Abstract
ASH1 encodes a protein that is localized specifically to the daughter cell nucleus, where it has been proposed to repress transcription of the HO gene. Using Ash1p purified from baculovirus-infected insect cells, we have shown that Ash1p binds specific DNA sequences in the HO promoter. DNase I protection analyses showed that Ash1p recognizes a consensus sequence, YTGAT. Mutation of this consensus abolishes Ash1p DNA binding in vitro. We have shown that Ash1p requires an intact zinc-binding domain in its C terminus for repression of HO in vivo and that this domain may be involved in DNA binding. A heterologous DNA-binding domain fused to an N-terminal segment of Ash1p functions as an active repressor of transcription. Our studies indicate that Ash1p is a DNA-binding protein of the GATA family with a separable transcriptional repression domain.
The budding yeast Saccharomyces cerevisiae divides to give rise to two cells with distinct developmental fates (1, 2). After cell division, the mother cell is competent to transcribe the HO gene and thus switch mating type, whereas the daughter cell is not competent to transcribe HO. The difference between mother and daughter cells appears to be due to the presence in daughter cells of a negative regulator, Ash1p, which turns off HO transcription in daughter cells. In mutants lacking Ash1p, daughter cells switch mating type; overexpression of ASH1 inhibits mating type switching in mothers (3, 4). Ash1p has 588 amino acid residues and is predicted to contain a zinc-binding domain related to those of the GATA family of transcriptional regulators. Ash1p is localized to the daughter cell nucleus in cells that have undergone nuclear division. Thus, Ash1p is a cell-fate determinant that is asymmetrically localized to the daughter cell nucleus, where it is a negative regulator of HO transcript level, presumably by repressing HO transcription.
HO is subject to many levels of transcriptional control; it is expressed only in haploid cells, mother cells, and the G1 phase of the cell cycle (5, 6). Consequently, appropriate expression of HO relies on the integration of signals from cell-type information, mother or daughter identity, and cell-cycle status and is controlled through regulatory sequences in the HO promoter. A large number of genes encoding activators and repressors of transcription, as well as chromatin remodeling factors, participate in HO expression. Recently, HO regulation was reported to depend on the highly ordered and temporally regulated recruitment and activities of Swi5p, Swi/Snf, SAGA, and SBF, in succession, for expression (7, 8).
Cell-cycle and mother/daughter information is integrated through distinct upstream regulatory sequences (URS) of the HO promoter, URS2 and URS1, respectively. Cell-cycle regulation of HO expression requires a transcriptional activator (SBF) that binds to many sites in the upstream regulatory sequence (URS2) of the HO promoter in a cell-cycle–controlled manner (9–11).
The SWI5 gene product is required for mother cell-specific transcription of HO and functions by binding to sites in URS1 after its cell cycle-regulated entry into the nucleus (12, 13). Biochemical studies have shown that Swi5p binds cooperatively at these sites with the product of the PHO2 gene, a coactivator of many genes (14–17). Overexpression of Swi5p allows daughter cells to switch mating type (18). Unlike Ash1p, Swi5p is found in the nuclei of both mother and daughter cells (19).
Our approach to understanding the mechanism of asymmetric expression of HO focused on the product of the ASH1 gene as a key determinant of asymmetric cell fate. We have undertaken biochemical and genetic studies to determine the mechanism of ASH1-mediated repression of HO. Here we report that Ash1p binds directly to sequences in the HO promoter DNA; DNase I footprinting studies have allowed identification of an Ash1p consensus recognition sequence, YTGAT. Mutation of this consensus abolishes DNA binding by Ash1p in vitro. We have also identified presumptive DNA-binding and transcriptional-repression domains of Ash1p that are required for its function in vivo.
Materials and Methods
Cells and Viruses.
Sf9 and High Five cells were purchased from Invitrogen. Sf9 cells were grown in supplemented Grace's medium with 10% FBS. High Five cells were grown in plates in Ex-Cell 405 medium (JRH Biosciences, Lenexa, KS). For protein expression, 2 × 107 High Five cells were seeded onto each 150 × 25-mm plate and infected at a multiplicity of infection of 10 with recombinant baculoviruses in 20 ml of medium. Cells were harvested after incubating for 72 h at 27°C.
Construction of Recombinant Baculoviruses.
Recombinant baculoviruses were constructed essentially as described in the Bac-To-Bac Baculovirus Expression System Instruction Manual (Life Technologies, Rockville, MD). ASH1 was subcloned into the pFastBac HTb donor plasmid and transformed into DH10BAc cells for transposition into the bacmid. Recombinant bacmid DNA was transfected into Sf9 cells by using CellFECTIN. Medium containing recombinant baculovirus was harvested 72 h after transfection. The virus stock was amplified by several rounds of infection of Sf9 cells. The virus stock titer was determined by an end-point dilution assay.
Purification of His-Ash1p from Infected Cells.
A total of 2 × 108 High Five cells infected as described above were harvested. Subsequent steps were performed at 4°C. Buffers contained Complete (EDTA-free) Protease Inhibitor mixture (Roche Molecular Biochemicals). Cell pellets were resuspended in buffer containing 8 M urea, 0.1 M NaH2PO4, and 0.01 M Tris⋅HCl (pH 8.0). The resuspension was centrifuged to remove any insoluble material, and the supernatant was applied to a nickel resin column (Qiagen, Chatsworth, CA). The bound material was renatured on the column by using a series of buffers containing 0.1 M NaCl, 10% glycerol, 0.02 M Tris⋅HCl (pH 8.0), and decreasing concentrations of urea (6 M, 4 M, 2 M, 1 M) followed by a buffer lacking urea. His-Ash1p was eluted by using elution buffer: 0.1 M NaCl/0.02 M Tris⋅HCl (pH 8.0)/0.25 M imidazole/10% glycerol. The eluted material was dialyzed into 0.5 mM DTT/0.1 M NaCl/0.02 M Tris⋅HCl (pH 8.0)/10% glycerol.
Anti-Ash1p Peptide Antibodies.
Antibodies specific to the N-terminal Ash1p sequence SSLYIKTPLHALSAGPDSHANSSYY (+GGC at the C terminus to promote solubility) were prepared from rabbits by using standard methods at Caltag Laboratories (Burlingame, CA). Antibodies were affinity-purified over resin conjugated to peptide antigen and dialyzed into buffer described above.
Genetic and Molecular Biological Methods.
Yeast genetic methods were performed as described (20). DNA manipulations were as described (21).
Electrophoretic Mobility-Shift Analysis.
Gel shifts were analyzed by using standard methods (14) with the following modifications. Reactions were run on an 8% polyacrylamide gel for 2 h at 4°C. An unknown but dilute concentration of 1–6 μl of His-Ash1p was added, depending on the experiment. Affinity-purified anti-Ash1p peptide antibodies (2 μl of unknown concentration) were added to gel-shift experiments.
DNase I Footprinting Experiments.
Footprint analysis was carried out by using standard techniques (14) and varied electrophoresis conditions to maximize resolution of bound regions. Probes were PCR products derived from the HO promoter by using oligonucleotides OM38 and OM39, subsequently labeled with [γ-32P]ATP or restriction fragments of PCR products (OM38 = GGCAAAGAAATCGATGCATACC and OM39 = AAGCACATCGATTATTTGATACCCC). Maxam–Gilbert sequencing (22) was performed by using a commercially available kit from Sigma to identify the consensus sequence bound at all sites.
Strains.
Yeast strains used were derivatives of W303 (see Table 1).
Table 1.
Strains
| Strain | Genotype | Plasmid |
|---|---|---|
| IH3964 | MATα ho∷HO-ADE2 can1∷HO-CAN1 ash1Δ∷TRP1 | None |
| MY158 | MATα ho∷HO-ADE2 can1∷HO-CAN1 ash1Δ∷TRP1 | pMM111 (ASH1) |
| MY159 | MATα ho∷HO-ADE2 can1∷HO-CAN1 ash1Δ∷TRP1 | pMM112 (ASH1C502R) |
| MY160 | MATα ho∷HO-ADE2 can1∷HO-CAN1 ash1Δ∷TRP1 | pRS316 (vector) |
| MY185 | MATα ho∷HO-ADE2 can1∷HO-CAN1 ash1Δ∷TRP1 | pAJ1621 pEG203 (pLexA) |
| MY186 | MATα ho∷HO-ADE2 can1∷HO-CAN1 ash1Δ∷TRP1 | pAJ1621 pMM94 (pLexA-ASH1) |
| MY208 | MATα ho∷HO-ADE2 can1∷HO-CAN1 ash1Δ∷TRP1 | pAJ1621 pMM129 (pLexA-ΔC90) |
| MY209 | MATα ho∷HO-ADE2 can1∷HO-CAN1 ash1Δ∷TRP1 | pAJ1621 pMM131 (pLexA-ΔN300) |
| MY210 | MATα ho∷HO-ADE2 can1∷HO-CAN1 ash1Δ∷TRP1 | pAJ1621 pMM134 (pLexA-ΔC288) |
| MY211 | MATα ho∷HO-ADE2 can1∷HO-CAN1 ash1Δ∷TRP1 | pAJ1621 pMM127 (pLexA-C502R) |
| MY212 | MATα ho∷HO-ADE2 can1∷HO-CAN1 ash1Δ∷TRP1 | pAS174 (2μ ASH1) |
| MY6 | MATα ho | None |
| MY115 | MATα ho ash1Δ∷LEU2 | None |
Strains used were derivatives of W303, whose complete genotype is ade2-1 trp1-1 can1-100 leu2-3,112 his3-11,13 ura3-52 GAL+ psi+, bar1-1.
Plasmid Construction.
pAJ1621 and pLexA (pEG203) were gifts from Sandy Johnson's and Erin O'Shea's laboratories, respectively (University of California, San Francisco). All pLexA constructs containing fragments of ASH1 were PCR derivatives of pMM62 (containing the ASH1 ORF) cloned into the XhoI site of pEG203. pLexA-Ash1p (pMM94) was generated by using oligonucleotides OM20 and OM21 (OM20 = GGCCGGCTCGAGATGTCAAGCTTA and OM21 = CCGGCCCTCGAGAGGATGACCAATCTATTGCGC). pLexA-ΔC90 (pMM129) was made by using OM58 and OM60 (OM58 = GGTTCCGCGTGGATCCTCAAGCTTATAC and OM60 = CCGGGATCCTTACACTCTTGTGGTGTGACG). pLexA-ΔN300 (pMM131) was made by using OM59 and OM62 (OM59 = GCGGATCCCCGTAGAATTAGACAAGTCC and OM62 = CCGGAATTCATGGAGCGCACGCTTAGAGG). LexA-ΔC288 (pMM134) was generated by using OM58 and OM63 (OM63 = CCGGAATTCTTATCCTCTAAGCGTCGTGCGCTCCAT). LexA-C502R (pMM127) was made by using OM20 and OM21 on the pASH1C502R plasmid template (pMM83) described below.
Construction of the ASH1 C502R Mutant (pMM83).
The zinc-binding domain of Ash1p was mutated at residue 502 by using two rounds of sequential PCR (products of OM34 + OM35 added to the products of OM36 + OM37 and amplified in a second reaction after gel purification with OM34 + OM37). Primer sequences used were as follows: OM34 = GGCCGGAGGCCTCTTTTTTTGAGGGG; OM35 = ATCACTCGAATGTCGCGACACGCACAC; OM36 = GTGTGCGTGTCGCGACATTCGAGTGAT; and OM37 = CCGGGTACCCTTCAATTTCGC.
The PCR product was gel-purified, digested with StuI and KpnI, and used to replace the wild-type 790-bp StuI/KpnI fragment of ASH1 to generate the C502R allele.
Construction of Consensus Binding Site Mutations.
The YTGAT consensus was mutated at −1730 (G to T) by using PCR mutagenesis and oligonucleotides OM96 and OM97. The mutated product was subcloned into the Invitrogen pCR2.1 vector and used in successive PCR experiments to generate pMM160, (a triple mutant with G-to-T mutations in Ash1p consensus binding sites at −1784, −1730, and −1321) subcloned into pCR2.1. The −1784 mutation was made by using OM98 and OM99. The −1321 mutation was made by using OM110 and OM111. A mutation at −1733 outside the YTGAT consensus was generated by using oligonucleotides OM79 and OM80. The oligonucleotide sequences show mutated bases as bold characters and are as follows: OM79 = GGAACTAAACGGTAAAGATAAAATATCACC; OM80 = GGTGATATTTTATCTTTACCGTTTAGTTCC; OM96 = CAAAAAAAGGCGGATAAAGATGTATG; OM97 = CATACATCTTTATCCGCCTTTTTTTG; OM98 = CTCTTTATTTTTCCAAATAAGAAAAATTAATATG; OM99 = CATATTAATTTTTCTTATTTGGAAAAATAAAGAG; OM110 = GCCTGCGATGAGATACATAAATTTAAAAAAAAAACCAGC; and OM111 = GCTGGTTTTTTTTTTAAATTTATGTATCTCATCGCAGGC.
Immunoblot Analysis.
Immunoblot analysis for Ash1p was performed by using standard techniques (23) with the following modifications. Urea extracts were prepared from ash1Δ strains transformed with either empty vector or plasmids carrying a wild-type or C502R mutant ASH1 allele. Extracts were prepared by using buffer containing 8 M urea and six bead-beater pulses of 30 sec each at 4°C. Anti-Ash1p peptide antibodies were diluted 1:200 (vol/vol) in Tris-buffered saline and incubated for 12 h at 4°C.
Canavanine Resistance Assays.
In vivo repression analysis of the integrated HO-CAN1 reporter was performed on glucose plates lacking histidine (to select for ASH1 plasmids) in the presence of 60 μg/ml canavanine (Sigma).
β-Galactosidase Assays.
Quantitative β-galactosidase measurements were performed by using standard techniques (24).
Results
Ash1p Is a Site-Specific DNA-Binding Protein.
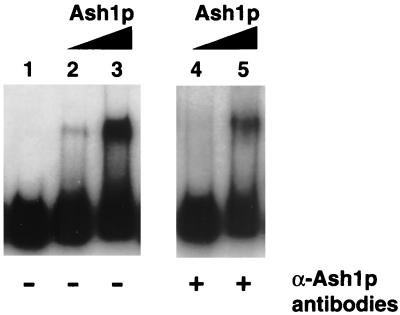
To determine the mechanism by which Ash1p negatively regulates expression of HO, a recombinant version of Ash1p tagged with six histidine residues (His-Ash1p) was purified from baculovirus-infected cells and assayed for ability to bind DNA by gel-shift analysis. On a 705-bp HO promoter fragment (Fig. 1, probe 3) that spans the region from −1915 to −1220, His-Ash1p caused a shift of the radiolabeled DNA [Fig. 2, lanes 2 and 4 (1 μl of His-Ash1p) and 3 and 5 (6 μl of His-Ash1p)], whereas a column eluate from uninfected cells did not (data not shown). Anti-Ash1p antibodies inhibited His-Ash1p gel-shift activity (Fig. 2, lanes 4 and 5). Inhibition by anti-Ash1p antibodies was specific; anti-Ash1p antibodies did not inhibit the gel-shift ability of bacterial recombinant Swi5p, which also binds probe 3 (data not shown).
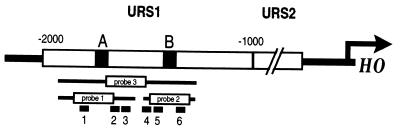
Figure 1.
Schematic representation of the HO promoter. Swi5p-binding sites are designated A and B and are located at −1800 and −1300, respectively. DNA probes used for electrophoretic mobility-shift and DNase I footprint analyses are indicated as probe 1 (approximately −1915 to −1350), probe 2 (−1350 to −1220), and probe 3 (−1915 to −1220). Ash1p-binding sites as determined by in vitro DNase I footprint analysis are illustrated as black boxes numbered 1 through 6.
Figure 2.
His-Ash1p binds HO promoter DNA. His-Ash1p shifted a radiolabeled HO probe corresponding to −1915 to −1220 of the HO promoter (lanes 2–5). The addition of anti-Ash1p peptide antibodies inhibited binding of His-Ash1p to DNA (lanes 4 and 5).
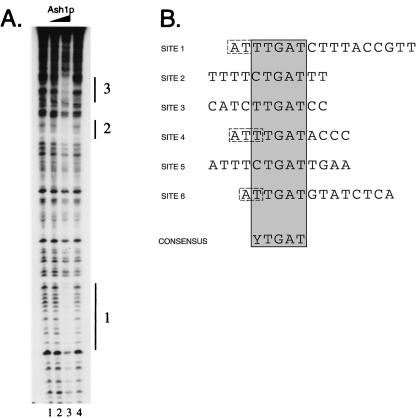
To identify the site(s) bound by His-Ash1p, DNase I footprinting experiments were performed by using fragments of the HO promoter. On a fragment of URS1 that spans −1915 to −1350 (Fig. 1, probe 1), the addition of His-Ash1p (1 μl in lane 2, 5 μl in lane 3) resulted in protection of three distinct regions of the promoter at approximately −1870 (region 1), −1785 (region 2), and −1730 (region 3) (Fig. 3A, lane 3). Various electrophoresis conditions were used to increase resolution of the protected regions, and Maxam-Gilbert DNA sequencing reactions were performed to identify the DNA sequences protected by His-Ash1p (M.E.M., unpublished results). Similar experiments were carried out with probe 2 (Fig. 1). A total of six protected regions of HO were characterized (Fig. 1). From the sequences of the protected regions, the consensus YTGAT was determined (Fig. 3B). His-Ash1p appears to protect a region of DNA larger than the YTGAT consensus at all sites analyzed. The HO upstream region (approximately −2000 to + 1) predicts a total of 20 matches to the YTGAT consensus sequence.
Figure 3.
(A) His-Ash1p binds to specific sequences in HO promoter DNA. The addition of increasing amounts of His-Ash1p (lanes 2 and 3) resulted in the specific protection of sites at approximately −1870 (region 1), −1785 (region 2), and −1730 (region 3). Lanes 1 and 4 show the DNase I cleavage pattern in the absence of added recombinant protein. (B) Ash1p binding site identification. DNase I-protected regions from probe 1 and probe 2 were analyzed for consensus sequences. All bases shown were protected from DNase I cleavage; bases shown in dashed line boxes were assumed with confidence to be protected based on their positions in cleavage site-poor regions of the probe. The consensus is shown at the bottom.
The DNase I footprint patterns of Swi5p, Pho2p, and the Swi5/Pho2p heteromer have been characterized on fragments of the HO promoter (14, 15, 17). We tested the ability of Ash1p to disrupt or alter the DNase I footprints of Swi5p, Pho2p, or Swi5p/Pho2p and found that addition of Ash1p did not affect any of these footprint patterns (data not shown).
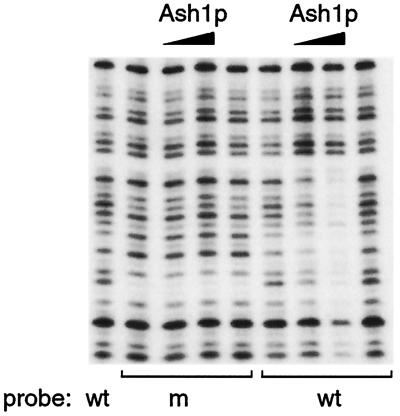
To analyze further the specificity of His-Ash1p DNA binding, a single base alteration of the consensus sequence at −1730 (site 3) from TTGAT to TTTAT was constructed by PCR mutagenesis, and the resulting mutant DNA fragment was analyzed for His-Ash1p binding. As shown in Fig. 4, alteration of the consensus rendered His-Ash1p unable to bind this site (2 μl and 6 μl of His-Ash1p were tested on each probe). Alteration of a base outside the YTGAT consensus but within the area bound by His-Ash1p, for example at −1730 from TTGATC to TTGATT, did not affect His-Ash1p DNA binding (data not shown).
Figure 4.
His-Ash1p binds specifically to YTGAT. A single base alteration of the consensus at −1730 from TTGAT to TTTAT was tested for effects on His-Ash1p DNA binding. The addition of His-Ash1p to wild-type (wt) and mutant (m) probes showed that His-Ash1p bound only to the probe containing the wild-type DNA sequence.
To assess effects of consensus mutations in vivo, similar point mutations in three Ash1-binding sites in URS1 at −1784, −1730, and −1321 were generated by using successive rounds of PCR mutagenesis and introduced by replacement into the HO promoter. Mutation of these three Ash1p-binding sites had no significant effect on mating type switching frequencies of haploid daughter or mother cells as determined by pedigree analysis (data not shown).
The Zinc-Binding Motif of Ash1p Is Required for Function.
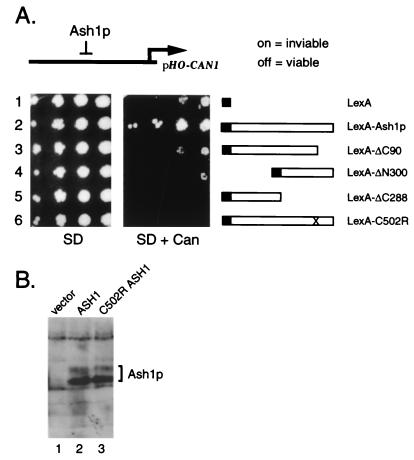
Ash1p contains a region similar to the zinc-binding domain of the GATA-1 transcription factor, a domain known to play a role in DNA binding (25). To determine whether this region is required for Ash1p function, we changed a cysteine residue predicted to participate in zinc coordination to arginine (C502R) and assayed the activities of the mutant allele. Ash1p activity was tested in vivo by monitoring growth in the presence of the amino acid analog canavanine by using an integrated pHO-CAN1 reporter in an ash1Δ strain (3, 26). In this assay, activation of the HO promoter causes cells to be sensitive to canavanine. If the HO promoter is repressed by Ash1p, the cell survives in the presence of canavanine. As shown in Fig. 5A, a strain with a high-copy plasmid carrying wild-type ASH1 grew robustly on canavanine medium (row 2), whereas a strain with the vector control did not (row 1). The strain carrying the C502R mutation did not grow in the presence of the drug, indicating that HO was not repressed under these conditions (row 6). To determine whether the mutant Ash1 protein was expressed and stable in yeast, a Western blot using anti-Ash1p peptide antibodies was performed on extracts made from strains transformed with plasmids carrying an empty vector, wild-type, or mutant ASH1. The Ash1p C502R mutant protein resembled the wild-type protein in both relative amounts and molecular weight (Fig. 5B, lane 3).
Figure 5.
(A) In vivo repression analysis of constructs carrying wild-type and mutant versions of ASH1. Wild-type and mutant versions of ASH1 were fused to the LexA DNA-binding domain and were tested for repression activity of HO-CAN1 in an ash1Δ strain in the presence or absence of canavanine as indicated. Cultures growing in liquid selective medium were diluted and spotted on plates from highest dilution to lowest (from left to right). SD, synthetic dextrose. (B) ASH1 C502R encodes a full-length protein. An ash1Δ strain (IH3964) was transformed with plasmids carrying a vector control (MY160, lane 1), wild-type ASH1 (MY158, lane 2), or ASH1 C502R (MY159, lane 3) and assayed for Ash1p by using anti-Ash1p peptide antibodies.
In addition, a purified recombinant version of the Ash1p C502R protein did not demonstrate in vitro gel-shift activity (data not shown), in further support of the notion that the zinc-binding domain is critical for normal protein function.
Ash1p Is a Transcriptional Repressor.
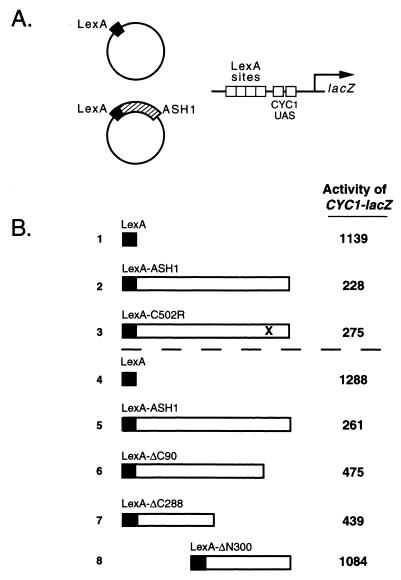
Transcriptional repression in eukaryotic cells occurs by active and passive mechanisms (27–31). To identify the means by which Ash1p negatively regulates HO expression, in vivo repression assays were performed (32). The ASH1 coding sequence was fused to the LexA-DBD (LexA-ASH1) and tested for its ability to repress transcription of the synthetic pCYC1-lacZ reporter plasmid carrying multiple LexA operators (Fig. 6A). The fusion of full-length, wild-type Ash1p to LexA-DBD repressed transcription from the reporter 2- to 5-fold (Fig. 6B, compare row 1 with row 2 and row 4 with row 5). Deletion derivatives of the ASH1 coding sequence were fused to the LexA-DBD to roughly map the domain structure of Ash1p. Derivatives lacking the C-terminal 90 amino acid residues (ΔC90) or C-terminal half of the molecule (ΔC288) were also able to repress the reporter construct (Fig. 6B, compare row 4 with rows 6 and 7, respectively). In contrast, a derivative lacking the N-terminal half of Ash1p (ΔN300) was unable to repress the reporter, indicating that the N-terminal region of Ash1p is responsible for transcriptional repression activity. These deletion derivatives were also tested for the ability to repress pHO-CAN1 in the canavanine sensitivity assay (Fig. 5A). None of the deletion derivatives [including ΔC90 and ΔC288, which exhibited repression activity in the LexA reporter assay (Fig. 6B)], conferred canavanine resistance to the test strain, suggesting that all of the deletion derivatives lacked the ability to repress.
Figure 6.
(A) Diagram of the in vivo heterologous transcription repression assay. The LexA-DBD plasmid and LexA-ASH1 plasmid are shown as examples. In this assay, the lacZ reporter provides a readout of active transcriptional repression when a repressor is targeted to the promoter by specific DNA interactions mediated by the LexA-DBD. (B) LexA-Ash1p represses CYC1-lacZ when recruited to the promoter via the LexA-DBD. β-Galactosidase measurements were performed in duplicate in two representative transcriptional repression experiments on three independent transformants. LexA-Ash1p and LexA-C502R repressed transcription of the heterologous promoter approximately 4- to 5-fold. Deletion derivatives of Ash1p indicated that the N terminus of Ash1p is required for repression of the reporter.
The potential contribution of Swi5p to Ash1p-mediated transcriptional repression was analyzed by using the pCYC1-lacZ reporter in a swi5Δ strain. Deletion of SWI5 had no effect on the ability of LexA-Ash1p to repress, indicating that Swi5 is not required for Ash1p-mediated repression (data not shown). Similarly, we tested known repressors of HO—TUP1 and RPD3—for their roles in facilitating Ash1p-mediated repression, but found that deletion of neither gene affected the ability of Ash1p to repress pCYC1-lacZ (data not shown).
To characterize further the in vivo defect of the zinc-binding domain mutant, the ASH1 ORF carrying the C502R mutation was fused to the LexA-DBD and assayed for repression of the pCYC1-lacZ reporter. Like LexA-ASH1, LexA-C502R repressed transcription of the reporter (Fig. 6B, row 3). This observation suggests that failure of this mutant to repress HO as defined by the pHO-CAN1 reporter was not due to inability to repress transcription per se but presumably was due to a defect in specific DNA binding at the HO promoter.
To identify genes in addition to HO that might be regulated by Ash1p, we searched the yeast genome for promoters containing the Ash1-binding site consensus. By using the PatMatch function on the Saccharomyces Genome Database web site, YTGAT was compared against 5′ untranslated regions within 2,000 bp upstream of predicted and known ORFs. Nearly all of the predicted ORFs contain one or more YTGAT sequences in this region: 487 genes have at least 10 consensus binding sites for Ash1p within 1,000 bp upstream of their translational starts.
The frequent occurrence of YTGAT in the yeast genome presents a challenge for identifying unknown targets of Ash1p by pattern matching. In collaboration with the laboratory of Patrick Brown at Stanford University, we performed DNA microarray analysis with mRNA prepared from asynchronous yeast strains to identify genes in addition to HO that might be regulated by Ash1p (33). In two independent array experiments [one comparing an ash1Δ strain (MY115) with a wild-type strain (MY6) and another comparing an ash1Δ strain (IH3964) with a strain carrying ASH1 on a 2μ plasmid (MY212)], we observed four genes in which the expression ratio (ash1Δ/ASH1) was 2–4. These genes—LYS9, ARG3, ARG5,6 and UGA3—contain 2, 3, 4, and 1 Ash1p consensus binding sites within 2 kb of their translational starts, respectively. Contrary to our expectation, HO was not significantly induced in the absence of ASH1 in these experiments. The functional significance of these results remains to be determined. Identification of target genes by this approach may be difficult if transcriptional regulation by Ash1p is less than 2- or 3-fold.
Discussion
Ash1p Is a Bipartite Protein with DNA-Binding and Repression Functions.
We have focused on the daughter cell-specific transcription factor Ash1p and its action on the HO promoter to determine the mechanism of repression of HO in daughter cells. Our experiments indicate that Ash1p is a modular transcription factor that acts at the DNA level to control transcription of HO. In vitro gel-shift and DNase I footprint experiments showed that Ash1p bound specifically to the HO promoter. In vivo repression assays using both HO (pHO-CAN1) and heterologous (pCYC1-lacZ) promoter constructs revealed that Ash1p is bifunctional, containing DNA-binding and repression functions at its C and N termini, respectively.
Use of a reporter in which LexA operators are upstream of a functional upstream activating sequence (UAS) allowed us to test Ash1p for active repression activity. Proteins with active repressor activity can negatively influence transcription when positioned upstream of the UAS, whereas proteins lacking repressor capability cannot function to decrease transcription of the reporter. We found that the LexA-DBD–Ash1p fusion repressed transcription when recruited to this reporter by the LexA DNA-binding domain, suggesting that Ash1p functions via an active mechanism for transcriptional repression.
We analyzed the role of the predicted GATA-type zinc-binding domain in Ash1p activity by creating and characterizing a mutant Ash1p with an altered zinc-binding domain. Our in vivo repression experiments using the pHO-CAN1 reporter indicated that the zinc-binding domain was required for Ash1p function at HO because pHO-CAN1 was not repressed in this assay. Immunoblot experiments ruled out the possibility that the mutant Ash1 protein was not expressed or was unstable in yeast. Interestingly, when the zinc-binding domain mutant was fused to the LexA-DBD, the mutant fusion protein was fully competent for transcriptional repression of the pCYC1-lacZ reporter, revealing that the zinc-binding domain is not required for transcriptional repression but may be specifically required for DNA binding. In vivo repression assays using a pCYC1-lacZ reporter and LexA-DBD/Ash1p fusions allowed us to crudely define a domain required for repression. We found that all fusions tested except a construct lacking the N-terminal half of ASH1 were competent to repress the pCYC1-lacZ reporter. These results suggest that Ash1p is recruited to the HO promoter by the C-terminal DNA-binding domain defined by a zinc-coordinating structure and functions to repress transcription by an active mechanism dependent on the N-terminal half of the protein.
Ash1p Binds to YTGAT Sequences Upstream of HO.
Ash1p binds specifically to sites containing a YTGAT consensus sequence in the URS1 region of the HO promoter, as determined by DNase I footprinting and Maxam–Gilbert sequencing analysis. In addition, we have shown that a single point mutation in one base pair of the consensus prevents recognition of the site by Ash1p in vitro. Consensus search analysis reveals a total of 20 matches to the consensus in the HO promoter, 13 in URS1, and 7 in URS2. Site-specific mutation of three Ash1p-binding sites adjacent to the two Swi5p-binding sites upstream of HO in URS1 had no demonstrable effect on the frequency of mother or daughter cell mating type switching in vivo. These findings suggest that repression of HO by Ash1p in daughter cells may require the occupation of many Ash1p-binding sites.
The zinc-binding domain of Ash1p has some similarity to the first of two such domains found in the GATA-1 protein and recognizes a sequence, YTGAT, that is related to the canonical (A/T)GATA(A/G) sequence bound by GATA-1 and most other GATA factors. GATA-1 has a characteristic DNA-binding domain with two zinc-binding coordination sites of the configuration Cys-Xaa2-Cys-Xaa17-Cys-Xaa2-Cys (25). Fungal GATA transcription factors such as the well-studied Aspergillus nidulans AreA generally possess one such domain instead of the two typically found in metazoans but maintain the ability to bind the canonical consensus site (34). The primary sequence of Ash1p suggests at least two differences from other GATA proteins that may be responsible for recognizing the variant GATA site: an altered residue at position 22 of the zinc-binding domain and extra amino acids in the presumptive DNA-binding loop. It was recently reported that a change from leucine to valine at position 22 of the AreA zinc-binding domain results in a preference for TGATAG sites over the (A/C)GATAG sites bound by wild-type AreA (34). Thus, position 22 of the zinc-binding domain may be important for DNA site selection. Ash1p has a cysteine residue at position 22 of its zinc-binding domain, unlike fungal GATA factors (AreA and Gln3p) and GATA factors of Caenorhabditis elegans and humans (elt-1 and GATA-1, respectively). The second difference is that the latter GATA factors have the typical Cys-Xaa2-Cys-Xaa17-Cys-Xaa2-Cys configuration in the DNA-binding loop, whereas Ash1p has three additional amino acids in this position (X17 → X20) at this position. These differences may be responsible for ability of Ash1p to recognize a variant GATA site.
A Model for Repression of HO by Ash1p.
Ash1p may repress transcription at the DNA level by a mechanism independent of Swi5p function and dependent on the N-terminal half of the Ash1 protein. Our genetic and biochemical experiments indicate that Ash1p may repress transcription of HO directly by first binding to specific sequences throughout the HO promoter and then repressing transcription by an active mechanism after DNA binding. This activity appears to be independent of the general repressor TUP1 and the histone deacetylase RPD3, negative regulators shown to play a role in active repression of HO.
Ash1p may function to repress HO by a mechanism that does not require the displacement of Swi5p on HO promoter DNA. Two of our observations support this notion. First, in vitro footprinting experiments suggest that Ash1p does not require Swi5p to bind DNA nor does it displace Swi5p bound to HO DNA. Second, in vivo repression assays suggest that Ash1p does not require Swi5p to repress transcription on a heterologous promoter, indicating that a conformational change induced by any Ash1p/Swi5p contact is not required for repressor activity. In addition, the binding of Swi5p to HO promoter DNA in vivo is unaffected by the inappropriate localization of Ash1p in both mother and daughter cells, suggesting that Ash1p does not prevent recognition of HO by Swi5p (7).
Ash1p may prevent chromatin remodeling at HO. Recently, Cosma et al. (7) have shown that transcription and chromatin remodeling factors are recruited to the HO promoter in a temporally ordered manner. The binding of Swi5p to URS1 at the end of anaphase was proposed to allow the binding of the Swi/Snf complex to URS1 and URS2. After Swi/Snf binding, SAGA binds URS2 and allows the subsequent recruitment of SBF to URS2. Ash1p was determined to bind within URS1 transiently and after the binding of Swi5p. Binding of Ash1p to HO was found to prevent Swi/Snf recruitment by Swi5p, a requirement for HO expression.
Our studies suggest that Ash1p may bind at several sites on the HO promoter. There are many models for Ash1p-mediated repression of HO. Promoter occupancy by Ash1p may prevent an essential recruitment function of Swi5p for Swi/Snf on HO, or it may function to inhibit Swi/Snf recruitment directly. Another possibility is that Ash1p represses transcription through interactions with chromatin components directly, inducing a structure refractory to Swi/Snf binding.
Ash1p May Regulate Genes in Addition to HO.
Ash1p binds to the consensus YTGAT and can repress transcription in the absence of HO-specific regulators such as Swi5p. These observations suggest that Ash1p may function at other promoters in addition to HO. A role for ASH1 in pseudohyphal growth (35) also supports a role for Ash1p in regulating genes in addition to HO. Our DNA microarray experiments identified several genes for which transcription was apparently induced in the absence of ASH1; these genes are candidates for regulation by Ash1p.
The identification of an Ash1p-binding site enables an affinity method to isolate proteins and protein complexes that bind with specificity to this consensus. Such an approach is expected to reveal whether alternative binding partners contribute to DNA-binding specificity in the recognition of sites controlling HO and other target genes such as those relevant to pseudohyphal growth and other aspects of mother/daughter asymmetry.
Acknowledgments
We thank John Anzola for help with the production of recombinant baculovirus-expressed Ash1p, Helen McBride and David Stillman for help in purifying recombinant Swi5p and Pho2p, and David King for synthesis of the N-terminal ASH1 peptide used for antibody production. Joe DeRisi is acknowledged for his generosity and help in performing DNA microarray analysis. We thank Anita Sil and members of the Herskowitz, O'Farrell, O'Shea, and Johnson laboratories for strains, plasmids, and reagents. We thank Anita Sil, Pat O'Farrell, Sandy Johnson, and members of the Herskowitz laboratory for advice and useful discussions. We are grateful to Charles Girdham for help with the figures and Thea Norman for her critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (AI18738) to I.H. and the Helen Hay Whitney Foundation and the American Cancer Society (to M.E.M.).
Abbreviation
- URS
upstream regulatory sequences
References
- 1.Strathern J N, Herskowitz I. Cell. 1979;17:371–381. doi: 10.1016/0092-8674(79)90163-6. [DOI] [PubMed] [Google Scholar]
- 2.Nasmyth K. Nature (London) 1983;302:670–676. doi: 10.1038/302670a0. [DOI] [PubMed] [Google Scholar]
- 3.Bobola N, Jansen R P, Shin T H, Nasmyth K. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- 4.Sil A, Herskowitz I. Cell. 1996;84:711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 5.Herskowitz I, Rine J, Strathern J. In: The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae: Gene Expression. Jones E W, Pringle J R, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 583–656. [Google Scholar]
- 6.Nasmyth K. Curr Opin Genet Dev. 1993;3:286–294. doi: 10.1016/0959-437x(93)90036-o. [DOI] [PubMed] [Google Scholar]
- 7.Cosma M P, Tanaka T, Nasmyth K. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 8.Krebs J E, Kuo M-H, Peterson C. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breeden L N K. Nature (London) 1987;329:651–654. doi: 10.1038/329651a0. [DOI] [PubMed] [Google Scholar]
- 10.Andrews B J, Herskowitz I. Nature (London) 1989;342:830–831. doi: 10.1038/342830a0. [DOI] [PubMed] [Google Scholar]
- 11.Harrington L A, Andrews B J. Nucleic Acids Res. 1996;24:558–565. doi: 10.1093/nar/24.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stillman D J, Bankier A T, Seddon A, Groenhout E G, Nasmyth K. EMBO J. 1988;7:485–494. doi: 10.1002/j.1460-2075.1988.tb02836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tebb G, Moll T, Dowzer C, Nasmyth K. Genes Dev. 1993;3:517–528. doi: 10.1101/gad.7.3.517. [DOI] [PubMed] [Google Scholar]
- 14.Brazas R M, Stillman D J. Mol Cell Biol. 1993;9:5524–5537. doi: 10.1128/mcb.13.9.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brazas R M, Stillman D J. Proc Natl Acad Sci USA. 1993;90:11237–11241. doi: 10.1073/pnas.90.23.11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brazas R M, Bhoite L T, Murphy M D, Yu Y, Chen Y, Neklason D W, Stillman D J. J Biol Chem. 1995;270:29151–29161. doi: 10.1074/jbc.270.49.29151. [DOI] [PubMed] [Google Scholar]
- 17.McBride H J, Brazas R M, Yu Y, Nasmyth K, Stillman D J. Mol Cell Biol. 1997;17:2669–2678. doi: 10.1128/mcb.17.5.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasmyth K. EMBO J. 1987;6:243–248. doi: 10.1002/j.1460-2075.1987.tb04745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nasmyth K, Adolf G, Lydall D, Seddon A. Cell. 1990;62:631–647. doi: 10.1016/0092-8674(90)90110-z. [DOI] [PubMed] [Google Scholar]
- 20.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 22.Maxam AM, Gilbert W. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 23.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 24.Stern M, Jensen R, Herskowitz I. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 25.Simon M. Nat Genet. 1995;11:9–11. doi: 10.1038/ng0995-9. [DOI] [PubMed] [Google Scholar]
- 26.Jansen R P, Dowzer C, Michaelis C, Galova M, Nasmyth K. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- 27.Johnson A D. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- 28.Tyler J, Kadonaga J T. Cell. 1999;99:443–446. doi: 10.1016/s0092-8674(00)81530-5. [DOI] [PubMed] [Google Scholar]
- 29.Knoepfler P S, Eisenmann R N. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 30.Bird A P, Wolffe A P. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 31.Maldonado E, Hampsey M, Reinberg D. Cell. 1999;99:455–458. doi: 10.1016/s0092-8674(00)81533-0. [DOI] [PubMed] [Google Scholar]
- 32.Keleher C A, Redd M J, Schultz J, Carlson M, Johnson A D. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 33.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 34.Starich M, Wikstrom M, Schumacher S, Arst H N, Jr, Gronenborn A M, Clore G M. J Mol Biol. 1998;277:621–634. doi: 10.1006/jmbi.1997.1626. [DOI] [PubMed] [Google Scholar]
- 35.Chandarlapaty S, Errede B. Mol Cell Biol. 1998;18:2884–2891. doi: 10.1128/mcb.18.5.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]