Abstract
Changes in gene expression levels determine differentiation of tissues involved in development and are associated with functional decline in aging. Although development is tightly regulated, the transition between development and aging, as well as regulation of post-developmental changes, are not well understood. Here, we measured messenger RNA (mRNA), microRNA (miRNA), and protein expression in the prefrontal cortex of humans and rhesus macaques over the species' life spans. We find that few gene expression changes are unique to aging. Instead, the vast majority of miRNA and gene expression changes that occur in aging represent reversals or extensions of developmental patterns. Surprisingly, many gene expression changes previously attributed to aging, such as down-regulation of neural genes, initiate in early childhood. Our results indicate that miRNA and transcription factors regulate not only developmental but also post-developmental expression changes, with a number of regulatory processes continuing throughout the entire life span. Differential evolutionary conservation of the corresponding genomic regions implies that these regulatory processes, although beneficial in development, might be detrimental in aging. These results suggest a direct link between developmental regulation and expression changes taking place in aging.
The human brain changes dramatically during postnatal development, both structurally and histologically (de Graaf-Peters and Hadders-Algra 2006; Marsh et al. 2008). Some developmental processes, such as cortical axon myelinization, extend far into adulthood, concluding at ∼40 yr of age (Sowell et al. 2004). Whereas between birth and adulthood, human cognitive abilities undergo remarkable remodeling (Marsh et al. 2008), in later life, the brain begins to change in a destructive manner. Such changes include a decrease in brain volume, loss of synapses, cognitive decline, and a rise in the frequency of neurological disorders (Courchesne et al. 2000; Sowell et al. 2004; Peters et al. 2008; Salthouse 2009). Although developmental and aging-related changes are clearly observed in histology and cognitive function, their molecular bases are still not well understood.
During the last decade, messenger RNA (mRNA) expression profiling has been widely used to investigate changes in gene expression levels in aging human and mammalian brains (Lee et al. 2000; Lu et al. 2004; Erraji-Benchekroun et al. 2005; Xue et al. 2007; Zahn et al. 2007; Loerch et al. 2008). These studies have identified several mRNA expression patterns in the aging brain, including an up-regulation of stress and immune response pathways, as well as a decrease in expression of genes involved in energy metabolism and neuronal functions. Many of the observed changes in expression levels were attributed to oxidative stress and associated accumulation of DNA damage, especially in gene promoter regions (Lu et al. 2004). Interestingly, at 13 yr of age, some of the gene expression changes observed in human brain aging are already detectable (Erraji-Benchekroun et al. 2005). This either reflects the early effects of DNA damage or indicates that some of the expression changes seen in aging reflect regulatory patterns established in brain development (Finch 1976; de Magalhaes and Church 2005; de Magalhães et al. 2009). Although the latter notion is not well recognized, recent studies in Caenorhabditis elegans have identified several developmental regulatory patterns that persist into aging, effectively limiting the life span of the animals studied (Boehm and Slack 2005; Budovskaya et al. 2008). Notably, such regulation involved both microRNA (miRNA) and transcription factors (TFs).
In mammals, development and aging are commonly studied separately. Thus, whereas many developmental changes, including those involved in postnatal neural development, are known to be controlled by regulatory programs (Polleux et al. 2007; Stefani and Slack 2008; Schratt 2009), little is known about when and whether these programs conclude or when aging-related changes commence. To address these questions, here we survey mRNA, miRNA, and protein expression changes in the prefrontal cortex of humans and rhesus macaques throughout the bulk of each species' life span.
Results
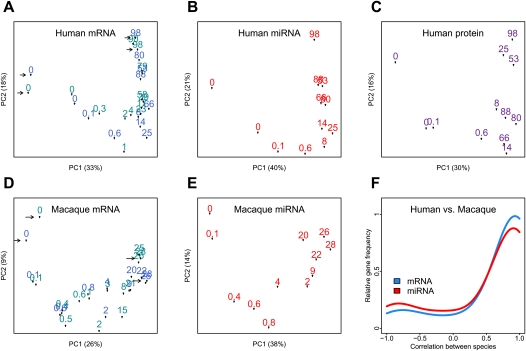
Using Affymetrix Human Gene 1.0 ST microarrays, we measured mRNA expression in postmortem samples from the superior frontal gyrus of the prefrontal cortex, from 23 cognitively healthy humans and 24 healthy macaques, with ages ranging from 2 d to 98 yr and from 16 d to 28 yr, respectively (Methods; Supplemental Tables S1, S2). In humans, we reliably detect expression signal from a total of 12,396 genes. For these genes, technical replicates indicated little experimental variation (Fig. 1A), and other factors, such as sex, postmortem delay, or RNA quality, did not explain a significant amount of expression variation among individuals (Supplemental Text S1). In contrast, ∼60% of the total variation in gene expression can be attributed to the age of individuals (permutation test [PT], P < 0.01) (Supplemental Fig. S1). The effect of age on total variation can also be observed from a Principal Components Analysis (Fig. 1A). Consequently, we find that for 33% of expressed genes (n = 4084), referred to as “age-related genes,” age explains a significant proportion of the total variation (median 83%, F-test, P < 1 × 10−4, false discovery rate [FDR] < 0.1%). Comparing age-related changes identified in this study with two published human brain data sets (0–83 yr, n = 44 [Somel et al. 2009], and 26–106 yr, n = 30 [Lu et al. 2004]), we find that expression trajectories are highly consistent across the studies (median [Pearson correlation] r = 0.98 and r = 0.68, respectively) (Supplemental Fig. S1).
Figure 1.
mRNA, protein, and miRNA expression changes during life span. (A–E) The first two principal components of mRNA, miRNA, and protein expression in human and rhesus macaque brains. The analysis was performed by singular value decomposition, using the “prcomp” function in the R “stats” package, with each gene scaled to unit variance before analysis. The numbers represent each individual's age in years. The proportion of variance explained by each principal component is shown in parentheses. For mRNA, arrows indicate pairs of technical replicates, and shades of blue represent two experimental batches. Individuals group according to their age, indicating a substantial influence of age on total expression variation. (F) Distribution of Pearson correlation coefficients between human and macaque expression time series, calculated for 3233 orthologous mRNAs (blue) or 98 orthologous miRNAs (red), showing significant expression change with age in human (Supplemental Text S1). The y-axis shows the relative frequency (the Gaussian kernel density estimate, calculated with the R “density” function) of genes showing a certain Pearson correlation coefficient.
Similarly, using only probes that match the rhesus macaque genome perfectly, we detect expression of 9628 genes in macaque brains (Fig. 1D), 21% of which show significant change with age. Notably, we find strong positive correlation between human and macaque age-related expression changes (median r = 0.94; PT, P < 0.01) (Fig. 1F). This observation is important because the macaques used in this study, as opposed to the humans, shared the same environment throughout their life, as well as the same postmortem conditions, thus excluding a potential source of non-age-related artifacts. The high reproducibility of the identified age-related gene expression changes, across human and macaque species and across several human data sets, indicates that technical errors or biological factors unrelated to age do not have a large impact on our data.
Aging as the reversal or extension of development
We then investigated the major patterns of expression change with age in the human prefrontal cortex using the 4084 age-related genes. To analyze expression changes in the classical framework, we separated human life span into “development” (0–20 yr) and “aging” (20–100 yr), following the commonly used age intervals for pre- and post-adulthood (e.g., Lu et al. 2004; Rodwell et al. 2004).
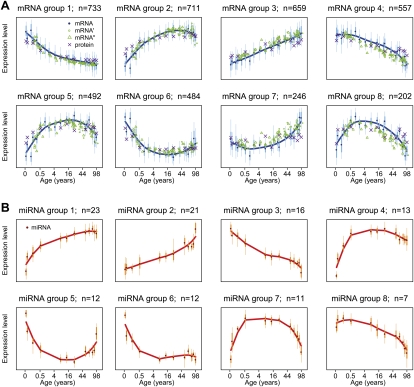
Four important observations emerged. First, for 64% of age-related genes, there was a reversal in the expression change trends observed between development and aging (Supplemental Fig. S1C). This can be seen visually by clustering gene expression profiles into groups of genes with similar trajectories (Fig. 2; Supplemental Fig. S2) (we use eight groups in the following analysis, but using different numbers of groupings yields consistent results; see Supplemental Figs. S3, S4). Figure 2 shows that for genes in five out of eight coexpressed groups (groups 2, 5, 6, 7, and 8) there is reversal in the trend of expression change between development and aging. The remaining genes either continue the developmental trend (group 3), show limited or no change during aging (group 1), or show limited change during the first years of postnatal development (group 4).
Figure 2.
Major patterns of mRNA and miRNA changes with age. (A) Shows the average expression levels in eight coexpressed gene groups (see Methods). Dark blue points represent the mean expression level of all genes in a group per individual. The y-axis shows standardized expression levels, where each unit indicates one standard deviation difference from the mean. The x-axis shows age of individuals on (age)¼ scale (i.e., the fourth root scale, which provided optimal resolution of expression changes during both developmental and adulthood periods). Bold vertical bars indicate the 25%–75% quantile range, and thin bars indicate the 2.5%–97.5% quantile range. Mean expression change with age within each group is summarized by spline curves. Green circles, green triangles, and purple crosses show the mean expression levels for the same genes among individuals from two published mRNA data sets (Lu et al. 2004; Somel et al. 2009) (GEO data set accession nos. GSE1572 and GSE11512), and the protein data set from the present study, respectively. (B) Major patterns of miRNA changes with age. Labels are as in A.
Second, there are no prominent expression change patterns that can be exclusively related to aging. In fact, 0.5% of all genes expressed in brain show substantial change during aging, but no change during development (Supplemental Fig. S5). Importantly, these genes cannot be functionally discerned from other age-related gene groups. In addition, expression changes in early development have substantially larger amplitude than changes during aging (Supplemental Fig. S6). This is consistent with the rate of anatomical change in the brain during postnatal life (Courchesne et al. 2000).
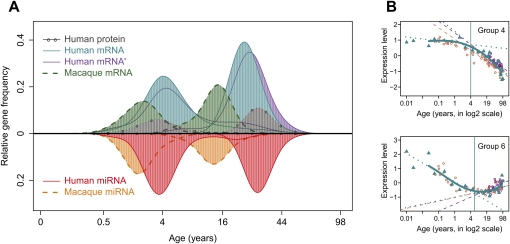
Third, we investigated the transitions between early- and late-life expression trends. Here, for each age-related gene, we determined when one trend finishes and another begins (Methods). We find that in humans, most transitions occur at two particular time intervals: ∼3–4 yr and/or 20–25 yr of age (Fig. 3A; Supplemental Table S3). Interestingly, many expression changes that had been previously associated with aging, such as down-regulation of genes involved in neural functions (Lu et al. 2004; Loerch et al. 2008), initiate in early childhood (Fig. 3B; discussed below).
Figure 3.
Transition points of expression change with age for mRNA, miRNA, and proteins. (A) The age distribution of expression transition points determined on gene-by-gene basis. The y-axis shows the relative frequency (the Gaussian kernel density estimate, calculated with the R “density” function) of genes showing a certain transition point. The x-axis shows transition ages on the (age)¼ scale. (Blue) Human mRNA; (green) macaque mRNA; (purple) a published human mRNA data set (Somel et al. 2009); (gray) human protein; (red) human miRNA; (orange) macaque miRNA. Only age-related genes following nonlinear trajectories and showing significant transition points are represented (Supplemental Table S3). (B) The transition point identification procedure illustrated using genes in groups 4 and 6 (as shown in Fig. 2A). The y-axes indicate mean normalized expression levels of genes in the group. The x-axes show individuals' ages in log2 scale, allowing improved resolution of developmental changes (Methods; Supplemental Fig. S14). Blue points represent expression levels from the human mRNA data set; blue solid lines show spline curves fit to these data. Blue vertical lines show the transition points. Dotted blue lines show linear regression of expression on age before and after the transition point. Purple and brown points/lines represent mean expression levels/linear regression lines from two published data sets (Lu et al. 2004; Somel et al. 2009), respectively. Note that the results shown in A are calculated per gene, and in B using the means of gene groups.
Finally, we find that the two transition intervals in macaques are shifted to earlier ages compared to humans (Fig. 3A). This shift is consistent with life-history differences such as time of reproductive maturation and maximum life span between the two species (de Magalhães and Costa 2009).
To test whether these observations apply to protein expression changes with age, we measured protein expression in 12 of the human samples with shotgun mass spectrometry (Methods; Fig. 1C). We used a label-free proteomic approach, which allows quantification of protein expression based on peptide counting (Old et al. 2005; Fu et al. 2009). Although both the sample size and the number of reliably detected proteins (n = 2229, with total peptide count ≥ 20, FDR = 3%) are limited compared to the mRNA data set, we find that age-related changes in protein expression are strongly correlated with mRNA changes across all 901 age-related genes (median r = 0.75) (Supplemental Fig. S1D). Accordingly, both the reversal of expression change between development and aging as well as the timing of these transitions are reflected at the protein level (Fig. 2; Supplemental Fig. S3). Thus, at both mRNA and protein levels, gene expression changes in the aging brain are connected to developmental patterns.
miRNAs regulate expression changes in both development and aging
To investigate whether some of the transitions between developmental and aging expression trends can be explained by the regulatory effects of miRNA, we analyzed miRNA expression in 12 humans and 12 rhesus macaques, selected among the individuals studied at the mRNA level, using Illumina high-throughput sequencing (Methods). In humans and macaques, we obtained a total of 56,661,685 and 69,883,506 sequence reads, respectively, that map perfectly to the corresponding genomes. Ninety-three percent and 95% of the sequence reads match annotated human miRNA and their macaque orthologs, respectively (Methods). Principal component analyses indicate that, in both species, miRNA expression is highly influenced by age (Fig. 1B,E). Out of 373 miRNAs represented by a total of ≥100 sequence reads in humans, 115 (31%) show significant expression changes with age (FDR < 0.1). Comparing these results to published miRNA studies on postnatal mouse cortex development (Smirnova et al. 2005; Dogini et al. 2008), we find consistent patterns of change for all reported cases (Supplemental Table S4). Similarly, we find good positive correlation between humans and macaques in miRNA expression changes with age (median r = 0.89; PT, P < 0.02) (Fig. 1F).
We find that age-related miRNA expression changes closely resemble those of mRNA. This includes the overall pattern of expression variation, the expression patterns of miRNA groups, and the timing of transitions (Figs. 1–3). This correspondence suggests that age-related mRNA changes might be directly shaped by miRNA regulation. In this case, we expect that coexpressed and, presumably, coregulated mRNA would be enriched for particular miRNA binding sites. Furthermore, we expect to find negative correlation between expression levels of these putative target mRNAs and the corresponding miRNAs.
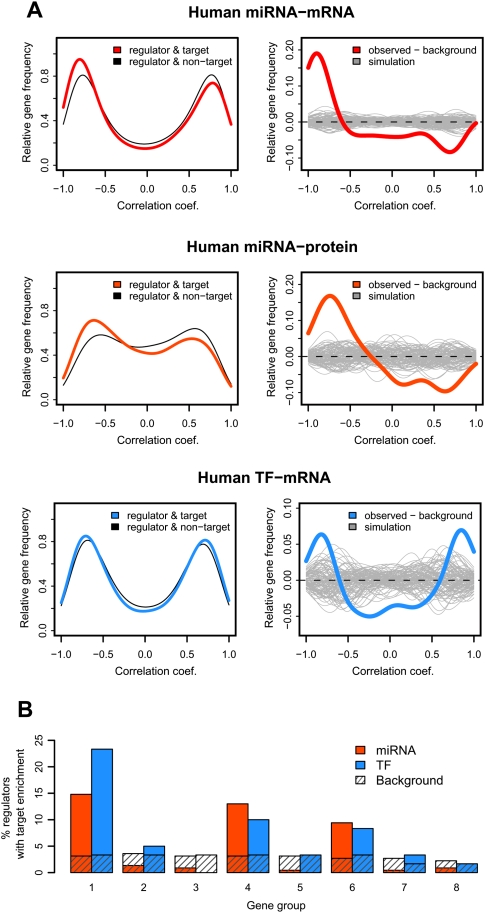
Using the TargetScan database (Lewis et al. 2005) to predict miRNA-binding sites, we show that age-related miRNA expression profiles are more negatively correlated with their targets' expression profiles compared to randomly chosen miRNA–target constellations (Fig. 4A). This holds for both mRNA and protein expression. We then separately tested the eight coexpressed gene groups for target site enrichment. This yields 90 cases of miRNAs with significant target enrichment in a gene group (at hypergeometric test [HT], P < 0.05; PT, P < 0.001) (Fig. 4B). Notably, this enrichment is not uniform among the groups, but found mainly in groups 1, 4, and 6 (Fig. 4B). We next examined the expression profiles of the miRNAs targeting these three groups. As predicted, these miRNAs show a significantly greater extent of negative correlation with the targets' expression profiles, compared to randomly chosen miRNA–target constellations (49% vs. 20%; HT, P = 5 × 10−67). Hence, both the overall miRNA–target expression relationship and the correlations between miRNA–target pairs identified, based on target site enrichment (i.e., targets in groups 1, 4, and 6), support the regulatory effect of miRNA on mRNA expression changes over the human life span.
Figure 4.
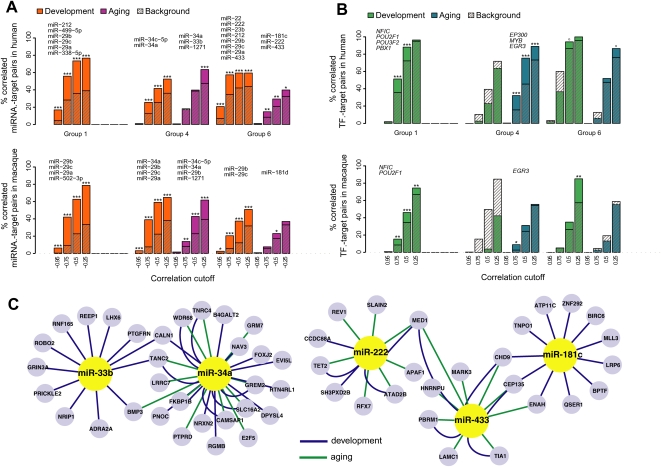
miRNA and TF regulation of expression changes with age. (A) Excess of negative correlations between miRNA/TF–target pairs. (Left panels) The y-axes show the relative frequency (the Gaussian kernel density estimate, calculated with the R “density” function) of Pearson correlation coefficients. Shown are correlations between expression profiles of age-related regulators (miRNA or TF) and their age-related target genes (Methods). Colored curves represent the distribution of regulator–target correlations for miRNA–mRNA (red), miRNA–protein (orange), and TF–mRNA (blue). Gray curves show the background distribution: correlations between regulators and non-targets (genes with no evidence of being targeted by the respective regulators). (Right panels) The difference between the kernel density distributions of regulator–target correlations and the background. The gray lines represent 100 simulation results, generated by randomly selecting the same number of background pairs, as regulator–target pairs. The bimodality of the correlation coefficient distributions is because we calculate correlations between age-related regulators and targets only; so, each pair shows some degree of correlation, positive or negative. We therefore test the excess of negative correlations for predicted miRNA–target pairs, relative to randomly paired age-related miRNA and mRNA. Similarly, we test the excess of strong positive and negative correlations for the predicted TF–target pairs (given the dual role of TFs as activator and/or repressor of transcription), relative to randomly paired age-related TFs and mRNA. (B) The proportion of expressed miRNA or TFs showing target enrichment among eight coexpressed gene groups (at HT, P < 0.05).
Importantly, when we separately analyze expression changes in “development” and “aging” (0–20 and 20–100 yr, respectively), we find putative regulatory effects of miRNA in both periods (Fig. 5A). For genes in groups 1, 4, and 6, we identify 16 miRNAs with a significant excess of binding sites and a significant negative expression correlation in development. Six miRNAs show a similar excess of enrichment and negative correlation during aging (Supplemental Table S5). Three of these—miR-34a, miR-222, and miR-433—are correlated with their targets in both development and aging and thus may regulate gene expression changes in both periods (Fig. 5C). Notably, this result does not depend on the cutoff used for dividing life span into two phases (Supplemental Fig. S7), indicating that molecular changes in early and late phases of life span are not distinct but form a continuum.
Figure 5.
miRNA and TF regulation in development and aging. (A) Excess of negative correlations among selected miRNA–target pairs in three coexpressed gene groups in human or macaque cortex. The colored bars show the proportions of negative correlations among miRNA with significant target enrichment within a gene group (at HT, P < 0.05) and their targets in that group at different correlation cutoffs. Hatched bars indicate the proportions of negative correlations among miRNA without target enrichment in a gene group and their targets in that group (the background). The asterisks indicate support for observed–background difference, calculated by bootstrapping the background set 1000 times; ***P < 0.001; **P < 0.01; *P < 0.05; o P < 0.10. Both observed and background correlations are calculated separately for developmental and aging periods. The names of identified putative regulatory miRNA are shown above each gene group. Genes in group 1 show limited expression change during aging; therefore, we do not estimate regulators for this group at this period. For macaque, regulators shown in the figure are predicted based on the macaque data and independently of human analysis results, using the same significance levels. Additionally, ∼80% of regulators predicted in humans show a tendency for negative correlation with their targets in macaques (Supplemental Table S5). (B) Excess of negative and positive correlations among TF–target pairs in three gene groups. The colored and hatched bars represent proportions of observed and background TF–target gene correlation pairs, as in B, but the x-axis shows the absolute Pearson correlation cutoff. The names of identified putative regulatory TFs are shown above each gene group. (C) A network of regulatory interactions identified in groups 4 and 6. Only part of the full network, listed in Supplemental Table S5, is shown. The represented genes are those containing the specific miRNA binding site and that show significant negative correlation with that miRNA's expression profile, either in development (green edges) or aging (blue edges). The figure was drawn using Cytoscape software (v 2.6.3).
As our analyses depend on indirect assessment of regulatory interactions, we conducted six additional analyses to support or falsify the estimated regulatory relationships as follows:
For 15/16 and 5/6 of the miRNA–target gene pairs identified in the mRNA expression data set, we find support for putative regulatory effects at the protein expression level (Supplemental Table S5) (odd's ratio = 9.7; HT, P = 0.001).
For 12/16 and 5/6 of these miRNAs, we find support for negative miRNA–target correlations in macaque development (0–4 yr) and aging (4–28 yr), respectively (Supplemental Table S5) (odd's ratio = 4.4; HT, P = 0.03).
We tested whether miRNA expression differences between humans and macaques are reflected in expression of their putative target genes (Supplemental Fig. S8C,D).
We investigated whether mutations in miRNA binding sites present in the rhesus macaque genome, which are expected to disrupt the regulatory relationship between miRNAs and their target genes in macaques, indeed cause a loss of correlation between miRNAs and target gene expression profiles in this species (Supplemental Fig. S8E).
We tested whether negative correlations could be caused by factors other than age, by using interpolated points instead of the original expression values in the correlation tests (Supplemental Fig. S8A).
We compared our list of putative miRNA–target pairs to published lists of experimentally identified regulators (Supplemental Table S6).
Taken together, the results of these six tests (Fig. 5; Supplemental Fig. S8; Methods; Supplemental Text S1) indicate that the majority of the identified miRNA–target gene pairs likely reflect genuine regulatory relationships. Most importantly, this suggests that at least part of expression changes occurring during aging are driven by the same general mechanism and, in some cases, the same regulatory factors as in postnatal development.
Besides miRNA, other gene expression regulators, such as TFs, may influence mRNA expression during development and aging. Based on the TRANSFAC database (Kel et al. 2003), we identified 47 human age-related TFs with at least one target gene expressed in the human brain. Remarkably, TFs show significant binding site enrichment in the promoters of genes in coexpressed groups 1, 4, and 6, the very same gene groups preferentially targeted by miRNA (Fig. 4B). TF–target pairs show a significant excess of both positive and negative correlations compared to randomly selected pairs (Fig. 4A; Supplemental Fig. S9), indicating that TFs may play activator or repressor roles in the brain. Furthermore, we find a significant excess of TF–target correlations for both the development and aging periods (Fig. 5B; Supplemental Fig. S9). Notably, the putative regulatory relationships found in development and in aging in humans are also found in macaques (Fig. 5B; Supplemental Table S4). Hence, like miRNAs, TFs appear to regulate gene expression changes in brain cortex throughout postnatal life.
Functional characterization of lifetime expression changes
It is generally accepted that expression changes taking place during development are optimized by purifying selection to yield a functional and reproducing organism (de Magalhaes and Church 2005). In contrast, molecular changes during aging are typically explained by accumulating somatic damage. Two models have been proposed to account for aging-related gene expression level changes: (1) down-regulations due to oxidative promoter damage (Lu et al. 2004) and (2) responses to accumulating somatic damage (Zahn et al. 2007). While the first type of change is probably invariably detrimental, the second type is presumably an adaptive response and may even contribute to longevity. Meanwhile, a third model, the extension of developmental patterns into old age, has rarely been associated with gene expression profiles found in aging (de Magalhães et al. 2009). Here we inspect the identified expression changes during development and aging with respect to the described models, with a particular emphasis on gene groups showing miRNA and TF regulation.
Strictly developmental regulation
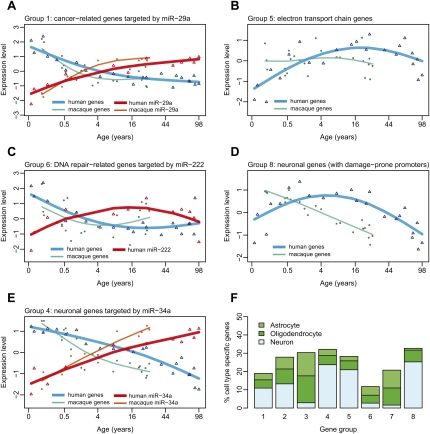
Group 1 genes show the following pattern: They are down-regulated throughout early development, but thereafter show no substantial change (Figs. 1, 6A). With respect to their function, these genes are mainly involved in regulation of transcription, nervous system development, and cell division (Supplemental Fig. S10; Supplemental Tables S7, S8). Among the candidate regulators of this group is the miR-29 family, shown to have tumor suppressor roles in multiple tissues (Fig. 6A; Fabbri et al. 2007; Wang et al. 2008). Interestingly, miR-29 loss has been linked to sporadic Alzheimer's disease (Hebert et al. 2008). Thus, miR-29-driven down-regulation of cell proliferation-associated genes may exemplify developmental changes protecting against late-onset disease.
Figure 6.
Functions, regulation, and specificity of coexpressed gene groups. Shown are mean expression profiles of selected genes within coexpressed gene groups, and their putative miRNA regulators. The empty triangles show mean standardized (z-transformed) human mRNA (blue) and miRNA (red) expression levels, while empty circles show mean standardized macaque mRNA (green) and miRNA (orange) expression levels (note the differences in timing of expression changes between human and macaque, for both mRNA and miRNA expression). The x-axis shows age of individuals on the (age)¼ scale. The lines correspond to cubic spline curves. The depicted genes are associated with specific Gene Ontology functional terms significantly enriched within the given coexpressed group. For A, C, and E, the genes are further targeted by specific miRNAs. (A) miR-29a and its four cancer-related targets in group 1 (MMP2, TRAF4, COL4A2, COL4A1). (B) Seventeen genes involved in electron transport in group 5. (C) miR-222 and its target in group 6, REV1, involved in DNA damage repair. (D) Fifty-seven neuronal genes in group 8. (E) miR-34a and its seven target neuronal genes in group 4 (GREM2, CAMSAP1, TANC2, CALN1, RGMB, FKBP1B, RTN4RL1). (F) Cell-type specificity of gene groups. The y-axis shows the percentage of cell-type-specific genes among the eight coexpressed age-related gene groups (based on Cahoy et al. 2008; see Methods).
Reversal of development as a possible response to damage
Group 6 genes show development-aging reversal in their expression profiles: In old age, they are up-regulated to levels seen in infants. The reason is uncertain, but the answer may lie with groups 2 and 5. These are the mirror images of group 6 and contain energy metabolism-related genes (Figs. 2, 6B; Supplemental Tables S7, S8). For instance, 39/46 of age-related electron transport chain genes fall in groups 2 and 5 (odd's ratio > 4; HT, P < 0.001). This indicates that brain energy production peaks around 20 yr of age and then subsides. In fact, reduced electron transport chain activity is a hallmark of aging-related expression changes in a wide range of tissues and species (Zahn et al. 2007; de Magalhães et al. 2009). The hypothesized reason is that energy production results in accumulating oxidative damage, and at old age, the soma reduces energy production so as to limit further harm (Zahn et al. 2007).
If attenuated energy metabolism at old age is triggered by somatic damage accumulation from the energy production peak at reproductive maturity, we expect this peak to also trigger damage response pathways. Indeed, genes involved in DNA damage response are enriched in group 6 only (20/68 age-related genes, odd's ratio = 3.2; HT, P < 0.001). One of these genes, REV1, a deoxycytidyl transferase involved in DNA repair (Lin et al. 1999), is down-regulated by miR-222 during development and up-regulated during aging (Fig. 6C). Thus, DNA damage response genes might be actively up-regulated in aging to curb further accumulation of oxidative damage. The reason for down-regulation of damage repair pathways in the developmental period, however, is less clear.
Reversal of development as a possible consequence of damage
Another common pattern seen in studies of brain aging is decreased neuronal gene expression (Lu et al. 2004; Loerch et al. 2008). This molecular trend might be associated with phenotypic trends of age-related synapse loss and cognitive decline (Peters et al. 2008; Salthouse 2009). In our data set, genes involved in neuronal function are enriched in two gene groups, 4 and 8 (52/170 age-related genes, odd's ratio > 1.7; HT, P < 0.001). Supporting this, groups 4 and 8 are significantly enriched in genes showing neuron-specific expression (Fig. 6F). Both gene groups' expression levels decrease during aging. The timing, however, is different: Expression of neuronal genes in group 8 starts decreasing around young adulthood, whereas group 4 genes' expression levels start decreasing already in early childhood (Fig. 6D,E). Furthermore, for group 4, but not for group 8, we find indication that this expression trend is actively regulated (discussed below). This implies that an alternative mechanism is involved in the down-regulation of group 8 genes. One possibility is that these genes show high levels of oxidative damage in their promoters, possibly due to higher GC content of their promoter sequences (Lu et al. 2004). Indeed, gene groups decreasing in expression with age have higher promoter GC content than other genes (Supplemental Fig. S11). Group 8 gene promoters are also more GC-rich than group 4 promoters (one-sided Wilcoxon test, P = 0.015). Thus, the expression levels of neuronal genes in group 8 might decline in aging due to accumulating promoter damage, rather than trans regulation.
Aging as extended development
Group 4 genes are related to neural development and function (e.g., axon guidance and long-term depression), as well as cell communication and cell–cell adhesion (Supplemental Tables S7, S8). Expression levels of these genes are constant during the first few years of life, but start decreasing in early childhood and continue decreasing during aging (Fig. 6E). Thus, for these genes, expression changes found in aging arguably represent the continuation of a developmental trend. Our results, from both humans and macaques, suggest that a number of regulators, including the TF early growth response protein 3 (EGR3) involved in sympathetic neuron development (Eldredge et al. 2008), and miR-34a, involved in the apoptosis (Yamakuchi et al. 2008) and tumor suppression in neuroblastoma (Cole et al. 2008), might be responsible for at least some of these continuous expression changes (Fig. 6E). What could the consequences of such extended regulation be? Parallel to increased expression of miR-34a during normal aging, a recent study showed that miR-34a is also up-regulated in a mouse model of Alzheimer's disease (Wang et al. 2009). Meanwhile, its knockdown attenuates the disease phenotype. Hence, we speculate that the continuous up-regulation of miR-34a might be a detrimental factor in human brain aging.
Discussion
Here we analyzed gene expression changes in the human and macaque brains across life span at the mRNA, protein, and miRNA levels. Our results indicate that in both species, gene expression regulation does not finalize at maturity but persists into old age. This pattern is apparent at both mRNA and protein expression levels and pertains to both miRNA and TF regulation.
This said, our work has a number of limitations. First, we have studied expression changes in one specific brain region, the superior frontal gyrus. Patterns of expression changes with age are known to diverge among tissues within the same organism (Zahn et al. 2007). It therefore remains to be shown whether other brain regions, or tissues, also show continuity between developmental and aging expression profiles.
Second, given the limitations of the current miRNA and TF target gene annotation (Chi et al. 2009), we expect that our analysis might miss many actual regulatory interactions. In addition, our estimation of regulator effects is based on statistical evidence, enrichment of binding sites, and correlation between regulator and target gene expression, rather than direct experimental testing. Thus, the identified interactions might contain false positives (Methods). Nonetheless, a number of observations corroborate the general validity of our predictions. These include the reproducibility of the regulator–target pair estimations in the protein data and in rhesus macaque species and coordinated shifts in expression timing of regulators and their targets between humans and macaques, as well as overlap with experimentally verified miRNA targets.
In fact, expression regulation by TFs and miRNAs in postnatal central nervous system development is well recognized (Schratt et al. 2006; Polleux et al. 2007; Schratt 2009). The novel finding here is that gene regulation by miRNAs and TFs occurs to a similar extent in both developmental and aging periods. Although the functional significance of this regulation during aging remains uncertain, some of our observations provide clues to its origin and its roles.
Transition points
In humans, age-related expression changes first alter their direction around 4 yr of age. This may be linked to key events in neural development. Indeed, synaptic density peaks around early/mid-childhood in the human prefrontal cortex (Huttenlocher and Dabholkar 1997; Glantz et al. 2007; for review, see Toga et al. 2006), and in our data, genes associated with neural function (groups 4 and 8) show transition at this point (Fig. 2A). The second transition point takes place in humans at ∼25 yr of age and might correspond to conclusion of developmental processes such as myelinization, known to continue past adolescence (Toga et al. 2006). It may also coincide with the onset of senescence. For instance, genes associated with energy production, which decrease their expression during aging across various tissues and species (Zahn et al. 2006, 2007; de Magalhães et al. 2009), start decreasing at this transition point in our data (group 5; Fig. 2A). Hence, 25 yr of age in humans may mark the beginning of systemic change associated with certain senescence processes.
Conservation of expression changes with age
We observe that both developmental and aging expression profiles, as well as predicted regulatory interactions, are largely conserved between humans and macaques (Figs. 1, 4). This result is consistent with a previous study reporting similar expression profiles in human and rhesus macaque aging, but not between primates and mice (Loerch et al. 2008). Meanwhile, the most prominent difference between the human and macaque species is the delay in the timing of expression changes in humans compared to macaques (Figs. 3A, 6; Supplemental Fig. S8). This delay is in line with previous observations of developmental timing differences in gene expression among primates (Somel et al. 2009) and approximately corresponds to differences in life-history traits between the two species (Supplemental Fig. S12). The coordinated delay of molecular changes across the entire human life span, coupled with our observation that some regulatory changes found in aging are initiated in development, supports a proposed relationship between the extension of human development and human longevity (Kaplan et al. 2000). Human longevity, extreme among primates, may have evolved as a consequence of regulatory changes underlying human neotenic development (Somel et al. 2009).
In addition to timing differences, a small proportion of genes (10%–15%) exhibit opposite trends of expression changes with age in humans and macaques (Supplemental Fig. S13). Interestingly, such differences are ∼1.5 times more common in aging than in development, an observation consistent with the lower strength of purifying selection on the gene regulation at old age (discussed below). These differences could also reflect extreme shifts in developmental timing between species, as well as technical artifacts. Future studies, using additional species and alternative methodology, are needed to address this issue.
Aging as ‘runaway’ development
Our results suggest that gene expression changes in human brain aging are best explained as a combination of multiple models: (1) responses to damage accumulation, (2) direct products of damage accumulation, and (3) uncontrolled or “runaway” continuation of developmental regulatory traits. We cannot yet deduce the relative influences of these mechanisms. Nevertheless, the paucity of expression patterns specific to aging, pervasive extension of developmental expression trends into aging among neuronal genes, and persistence of some developmental regulatory effects into aging, collectively indicate that “runaway” developmental regulation might be a major factor shaping the aging brain.
In fact, a likely link between expression changes in development and aging was previously suggested (Finch 1976; de Magalhaes and Church 2005) and recently noted in the context of lifelong up-regulation of blood coagulation potential (de Magalhães et al. 2009). Such a connection between development and aging has two implications. First, if the two periods constitute a continuum, it should be preferable to study senescence-related molecular changes across life span, rather than restricting analyses to adulthood. Second, this model may explain certain phenotypic changes during aging. Specifically, following the antagonistic pleiotropy hypothesis of aging (Williams 1957), such “runaway” regulatory processes might be beneficial in development but have negative consequences with increased age. Observations of antagonistic regulatory relationships between developmental and aging changes have already been made in nematodes (Boehm and Slack 2005; Budovskaya et al. 2008). Meanwhile, in the context of brain development, this model could predict a connection between synaptic pruning in early life, required for efficient maturation of cognitive processes (Toga et al. 2006; Gonzalez-Burgos et al. 2008), and synaptic loss in later life, associated with cognitive decline (Peters et al. 2008).
Decrease in evolutionary conservation with age
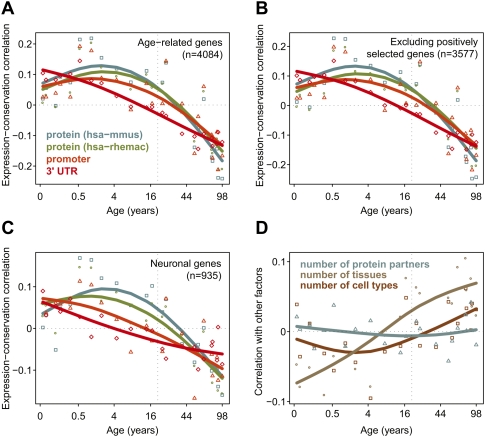
Supporting the notion of a development-aging antagonism, we observe that the extent of stabilizing/purifying selection, as reflected by sequence conservation in protein-coding and regulatory regions, decreases over lifetime (Fig. 7A; Methods). Specifically, we find that genes highly expressed at old age are substantially less conserved than genes highly expressed in early postnatal life. This observation is consistent with theories postulating a decrease in stabilizing selection pressure past optimal reproductive age (Rose 1991). This decrease in conservation remains after controlling for other factors correlated with sequence conservation (Duret and Mouchiroud 2000), such as expression breadth, cell type specificity, number of interaction partners, or positive selection (Fig. 7B–D). Similarly, expression changes taking place in aging are less conserved between humans and macaques, compared to developmental changes (Supplemental Fig. S13A,B). Evolutionarily, relaxation of stabilizing selection pressure on regulatory motifs of genes expressed at higher levels in old age would facilitate fixation of regulatory traits detrimental in aging.
Figure 7.
Diminishing stabilizing selection pressure with age. (A) The symbols and fitted spline curves show the stabilizing selection scores (SSS) calculated for protein coding (blue and green), promoter (orange), and 3′-untranslated (UTR) (red) regions (Methods). The SSS indicate correlation between conservation values and standardized expression levels per individual, across 4084 age-related genes. Conservation scores are corrected for variation in mutation rates. The x-axis shows age of individuals on the (age)¼ scale. Positive SSS indicate above-average correlation between expression levels and sequence conservation among genes, at a certain age. The dashed vertical line indicates 20 yr of age, when brain maturation is largely complete (de Graaf-Peters and Hadders-Algra 2006). (B) Same as A, but excluding genes possibly under positive selection (Methods). (C) Same as A, but only using genes with enriched expression levels in neurons. (D) Correlation between standardized expression levels and potential confounding factors across age-related genes: number of protein–protein interaction partners (blue), number of tissues (gray) or cell types (brown) a gene is expressed in (i.e., expression breath).
Taken together, our results indicate that the expression dynamics and regulatory interactions during development and aging are not independent. Transcriptional regulation through miRNAs and TFs appears as yet another mode of senescence, confirming the complex nature of this biological phenomenon. Further studies should determine the full set of regulatory interactions occurring over lifetime in the brain and other tissues, and reveal the exact role of developmental regulatory processes in determining the onset and the progression of aging.
Methods
Analyses were conducted in the R environment (http://www.r-project.org/). All miRNA names are based on miRBase (v. 11; http://www.mirbase.org/). We use HGNC gene symbols obtained from Ensembl (v. 54; http://www.ensembl.org).
Sample collection
We collected superior frontal gyrus samples from postmortem brains of healthy humans and rhesus macaques (Supplemental Tables S1, S2; for details, see Supplemental Text S1).
mRNA isolation, hybridization to microarrays, and data preprocessing
The experimental procedures followed Khaitovich et al. (2005) and Somel et al. (2009) and are described in Supplemental Text S1. Briefly, samples prepared from 2 μg of total RNA were processed and hybridized to Affymetrix Human Gene 1.0 ST arrays following the standard Affymetrix protocol. We used the R Bioconductor “affy” library (Gautier et al. 2004) and in-house code to extract, background-correct, quantile normalize, and summarize probe intensities. For macaque data preprocessing, we only used probes that perfectly and uniquely match the macaque genome (rhemac2).
miRNA isolation, sequencing, and quantification
The miRNA experiments and data preprocessing followed Hu et al. (2009) and are described in Supplemental Text S1. Briefly, low-molecular-weight RNA was isolated and sequenced following the Small RNA Sample Preparation Protocol (Illumina). For data preprocessing, trimmed sequences (18–26-nt long) were mapped to the human genome (hg18), requiring a perfect match. To annotate and quantify miRNAs, we used miRBase version 11 (Griffiths-Jones et al. 2006), only including sequences with copy number ≥ 2 and mapping within the proximity of mature miRNAs. We also included small RNA sequences mapping to the opposite arm of the precursor hairpin as novel miRNAs (Hu et al. 2009). For identifying rhesus macaque mature miRNA, we used reciprocal BLAST with human sequences from miRBase. This yielded 306 orthologous macaque miRNAs expressed in our data set and with high sequence similarity between species (Supplemental Fig. S8F).
Protein sample preparation, sequencing, and peptide identification
Protein expression measurements were performed using a label-free proteomic approach and peptide (spectral) counting, using a pH continuous online gradient (pCOG) 2D LC-MS/MS system (Fu et al. 2009). This method provides comparably accurate measurements for protein relative abundance (Old et al. 2005) and is widely used in large-scale comparative proteomic studies (e.g., Domon and Aebersold 2006; Nesvizhskii et al. 2007; Lu et al. 2009). We followed the procedure described in Fu et al. (2009), with a number of modifications (Supplemental Text S1). Briefly, starting from frozen prefrontal cortex tissue from 12 humans (also used in the mRNA data set; Supplemental Table S1), we extracted and trypsin-digested protein samples. These were loaded on ion exchange columns and eluted using a pH continuous gradient buffer. Fractions were automatically loaded on alternative trap columns and analyzed on an LTQ mass spectrometer (ThermoFinnigan). Peptides were identified by searching against the IPI human database (IPI human v3.61). We estimated FDR of peptide identification by reversed database searching. Peptide data were mapped to Ensembl genes, summarized per gene, and quantile-normalized (Supplemental Text S1).
Variance explained by age and other factors
We calculated the average expression variance explained by age by fitting a cubic polynomial formula (Somel et al. 2009), while transforming individual age to ranks (Supplemental Fig. S14; Supplemental Text S1). We estimated the significance of the proportion of variance explained by 300 random permutations of age. We calculated variance explained by sex, RIN, and PMI in the human cortex data set using the same procedure (Supplemental Text S1).
Age test
We tested the effect of age on expression level using polynomial regression models, following Somel et al. (2009). For each gene, we choose the best regression model with age as predictor and expression level as response, using families of cubic polynomial regression models and the “adjusted r 2” criterion (Faraway 2002). The significance of the chosen regression model was estimated using the F-test, and the FDR was calculated by 1000 random permutations of age. The median of the permutation distribution was used as the null expectation. For the mRNA and miRNA data sets, genes with an age-test FDR < 0.1% were termed “age-related.”
Clustering genes in groups
We used a modified version of the k-means algorithm to cluster age-related genes into groups with similar expression profiles (Supplemental Text S1).
Transition point analysis
We estimated the age when an early-life expression change trend gives way to a late-life trend (e.g., in group 1 in Fig. 2A, when the trend of a decrease in expression in childhood ceases at early adulthood). The procedure is based on segmental linear regression, which compares the fit of two linear regression models (for early and late life) to a single linear regression model (for the full life span). We used the log2 age scale (which allows resolution of developmental changes) for identifying transition points in early life, and the linear age scale for transition points in late life (Supplemental Table S3; Supplemental Text S1).
miRNA/TF binding site estimation
We used TargetScan5.0 (Lewis et al. 2005) for miRNA binding site identification and the MATCH algorithm based on TRANSFAC (Kel et al. 2003) for conserved TF binding site identification. For the latter, we used 2000 bp +/− around the transcription start site as proximal promoter, and 17-way vertebrate phastCons scores (Siepel et al. 2005) for conservation (Supplemental Text S1).
Regulator miRNA/TF identification
The identification of miRNA regulators of brain development/aging is described in Supplemental Figure S15 and Supplemental Text S1. For identifying an miRNA as a regulator, we required two conditions: (1) enrichment of targets in a coexpressed gene group, compared to other miRNAs and all other gene groups (at one-sided HT, P < 0.05); and (2) excess of negative correlation (in expression profiles) with its targets in the enriched gene group, compared to non-enriched miRNA (at one-sided binomial tests [BT], P < 0.05). We calculated miRNA–target correlations separately for development and aging periods, using 20 yr and 5 yr as borderlines for human and macaque, respectively, representing approximate times of age at first reproduction (Walker et al. 2006a,b). The same test was applied to identify putative regulatory TFs. As TFs (in contrast to miRNAs) showed both positive and negative correlations with their targets, we tested excess of both positive and negative correlations.
Testing predicted regulators
Additional tests to confirm the predicted target–regulator pairs are described in Supplemental Text S1. Briefly:
For testing possible age-independent effects on miRNA–target correlation, we used interpolated miRNA and mRNA profiles, rather than individual expression levels (Supplemental Fig. S8A).
We identified regulators conserved between macaques and humans by preselecting miRNAs with the same direction of expression change with age as in humans and testing the excess of negative miRNA–target correlations (using miRNAs enriched among human coexpressed gene groups; see above). We also checked if putative regulator miRNAs identified in humans show a tendency for excess negative correlations (mean r < 0) with their targets in macaques (Supplemental Table S5). Note that the macaque data, presumably due to their shorter age range, showed less age-related change and weaker correlations than human (both in miRNAs and mRNAs).
We tested whether miRNAs and their putative targets show coordinated divergence between humans and macaques (negative correlation in human–macaque differences between miRNAs and mRNAs) compared to random pairs of miRNAs and genes (Supplemental Fig. S8C,D).
We tested if among 298 miRNA–target pairs, for which macaque (but not human) has a mutation in the miRNA binding site, we see miRNA–target negative correlation in humans, but not in macaques (Supplemental Fig. S8E).
We compared our miRNA–target predictions with experimentally verified miRNA target gene sets: Tarbase (http://diana.cslab.ece.ntua.gr/tarbase/tarbase_download.php; Papadopoulos et al. 2009), Mirwalk (http://www.ma.uni-heidelberg.de/apps/zmf/mirwalk/contact.html), a published collection of results from multiple experiments (Khan et al. 2009), and an miR-181 overexpression experiment (Baek et al. 2008).
We estimated the FDR of the binding site enrichment and the regulator–target correlation tests, using permutations.
Functional analysis
We used the Gene Ontology (GO) (Ashburner et al. 2000) and KEGG (Kanehisa et al. 2008) databases for testing functional enrichment among gene groups. For identifying cell-type-specific expression, we used expression levels measured from purified mouse neurons, astrocytes, and oligodendrocytes from a published study (Cahoy et al. 2008). For details see Supplemental Text S1.
Evolutionary conservation analysis
For each human gene, we calculated conservation scores for 3′-untranslated and proximal promoter regions using the phastCons 18-way Placental Mammal Conservation Track (Siepel et al. 2005). These were corrected for variance in mutation rates among genes, using intronic conservation. We used dN/d S ratios from Ensembl as estimates for amino acid conservation. The scores shown in Figure 7 were calculated as the Pearson correlation coefficient between sequence conservation levels and the standardized expression levels of an individual, across age-related genes. Because the score is calculated using expression levels standardized per gene (z-transformed to mean = 0 and SD = 1) across the 23 humans, it is independent of the general positive correlation between mean expression levels and conservation. For details see Supplemental Text S1.
Acknowledgments
We thank the NICHD Brain and Tissue Bank for Developmental Disorders, the Chinese Brain Bank Center, and, in particular, H. Ronald Zielke and Jiapei Dai for providing the samples; Fenny Xue, Lin Tang, Xiling Liu, Yi Huang, Jia Jia Xu, Kai Weng, and Ningyi Shao for assistance; and R. Ed Green, Martin Vingron, Brian Cusack, Dan Rujescu, Jing Dong Jackie Han, Jennifer E. Dent, Joao Pedro de Magalhães, and two anonymous reviewers, all members of the Comparative Biology Group in Shanghai, for helpful discussions and suggestions. We thank the Ministry of Science and Technology of the People's Republic of China (grant no. 2007CB947004), the Chinese Academy of Sciences (grant nos. KSCX2-YW-R-094 and KSCX2-YW-R-251), the Shanghai Institutes for Biological Sciences (grant no. 2008KIT104), the Max Planck-Society, and the Bundesministerum fuer Bildung und Forschung for financial support.
Footnotes
[Supplemental material is available online at http://www.genome.org. All mRNA, miRNA, and protein expression data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under series accession no. GSE18069.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.106849.110.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: Tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP 2008. The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M, Slack F 2005. A developmental timing microRNA and its target regulate life span in C. elegans. Science 310: 1954–1957 [DOI] [PubMed] [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK 2008. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell 134: 291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA, et al. 2008. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci 28: 264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Chi SW, Zang JB, Zang JB, Mele A, Mele A, Darnell RB, Darnell RB 2009. Argonaute hits-clip decodes microRNA–mRNA interaction maps. Nature 460: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM, Maris JM 2008. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res 6: 735–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Chisum HJ, Townsend J, Cowles A, Covington J, Egaas B, Harwood M, Hinds S, Press GA 2000. Normal brain development and aging: Quantitative analysis at in vivo MR imaging in healthy volunteers. Radiology 216: 672–682 [DOI] [PubMed] [Google Scholar]
- de Graaf-Peters VB, Hadders-Algra M 2006. Ontogeny of the human central nervous system: What is happening when? Early Hum Dev 82: 257–266 [DOI] [PubMed] [Google Scholar]
- de Magalhães JP, Church GM 2005. Genomes optimize reproduction: Aging as a consequence of the developmental program. Physiology 20: 252–259 [DOI] [PubMed] [Google Scholar]
- de Magalhães J, Costa J 2009. A database of vertebrate longevity records and their relation to other life-history traits. J Evol Biol 22: 1770–1774 [DOI] [PubMed] [Google Scholar]
- de Magalhães JP, Curado J, Church GM 2009. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25: 875–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogini DB, Ribeiro PA, Rocha C, Pereira TC, Lopes-Cendes I 2008. MicroRNA expression profile in murine central nervous system development. J Mol Neurosci 35: 331–337 [DOI] [PubMed] [Google Scholar]
- Domon B, Aebersold R 2006. Mass spectrometry and protein analysis. Science 312: 212–217 [DOI] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D 2000. Determinants of substitution rates in mammalian genes: Expression pattern affects selection intensity but not mutation rate. Mol Biol Evol 17: 68–70 [DOI] [PubMed] [Google Scholar]
- Eldredge LC, Gao XM, Quach DH, Li L, Han X, Lomasney J, Tourtellotte WG 2008. Abnormal sympathetic nervous system development and physiological dysautonomia in EGR3-deficient mice. Development 135: 2949–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erraji-Benchekroun L, Underwood MD, Arango V, Galfalvy H, Pavlidis P, Smyrniotopoulos P, Mann JJ, Sibille E 2005. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry 57: 549. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. 2007. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3a and 3b. Proc Natl Acad Sci 104: 15805–15810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraway J 2002. Practical regression and ANOVA using R. http://cran.r-project.org/doc/contrib/Faraway-PRA.pdf
- Finch CE 1976. The regulation of physiological changes during mammalian aging. Q Rev Biol 51: 49–83 [DOI] [PubMed] [Google Scholar]
- Fu X, Fu N, Guo S, Yan Z, Xu Y, Hu H, Menzel C, Chen W, Li Y, Zeng R, et al. 2009. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics 10: 161 doi: 10.1186/1471-2164-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA 2004. affy––analysis of Affymetrix Genechip data at the probe level. Bioinformatics 20: 307–315 [DOI] [PubMed] [Google Scholar]
- Glantz LA, Gilmore JH, Hamer RM, Lieberman JA, Jarskog LF 2007. Synaptophysin and postsynaptic density protein 95 in the human prefrontal cortex from mid-gestation into early adulthood. Neuroscience 149: 582–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Burgos G, Kroener S, Zaitsev AV, Povysheva NV, Krimer LS, Barrionuevo G, Lewis DA 2008. Functional maturation of excitatory synapses in layer 3 pyramidal neurons during postnatal development of the primate prefrontal cortex. Cereb Cortex 18: 626–637 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B 2008. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci 105: 6415–6420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Yan Z, Xu Y, Hu H, Menzel C, Zhou YH, Chen W, Khaitovich P 2009. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics 10: 413 doi: 10.1186/1471-2164-10-413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher PR, Dabholkar AS 1997. Regional differences in synaptogenesis in human cerebral cortex. J Comp Neurol 387: 167–178 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan H, Hill K, Lancaster J, Hurtado AM 2000. A theory of human life history evolution: Diet, intelligence, and longevity. Evol Anthropol 9: 156–185 [Google Scholar]
- Kel AE, Gössling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E 2003. MATCH: A tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res 31: 3576–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Hellmann I, Enard W, Nowick K, Leinweber M, Franz H, Weiss G, Lachmann M, Paabo S 2005. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309: 1850–1854 [DOI] [PubMed] [Google Scholar]
- Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS 2009. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol 27: 549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Weindruch R, Prolla TA 2000. Gene-expression profile of the ageing brain in mice. Nat Genet 25: 294–297 [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Lin W, Xin H, Zhang Y, Wu X, Yuan F, Wang Z 1999. The human REV1 gene codes for a DNA template-dependent DCMP transferase. Nucleic Acids Res 27: 4468–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerch PM, Lu T, Dakin KA, Vann JM, Isaacs A, Geula C, Wang J, Pan Y, Gabuzda DH, Li C, et al. 2008. Evolution of the aging brain transcriptome and synaptic regulation. PLoS ONE 3: e3329 doi: 10.1371/journal.pone.0003329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Pan Y, Kao S, Li C, Kohane I, Chan J, Yankner BA 2004. Gene regulation and DNA damage in the ageing human brain. Nature 429: 883–891 [DOI] [PubMed] [Google Scholar]
- Lu A, Wiśniewski JR, Mann M 2009. Comparative proteomic profiling of membrane proteins in rat cerebellum, spinal cord, and sciatic nerve. J Proteome Res 8: 2418–2425 [DOI] [PubMed] [Google Scholar]
- Marsh R, Gerber AJ, Peterson BS 2008. Neuroimaging studies of normal brain development and their relevance for understanding childhood neuropsychiatric disorders. J Am Acad Child Adolesc Psychiatry 47: 1233–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Vitek O, Aebersold R 2007. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat Methods 4: 787–797 [DOI] [PubMed] [Google Scholar]
- Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG 2005. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics 4: 1487–1502 [DOI] [PubMed] [Google Scholar]
- Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG 2009. The database of experimentally supported targets: A functional update of Tarbase. Nucleic Acids Res 37: D155–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Sethares C, Luebke JI 2008. Synapses are lost during aging in the primate prefrontal cortex. Neuroscience 152: 970–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polleux F, Ince-Dunn G, Ghosh A 2007. Transcriptional regulation of vertebrate axon guidance and synapse formation. Nat Rev Neurosci 8: 331–340 [DOI] [PubMed] [Google Scholar]
- Rodwell GEJ, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, Xiao W, Mindrinos M, Crane E, Segal E, et al. 2004. A transcriptional profile of aging in the human kidney. PLoS Biol 2: e427 doi: 10.1371/journal.pbio.0020427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MR 1991. Evolutionary biology of aging. Oxford University Press, New York [Google Scholar]
- Salthouse TA 2009. When does age-related cognitive decline begin? Neurobiol Aging 30: 507–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G 2009. MicroRNAs at the synapse. Nat Rev Neurosci 10: 842–849 [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME 2006. A brain-specific microRNA regulates dendritic spine development. Nature 439: 283–289 [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova L, Gräfe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG 2005. Regulation of miRNA expression during neural cell specification. Eur J Neurosci 21: 1469–1477 [DOI] [PubMed] [Google Scholar]
- Somel M, Franz H, Yan Z, Lorenc A, Guo S, Giger T, Kelso J, Nickel B, Dannemann M, Bahn S, et al. 2009. Transcriptional neoteny in the human brain. Proc Natl Acad Sci 106: 5743–5748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell ER, Thompson PM, Toga AW 2004. Mapping changes in the human cortex throughout the span of life. Neuroscientist 10: 372–392 [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ 2008. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 9: 219–230 [DOI] [PubMed] [Google Scholar]
- Toga AW, Thompson PM, Sowell ER 2006. Mapping brain maturation. Trends Neurosci 29: 148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker R, Burger O, Wagner J, Von Rueden CR 2006a. Evolution of brain size and juvenile periods in primates. J Hum Evol 51: 480–489 [DOI] [PubMed] [Google Scholar]
- Walker R, Gurven M, Hill K, Migliano A, Chagnon N, De Souza R, Djurovic G, Hames R, Hurtado A, Kaplan H, et al. 2006b. Growth rates and life histories in twenty-two small-scale societies. Am J Hum Biol 18: 295–311 [DOI] [PubMed] [Google Scholar]
- Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, et al. 2008. NF-κB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell 14: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu P, Zhu H, Xu Y, Ma C, Dai X, Huang L, Liu Y, Zhang L, Qin C 2009. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer's disease, inhibits BCL2 translation. Brain Res Bull 80: 268–273 [DOI] [PubMed] [Google Scholar]
- Williams GC 1957. Pleiotropy, natural-selection, and the evolution of senescence. Evolution 11: 398–411 [Google Scholar]
- Xue H, Xian B, Dong D, Xia K, Zhu S, Zhang Z, Hou L, Zhang Q, Zhang Y, Han JJ 2007. A modular network model of aging. Mol Syst Biol 3: 147 doi: 10.1038/msb4100189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Ferlito M, Lowenstein CJ 2008. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci 105: 13421–13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, Sonu R, Vogel H, Crane E, Mazan-Mamczarz K, Rabkin R, Davis RW, Becker KG, Owen AB, Kim SK 2006. Transcriptional profiling of aging in human muscle reveals a common aging signature. PLoS Genet 2: e115 doi: 10.1371/journal.pgen.0020115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn JM, Poosala S, Owen AB, Ingram DK, Lustig A, Carter A, Weeraratna AT, Taub DD, Gorospe M, Mazan-Mamczarz K, et al. 2007. AGEMAP: A gene expression database for aging in mice. PLoS Genet 3: e201 doi: 10.1371/journal.pgen.0030201 [DOI] [PMC free article] [PubMed] [Google Scholar]