Detailed results are presented in Supplemental Table S4. The incidence of spontaneous activity was almost double in the stg/stg mice compared to the stg/+ mice, 12.2% versus 6.9%, respectively. The average of the two +/+ mice was 8.4%. The difference between stg/stg and stg/+ was statistically significant (Mann-Whitney U-test, P = 0.05). Since stg/+ and +/+ mice are phenotypically similar (low), we also analyzed them jointly as controls compared to stg/stg resulting in P = 0.02, or P = 0.01 for a one-tailed test, justified by our a priori prediction. These statistical tests were performed in a conservative manner, assigning a percentage value to each mouse (percent of axons that exhibited spontaneous firing out of all the axons sampled in that mouse) and then using the number of mice as the sample size of our analysis. A more relaxed analysis would be a logistic regression assigning 1 or 0 to each afferent axon exhibiting some or no spontaneous firing, respectively. Using this approach, the genotype effect on spontaneous ectopic firing had a statistical significance of P = 3.4 × 10−7. Based on spike waveform characteristics (Liu et al. 2000), the bulk of the ectopia was carried in afferent A-fibers. Firing patterns and rates of afferents that discharged spontaneously did not differ among the genotypes (data not shown).
It is important to note that the hypomorphic Cacng2 mutation occurred in an A/J background. Following the stargazer breeding scheme, the wild-type chromosome has C57BL/6J (B6) alleles in that region. Consequently, one cannot rule out the possibility that the phenotypic results for the stargazer F2 population are (partially or entirely) due to background allele effect.
Human CACNG2 and chronic pain
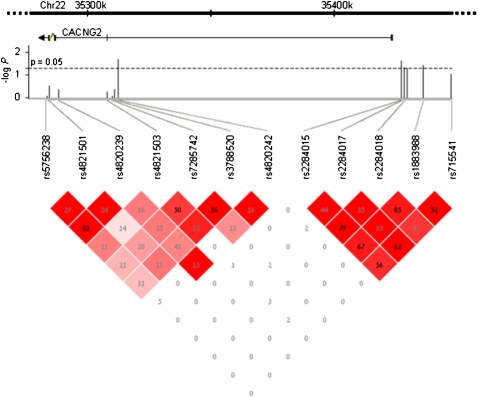
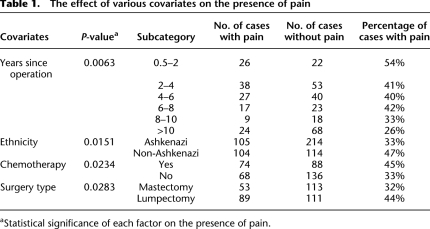
Having identified Cacng2 as an important gene for the pain phenotype in mice, we analyzed the homologous human gene CACNG2 in a human neuropathic pain cohort. The cohort comprised 549 breast cancer patients who had undergone unilateral surgical removal (or partial removal) of a breast. Some women develop chronic pain following mastectomy (Rothemund et al. 2004; Bjorkman et al. 2008). Of the 549 women, 215 reported chronic pain and 334 did not. Based on HapMap information (http://www.hapmap.org), we selected 12 SNPs covering CACNG2 exons and regulatory regions (Fig. 6). All genotype and phenotype raw data are presented in Supplemental Table S5. We first performed a single 12-SNP global haplotype test for association with the pain phenotype, which was found to be significant (P = 0.0415). This and subsequent analyses used ethnicity, number of years since operation, chemotherapeutic treatment, and surgery type (mastectomy or lumpectomy) as covariates. Descriptive statistics and the statistical significance of these variables are presented in Table 1. Briefly, less pain was reported in (1) patients with shorter times since operation; (2) patients not treated with chemotherapy; (3) patients who underwent complete mastectomy; and (4) patients of Ashkenazi Jewish ancestry. Of the 12 SNPs (see haplotype structure in Fig. 6), five showed significant association with chronic pain individually. One SNP is located near exon 3 (rs4820242, P = 0.02), and the four others are near exon 4 in an independent haplotype block (rs2284015, P = 0.02; rs2284017, P = 0.04; rs2284018, P = 0.05; rs1883988, P = 0.03), possibly suggesting the presence of two independent effects. Considering multiple testing, we ran a permutation test by reshuffling the phenotypes 10,000 times. This yielded a probability of P = 0.018 to obtain five or more SNPs each with P < 0.05 on independent testing of the 12 SNPs, which is the result we obtained. The single SNP effects as well as the 12-SNP haplotype effect can all be explained by a core of three SNPs: rs4820242, rs2284015, and rs2284017. The homozygous A-C-C haplotype of these SNPs significantly increased susceptibility to pain (odds ratio [OR] = 1.65; P = 0.001). Of the 549 women, 319 were of Ashkenazi Jewish (AJ) ancestry. As noted, we accounted for ethnicity in our analysis. Nonetheless, we also performed the analysis on the ethnically homogeneous subgroup of AJ women alone and found that the A-C-C haplotype remained statistically significant (OR = 1.51, P = 0.031). The P-values reported here are all significant, but only marginally so. Nevertheless, the results are meaningful in light of the fact that only one gene candidate was tested, and in the context of the robust corresponding findings obtained in the mice. Future analysis of other independent human populations will definitely be of interest.
Figure 6.
Haplotype analysis of human CACNG2. The 12 SNPs used for genotyping human CACNG2 are listed, along with their map locations relative to the gene on human chromosome 22 (exons and gene orientation are indicated). The statistical significance of allele association with the pain phenotype (in −log P units) is plotted for each SNP. The dashed horizontal line is drawn at P = 0.05 (−log P = 1.3). The bottom part of the figure presents linkage disequilibrium (LD) patterns for the 12 SNPs. In each square, the LD r 2 value of the two SNPs that the square intersects is shown. The intensity of the color represents D′ (the higher D′, the darker the color of the square).
Table 1.
The effect of various covariates on the presence of pain
aStatistical significance of each factor on the presence of pain.
Discussion
In this study we applied an integrative approach to identify the gene underlying Pain1. Advanced fine-mapping strategies allowed us to locate the relevant QTL to a small interval containing only 155 genes. These candidate genes were selected in a hypothesis-free manner on the basis of genetic linkage only, and therefore benefitted from the unbiased discovery potential of the genome-wide association strategy. We did not depend on prior knowledge of pain physiology to select them. We then applied bioinformatics and whole-genome expression analysis to prioritize the candidate genes. Integrating the various sources of information yielded one candidate that stood out among the rest, the Cacng2 gene. Obviously, the screening criteria can serve only to prioritize candidate genes. Follow-up functional tests are essential. For example, a PubMed “pain hit” strengthens the candidacy of a gene, but genes that have not (yet) been associated with pain can by no means be excluded. Doubtless our screening criteria might have missed the gene underlying Pain1 while meeting all of our criteria is no guarantee of positive identification. Nevertheless, the prioritization process guided us through the daunting task of identifying the gene. Applying it yielded a single outstanding candidate that was verified functionally. The follow-up analysis using stargazer mice provided behavioral and electrophysiological support for the relevance of Cacng2 to pain. Finally, to complete the broad base of evidence, CACNG2 SNPs were found to associate with a relevant neuropathic pain phenotype in humans, chronic pain following complete or partial breast amputation in cancer patients.
Identification of CACNG2 as a pain susceptibility gene is significant for several reasons. First, it provides additional support for the notion that susceptibility to chronic pain following nerve injury, including diagnoses that feature spontaneous pain such as anesthesia dolorosa and phantom pain, is subject to genetic control. This conclusion can be of immediate social benefit. Patients who report more pain than might be expected given the extent of their injuries are frequently stigmatized, with the suspicion that they are exaggerating their symptoms in the hopes of obtaining secondary gains (Fishbain et al. 2002; Etherton et al. 2006). Although knowledge of the existence of genetic factors controlling pain susceptibility does not exclude the possibility of fraud and malingering, it ought to reduce unwarranted stigmatization of patients with severe pain due to no fault of their own. Second, although abundant functional evidence indicates that autotomy behavior in the Neuroma model indeed reflects spontaneous neuropathic pain (Devor 2007; Minert et al. 2007), the documentation that a pain susceptibility gene discovered using this model also affects a corresponding pain phenotype in humans lends further support to the model's validity. Hence, it supports the potential usefulness of this and related animal models for the study of pain and for the screening of potential therapeutic agents. Finally, the mechanism(s) whereby CACNG2 polymorphisms affect spontaneous pain can be expected to inform our understanding of pain physiology. Several pain susceptibility genes have been identified previously based on family linkage studies or candidate gene approaches, including one remarkable example in which a genetic polymorphism directly related to mechanisms that regulate the electrical excitability of DRG neurons (Estacion et al. 2009).
To date, CACNG2 has mostly been studied within the context of epilepsy (Moss et al. 2003; Payne et al. 2006; Ryu et al. 2008). In this regard, the potential relationship between mechanisms of epilepsy and neuropathic pain is noteworthy (Devor 2006b). An association to formalin-induced pain in a cutaneous inflammation model has also been noted (Tao et al. 2006). Considering the known information about CACNG2 and related genes, the discovery of its role in neuropathic pain is striking. For example, mutations in another Ca2+ channel subunit, CACNA1A, are a known cause of chronic pain in familial hemiplegic migraine (de Vries et al. 2009). Likewise, the neuropathic pain-suppressing drugs gabapentin and pregabalin act by binding to yet another Ca2+ channel subunit, CACNA2D (a2δ). This auxiliary protein, like stargazin, is thought to play a role in ion channel trafficking (Taylor 2009).
CACNG2 itself encodes a transmembrane protein that is a putative subunit of neuronal voltage-sensitive Ca2+ channels Cav2.1 and Cav2.2 (Kang et al. 2001; Sandoval et al. 2007). It is also known to be a TARP, intimately involved in the trafficking of AMPA-type glutamate receptors to the neuronal membrane at the cell soma and synaptic cleft (Bats et al. 2007; Cokic and Stein 2008; Milstein and Nicoll 2009), and to modulate the ion channel kinetics of AMPA receptors (Priel et al. 2005). Both functions, ionic conductance modulation and membrane trafficking, are relevant to pain. The contribution of Ca2+ inflow to electrical excitability in afferent neurons is complex. Ca2+ conductances play a small excitatory role in spike generation in primary afferents, but they also contribute to membrane hyperpolarization (hence pain suppression) by activation of the Ca2+-activated K+ conductance (gK+ Ca2+). The balance between these two factors appears to favor suppression as our electrophysiological recordings revealed that mice with the Cacng2 mutation, and hence depleted stargazin, develop enhanced levels of spontaneous afferent discharge in their injured nerves, nearly double the incidence of control mice. It is likely that this enhanced discharge directly drives spontaneous pain sensation in these animals and enhanced spontaneous pain behavior in the form of autotomy (Devor 2006a).
AMPA receptors on postsynaptic neurons in the spinal cord are important for excitatory neurotransmission. Their activation by intrathecal drug administration provokes pain, and their blockade is analgesic (Nishiyama 2000). Reduced stargazin might therefore be expected to suppress pain. However, reduced trafficking of AMPA receptors to the presynaptic terminals of primary afferents in stg/stg mice is expected: (1) to reduce glutamate-induced presynaptic inhibition and (2) to increase the potential across the terminal's membrane, hence increasing spike-induced neurotransmitter release (Engelman and MacDermott 2004). Both effects are expected to enhance pain.
Further research is needed to define the specific role played by CACNG2 in neuropathic pain and, in particular, whether both enhanced ectopic firing of afferents due to altered membrane excitability and altered synaptic function at their central terminal play equal roles in promoting pain. It is also important to determine why modifications of different Ca2+ channel subunits appear to associate with different chronic pain syndromes. Answers to these questions are expected to advance our understanding of pain processing and may ultimately pay dividends in terms of the development of better therapeutic options.
Methods
Animals
All progenitor mice were purchased from The Jackson Laboratory (Bar Harbor, ME) except for the C3H/HeN mice, which were purchased from Harlan Laboratories Ltd. Mice were handled in a standard manner as previously described (Devor et al. 2007). All procedures were carried out with the approval of the Institutional Animal Care and Use Committees of the Institute for Life Sciences of the Hebrew University of Jerusalem, of the Karolinska Institutet, or in accordance with the German Animal Protection Law as overseen by the local Ethics Committee for Research on Laboratory Animals at Sanofi-Aventis Deutschland GmbH.
Recombinant progeny test (RPT)
For the RPT experiment, 75 [C3H/HeN × C58/J] × C3H/HeN BC males were genotyped with 17 SNPs, and one of the recombinant males was genotyped with two additional SNPs as described in the Results section. All SNPs are listed in Supplemental Table S1. Eight male mice were selected in which a recombination event was detected within the ∼25-cM interval of interest in chromosome 15 (Devor et al. 2005). Each of these male mice was mated with three C3H/HeN inbred females. Female offspring of these matings were screened with the same 17 SNPs to assess whether they received the recombinant chromosome (within the 25-cM interval) from their fathers. A total of 114 females that received the recombinant chromosome were subsequently phenotyped for autotomy.
Recombinant inbred segregation test (RIST)
For the RIST experiment, BXA7 and BXA8 recombinant inbred (RI) strains were each separately crossed to the A/J or C57BL/6J parental inbred strains. Each cross consisted of one male from the RI strain and two females from one of the standard inbred strains. F1 males were then mated (backcrossed, BC) with two A/J females creating four separate BC populations, according to the RIST design (Darvasi 1998). Specifically, for each RI strain (BXA7 and BXA8), two BC populations were created, one using F1 individuals generated by crossing the RI with A/J and the other using F1 individuals generated by crossing the RI with C57BL/6J. Only female BC offspring, totaling 97 mice, were phenotyped for autotomy.
Gene expression experiment
For the gene expression analysis, adult males of five inbred strains were used: AKR/J, CBA/J, C3H/HeJ, C57BL/6J, and C58/J. Mice obtained from the Jackson Laboratory were used directly without any further in-house breeding (Persson et al. 2009).
Stargazer mice
In this experiment, B6C3Fea/a-Cacng2stg/+ (stg/+) heterozygous mice were purchased from the Jackson Laboratory. The initial F2 population was created by mating one male with two females. They produced 10 litters and a total of 70 F2 offspring. The F2 offspring were classified by their Cacng2 genotype. The mutant homozygote stg/stg (“Stargazer”) is clearly recognizable at 14 d of age by its smaller body size and slightly ataxic gait. The other two possible genotypes, stg/+ and +/+ were identified by DNA analysis as previously described (Letts et al. 1998). The initial F2 population consisted of four female and four male stg/stg, 23 female and 19 male stg/+, and eight female and 12 male +/+ mice. All mice were phenotyped for autotomy. The second F2 population was produced from new stg/+ mice purchased from the Jackson Laboratory. As with the initial F2 population, each male was mated with two females. Seventy-one F2 offspring, having reached adulthood, were operated on but only 58 (13 stg/stg, 16 stg/+, and 29 +/+) completed phenotyping (13 died, primarily due to fights with C3H/HeN mice present in the cage during phenotyping). The C3H/HeN cagemate was also operated on. Cages in which the C3H/HeN mouse did not exhibit high autotomy were excluded from the experiment. Thus, the phenotype in this second experiment was based on observations on 47 F2 mice (five female and six male stg/stg, eight female and five male stg/+, and nine female and 14 male +/+).
Human cohort
A cohort of 549 Jewish breast cancer patients who had undergone breast surgery was recruited to the study. Ethical committee approval was obtained from the participating institutions, and all subjects signed an informed consent form. In this study, inclusion criteria were as follows: (1) All women were Israeli of Jewish origin, having both parents of the same ethnic origin, either Ashkenazi or non-Ashkenazi. (2) The age breast cancer was diagnosed was ≥18 yr. (3) Diagnosis and treatment were done in an Israeli hospital, to reduce the variability in diagnostic criteria and surgical and adjuvant anti-cancer treatments (i.e., radio-, chemo- or hormonal therapies). (4) Breast cancer was unilateral, malignancy stage was <3, and there were no metastases or recurrences. (5) They were >6 mo after the surgery, which included unilateral mastectomy (i.e., removal of the whole breast but not including the underlying chest muscles) or breast-conserving surgery that excised the tumor (“lumpectomy”). (6) Both surgical approaches were accompanied by axillary lymph node dissection on the side of the breast surgery. (7) At least 3 mo had passed since the last adjuvant treatment. The participants were recruited to the study from 1998 to 2002, in two major hospitals: Sheba Medical Center (Ramat Gan) and Hadassah Medical Center (Jerusalem). The participants ranged in age from 22 to 80 yr (average = 52.9 yr), 60.4% were Ashkenazi, and 39.6% were non-Ashkenazi. They were recruited to the study on average 6.6 yr (6 mo minimum) after the surgery; 54.3% had mastectomy, 45.7% lumpectomy, and 100% had axillary lymph node dissection; 51.4% had chemotherapy, 38.8% radiotherapy, and 32.1% hormonal therapy.
Pain phenotyping
Surgery
Mice were 8–19 wk of age at the time of surgery, weighing 25–35 g (males) and 20–30 g (females). For induction of autotomy behavior, mice were anesthetized (pentobarbital 50 mg/kg i.p. or chloral hydrate 350 mg/kg i.p.), and unilateral total hindpaw denervation was produced by tightly ligating and severing distally the sciatic nerve at mid-thigh and the saphenous nerve at mid-calf (Seltzer et al. 2001). Mice used for expression analysis and electrophysiological recording were operated on similarly, but with nerves transected closer to the DRG as we previously described (Persson et al. 2009). Sham operated control mice underwent the identical surgical exposures, but nerves were not ligated or transected.
The mouse Neuroma model (autotomy scoring)
Autotomy phenotyping began with surgical denervation of one hindpaw as described above. Over the ensuing weeks, some operated animals began to lick, scratch, and bite their numb paw. This behavior, “autotomy,” was scored on a 0–11 scale at 7-d intervals for 35 d post-surgery, as previously described (Wall et al. 1979). Briefly, one point was given for loss of one or more toenails, and an additional point was given for injury that extended onto the proximal or distal half of each phalanx for a maximum total score of 11. In the RPT experiment, adult females were phenotyped in caging groups of two or three, where all mice in a cage were offspring of the same father. In the RIST experiment, females were phenotyped, one per cage. In the stargazer experiments, both females and males were phenotyped, in cages containing one or two mice of the same sex and the same Cacng2 genotype. In the second F2 stargazer experiment, each cage also contained an operated C3H/HeN mouse of the same sex as the other mice in that cage. Only cages where the C3H/HeN mouse exhibited autotomy behavior (26 out of 33) were included in the experiment.
Pain assessment in breast cancer patients
To assess pain in the human cohort, subjects were interviewed by a study nurse. We divided the patients into two discrete groups; those with pain and those without. Assigning a patient to a specific group was based, in this study, on answers to a single question from the questionnaire: “Have you experienced pain related to your breast surgery during the course of the last year?” (question phrased in Hebrew). Note that patients were on average 6.6 yr after surgery. Those who answered “no” were assigned a “no pain” phenotype, and those who answered “yes” were assigned a “pain” phenotype. Obviously, our pain definition includes some noise, and it may bulk together different kinds of pain (e.g., scar pain, phantom breast pain). This broad pain definition, however, allows efficient sample collection and statistical analysis. The downside is a decrease in statistical power, but this should not increase the false-positive rates since the noise is not expected to carry a genetic bias. An assessment of the specific pain phenotype is of considerable interest as it might indicate genetic association for one type of pain and not another. However, this level of analysis will probably require a much larger patient cohort, and it is beyond the scope of the present study. Additional data were also collected at the time of the patient interview and from the patients' medical records. These included age at operation, years since operation, treatment type, and surgery type. A detailed description of all descriptive parameters is presented in Table 1.
DNA extraction, SNP selection, and genotyping
Mice
Genomic DNA was extracted from earpieces using the DirectPCR Lysis Reagent (Ear) kit (Viagen Biotech, Inc.). Polymorphic SNPs were selected from the Wellcome Trust Center mouse strain database (http://www.well.ox.ac.uk/mouse/INBREDS/). SNP genotyping was carried out by restriction fragment length polymorphism (RFLP) analysis (Kwok 2001). Primers were designed using the PRIMER3 online program (Rozen and Skaletsky 2000) and purchased from IDT Inc. Restriction enzymes were purchased from Fermentas UAB and from New England Biolabs, Inc. SNPs used in the RPT and RIST experiments and their details including primers and restriction enzymes used are presented in Supplemental Table S1. Mouse genomic DNA (4 ng/μL) was PCR-amplified in an M/J Research PTC-200 instrument (10 min at 95°C, 20 sec at 94°C, 30 sec at 55°C, and 30 sec at 72°C, for a total of 45 cycles). The PCR products were subsequently digested according to the vendor's protocols. The digested PCR products were separated on a 3% low EEO agarose (Hispanagar). Genotypes were assessed manually.
Humans
Genomic DNA was extracted from a blood sample (30 mL) using the QIAampTM 96 DNA Blood Kit (QIAGEN). All DNA samples were checked for quality on agarose gel electrophoresis, and intact samples were selected for genotyping. DNA concentration was determined with the Nanodrop spectrophotometer and diluted to a final concentration of 50 ng/μL. Twelve SNPs were manually selected to cover exons and regulatory regions of CACNG2 using HapMap information (http://www.hapmap.org). The selected SNPs were genotyped in one reaction using the iPLEX assay (Sequenome). Briefly, DNA was amplified using the following PCR conditions: initial denaturation for 15 min at 94°C, followed by 45 cycles of 20 sec at 94°C, 30 sec at 56°C, and 1 min at 72°C, and a final step of 3 min at 72°C. To decrease contamination, unincorporated dNTPs were dephosphorylated with SAP buffer and enzyme for 40 min at 37°C, 5 min at 85°C, and kept at 4°C. Primer extension followed by using iPLEX buffer plus terminator, enzyme, and a primer mix (adjusted for high and low mass primer concentrations), then cycled using a two-step 200 short cycle program that used two cycling loops, one of five cycles (annealing for 5 sec at 52°C and extension for 5 sec at 80°C) that sits inside a loop of 40 cycles (denaturation for 5 sec at 94°C followed by the five-cycle annealing and extension steps). A final extension was done for 3 min at 72°C, and then the sample was cooled to 4°C. The MassEXTENDED iPLEX reaction products (desalted with 6 mg of resin) were dispensed using a nanodispenser onto a 384-element SpectoCHIP bioarray, and processed with TYPER software.
Expression analysis
RNA extraction
Three days after the surgery mice were killed by CO2 inhalation, and the lumbar spine was divided longitudinally. L5 DRGs were removed into PBS, frozen on dry ice, and stored at −70°C. After DRGs from all groups were collected, total RNA was isolated from the individual ganglia using the PicoPure RNA Isolation Kit following the manufacturer's protocol (Arcturus Bioscience Inc.). The average RNA yield was ∼500 ng/DRG. The high quality of the RNA extracted was verified using the Agilent RNA 6000 Pico Kit (Agilent Technologies).
Microarray expression profiling
First-strand cDNA synthesis was performed using 500 ng of total RNA with a 100pM T7-(dT)24 oligomer (GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGG-dT24) according to Baugh et al. (2001) and SuperScript II reverse transcriptase following the manufacturer's instructions. Double-stranded cDNA was synthesized and then extracted using phenol-chloroform followed by an ethanol precipitation step. An in vitro transcription reaction was performed with the double-stranded cDNA sample using the BioArray High Yield RNA Transcription Labeling kit (Enzo Life Sciences) according to the manufacturer's instructions. This linearly amplifies mRNA quantity. Transcription reactions were incubated for 16 h at 37°C. cRNA was purified using the RNeasy Mini kit protocol for RNA cleanup (QIAGEN GmbH) and quantified spectrophotometrically. The biotin-labeled cRNA was fragmented using an RNA fragmentation buffer (200 mM tris-acetate, 500 mM KOAc, 150 mM MgOAc at pH 8.1). Hybridization and staining of mouse MG430_2 GeneChipsTM (Affymetrix Inc.) was performed according to the manufacturer's instructions. The microarrays were scanned using a GeneChip 3000 Scanner and analyzed using Resolver v5.1 expression data analysis software (Rosetta Biosoftware). Negative expression levels estimated by the software were truncated to zero. We also set a threshold whereby transcripts with 10 or more mice exhibiting zero expression were excluded. Expression values were further standardized by dividing the value of each transcript by the mean expression of that transcript across all mice. Subsequently, within each group of mice of a given strain subjected to the same surgical procedure, observations that were above or below two standard deviations from the within-group mean were excluded from the analyses (∼1.5% of all mice).
Immunohistochemistry
Male CBA/J mice obtained from the Jackson Laboratory were deeply anaesthetized with chloral hydrate (350 mg/kg i.p.) and perfused transcardially first with 20 mL of Tyrode's solution, and then with 40 mL of 4% formaldehyde in a 0.1 M phosphate buffer containing 0.4% picric acid (4°C). L5 DRGs were quickly dissected out and post-fixed in the perfusion fixative for 90 min, immersed in 0.1 M buffer with 30% sucrose overnight (4°C), and rapidly frozen. The specimens were then cryosectioned at 12 μm and thaw-mounted onto Super Frost/Plus slides (Menzel GmbH). Sections were incubated for 24 h in a humid atmosphere at 4°C with rabbit polyclonal antibodies against stargazin (Chemicon, cat. # AB5474; dilution 1:400). After rinsing with 0.01 M PBS, sections were incubated with Cy2-conjugated donkey anti-rabbit antiserum (diluted 1:250; Jackson ImmunoResearch) in a humid atmosphere for 1 h at room temperature. All reagents were diluted in 0.01 M PBS containing 0.3% Triton X-100, 5% bovine serum albumin (BSA), 3% normal donkey serum, and 0.1% sodium azide. Control sections were incubated as above but without a primary antibody. This gave no immunostaining. Subsequently the sections were rinsed in 0.01 M PBS, mounted in a mixture of glycerol and 0.01 M PBS (3:1), coverslipped, and examined in a Nikon E600 fluorescence microscope equipped with appropriate filter combinations. Photographs were taken with a Nikon DXM 1200 digital camera and processed using Adobe Photoshop CS4 11.0.
Electrophysiology
Ectopic spontaneous impulse discharges generated in nerve-end neuromas and the associated DRGs were assessed 2–3 d following L4 or L5 spinal nerve transection. This is the time of peak activity from previous studies. Mice were anesthetized with chloral hydrate (400 mg/kg, followed by ∼60 mg/kg/h, i.p.), and the spinal canal was opened by laminectomy to expose the lower lumbar dorsal roots. The teased fiber recording method was used to quantitatively evaluate spontaneous ectopic discharge. Briefly, small bundles of axons (microfilaments) were teased from the L4 or L5 dorsal root, cut centrally but in continuity with the DRG and neuroma peripherally, and adhered to an Ag/AgCl recording electrode referenced to a nearby indifferent electrode. Each microfilament was observed passively for at least 1 min, and a spike-triggered delay-line and a multispike detector system (MSD 308; Alpha-Omega Engineering) were used to determine the presence of distinct, all-or-none axons in each microfilament that fired spontaneously. If any spontaneous activity was detected, observations were extended to 4–5 min to enable more detailed characterization. We confirmed that microfilaments ended in the neuroma by verifying that no axons responded to mechanical stimulation of the hindpaw. The percent activity per microfilament was derived by dividing the number of spontaneously active axons observed by 15, an estimate from prior studies of the number of resolvable A- and C-fiber afferents present, on average, in microfilaments of the size used. The percent of axon activity for a given mouse was obtained by averaging all microfilaments analyzed in that mouse. We also evaluated spike waveform, firing frequency, and discharge pattern (tonic, on/off, and irregular) and used this information to identify active afferents as A- or C-fibers using the criteria given in Liu et al. (2000). All axon-firing values are presented in Supplemental Table S4.
Bioinformatics analysis
Detailed SNP information for various mouse inbred strains was obtained from the Mouse Phenome Database (MPD; http://www.jax.org/phenome, build 37.1). The list of the 155 genes in the 4.2-Mb interval of interest on chromosome 15 was initially obtained also from MPD. The list was then curated from duplicate gene entries using the NCBI database (http://www.ncbi.nlm.nih.gov).
Statistical analyses
Group differences in the proportion of mice exhibiting autotomy behavior (autotomy score > 1, except as noted) were tested using a Fisher's exact test. The effect of genotype in the electrophysiological experiments was analyzed using the nonparametric Mann-Whitney U-test and logistic regression. Differences in gene expression were tested using ANOVA or a t-test as noted. Haplotype analysis of the human cohort was done using the R haplo.stats package (the R haplo.score command) (Schaid et al. 2002), where all Table 1 parameters were included as covariates and a multiplicative model was employed. This package handles missing values with the expectation maximization (EM) algorithm. Global and specific haplotype P-values were obtained through 10,000 simulations. Single SNP P-values were obtained using χ2 tests. The P-values for the logistic regression analysis presented in Table 1 were obtained using the glm function of the R stats package (Hastie and Pregibon 1992).
Acknowledgments
This work was supported by the Israel Science Foundation (ISF), The Hebrew University Center for Research on Pain, The Canada Research Chair Program (ZS), and the European Community's 6th Framework Program (project LSHM-CT-2004-502800 PainGenes). The manuscript reflects only the authors' views. The European Community is not liable for any use that may be made of the information contained therein. We thank Danping Ding-Pfennigdorff and Anke M. Schulte for their contribution to the expression experiments; Yoram Shir, Jean-Jacques Vatine, Tamar Peretz-Jablonsky, Raphael Pfeffer, Edith Gershon, and Jessica Livneh-Fuchs for phenotyping and DNA collection of the human cohort; and Pnina Raber for phenotyping and managing mouse colonies.
Footnotes
[Supplemental material is available online at http://www.genome.org. The human data from this study have been submitted to the Human Genome Variation Database of Genotype-to-Phenotype (http://www.hgvbaseg2.org) under accession no. HGVST514.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.104976.110.
References
- Bats C, Groc L, Choquet D 2007. The interaction between Stargazin and PSD-95 regulates AMPA receptor surface trafficking. Neuron 53: 719–734 [DOI] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Brown EL, Hunter CP 2001. Quantitative analysis of mRNA amplification by in vitro transcription. Nucleic Acids Res 29: e29 doi: 10.1093/nar/29.5.e29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkman B, Arner S, Hyden LC 2008. Phantom breast and other syndromes after mastectomy: Eight breast cancer patients describe their experiences over time: A 2-year follow-up study. J Pain 9: 1018–1025 [DOI] [PubMed] [Google Scholar]
- Bouhassira D, Lanteri-Minet M, Attal N, Laurent B, Touboul C 2008. Prevalence of chronic pain with neuropathic characteristics in the general population. Pain 136: 380–387 [DOI] [PubMed] [Google Scholar]
- Breivik H, Collett B, Ventafridda V, Cohen R, Gallacher D 2006. Survey of chronic pain in Europe: Prevalence, impact on daily life, and treatment. Eur J Pain 10: 287–333 [DOI] [PubMed] [Google Scholar]
- Brown AM, Sease KL, Robey JL, Shofer FS, Hollander JE 2007. The impact of B-type natriuretic peptide in addition to troponin I, creatine kinase-MB, and myoglobin on the risk stratification of emergency department chest pain patients with potential acute coronary syndrome. Ann Emerg Med 49: 153–163 [DOI] [PubMed] [Google Scholar]
- Cokic B, Stein V 2008. Stargazin modulates AMPA receptor antagonism. Neuropharmacology 54: 1062–1070 [DOI] [PubMed] [Google Scholar]
- Dai G, Haedo RJ, Warren VA, Ratliff KS, Bugianesi RM, Rush A, Williams ME, Herrington J, Smith MM, McManus OB, et al. 2008. A high-throughput assay for evaluating state dependence and subtype selectivity of Cav2 calcium channel inhibitors. Assay Drug Dev Technol 6: 195–212 [DOI] [PubMed] [Google Scholar]
- Darvasi A 1998. Experimental strategies for the genetic dissection of complex traits in animal models. Nat Genet 18: 19–24 [DOI] [PubMed] [Google Scholar]
- Devor M 2006a. Response of nerves to injury in relation to neuropathic pain. In Wall and Melzack's textbook of pain (ed. McMahon S, Koltzenburg M), pp. 905–927 Churchill Livingstone, London, UK [Google Scholar]
- Devor M 2006b. Sodium channels and mechanisms of neuropathic pain. J Pain 7: S3–S12 [DOI] [PubMed] [Google Scholar]
- Devor M 2007. Anesthesia dolorosa model, autotomy. In Encyclopedia of pain (ed. Schmidt R, Willis W), pp. 84–87 Springer-Verlag, Berlin, Germany [Google Scholar]
- Devor M, Raber P 1990. Heritability of symptoms in an experimental model of neuropathic pain. Pain 42: 51–67 [DOI] [PubMed] [Google Scholar]
- Devor M, Gilad A, Arbilly M, Yakir B, Raber P, Pisante A, Darvasi A 2005. pain1: A neuropathic pain QTL on mouse chromosome 15 in a C3HxC58 backcross. Pain 116: 289–293 [DOI] [PubMed] [Google Scholar]
- Devor M, Gilad A, Arbilly M, Nissenbaum J, Yakir B, Raber P, Minert A, Pisante A, Darvasi A 2007. Sex-specific variability and a ‘cage effect’ independently mask a neuropathic pain quantitative trait locus detected in a whole genome scan. Eur J Neurosci 26: 681–688 [DOI] [PubMed] [Google Scholar]
- de Vries B, Frants RR, Ferrari MD, van den Maagdenberg AM 2009. Molecular genetics of migraine. Hum Genet 126: 115–132 [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Nackley AG, Tchivileva IE, Shabalina SA, Maixner W 2007. Genetic architecture of human pain perception. Trends Genet 23: 605–613 [DOI] [PubMed] [Google Scholar]
- Engelman HS, MacDermott AB 2004. Presynaptic ionotropic receptors and control of transmitter release. Nat Rev Neurosci 5: 135–145 [DOI] [PubMed] [Google Scholar]
- Estacion M, Harty TP, Choi JS, Tyrrell L, Dib-Hajj SD, Waxman SG 2009. A sodium channel gene SCN9A polymorphism that increases nociceptor excitability. Ann Neurol 66: 862–866 [DOI] [PubMed] [Google Scholar]
- Etherton JL, Bianchini KJ, Ciota MA, Heinly MT, Greve KW 2006. Pain, malingering and the WAIS-III Working Memory Index. Spine J 6: 61–71 [DOI] [PubMed] [Google Scholar]
- Fishbain DA, Cutler RB, Rosomoff HL, Steele-Rosomoff R 2002. Does the conscious exaggeration scale detect deception within patients with chronic pain alleged to have secondary gain? Pain Med 3: 39–46 [DOI] [PubMed] [Google Scholar]
- Hastie TJ, Pregibon D 1992. Generalized linear models. In Statistical models in S (ed. Chambers JM, Hastie TJ), pp. 195–246 Wadsworth & Brooks/Cole, Pacific Grove, CA [Google Scholar]
- Kang MG, Chen CC, Felix R, Letts VA, Frankel WN, Mori Y, Campbell KP 2001. Biochemical and biophysical evidence for gamma 2 subunit association with neuronal voltage-activated Ca2+ channels. J Biol Chem 276: 32917–32924 [DOI] [PubMed] [Google Scholar]
- Kwok PY 2001. Methods for genotyping single nucleotide polymorphisms. Annu Rev Genomics Hum Genet 2: 235–258 [DOI] [PubMed] [Google Scholar]
- LaCroix-Fralish ML, Mogil JS 2008. Progress in genetic studies of pain and analgesia. Annu Rev Pharmacol Toxicol 49: 97–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCroix-Fralish ML, Mo G, Smith SB, Sotocinal SG, Ritchie J, Austin JS, Melmed K, Schorscher-Petcu A, Laferriere AC, Lee TH, et al. 2009. The β3 subunit of the Na+,K+-ATPase mediates variable nociceptive sensitivity in the formalin test. Pain 144: 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts VA, Felix R, Biddlecome GH, Arikkath J, Mahaffey CL, Valenzuela A, Bartlett FS II, Mori Y, Campbell KP, Frankel WN 1998. The mouse stargazer gene encodes a neuronal Ca2+-channel γ subunit. Nat Genet 19: 340–347 [DOI] [PubMed] [Google Scholar]
- Liu CN, Wall PD, Ben-Dor E, Michaelis M, Amir R, Devor M 2000. Tactile allodynia in the absence of C-fiber activation: Altered firing properties of DRG neurons following spinal nerve injury. Pain 85: 503–521 [DOI] [PubMed] [Google Scholar]
- Ma L, Uchida H, Nagai J, Inoue M, Aoki J, Ueda H 2010. Evidence for de novo synthesis of lysophosphatidic acid in the spinal cord through phospholipase A2 and autotaxin in nerve injury-induced neuropathic pain. J Pharmacol Exp Ther 333: 540–546 [DOI] [PubMed] [Google Scholar]
- McGraw J, Gaudet AD, Oschipok LW, Steeves JD, Poirier F, Tetzlaff W, Ramer MS 2005. Altered primary afferent anatomy and reduced thermal sensitivity in mice lacking galectin-1. Pain 114: 7–18 [DOI] [PubMed] [Google Scholar]
- Milstein AD, Nicoll RA 2009. TARP modulation of synaptic AMPA receptor trafficking and gating depends on multiple intracellular domains. Proc Natl Acad Sci 106: 11348–11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minert A, Gabay E, Dominguez C, Wiesenfeld-Hallin Z, Devor M 2007. Spontaneous pain following spinal nerve injury in mice. Exp Neurol 206: 220–230 [DOI] [PubMed] [Google Scholar]
- Mitsukawa K, Lu X, Bartfai T 2008. Galanin, galanin receptors and drug targets. Cell Mol Life Sci 65: 1796–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogil JS, Miermeister F, Seifert F, Strasburg K, Zimmermann K, Reinold H, Austin JS, Bernardini N, Chesler EJ, Hofmann HA, et al. 2005. Variable sensitivity to noxious heat is mediated by differential expression of the CGRP gene. Proc Natl Acad Sci 102: 12938–12943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss FJ, Dolphin AC, Clare JJ 2003. Human neuronal stargazin-like proteins, γ2, γ3 and γ4; an investigation of their specific localization in human brain and their influence on CaV2.1 voltage-dependent calcium channels expressed in Xenopus oocytes. BMC Neurosci 4: 23 doi: 10.1186/1471-2202-4-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolajsen L, Jensen TS 2001. Phantom limb pain. Br J Anaesth 87: 107–116 [DOI] [PubMed] [Google Scholar]
- Nishiyama T 2000. Interaction among NMDA receptor-, NMDA glycine site- and AMPA receptor antagonists in spinally mediated analgesia. Can J Anaesth 47: 693–698 [DOI] [PubMed] [Google Scholar]
- Nissenbaum J, Shpigler H, Pisante A, DelCanho S, Minert A, Seltzer Z, Devor M, Darvasi A 2008. pain2: A neuropathic pain QTL identified on rat chromosome 2. Pain 135: 92–97 [DOI] [PubMed] [Google Scholar]
- Nordin M, Nystrom B, Wallin U, Hagbarth KE 1984. Ectopic sensory discharges and paresthesiae in patients with disorders of peripheral nerves, dorsal roots and dorsal columns. Pain 20: 231–245 [DOI] [PubMed] [Google Scholar]
- Ochoa JL, Campero M, Serra J, Bostock H 2005. Hyperexcitable polymodal and insensitive nociceptors in painful human neuropathy. Muscle Nerve 32: 459–472 [DOI] [PubMed] [Google Scholar]
- Payne HL, Donoghue PS, Connelly WM, Hinterreiter S, Tiwari P, Ives JH, Hann V, Sieghart W, Lees G, Thompson CL 2006. Aberrant GABAA receptor expression in the dentate gyrus of the epileptic mutant mouse stargazer. J Neurosci 26: 8600–8608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson AK, Gebauer M, Jordan S, Metz-Weidmann C, Schulte AM, Schneider HC, Ding-Pfennigdorff D, Thun J, Xu XJ, Wiesenfeld-Hallin Z, et al. 2009. Correlational analysis for identifying genes whose regulation contributes to chronic neuropathic pain. Mol Pain 5: 7 doi: 10.1186/1744-8069-5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priel A, Kolleker A, Ayalon G, Gillor M, Osten P, Stern-Bach Y 2005. Stargazin reduces desensitization and slows deactivation of the AMPA-type glutamate receptors. J Neurosci 25: 2682–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothemund Y, Grusser SM, Liebeskind U, Schlag PM, Flor H 2004. Phantom phenomena in mastectomized patients and their relation to chronic and acute pre-mastectomy pain. Pain 107: 140–146 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Ryu MJ, Lee C, Kim J, Shin HS, Yu MH 2008. Proteomic analysis of stargazer mutant mouse neuronal proteins involved in absence seizure. J Neurochem 104: 1260–1270 [DOI] [PubMed] [Google Scholar]
- Saban R, Simpson C, Vadigepalli R, Memet S, Dozmorov I, Saban MR 2007. Bladder inflammatory transcriptome in response to tachykinins: Neurokinin 1 receptor-dependent genes and transcription regulatory elements. BMC Urol 7: 7 doi: 10.1186/1471-2490-7-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval A, Andrade A, Beedle AM, Campbell KP, Felix R 2007. Inhibition of recombinant N-type CaV channels by the γ2 subunit involves unfolded protein response (UPR)-dependent and UPR-independent mechanisms. J Neurosci 27: 3317–3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA 2002. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 70: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltzer Z, Wu T, Max MB, Diehl SR 2001. Mapping a gene for neuropathic pain-related behavior following peripheral neurectomy in the mouse. Pain 93: 101–106 [DOI] [PubMed] [Google Scholar]
- Sherman R, Devor M, Casey Jones DE, Katz J, Marbach JJ 1996. Phantom pain. Plenum, New York [Google Scholar]
- Tao F, Skinner J, Su Q, Johns RA 2006. New role for spinal Stargazin in α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor-mediated pain sensitization after inflammation. J Neurosci Res 84: 867–873 [DOI] [PubMed] [Google Scholar]
- Taylor CP 2009. Mechanisms of analgesia by gabapentin and pregabalin–calcium channel α2-δ [Cavα2-δ] ligands. Pain 142: 13–16 [DOI] [PubMed] [Google Scholar]
- Tegeder I, Costigan M, Griffin RS, Abele A, Belfer I, Schmidt H, Ehnert C, Nejim J, Marian C, Scholz J, et al. 2006. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nat Med 12: 1269–1277 [DOI] [PubMed] [Google Scholar]
- Thorsen TS, Madsen KL, Rebola N, Rathje M, Anggono V, Bach A, Moreira IS, Stuhr-Hansen N, Dyhring T, Peters D, et al. 2010. Identification of a small-molecule inhibitor of the PICK1 PDZ domain that inhibits hippocampal LTP and LTD. Proc Natl Acad Sci 107: 413–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tselniker I, Tsemakhovich VA, Dessauer CW, Dascal N 2010. Stargazin modulates neuronal voltage-dependent Ca2+ channel Cav2.2 by a Gβγ-dependent mechanism. J Biol Chem 285: 20462–20471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytherleigh MG, Koshy CE, Evans J 1998. Phantom breast pain. Plast Reconstr Surg 102: 921. [DOI] [PubMed] [Google Scholar]
- Vadivelu N, Schreck M, Lopez J, Kodumudi G, Narayan D 2008. Pain after mastectomy and breast reconstruction. Am Surg 74: 285–296 [PubMed] [Google Scholar]
- Wall PD, Devor M, Inbal R, Scadding JW, Schonfeld D, Seltzer Z, Tomkiewicz MM 1979. Autotomy following peripheral nerve lesions: Experimental anaesthesia dolorosa. Pain 7: 103–111 [DOI] [PubMed] [Google Scholar]
- Wynn Parry CB 1980. Pain in avulsion lesions of the brachial plexus. Pain 9: 41–53 [DOI] [PubMed] [Google Scholar]
- Yao MZ, Gu JF, Wang JH, Sun LY, Lang MF, Liu J, Zhao ZQ, Liu XY 2002. Interleukin-2 gene therapy of chronic neuropathic pain. Neuroscience 112: 409–416 [DOI] [PubMed] [Google Scholar]