Abstract
The centromere is essential for faithful chromosome segregation by providing the site for kinetochore assembly. Although the role of the centromere is conserved throughout evolution, the DNA sequences associated with centromere regions are highly divergent among species and it remains to be determined how centromere DNA directs kinetochore formation. Despite the active use of chicken DT40 cells in studies of chromosome segregation, the sequence of the chicken centromere was unclear. Here, we performed a comprehensive analysis of chicken centromere DNA which revealed unique features of chicken centromeres compared with previously studied vertebrates. Centromere DNA sequences from the chicken macrochromosomes, with the exception of chromosome 5, contain chromosome-specific homogenous tandem repetitive arrays that span several hundred kilobases. In contrast, the centromeres of chromosomes 5, 27, and Z do not contain tandem repetitive sequences and span non-tandem-repetitive sequences of only ∼30 kb. To test the function of these centromere sequences, we conditionally removed the centromere from the Z chromosome using genetic engineering and have shown that that the non-tandem-repeat sequence of chromosome Z is a functional centromere.
The contribution of DNA sequences to centromere formation remains controversial (Marshall et al. 2008). While there exist large arrays of repetitive sequences in human centromere regions (alphoid sequences; alpha satellite DNA) that contribute to the efficient construction of human artificial chromosomes (Harrington et al. 1997; Ikeno et al. 1998; Ohzeki et al. 2002), analysis of human neocentromeres suggests that these repetitive sequences are not absolutely essential for centromere formation (Marshall et al. 2008). To understand the molecular basis of centromere formation, it is necessary to define both the protein complexes that associate with these regions and their underling centromere DNA sequences. We have previously isolated and analyzed multiple centromere-localized proteins from chicken DT40 cells (Okada et al. 2006; Fukagawa 2004, 2008; Hori et al. 2008; Amano et al. 2009). Due to high rates of homologous recombination, DT40 cells provide the unique ability to conduct genetic modifications to analyze the consequences of specific changes to centromere DNA (CenDNA) sequences to kinetochore formation. In addition, while mammalian centromere sequences have been defined, the analysis of centromeres in a distant vertebrate would provide an important evolutionary view of CenDNA. While previous studies have analyzed the chicken whole genome, sequence information from the chicken centromere was not defined (International Chicken Genome Sequencing Consortium 2004). Here, we report the comprehensive analysis of centromere DNA sequences in chicken DT40 cells.
Results
Identification of chicken centromeric DNA
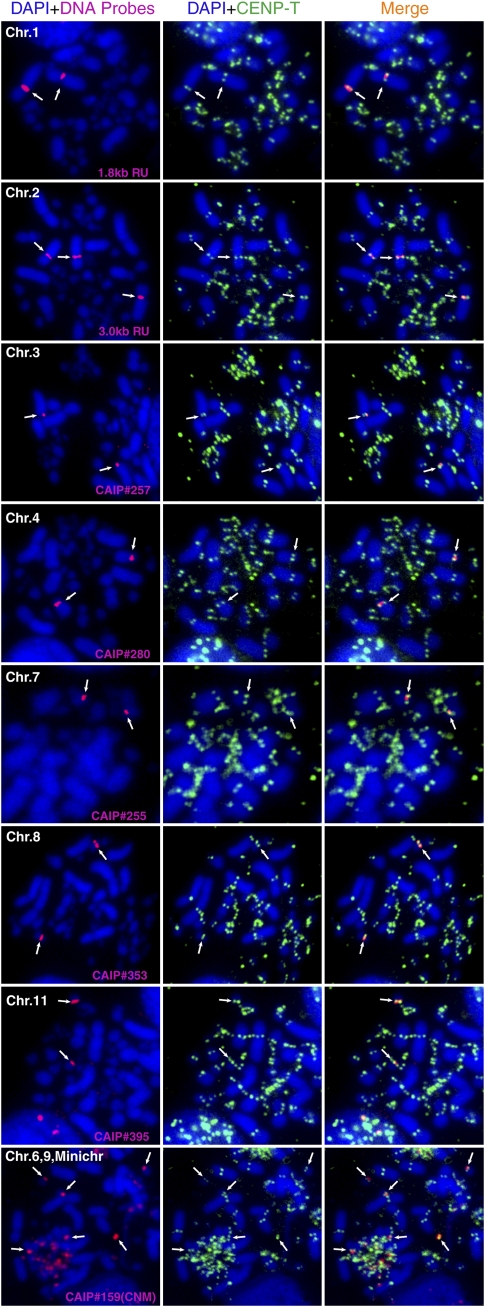
To identify and characterize CenDNA from chickens, we used the centromere-specific histone H3 variant CENPA (Palmer and Margolis 1987). CENPA is a defined marker for active centromeres including neocentromeres (Marshall et al. 2008), and thus DNA associated with CENPA represents bona fide functional CenDNA. To characterize chicken CenDNAs, we created a DT40 cell line stably expressing CENPA-Flag. We next performed chromatin immunoprecipitation (ChIP) experiments in the CENPA-Flag DT40 cell line with anti-Flag antibodies and isolated and cloned the DNAs from the immunoprecipitates. In total, we sequenced 292 clones (CAIP clones, Supplemental Table S1). To confirm that these DNA clones represent centromere sequences, we performed FISH analysis for ∼100 of these clones to examine the centromere localization of these sequences (Fig. 1; Supplemental Table S1).
Figure 1.
Identification of centromere DNA in chicken. Using DNAs precipitated with CENPA, FISH analysis were performed. Probes are indicated in each panel. FISH signals are shown in red. Centromeres are stained with anti-CENPT antibodies (green). CenDNAs of chromosomes 1, 2, 3, 4, 7, 8, and 11 are identified. These sequences are chromosome-specific. The clone containing CNM repeats hybridized with microchromosomes and chromosomes 6 and 9.
Chicken cells contain 10 pairs of macrochromosomes (chromosomes 1–10), 28 pairs of small chromosomes called microchromosomes, and Z/W sex chromosomes (Masabanda et al. 2004). Previous reports identified a 42-bp tandem repeat (CNM sequence) that localizes to centromeres from some of microchromosomes and on chromosomes 6 and 9 (Matzke et al. 1990; Wang et al. 2002; Krasikova et al. 2006). Consistent with these reports, we found CNM sequences in 15 of our CENPA-associated DNA clones (Supplemental Table S1). FISH analysis with a clone containing the CNM sequence confirmed its localization to a subset of microchromosomes and to chromosomes 6 and 9 (Fig. 1). Although the CenDNA sequences of the other macrochromosomes were previously unclear, our FISH analysis revealed distinct centromere sequences for chromosomes 1, 2, 3, 4, 7, 8, and 11 (Fig. 1). Centromere sequence of chromosome 5 will be described later. Unfortunately, we have not identified a specific sequence for chromosome 10 among the 292 clones. Interestingly, each CenDNA from chromosomes 1, 2, 3, 4, 7, 8, and 11 has a specific sequence and we did not detect cross hybridization with other centromeres (see Methods for hybridization condition). For each of these macrochromosomes, we isolated approximately three independent clones. As chicken have 39 chromosome pairs, we calculate that ∼117 (39 × 3) of the isolated clones were from centromere, suggesting that centromere DNA was strongly enriched in the CENPA ChIP-based cloning experiment (see also Supplemental Fig. S1C).
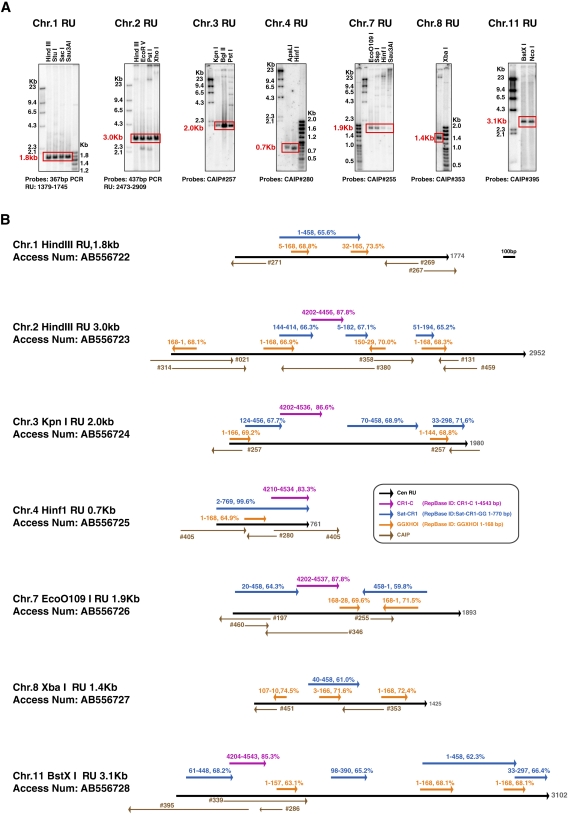
We next analyzed the detailed sequence of each chicken CenDNA. First, genomic DNA from DT40 cells was digested with several restriction enzymes and Southern hybridization analysis was performed with each CenDNA as a probe to determine whether it contained repetitive elements. As shown in Figure 2A, centromeres from several chromosomes have distinct repeated units with chromosomes 1, 2, 3, 4, 7, 8, and 11 having 1.8-, 3.0-, 2.0-, 0.7-, 1.9-, 1.4-, and 3.2-kb repeats, respectively. The sequences from each repeat-unit have been deposited in the DDBJ/EMBL/GenBank DNA database, and the corresponding accession numbers are shown in Figure 2. While global homology between repeat-unit sequences is ∼50%, we found common features in repeat-unit sequences. An important feature of these repeat sequences is that all these sequences contain a homology region (>60% homology) with Satellite CR-1 sequences (Fig. 2B). In addition, a limited region of the CR1-C retrotransposon (4200–4500 bp) shows >80% homology with the repeat-unit sequences of Cen2, Cen3, Cen4, Cen7, and Cen11 (Fig. 2B), suggesting that Cen2, Cen3, Cen4, Cen7, and Cen11 are at least partially derived from a common ancestral sequence.
Figure 2.
DNA sequence of centromere of each macro-chromosome is specific. (A) Using DNA clones, which hybridized with centromere region by FISH, DNA sequence of each repeat-unit was characterized. Each DNA sequence was deposited in DDBJ/EMBL/GenBank DNA database. Accession numbers of Cen1, Cen2, Cen3, Cen4, Cen7, Cen8, and Cen11 are AB556722, AB556723, AB556724, AB556725, AB556726, AB556727, and AB556728, respectively. Length of each repeat-unit was determined by Southern hybridization. Probes are indicated in each panel. (B) Homology region in each repeat-unit sequence with known interspersed repeats is shown. Percentage of identity and position of repeats are also shown. Sequences of interspersed repeats are from RepBase. RepBase IDs of CR1-C, Satellite CR1, and GGXHOI sequence are CR1-C, Sat-CR1-GG, and GGXHOI, respectively. Li and Leung (2006) have previous shown that Cen4 contains a CR-1 repeat.
The repeat-unit from chromosome 8 is reported as a PIR repeat (Wang et al. 2002; Li et al. 2007) which contains a 21-bp consensus sequence in which clusters of (A)3-5 and (T)3-5 are separated by 5–7 bp (for a consensus sequence of gaaaataccnctttttctccc). As the CNM repeat also contains a sequence related to this 21-bp PIR repeat, we examined whether CenDNAs identified in macrochromosomes show homology with this 21-bp sequence (RepbaseID: GGXHOI). We found that all repeat sequences have regions with homology with this GGXHOI 21-bp sequence (>65% homology) (Fig. 2B). We also found the other 21-bp satellite sequence PO41 (Wicker et al. 2005) in CAIP clones (CAIP #217, #258, #381; Supplemental Table S1), but these clones did not localize to centromeres of macrochromosomes by FISH analysis, while signals were detected in mini chromosomes (Supplemental Table S1). However, it is very hard to conclude whether PO41 really colocalizes to centromere on mini-chromosomes due to limit of resolution of the FISH experiments.
In total, the Satellite CR-1/CR1-C/GGXHOI motifs mentioned above cover 41% (Cen1), 47%(Cen2), 82% (Cen3), 100% (Cen4), 81% (Cen7), 50% (Cen8), and 66% (Cen11) of the repeat-units (Fig. 2B). These data suggest that all the centromere repeat-unit sequences may originate from one or two progenitor sequences through cycles of interchromosomal transfer, divergence, rearrangement, and amplification, as has been suggested for human alpha satellite DNA (Shepelev et al. 2009).
Long-range organization of centromeres of chicken chromosomes 1 and 2
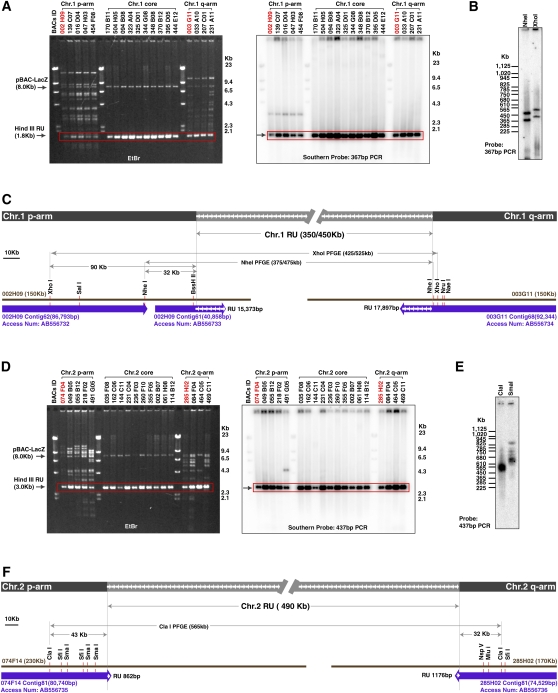
Our results demonstrate that centromeres from most chicken macrochromosomes contain tandem repetitive DNA sequences. To examine the long-range organization of these centromere regions, we focused on the centromeres from chromosomes 1 and 2 (Cen1 and Cen2) and isolated bacteria artificial chromosome (BAC) clones (Hori et al. 2000) that cover these regions (Fig. 3). We characterized BAC clones that contain both repetitive and flanking unique sequences (Fig. 3A,D) and defined the exact boundary between these sequences (Fig. 3; accession nos. AB556732–AB556736). Each human centromere contains a different homogeneous higher-order repeated unit (HOR) with a length ranging from 0.5 to 6.5 kb (Alexandrov et al. 2001; Shepelev et al. 2009). The kinetochore is formed at the HOR region in the long alphoid array (Schueler et al. 2001; Fukagawa 2004; Shepelev et al. 2009). In the case of chicken Cen1 and Cen2, we found that repeat-unit sequences isolated from BAC clones show high similarity (∼99%) with each other, suggesting regions containing high homology repeat-units from Cen1 and Cen2 correspond to the HOR region of human alphoid sequences and form the active centromere. In human centromere, there is a region of heterochromatin that contains divergent alphoid sequences. These adjacent divergent regions in human centromere are inactive centromeres of primate ancestors and have a clearly distinct origin from HOR sequences of alphoid DNA (Kazakov et al. 2003; Schueler and Sullivan 2006; Shepelev et al. 2009). In contrast, we did not find any evidence for such “old” centromere sequence near repeat-unit sequences on chicken Cen1 and Cen2.
Figure 3.
Genome organization of centromere region of chromosome 1 and chromosome 2. (A) DNAs from BAC clones hybridized with Cen1 were digested with HindIII and characterized by Southern analysis. Various BAC clones containing unique sequences and the 1.8-kb repeat-unit sequence were obtained. (B) A pulsed-field gel electrophoresis (PFGE) analysis was performed to determine the length consisting of the repeat-unit. Information of restriction enzyme sites is shown in C. (C) Genome organization around Cen1 region. Sequences of contigs from BAC002H09 and BAC003G11 are deposited in the DDBJ/EMBL/GenBank database with accession numbers AB556732–AB556734. Sequencing of BAC clones identified the exact boundary between unique region and repeat region. The length consisting of the repeat-unit was determined by a PFGE analysis. (D) DNAs from BAC clones hybridized with Cen2 were digested with HindIII and characterized by Southern analysis. Various BAC clones containing unique sequences and the 3.0-kb repeat-unit sequence were obtained. (E) A PFGE analysis was performed to determine the length consisting of the repeat-unit. Information of restriction enzyme sites is shown in F. (F) Genome organization around Cen2 region. Sequences of contigs from BAC074F14 and BAC285H02 are deposited in the DDBJ/EMBL/GenBank database with accession numbers AB556735 and AB556736, respectively. Sequencing of BAC clones identified the exact boundary between unique region and repeat region. The length consisting of the repeat-unit was determined by a PFGE analysis.
To determine the exact length of the entire repetitive arrays, we performed pulsed-field gel electrophoresis (PFGE) analysis with DT40 genomic DNA. We identified a length polymorphism for Cen1 with one homolog containing a repetitive array of 350 kb and the other 450 kb. These correspond to ∼190 and ∼250 copies of the Cen1 repeat sequence (Fig. 3B,C). The length of the repetitive array for Cen2 is 490 kb corresponding to ∼160 copies of the Cen2 repeat sequence (Fig. 3E,F). The long-range organization around the centromere region of chicken chromosomes 1 and 2 is summarized in Figure 3C,F, respectively. Both Cen1 and Cen2 contain several hundred kilobases of repetitive arrays. However, based on our studies of the other chicken centromeres described below, we do not believe that the kinetochore is formed along the entire region of these long repetitive arrays.
Centromeres have non-tandem-repetitive sequences in several chicken chromosomes
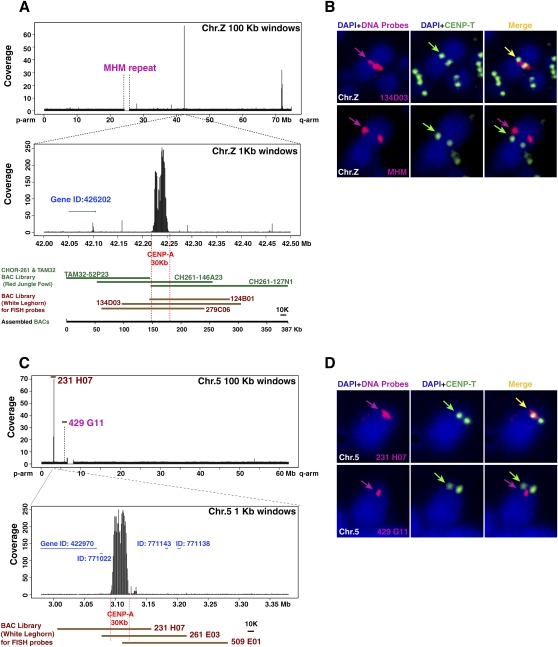
Although we identified CenDNAs for most of the chicken chromosomes, sequence information for chromosomes 5 and Z was lacking. To further define the chicken centromeres, we used next generation deep sequencing to analyze CENPA-associated DNAs (ChIP with massively parallel sequencing [ChIP-seq] analysis). As the chicken genome information deposited in the NCBI database does not contain most of the centromeric repetitive sequences (International Chicken Genome Sequencing Consortium 2004) that we identified above, we could not map CENPA-associated DNAs to the majority of chromosomes. However, we identified a strong enrichment of sequences that mapped to the 42-Mb position of the Z chromosome and the 3.1-Mb position of chromosome 5 (Fig. 4). Both regions lack tandem repetitive sequences. Chromosome Z contains a long-range repetitive array, termed MHM repeats, and spans a ∼2-Mb region (Fig. 4A; Teranishi et al. 2001). Chromosome 5 also may contain a long-range repeat sequence. To confirm which region colocalizes with centromere proteins, we isolated BAC clones containing both of these repeat sequences and the non-tandem-repetitive CENPA-associated DNAs and performed FISH analysis. As shown in Figure 4B,D, a BAC clone containing the corresponding CENPA-associated DNA, but not the long-range repeat sequences, colocalized with each centromere, indicating that the long-range repetitive arrays in chromosomes Z and 5 do not function as centromere DNA. Although we observed a second peak in Figure 4A (72-Mb region of Z chromosome), we did not observe colocalization of the region with CENPT, a second marker for a functional centromere (Hori et al. 2008). High-resolution DNA mapping indicates that the major CENPA-associated region spans ∼30 kb in both chromosomes 5 and Z (Fig. 4A,C). In addition, we also found a CENPA-associated sequence on one microchromosome, which is devoid of tandem repeat (chromosome 27; Supplemental Fig. S2). This sequence also spans ∼30 kb. These 30-kb centromere sequences were deposited in the DDBJ/EMBL/GenBank database with accession numbers AB556729–AB556731. We examined sequences of the 292 CENPA associated clones analyzed in Supplemental Table S1 and found that several clones contain these sequences (Supplemental Table S1). We compared the sequence homology between these three non-tandem-repetitive centromeres and found that there is an enrichment of retrotransposon sequences in CenZ and Cen27: Regions with homology with CR-1 and LTR elements (>70% homology) cover 55% of CenZ and 37% of Cen27. However, we did not detect further common features between these non-tandem-repetitive centromere sequences (Supplemental Fig. S3).
Figure 4.
Centromere region is devoid of tandem-repeated sequences in chromosomes 5 and Z. (A) A distribution of DNA precipitated with CENPA in entire chromosome Z. Major peak is around position of the 42 Mb. High-resolution profile in 1000-bp windows indicates that the binding region of CENPA is restricted in the 30-kb region. Detail sequence information is available in several BAC contigs. As part of sequence information around the 42-Mb position was absent in the NCBI database, we remapped the DNA against BAC contig (387 kb) covering these genomic region and the CENPA associated-DNA was mapped in the 30-kb region, which does not contain tandem-repetitive sequence. Gene ID around CENPA-associated DNA is shown. (B) FISH analysis using a BAC clone (134D03) containing CENPA binding region of chromosome Z (red in top panel). Centromeres are immunostained with anti-CENPT antibodies (green). FISH signals of the 134D03BAC colocalized with centromeres, while FISH signals of the MHM repeat sequence (BamHI 2.2-kb unit) (red in bottom panel) did not localize with CENPT. (C) A distribution of DNA precipitated with CENPA in entire chromosome 5. Major peak is around position of the 3 Mb. High-resolution profile in 1000-bp windows indicates that the binding region of CENPA is restricted in the 30-kb region. Gene IDs around CENPA associated DNA are shown. Clone numbers of BACs are indicated. (D) FISH analysis using a BAC clone (231H07) containing CENPA binding region of chromosome 5 (red in top panel). Centromeres are immunostained with anti-CENPT antibodies (green). FISH signals of the 231H07 BAC colocalized with CENPT, while FISH signals of a BAC clone (429G11) around chromosome 5 satellite repeat sequence (red of bottom panel) did not localize with CENPT.
Conditional removal of centromere sequences by genetic engineering in DT40 cells
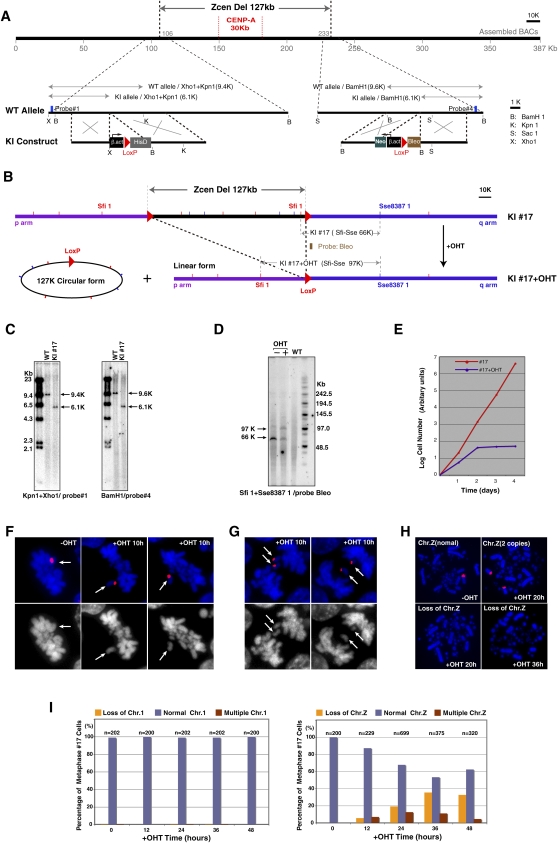
Chicken DT40 cells provide a powerful system for genetic manipulations creating the possibility to modify CenDNA efficiently. To analyze the function of the centromere sequences we identified, we sought to create a cell line in which CenDNA is conditionally removed. For these studies, we chose to focus on the non-tandem-repetitive centromere sequence from chromosome Z. To remove CenDNA, we inserted loxP sequences at both flanking sites of the Z chromosome centromere region using homologous recombination (Fig. 5A). Each target construct containing loxP and a distinct drug resistance gene was transfected sequentially into DT40 cells. Homologous recombination was confirmed by Southern hybridization (Fig. 5A,C). We then introduced a Mer-Cre-Mer construct (Verrou et al. 1999), which expresses an active form of Cre recombinase in the presence of hydroxytamoxifen (OHT), into cells containing loxP sequences at both centromere flanking sites. In this resulting cell line, addition of OHT should conditionally remove the complete centromere region (Fig. 5B). To test this, we performed PFGE analysis after addition of OHT in cells containing Mer-Cre-Mer and confirmed deletion of the centromere region (Fig. 5D). The Z sex chromosome is present in only a single copy in DT40 cells. To determine the consequences of removing the centromere region, we examined cell growth upon the addition of OHT. Cell growth ceased 2 d after addition of OHT and almost all cells had died 4 d after addition (Fig. 5E). We also examined chromosome behavior during mitosis after addition of OHT using FISH analysis with a probe for chromosome Z. As shown in Figure 5F, we frequently observed metaphase cells in which almost all chromosomes were aligned at the metaphase plate, but the chromosome Z had not aligned. During anaphase, sister Z chromosomes were not segregated into daughter cells and were left at the cell midzone (Fig. 5G). These observations indicate that cells with inserted loxP sequences completely lack centromere function after activation of Cre recombinase. We also investigated the stability of chromosome Z after deletion of the centromere region. We observed an increase in cells that lost chromosome Z or had two Z chromosomes, indicating that centromere deletion results in chromosome instability (Fig. 5H,I).
Figure 5.
Deletion of centromere region of chromosome Z results in chromosome instability. (A) To delete entire centromere region of chromosome Z, loxP sequences were inserted into both flanking sites of the centromere region using homologous recombination. CENPA binding region is also shown. (B) Expected genotype after recombination between both loxP sites upon activation of Cre recombinase. The 127-kb region should be removed after addition of OHT. (C) Confirmation of loxP insertions at both flanking sites of the centromere region. Information of restriction map and probes (#1 and #4) for Southern hybridization are shown in A. (D) Confirmation of recombination between both loxP sites. PFGE analysis was performed. Information for the restriction map and the probe (Bleo) for Southern hybridization is shown in B. After recombination a novel 97-kb band is observed. (E) Growth curve of DT40 cells after removal of centromere DNA. (F) Representative images of abnormal metaphase cells in which Z chromosome is not aligned at metaphase plate. Z chromosome was painted with a Z-specific macrosatellite DNA probe (red). (G) Representative images of abnormal anaphase cells in which sister Z chromosomes are left at midzone. Z chromosome was painted with a Z-specific macrosatellite DNA probe (red). (H) Representative images of FISH analysis with a Z-specific macrosatellite DNA probe (red). Cells with two copies of chromosome Z or lacking chromosome Z are observed. (I) Numbers of cells containing chromosome Z after removal of centromere sequence. As a control, numbers of cells containing chromosome 1 were scored. Total cell numbers (n) for the measurements are also shown.
Discussion
In this paper, we performed a comprehensive analysis of chicken CenDNA that demonstrates that (1) many chicken centromeres contain homogeneous repetitive arrays and the repeat-units of each centromere are chromosome-specific, (2) centromeres from three chromosomes (chromosomes 5, 27, and Z) span non-tandem-repetitive sequences of ∼30 kb, and (3) cells in which the identified centromere sequence is conditionally removed fail to segregate that chromosome.
Human alphoid DNA is chromosome-specific (Shepelev et al. 2009). Each active centromere contains a different higher-order repeated unit (HOR) that is related to each other, but unique for each chromosome. Sequence similarity between HORs is 70%–88% (Alexandrov et al. 2001; Shepelev et al. 2009). The copy number of HORs ranges from a hundred to thousands. This feature is similar to what we described here for the chicken centromere. The main difference between human and chicken centromeres is that the composition of chicken centromere repeat-units is much more complex and includes not only one satellite repeat, but a satellite (21 bp, GGXHOI) and pieces of CR1 (Fig. 2). The 21-bp sequence (GGXHOI) is also observed in W heterochromatin regions in chicken genome and shows unusual electrophoretic behavior, which is attributed to the occurrence of DNA curvatures (Kodama et al. 1987). It is possible that a specific DNA-associated protein recognizes these DNA curvatures and localize to centromeres and heterochromatin similar to the mammalian protein CENPB (Masumoto et al. 1989).
In many species, chromosomes contain satellite sequences (Fukagawa 2004; Rudd et al. 2006; Yan and Jiang 2007). Human and mouse active centromeres are composed exclusively of satellite DNA. There are no retrotransposons in active centromeres, although there are transposons in dead centromeres surrounding the active one (Schueler et al. 2001; Kazakov et al. 2003). On the other hand, in rice there are centromere-specific transposons in addition to the CentO repeat (Yan and Jiang 2007). As we also found retrotransposon-like sequences in chicken centromeres, chicken centromere is quite similar to that of rice. However, it appears in rice that there is no homogeneous core of defined repeat-units. In this respect, the chicken centromere is perhaps more similar to the human centromere. In Drosophila and Xenopus, chromosomes do not contain common centromere sequences (Sun et al. 1997; Edwards and Murray 2005), although the detailed sequence information from each centromere remains unclear. Extensive comparison with centromere sequences from various species will be important to understand centromere evolution.
We also identified non-tandem-repetitive sequences of ∼30 kb for three chromosomes. The complete Z chromosome (∼75 Mb) is 15 times bigger than that of chromosome 27 (∼5 Mb), suggesting that the size of the kinetochore is independent of chromosome size. Considering that similar levels of constitutive centromere proteins are present at each chicken chromosome (Jonston et al. 2010), we propose that the kinetochore region of each chromosome should occupy only ∼30 kb, even for chromosomes with long-range repetitive arrays. Therefore, most long-range repetitive sequences may serve as a mark for the centromere region within a chromosome rather than a specific functional element. In addition, for chicken chromosomes with repetitive centromere sequences, each of these repetitive sequences is quite divergent. We propose that centromeres without tandem-repetitive sequences represent a primitive form of these regions and that tandem-repetitive sequence were subsequently added during evolution. This idea is consistent with previous models regarding the formation of evolutionary new centromeres (ENC) (Ventura et al. 2007). Many ENCs have been identified in vertebrate and plant cells (Kasai et al. 2003; Carbone et al. 2006; Yan and Jiang 2007; Han et al. 2009). However, these ENCs have already acquired large arrays of satellite repeats. Very recently, an ENC without tandem-repetitive sequences was identified on chromosome 11 of domestic horse (Wade et al. 2009). Chickens contain three putative ENCs without tandem-repetitive array sequence that provide important insights into centromere evolution and function. Allshire and Karpen (2008) proposed that the centromere is formed by a sequence-independent epigenetic mechanism. Human neocentoromere and chicken putative ENCs would be important materials to understand centromere formation. We also found an enrichment of retrotransposons in putative chicken ENCs like human neocentromeres (Chueh et al. 2009). Retrotransposons may ultimately contribute to the continued evolution of the centromere (Carone et al. 2009).
Finally, we have established an experimental system by which the centromere can be conditionally removed. It is possible to perform such an experiment using yeast cells (Ishii et al. 2008). However, there is a strong advantage to using cytogenetics in vertebrate DT40 cells compared with yeast cells. Therefore, our conditional centromere knockout cell line represents a powerful system to investigate the specific requirements for centromere function in vertebrate cells.
Methods
Immunoprecipitation, cloning, and sequencing of centromere DNA
A CENPA-Flag expression construct under control of the CMV promoter was created. Flag was fused to the C-terminal end of CENPA. DT40 cells expressing CENPA-Flag grow similarly to wild-type DT40 cells. For immnoprecipitation of these cells with an Flag antibody, a nuclear fraction of cells expressing CENPA-Flag was collected and digested with excess MNase (3 U/mL) (Takara) at 37°C for 2 h. The samples were solubilized in 0.5 M NaCl. Anti-Flag M2-beads (Sigma) were incubated with the sample for 2–4 h at 4°C and washed four times with 1 mL of buffer B (20 mM Tris-HCl at pH 8.0, 5 mM EDTA, 500 mM NaCl, and 0.2% Tween 20) and eluted with buffer B in the presence of 3× Flag peptide (Sigma). DNA was extracted from immunoprecipitates and cloned into pBluescript. Alternatively, the DNA was subjected to Illumina DNA sequencing. Sequenced DNAs were mapped into a Chicken Genome database with the MAQ program (Li et al. 2008).
FISH and immunofluorescence
Two-hundred-ninety-two clones containing CENPA associated DNA was sequenced (Supplemental Table S1) and ∼100 clones were randomly selected to use for FISH analysis. DT40 metaphase spreads were prepared by the cytospin method and fixed in 4% paraformaldehyde for 15 min at room temperature. After a brief treatment with 0.5% NP-40 in PBS, glass slides were dehydrated in ethanol. Chromosome DNAs were heat-denatured for 5 min at 95°C. DNA probes were labeled with Biotin-16-dUTP (Roche) by nick translation procedure. Two hundred nanograms of probes were dissolved in a hybridization buffer (0.04 M phosphate buffer at pH 7.0, 2× standard saline citrate [SSC], 50% formamide, 10% dextran sulphate, 1× Denhart's solution, 100 μg/mL of salmon sperm DNA, and 0.5 mM EDTA) denatured for 10 min at 75°C. Hybridization was carried out at 37°C in a humidity chamber overnight. Slides were washed in 2× SSC, 50% formamide in 1× SSC, 0.1× SSC for 10 min at 42°C. Percentage base pair mismatches allowed in this hybridization were estimated from the difference in the melting temperature (T m) for the hybridization or washing. The T m was calculated for a perfectly matched hybrid having the average base pair composition of probes (G + C = 47.8%) and an average length of biotin-labeled 200-bp probes in the 2× SSC (0.39 M Na+), 50% formamide for the hybridization or 0.1× SSC (0.0195 M Na+), 50% formamide for the washing system. Using these parameters, T ms for hybridization and washing are 86°C and 62°C. It was assumed that the T m of a DNA–DNA hybrid decreased by 1°C for every 1% base pair mismatch present in the hybrid (Bonner et al. 1973), ∼20% mismatch was allowed in our FISH experiment. After washing out probes, Cy3-anti-avidin were added to slides and incubated at 37°C for 30 min. Rabbit-anti CENPT antibodies (Hori et al. 2008) were used as a centromere marker. All immunofluorescence and FISH images were collected with a cooled EM CCD camera (QuantEM; Roper Scientific Japan) mounted on an Olympus IX71 inverted microscope with a 100× objective lens together with a filter wheel and a DSU confocal unit. Z-sections (n = 15∼25) were acquired at 0.3-μm steps. All subsequent analysis and processing of images were performed using Metamorph software (Molecular Devices Japan). For detection of Z chromosome, Z-specific macrosatellite DNA (Hori et al. 1996) was used as a probe
Southern blot analysis
For Southern hybridization, 5–10 μg of genomic DNAs of DT40 cells were digested with various restriction enzymes as indicated in the figures and subjected to 0.7% agarose gel electrophoresis. Probes indicated in the figures were prepared. After blotting of gels to nylon membranes, hybridizations with probes were carried out in phosphate-SDS buffer (0.5 M Na-phosphate at pH 7.0, 1 mM EDTA, 7% SDS, 1% bovine serum albumin) and incubated overnight at 62°C. Membrane was sequentially washed in 2× SSC, 1× SSC, and 0.5× SSC buffer, all containing 0.5% SDS for 20 min at 65°C. Autoradiogram was scanned by a Storm 860 system (Molecular Dynamics).
Deletion of centromere DNA
Target constructs containing loxP sequence and several resistance cassettes under control of the beta-actin promoter were created by a standard method. Target constructs were transfected with a Gene Pulser II electroporator (Bio-Rad) into DT40 cells. All DT40 cells were cultured at 38°C in Dulbecco's modified medium supplemented with 10% fetal calf serum, 1% chicken serum, 2-mercaptoethanol, penicillin, and streptomycin. To activate the Cre recombinase, 4-hydroxytamoxifen (OHT; Sigma) was added to the culture medium to a final concentration of 100 nM.
Acknowledgments
We thank K. Suzuki, M. Takahashi, and K. Kita for technical assistance. We also thank I.M. Cheeseman for critical reading of the manuscript and M. Reth for providing us with a Mer-Cre-Mer plasmid. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan to T.F.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank) under accession nos. AB556430–AB556736.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.106245.110.
References
- Alexandrov I, Kazakov A, Tumeneva I, Shepelev V, Yurov Y 2001. Alpha-satellite DNA of primates: Old and new families. Chromosoma 110: 253–266 [DOI] [PubMed] [Google Scholar]
- Allshire RC, Karpen GH 2008. Epigenetic regulation of centromeric chromatin: Old dogs, new tricks? Nat Rev Genet 9: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M, Suzuki A, Hori T, Backer C, Okawa K, Cheeseman IM, Fukagawa T 2009. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J Cell Biol 186: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner TI, Brenner DJ, Neufield BR, Britten RJ 1973. Reduction in the rate of DNA reassociation by sequence divergence. J Mol Biol 81: 123–135 [DOI] [PubMed] [Google Scholar]
- Carbone L, Nergadze SG, Magnani E, Misceo D, Francesca Cardone M, Roberto R, Bertoni L, Attolini C, Francesca Piras M, de Jong P, et al. 2006. Evolutionary movement of centromeres in horse, donkey, and zebra. Genomics 87: 777–782 [DOI] [PubMed] [Google Scholar]
- Carone DM, Longo MS, Ferreri GC, Hall L, Harris M, Shook N, Bulazel KV, Carone BR, Obergfell C, O'Neill MJ, et al. 2009. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma 118: 113–125 [DOI] [PubMed] [Google Scholar]
- Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KH, Wong LH 2009. LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet 5: e1000354 doi: 10.1371/journal.pgen.1000354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards NS, Murray AW 2005. Identification of Xenopus CENP-A and an associated centromeric DNA repeat. Mol Biol Cell 16: 1800–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa T 2004. Centromere DNA, proteins and kinetochore assembly in vertebrate cells. Chromosome Res 12: 557–567 [DOI] [PubMed] [Google Scholar]
- Fukagawa T 2008. The kinetochore and spindle checkpoint in vertebrate cells. Front Biosci 13: 2705–2713 [DOI] [PubMed] [Google Scholar]
- Han Y, Zhang Z, Liu C, Liu J, Huang S, Jiang J, Jin W 2009. Centromere repositioning in cucurbit species: Implication of the genomic impact from centromere activation and inactivation. Proc Natl Acad Sci 106: 14937–14941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington JJ, Van Bokkelen G, Mays RW, Gustashaw K, Willard HF 1997. Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat Genet 15: 345–355 [DOI] [PubMed] [Google Scholar]
- Hori T, Suzuki Y, Solovei I, Saitoh Y, Hutchison N, Ikeda JE, Macgregor H, Mizuno S 1996. Characterization of DNA sequences constituting the terminal heterochromatin of the chicken Z chromosome. Chromosome Res 4: 411–426 [DOI] [PubMed] [Google Scholar]
- Hori T, Asakawa S, Itoh Y, Shimizu N, Mizuno S 2000. Wpkci, encoding an altered form of PKCI, is conserved widely on the avian W chromosome and expressed in early female embryos: Implication of its role in female sex determination. Mol Biol Cell 11: 3645–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, et al. 2008. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135: 1039–1052 [DOI] [PubMed] [Google Scholar]
- Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H 1998. Construction of YAC-based mammalian artificial chromosomes. Nat Biotechnol 16: 431–439 [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716 [DOI] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K 2008. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science 321: 1088–1091 [DOI] [PubMed] [Google Scholar]
- Jonston K, Joglekar A, Hori T, Suzuki A, Fukagawa T, Salmon ED 2010. Vertebrate kinetochore protein architecture: Protein copy number. J Cell Biol 189: 937–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai F, Garcia C, Arruga MV, Ferguson-Smith MA 2003. Chromosome homology between chicken (Gallus gallus domesticus) and the red-legged partridge (Alectoris rufa); evidence of the occurrence of a neocentromere during evolution. Cytogenet Genome Res 102: 326–330 [DOI] [PubMed] [Google Scholar]
- Kazakov AE, Shepelev VA, Tumeneva IG, Alexandrov AA, Yurov YB, Alexandrov IA 2003. Interspersed repeats are found predominantly in the “old” alpha satellite families. Genomics 82: 619–627 [DOI] [PubMed] [Google Scholar]
- Kodama H, Saitoh H, Tone M, Kuhara S, Sakaki Y, Mizuno S 1987. Nucleotide sequences and unusual electrophoretic behavior of the W chromosome-specific repeating DNA units of the domestic fowl, Gallus gallus domesticus. Chromosoma 96: 18–25 [DOI] [PubMed] [Google Scholar]
- Krasikova A, Deryusheva S, Galkina S, Kurganova A, Evteev A, Gaginskaya E 2006. On the positions of centromeres in chicken lampbrush chromosomes. Chromosome Res 14: 777–789 [DOI] [PubMed] [Google Scholar]
- Li J, Leung FC 2006. A CR1 element is embedded in a novel tandem repeat (HinfI repeat) within the chicken genome. Genome 49: 97–103 [DOI] [PubMed] [Google Scholar]
- Li J, Wang X, Leung FC 2007. The intragenomic polymorphism of a partially inverted repeat (PIR) in Gallus gallus domesticus, potential role of inverted repeats in satellite DNAs evolution. Gene 387: 118–125 [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res 18: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KH 2008. Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 82: 261–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masabanda JS, Burt DW, O'Brien PC, Vignal A, Fillon V, Walsh PS, Cox H, Tempest HG, Smith J, Habermann F, et al. 2004. Molecular cytogenetic definition of the chicken genome: The first complete avian karyotype. Genetics 166: 1367–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T 1989. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol 109: 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Varga F, Berger H, Schernthaner J, Schweizer D, Mayr B, Matzke AJ 1990. A 41-42 bp tandemly repeated sequence isolated from nuclear envelopes of chicken erythrocytes is located predominantly on microchromosomes. Chromosoma 99: 131–137 [DOI] [PubMed] [Google Scholar]
- Ohzeki J, Nakano M, Okada T, Masumoto H 2002. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol 159: 765–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR III, Desai A, Fukagawa T 2006. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol 8: 446–457 [DOI] [PubMed] [Google Scholar]
- Palmer DK, Margolis RL 1987. A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J Cell Biol 104: 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Sullivan BA 2006. Structural and functional dynamics of human centromeric chromatin. Annu Rev Genomics Hum Genet 7: 301–313 [DOI] [PubMed] [Google Scholar]
- Rudd MK, Wray GA, Willard HF 2006. The evolutionary dynamics of alpha-satellite. Genome Res 16: 88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF 2001. Genomic and genetic definition of a functional human centromere. Science 294: 109–115 [DOI] [PubMed] [Google Scholar]
- Shepelev VA, Alexandrov AA, Yurov YB, Alexandrov IA 2009. The evolutionary origin of man can be traced in the layers of defunct ancestral alpha satellites flanking the active centromeres of human chromosomes. PLoS Genet 5: e1000641 doi: 10.1371/journal.pgen.1000641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wahlstrom J, Karpen G 1997. Molecular structure of a functional Drosophila centromere. Cell 91: 1007–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teranishi M, Shimada Y, Hori T, Nakabayashi O, Kikuchi T, Macleod T, Pym R, Sheldon B, Solovei I, Macgregor H, et al. 2001. Transcripts of the MHM region on the chicken Z chromosome accumulate as non-coding RNA in the nucleus of female cells adjacent to the DMRT1 locus. Chromosome Res 9: 147–165 [DOI] [PubMed] [Google Scholar]
- Ventura M, Antonacci F, Cardone MF, Stanyon R, D'Addabbo P, Cellamare A, Sprague LJ, Eichler EE, Archidiacono N, Rocchi M 2007. Evolutionary formation of new centromeres in macaque. Science 316: 243–246 [DOI] [PubMed] [Google Scholar]
- Verrou C, Zhang Y, Zürn C, Schamel WW, Reth M 1999. Comparison of the tamoxifen regulated chimeric Cre recombinases MerCreMer and CreMer. Biol Chem 380: 1435–1438 [DOI] [PubMed] [Google Scholar]
- Wade CM, Giulotto E, Sigurdsson S, Zoli M, Gnerre S, Imsland F, Lear TL, Adelson DL, Bailey E, Bellone RR, et al. 2009. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326: 865–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li J, Leung FC 2002. Partially inverted tandem repeat isolated from pericentric region of chicken chromosome 8. Chromosome Res 10: 73–82 [DOI] [PubMed] [Google Scholar]
- Wicker T, Robertson JS, Schulze SR, Feltus FA, Magrini V, Morrison JA, Mardis ER, Wilson RK, Peterson DG, Paterson AH, et al. 2005. The repetitive landscape of the chicken genome. Genome Res 15: 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Jiang J 2007. Rice as a model for centromere and heterochromatin research. Chromosome Res 15: 77–84 [DOI] [PubMed] [Google Scholar]