Abstract
Aberrant DNA methylation frequently occurs at gene promoters during cancer progression. It is important to identify these loci because they are often misregulated and drive tumorigenesis. Bisulfite sequencing is the most direct and highest resolution assay for identifying aberrant promoter methylation. Recently, genomic capture methods have been combined with next-generation sequencing to enable genome-scale surveys of methylation in individual samples. However, it is challenging to validate candidate loci identified by these approaches because an efficient method to bisulfite sequence more than 50 differentially methylated loci across a large number of samples does not exist. To address this problem, we developed Bisulfite Patch PCR, which enables highly multiplexed bisulfite PCR and sequencing across many samples. Using this method, we successfully amplified 100% of 94 targeted gene promoters simultaneously in the same reaction. By incorporating sample-specific DNA barcodes into the amplicons, we analyzed 48 samples in a single run of the 454 Life Sciences (Roche) FLX sequencer. The method requires small amounts of starting DNA (250 ng) and does not require a shotgun library construction. The method was highly specific; 90% of sequencing reads aligned to targeted loci. The targeted promoters were from genes that are frequently mutated in breast and colon cancer, and the samples included breast and colon tumor and adjacent normal tissue. This approach allowed us to identify nine gene promoters that exhibit tumor-specific DNA methylation defects that occur frequently in colon and breast cancer. We also analyzed single nucleotide polymorphisms to observe DNA methylation that accumulated on specific alleles during tumor development. This method is broadly applicable for studying DNA methylation across large numbers of patient samples using next-generation sequencing.
Inappropriate CpG DNA methylation has been found in most types of cancer (Lyko and Brown 2005), and many genes involved in malignancy can acquire aberrant promoter methylation (Baylin et al. 1998). Tumor suppressor genes frequently exhibit promoter hypermethylation, an epimutation that is associated with inappropriate gene silencing (Baylin et al. 1998). A recent study has found that several key tumor suppressor genes exhibit promoter hypermethylation more often than genetic disruption, suggesting this mechanism is an important driver of tumorigenesis (Chan et al. 2008). Oncogenes can exhibit hypomethylation of their promoters, which is associated with inappropriate expression (Jun et al. 2009). More complicated misregulation of a gene can also be caused by aberrant methylation; a recent report found that hypermethylation of a p53 binding site blocked binding of the repressor, resulting in overexpression of the survivin oncogene (Nabilsi et al. 2009).
The identification of gene promoters that are aberrantly methylated during tumor development is valuable because it provides information about the biological pathways that are commonly disrupted during tumorigenesis (Klarmann et al. 2008; Suzuki et al. 2008). This knowledge may ultimately lead to new drug targets. Analysis of promoter methylation can also classify distinct subtypes of cancers that may have differential clinical characteristics in order to personalize treatment (Esteller et al. 2000; Widschwendter et al. 2004). Finally, loci that are hypermethylated in tumors are often detected in peripheral samples (e.g., blood or stool) and may serve as diagnostic or prognostic biomarkers (Laird 2005).
Many techniques have been developed to detect DNA methylation, including methods based on microarrays (Ushijima 2005), quantitative PCR (Eads et al. 2000), mass spectrometry (Ehrich et al. 2005), and DNA sequencing (Frommer et al. 1992). The method that is the most direct and has the highest resolution involves the treatment of genomic DNA with sodium bisulfite (which converts unmethylated cytosines to uracil, while leaving methylated cytosines intact) followed by the sequencing of single molecules. This method is capable of detecting any cytosine DNA methylation, including the non-CpG cytosine methylation found in stem cells (Lister et al. 2009). Not only does this method determine the methylation state at each cytosine across a single molecule, but it also detects single nucleotide polymorphisms (SNPs). This cis information makes it possible to distinguish allele-specific methylation (Frommer et al. 1992). This cis information is also valuable for quantifying rare densely methylated molecules in a background of unmethylated or sparsely methylated molecules.
The recent introduction of second-generation DNA sequencing technologies has significantly reduced the cost required to sequence DNA. This has led to several new approaches for studying aberrant methylation using bisulfite PCR and sequencing. Methods for genome-wide surveys of methylation have been developed, including whole-genome bisulfite sequencing (Cokus et al. 2008; Lister et al. 2009), bisulfite sequencing large fractions of restriction digested genomic DNA (Meissner et al. 2008), padlock probe-based strategies (Ball et al. 2009; Deng et al. 2009), and array-based hybridization capture (Hodges et al. 2009). These methods are powerful because they cast a wide net and can generate many novel hypotheses. However, in order to identify epimutations that are significantly associated with a disease, it is usually necessary to analyze hundreds of samples, something that remains cost-prohibitive with genome-scale methods. It is possible to analyze a few loci across many samples by amplifying each locus individually, labeling with sample-specific barcodes, and performing ultra-deep bisulfite sequencing (Taylor et al. 2007; Korshunova et al. 2008; Varley et al. 2009). However, these methods are limited to a small number of loci because the amplification of each locus separately is laborious and requires a significant amount of patient DNA per locus queried. Thus, there is a need for methods that enable the targeted multiplexed bisulfite PCR and sequencing of an intermediate number of loci (100–1000) across a large number of samples.
We sought to develop a method to perform highly multiplexed bisulfite sequencing across many patient samples simultaneously. We based our design on the Nested Patch PCR method that we developed for the multiplexed sequencing of loci to identify genetic mutations (Varley and Mitra 2008). Several key steps had to be modified to be compatible with the bisulfite treatment of genomic DNA. Bisulfite treatment significantly reduces the complexity of DNA sequence by converting most Cs to Ts. It also results in molecules from the same locus having different sequences depending on their methylation state. Therefore we perform the oligo hybridization and ligation-based selection of the targeted loci before the bisulfite treatment. The selection is highly sensitive and specific, and only one pair of oligos per locus is needed, even when selecting CpG-rich loci. The PCR amplification of selected loci is performed after the bisulfite. Therefore the universal primers used to amplify all loci simultaneously had to be designed to exclude Cs, so that they would remain unchanged through the bisulfite conversion. Since the major application of this method is likely to be in clinical specimens, we optimized the method so that it did not require large quantities of starting genomic DNA and was compatible with the DNA degradation inherit in the sodium bisulfite treatment.
We designed the method to be easy to implement in any laboratory with standard molecular biology techniques and reagents. We also tested that it would scale up well to process many patient samples in 96-well format. We integrated sample-specific DNA barcodes into the multiplexed amplification so that many patient samples can be pooled and sequenced simultaneously on second-generation sequencing machines. Here, we present a proof-of-principle experiment in which we amplified promoter regions from 94 targeted loci simultaneously and sequenced these loci across 48 samples, including colon and breast tumor and adjacent normal tissue samples. In this experiment, we characterized the promoter methylation of genes that are known to be frequently mutated in cancer. We identified several novel loci that undergo frequent tumor-specific promoter methylation, and we observed allele-specific methylation patterns that occur during tumor development. We demonstrated that this method uses the power of next-generation sequencing to study DNA methylation at many loci across many patient samples.
Results
Overview of Bisulfite Patch PCR
Bisulfite Patch PCR begins with a restriction digest of human genomic DNA to define the ends of the fragments that will be selected (Fig. 1A,B). Targeted loci are then selected from the genomic restriction fragments by annealing patch oligos to the ends of the targeted genomic fragments. These oligos serve as a patch between the correct fragments and universal primers (U1 and U2) (Fig. 1C). The universal primers are then ligated to the genomic fragments using a thermostable ligase (Fig. 1D). Unselected genomic DNA is then degraded with exonucleases to gain additional selectivity (Fig. 1E). Selected fragments are protected from degradation by a 3′ modification on the universal primer U2 (Fig. 1E). Next, the selected fragments are treated with sodium bisulfite to convert unmethylated cytosines to uracil, leaving the methylated bases intact (Fig. 1F). The universal primers do not contain cytosine bases so that the sequence remains unchanged through the bisulfite conversion. The bisulfite-treated selected fragments are then all amplified together simultaneously by PCR with the universal primers (U1 and U2′) (Fig. 1G). Sample-specific DNA barcodes are incorporated into the universal primers by tailing the 5′ end with a DNA sequence that is specific to each sample and the sequencing platform primers (454 Life Sciences [Roche] sequencing primers) (Fig. 1G). The final PCR amplicons from each of the samples can be pooled together for sequencing because the first few bases of each sequencing read will identify the sample from which that sequence originated.
Figure 1.
Bisulfite Patch PCR. (A,B) Genomic DNA restriction digest. (C) Anneal patch oligos and universal primers specifically to the ends of desired fragments. (D) Ligate universal primers (U1 and U2) to targeted fragments. (E) Degrade unselected DNA with exonucleases. Targeted loci are protected from exonuclease by 3′ modification on U2. (F) Treat with sodium bisulfite to convert unmethylated cytosine to uracil, leaving methylated cytosine intact. (G) PCR all loci simultaneously with universal primers tailed with sample-specific DNA barcodes and sequencing machine primers (454A and 454B). Pool PCR products from all samples together for sequencing.
Highly multiplexed bisulfite sequencing of CAN gene promoters in colon and breast cancer
To test the performance of Bisulfite Patch PCR, we analyzed the promoter methylation of 94 genes that are frequently mutated in breast and colon cancers (“CAN genes”) (Wood et al. 2007). We designed the patch oligos to select AluI restriction digest fragments containing at least three CpG positions within 700 bp upstream of the transcription start site (TSS). We chose 42 colon CAN gene promoters, 44 breast CAN gene promoters, four gene promoters that were identified as both colon and breast CAN genes, and four controls. The four controls include an imprinted locus, a housekeeping gene promoter, and two neutral loci that accumulate methylation with mitotic cell division (Kim et al. 2006). These targeted promoter regions ranged in length from 125–581 bp and totaled 25.4 kbp (Supplemental Data 1). To determine the amount of genomic DNA required for Bisulfite Patch PCR, we performed gel electrophoresis of the PCR products generated with different amounts of starting DNA. We observed DNA within the expected size range from reactions that started with as much as 1 μg and as little as 20 ng of human genomic DNA (Supplemental Fig. 1).
We performed Bisulfite Patch PCR on 250 ng of genomic DNA from each of the 48 samples in parallel in a 96-well plate. The genomic DNA was isolated from a panel of 12 colon tumors, 12 matched adjacent normal colon tissues, 12 breast tumors, and 12 matched adjacent normal breast tissues (Supplemental Table 2). We incorporated a 5-bp sample-specific DNA barcode in the final PCR, pooled the amplicons from all of the samples, and sequenced the pool using the 454 Life Sciences (Roche) FLX sequencer. We obtained 97,115 reads and aligned these to the in silico bisulfite-treated reference sequences of our targeted loci. We successfully amplified all 94 (100%) of the targeted loci, indicating that the method is highly sensitive. Ninety percent (87,458 reads) of all reads mapped to one of the targeted promoters, demonstrating that the method is highly specific. These results demonstrate that Bisulfite Patch PCR enables highly multiplexed bisulfite sequencing.
Coverage of promoters and reproducibility
To analyze the uniformity of the sequence coverage, we graphed the number of reads obtained for each targeted promoter versus the length of the targeted region. (Fig. 2A; Supplemental Table 1). The abundance of each promoter ranged from 10 to 5114 reads. We calculated that 93% of the promoters have coverage within 10-fold of the median coverage (444 reads). The Pearson's linear correlation coefficient between the amplicon length and the number of reads is −0.65, suggesting that longer amplicons are less abundant in the reaction. To determine if the observed correlation was statistically significant, we transformed the correlation to create a t-statistic having N − 2 degrees of freedom, and found that P < 1.3 × 10−12. If we had restricted our design to a maximum target length of 300 bp, then 92% (57/62) of those promoters would have coverage within fivefold of the median coverage (1051 reads), so approximately half of the variation in abundance of the loci is attributable to length bias. We suspect that this length bias is introduced by the bisulfite treatment, rather than by the multiplexed PCR, since we did not observe a correlation between amplification efficiency and length when performing the Nested Patch PCR (Varley and Mitra 2008), which does not use bisulfite treatment. Consistent with this hypothesis, others have observed that longer DNA fragments are more likely to be damaged during the bisulfite conversion (Munson et al. 2007).
Figure 2.
Method performance. (A) Number of sequencing reads per promoter for all 94 targeted promoters, order by length in base pairs (bp) on the x-axis. Longer promoter amplicons yield fewer sequencing reads (length bias), but 87 amplicons (93%) have coverage within 10-fold of the median coverage (444 reads). The abundance of each promoter ranged from 10 to 5114 reads. (B) Histogram of the pairwise squared correlation coefficients for the number of reads per promoter for all 48 samples. The mean correlation coefficient is 0.91, indicating that the number of reads per promoter is highly reproducible across patient samples.
To test if Bisulfite Patch PCR reproducibly amplifies selected loci, we calculated the number of reads per locus in each of the 48 samples that were prepared in parallel. We then calculated the correlation coefficient for the number of reads per locus between all possible pairs of samples. The histogram of correlation coefficients obtained for the pairwise correlations between all 48 samples is shown in Figure 2B. The mean correlation coefficient is 0.91, indicating that the number of reads per locus is highly reproducible across patient samples. This indicates that the abundance of each locus in the reaction is not stochastic but represents something intrinsic to the locus, including the length, as discussed above.
Methylation detection at control loci
To ensure that the multiplexed PCR aspect of Bisulfite Patch PCR did not diminish the ability to accurately detect methylation, we examined the bisulfite sequence at control loci. There was no DNA methylation detected at the negative control promoter of the housekeeping gene HSP90AB1 (NM_007355), indicating the sodium bisulfite conversion was effective. We did detect methylation at all three (100%) of the positive control loci, including the H19 imprinted promoter (AK311497) and two neutral loci that accumulated DNA methylation with mitotic division (NM_006941 Exon 2, and NM_004387 3′ untranslated region [UTR]) (Kim et al. 2006). These results led us to conclude the method could accurately detect methylation patterns, which agrees with previously published work in which methylation patterns obtained by bisulfite sequencing using 454 Life Sciences (Roche) sequencing were extensively validated (Taylor et al. 2007; Korshunova et al. 2008; Varley et al. 2009).
Next, we sought to determine if the methylated and unmethylated molecules from the same locus are amplified with similar efficiencies. This is required if the method is to be used to make quantitative measurements of promoter methylation. The imprinted region from the H19 locus (AK311497), which was included as a control, allows the direct comparison of the amplification efficiency of the methylated and unmethylated alleles. We identified nine patients in our panel who were heterozygous for a SNP (rs2251375) in the H19 locus. We used this SNP to identify allele-specific methylation and to quantify the number of sequencing reads obtained for each allele. Allele-specific methylation was observed, and both alleles were amplified with nearly equal efficiencies (Fig. 3). Imprinting methylation was observed on either allele in different individuals, consistent with the parent-of-origin determining which allele is methylated. Both alleles were represented at similar frequencies—on average 42% of the sequencing reads corresponded to the “G” allele; 58%, to the “T” allele. Thus, our method amplifies the methylated and unmethylated molecules from the same locus with nearly equal efficiency, which is crucial for quantifying heterogeneous methylation within tumors.
Figure 3.
Methylation at the H19 imprinted locus. Data from four patients who were germline heterozygous for a SNP (rs2251375) in this locus. The sequencing reads are aligned as rows in each panel. Each base in the read is color-coded to indicate the sequence: (yellow) a methylated cytosine; (blue) all other bases. The position of the SNP is indicated by the red and white column: (red base) reads from the G allele; (white base) reads from the T allele. The percentage of reads for each patient that are from the G allele is listed below the patient identifier for each sample. As expected for an imprinted locus, methylation is observed on one allele in both the tumor (left panels) and the adjacent normal tissue (right panels) for each patient. Both alleles and both methylated and unmethylated molecules were amplified and sequenced efficiently from this locus in all samples.
CAN gene promoter methylation
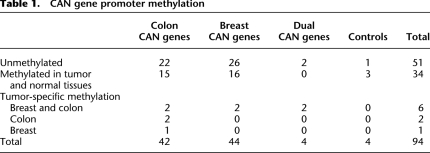
We next examined the methylation patterns found at the targeted CAN gene promoters to determine if they exhibited tumor-specific methylation. Since these genes were previously shown to be frequently mutated in colon and breast tumors (Wood et al. 2007), we hypothesized that the promoters of these genes might also be frequently hyper- or hypomethylated in these cancers. Promoter classification was straightforward, as the vast majority of sequencing reads were either fully methylated or completely unmethylated (Supplemental Fig. 2). We found that approximately half (51/94), of all the promoters were unmethylated in all the tissue types that we tested. Approximately one-third (34/94), of all promoters were methylated in both cancer and normal tissue. The remaining nine promoters exhibited tumor-specific methylation (for summary, see Table 1; for details, see Supplemental Table 1).
Table 1.
CAN gene promoter methylation
Tumor-specific promoter methylation
Of the nine promoters that exhibited tumor-specific methylation, five were promoters from colon CAN genes, two were promoters from breast CAN genes, and two were promoters from genes that were frequently mutated in both colon and breast cancer (“dual CAN genes”) (for summary, see Table 1; for details, see Supplemental Table 1).
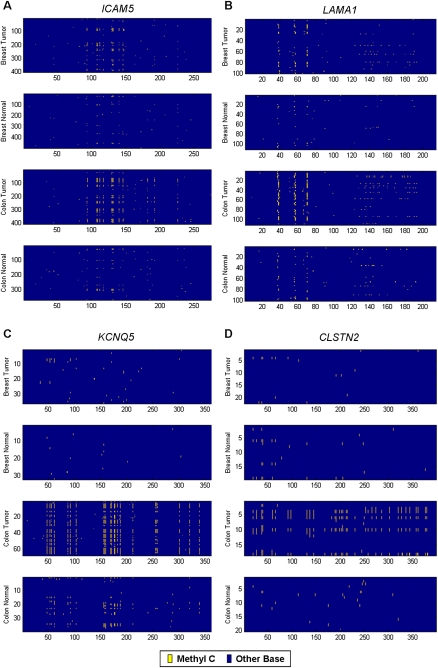
Five promoters exhibited tumor-specific hypermethylation in both breast and colon tumors (IGFBP3, UHRF2, LAMA1, ICAM5, PPM1E). One promoter (SORL1) exhibited tumor-specific hypomethylation in both types of cancer. The methylation patterns of ICAM5 and LAMA1 are shown in Figure 4, A and B, respectively. Tumor-specific promoter methylation of ICAM5 (Chan et al. 2008) and IGFBP3 (Tomii et al. 2007) was recently reported in different cohorts of breast and colon cancers. The other three loci are novel observations of aberrant tumor methylation. The frequent hypermethylation of these five loci in both types of tumors indicates that common molecular defects are shared between colon and breast cancer. The molecular defect could be an error in both types of tumors that directs methylation to these loci, or it could suggest that the inactivation of these genes is a key step in tumorigenesis in both tissues.
Figure 4.
Four promoters that exhibit tumor-specific methylation. Sequencing reads from all patients for each type of tissue are grouped together in panels; breast tumors, adjacent normal breast tissues, colon tumors, and adjacent normal colon tissues. The sequencing reads are aligned as rows in each panel and are grouped by patient. Each base in the read is color-coded to indicate the sequence: (yellow) a methylated cytosine; (blue) all other bases. (A,B) ICAM5 and LAMA1 promoters exhibit colon and breast tumor-specific methylation. (C,D) KCNQ5 and CLSTN2 promoters exhibit colon tumor-specific methylation.
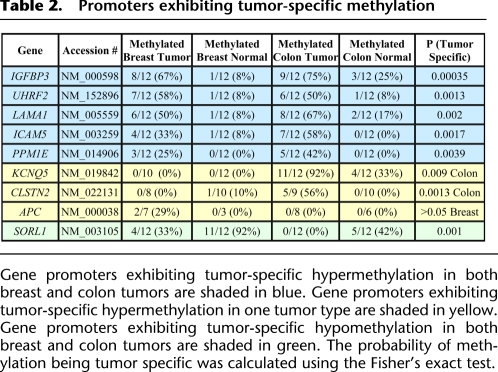
These five loci that are hypermethylated in both breast and colon cancer are methylated in 25%–75% of tumors (Table 2). Loci that exhibit frequent tumor-specific methylation are often useful as clinical biomarkers. A valuable biomarker would occur frequently in patients' tumors and would be easily distinguished from normal samples. We calculated the sensitivity and specificity of these loci across our samples. The presence of aberrant methylation at two or more of these five methylated markers is found in nine out of 12 breast tumors (75%), 11 out of 12 colon tumors (92%), one of 12 normal breasts (8%), and one of 12 normal colons (8%). These strong classifiers of cancer versus normal samples are good candidates for follow-up studies to evaluate their potential as biomarkers for stratifying disease subtypes or as diagnostic biomarkers that can be detected in peripheral specimens. The frequency of aberrant methylation at these loci approaches the significance of even the most common genetic mutations such as APC or TP53 mutations, which are reported to occur in 40%–80% of tumors (Sjoblom et al. 2006). This supports the previously proposed hypothesis that epigenetic defects at CAN genes may be more frequent than genetic mutations (Chan et al. 2008).
Table 2.
Promoters exhibiting tumor-specific methylation
Gene promoters exhibiting tumor-specific hypermethylation in both breast and colon tumors are shaded in blue. Gene promoters exhibiting tumor-specific hypermethylation in one tumor type are shaded in yellow. Gene promoters exhibiting tumor-specific hypomethylation in both breast and colon tumors are shaded in green. The probability of methylation being tumor specific was calculated using the Fisher's exact test.
Three of the CAN gene promoters show tumor-specific methylation in only one type of cancer. Colon tumor-specific methylation was found in the promoters of KCNQ5 (NM_019842) and CLSTN2 (NM_022131), and those methylation patterns are shown in Figure 4, C and D, respectively. Breast tumor-specific methylation was found in the promoter of APC (NM_000038). The frequency of these aberrant events in each tumor type is cataloged in Table 2 and suggests that these loci may represent frequent tumor-specific epimutations that merit follow-up investigation in a larger cohort of tumors and adjacent normal and cancer-free patient's tissue.
Allelic tumor methylation
The single molecule resolution of bisulfite sequencing allows us to simultaneously assess the methylation status and identify SNPs. As seen in Figure 5, we can distinguish whether tumor-specific methylation is occurring on one allele or on both alleles in individuals that are heterozygous for the SNP (rs2854744) in IGFBP3 (NM_000598). Although some aberrant promoter methylation events are known to always occur on both alleles, such as MLH1 promoter methylation (Veigl et al. 1998), we found examples in which aberrant methylation was observed on only one allele: Breast cancer patient 4 acquired tumor-specific methylation primarily on the A allele, while colon cancer patient 6 acquired tumor-specific methylation primarily on the C allele (Fig. 5). However, other patients acquired aberrant methylation on both alleles during tumorigenesis, such as breast cancer patient 6 and colon cancer patient 7 (Fig. 5). If associated with silencing, this biallelic methylation would indicate that both copies of the gene are inactive. Some patients exhibit different allelic methylation patterns between their tumor and the adjacent normal tissue: Colon cancer patient 12 has methylation on their A allele across all CpGs in both the tumor and the adjacent normal tissue, but as the tumor formed, the C allele acquired methylation, specifically in the region of the promoter most distal from the SNP. This suggests that the accumulation of methylation on each allele can occur in different regions of the locus and can occur at different times in tumor development. This type of allelic analysis is useful for resolving intratumor heterogeneity of DNA methylation, identifying heterozygous and homozygous epimutations, and understanding the accumulation of aberrant DNA methylation in different tumors.
Figure 5.
Allelic tumor-specific methylation. Data from six patients who are germline heterozygous for a SNP (rs2854744) in the IGFBP3 promoter. The sequencing reads are aligned as rows in each panel. Each base in the read is color-coded to indicate the sequence: (yellow) a methylated cytosine; (blue) all other bases. The position of the SNP is indicated by the red and white column: (red) reads from the A allele; (yellow) methylated C allele; (white) C allele, if unmethylated and converted to a T. Patient “breast 8” is unmethylated on both alleles in both the tumor (left column) and normal tissue (right column). Patients “breast 4” and “colon 6” display tumor-specific methylation on only one allele, and the methylated allele differs between them. Patients “colon 7” and “colon 12” display tumor-specific methylation on both alleles. Patient “colon 12” displays different patterns of methylation on each allele in the tumor.
Discussion
Several methods have been developed to perform the genome-wide profiling of DNA methylation in individual samples. These methods include genome-wide surveys of methylation such as whole-genome bisulfite sequencing (Cokus et al. 2008; Lister et al. 2009) and bisulfite sequencing large fractions of restriction digested genomic DNA (Meissner et al. 2008) or methods that target specific subclasses of loci such as padlock probe-based strategies (Ball et al. 2009; Deng et al. 2009) and array-based hybridization capture (Hodges et al. 2009). These methods are producing long lists of candidate loci that will need to be validated across many DNA samples. Bisulfite Patch PCR provides an efficient workflow to use second-generation sequencing to follow up and validate aberrant methylation at a moderate number of loci (50–100) across large numbers of samples. This method is highly sensitive and integrates DNA barcoding into the library construction process so that many samples can be pooled to fully utilize the capacity of next-generation sequencing. We designed the method so that it is easy to implement in any laboratory, has an easy workflow that does not involve shotgun library construction, and requires small amounts of sample DNA. The method specifically targets and selects loci before sodium bisulfite treatment, which increases the specificity and decreases the oligo cost and design failures compared with similar methods that select after bisulfite conversion (Hodges et al. 2007; Ball et al. 2009; Deng et al. 2009). Bisulfite Patch PCR should have broad utility for the investigation of DNA methylation across disciplines, including cancer and other disease research, stem cell research, aging research, population-based studies of natural variation, and epidemiological studies of environmental exposures.
There are a few constraints and unknown parameters of the method that are important to note. The boundaries of target loci are defined by restriction digest. This is a design constraint that limits the number of loci that can be targeted in a single reaction, either because the regions do not contain restriction sites or because the restriction cut occurs in a repetitive element and a unique patch oligo cannot be designed. We chose to use the restriction enzyme AluI because it is not sensitive to methylation, it is an efficient enzyme, it has a four-base recognition sequence so it cuts frequently in the genome, and it cuts the middle of its recognition sequence and leaves blunt fragments, so there is not concern about imprecise cut positions. We computationally determined that 59% (161/271) of CAN gene promoters could be targeted with the current design criteria using AluI restriction, and picked 94 of these for this experiment (see section Design of Patch Oligonucleotides and Universal Primers). We also computed the optimal restriction enzyme pair that produces the most restriction fragments from the CAN gene promoters. We determined that 71% of the CAN genes promoters could be targeted using a double digest of Tsp509I and HaeIII. To further improve the number of loci that can be targeted, separate restriction digests could be performed on aliquots of the sample gDNA, and the DNA could be repooled for the subsequent patch ligation and PCR. For example, if an AluI digest was performed in one tube, a Tsp509I/HaeIII double digest was performed in a second tube, and the reactions were pooled for the patch ligation, then 83% of CAN gene promoters could be targeted. The development of a way to specifically cleave the DNA at the boundaries of the targeted loci would significantly improve the design success rate of the method. One unknown parameter for the method is the number of loci that can be amplified in a single reaction. As the capacity of next-generation sequencing machines expands, it will be logical to expand the level of multiplexing achieved by Bisulfite Patch PCR. Since we successfully amplified all 94 of the targeted loci in this initial experiment, we have not determined the limit to increasing numbers of targeted loci. Additionally, as more loci are targeted, the cost of oligo synthesis becomes a concern, and methods besides standard oligo synthesis will need to be explored (Porreca et al. 2007).
In this proof-of-principle experiment, we applied this method to characterize the promoter methylation of genes that are frequently mutated in cancer. From the 94 gene promoters that we analyzed, we found that ∼10% showed tumor-specific DNA methylation in breast or colon cancer compared with the adjacent normal tissue. Our data support the previously proposed hypothesis that a relatively small set of genes that are important for tumorigenesis are disrupted in multiple ways in cancers, including frequent epigenetic defects (Chan et al. 2008). We found five loci that can be used to classify tumor and normal samples with high sensitivity. Follow-up studies that include larger cohorts, cancer-free control patients, and peripheral samples from patients with cancer will help determine if these molecular defects can be useful biomarkers in the clinic. We also used SNPs in the sequencing data to observe allele-specific methylation patterns that provide insights into the accumulation of aberrant DNA methylation during tumor development. This method would be valuable for comparing the allelic accumulation of methylation across tumors with different stages and grades to understand the timing of aberrant methylation.
Undoubtedly, the characterization of individual tumors will require an integrative approach, since key genes can be disrupted by numerous defects, including methylation, point mutation, and genetic instability (The Cancer Genome Atlas Research Network 2008; Chan et al. 2008). While, any individual method does not provide a complete picture of the spectrum of defects in a tumor, ideally we will move toward incorporating many different tools to obtain a comprehensive view of the expression differences, copy number variation, genome sequence, and epigenetic changes in each tumor. The method presented here fills a gap in the arsenal of tools for the characterization of aberrant DNA methylation. It provides the high resolution of bisulfite sequencing with the throughput of sampling many loci across many samples. This enables an experimental scale that promises to be useful in the effort to understand cancer.
Methods
Design of patch oligonucleotides and universal primers
Human promoter sequence between the TSS and 700 bp upstream of the TSS was downloaded from the March 2006 assembly on the UCSC Genome Browser (http://www.genome.ucsc.edu) for the RefSeq genes listed in Supplemental Data 1. These sequences were then scanned for the AluI restriction enzyme recognition sequences, and the AluI restriction fragments that were between 125 bp and 600 bp in length and containing at least three CpG positions were selected. A patch oligo was then designed by sequentially included base pairs from the AluI restriction site into fragment sequence until the Tm of Patch oligo was between 62°C and 67°C. Any fragment whose patch oligos contained repetitive elements according to the RepeatMasker track on the UCSC Genome Browser (http://www.genome.ucsc.edu) were excluded. The patch oligos were then appended with the complement universal primer sequences to result in the appropriate patch sequence. Patch oligonucleotides were synthesized by SigmaGenosys (http://www.sigmaaldrich.com/Brands/Sigma_Genosys.html). Ninety-four pairs of patch oligos were ordered in a 96-well plate. The patch oligos for two loci were duplicated on the plate so that when the equimolar portions were pooled from each well, these two loci were twice as concentrated in the pool. This was used to measure how the concentration of patch oligos affected amplification efficiency during protocol development. Two universal primer sequences were synthesized by IDT (http://www.idtdna.com), including U2, which has a 5′ phosphate and a three-carbon spacer on the 3′ end. Oligonucleotide sequences are listed in Supplemental Table 3.
Bar coded universal primers are used to PCR amplify the selected bisulfite converted loci from each sample. A different pair of universal primers is used to PCR amplify each sample, and they are distinguished by a 5-bp sample-specific DNA barcode that resides between the universal primer sequence and the 454 machine-specific sequence. The universal primer sequence is listed in Supplemental Table 3, and the barcode used for each patient sample is listed in Supplemental Table 2. There are 1024 possible 5-bp DNA sequences. We first excluded barcodes that contained homopolymers since 454 sequencing is known to produce more errors in this sequence context. We then calculated the number of sequence differences between all possible pairs of barcodes. We selected 48 sample-specific barcodes, one for each sample that had the least sequence similarity to each other, so that PCR error or sequencing error was least likely to turn one barcode sequence into another, resulting in the misassignment of a read to the wrong patient. We then ordered the 48 universal primer pairs containing barcodes from IDT (http://www.idtdna.com).
Bisulfite Patch PCR
Genomic DNA from cancer and adjacent normal tissue was obtained from Biochain (http://www.biochain.com) for both the breast and colon . Patient information and lot numbers are listed in Supplemental Table 2. Each patient sample was aliquoted into a well of a 96-well plate and digested with the AluI restriction endonuclease in 10 μL of total volume reaction containing 250 ng DNA, 10 U of AluI enzyme (NEB), and 1× NEBuffer 2 (NEB). This reaction was incubated for 1 h at 37°C, followed by heat inactivation of the enzyme for 20 min at 65°C, and was held at 4°C until the subsequent step.
Patch driven ligation of the universal primers to selected fragments was performed by addition of more reactants to the initial tube to result in the following final concentrations: 2 nM each Patch oligo, 200 nM U1 primer, 200 nM U2 primer (contains 5′ phosphate and 3′ three carbon spacer), 5 U of Ampligase (Epicentre), and 1× Ampligase Reaction Buffer (Epicentre) in a total volume of 25 μL. This reaction was incubated for 15 min at 95°C, followed by 30 sec at 94°C and 8 min at 65°C for 100 cycles, and was held at 4°C.
Incorrect products, template genomic DNA, and excess primer were degraded by the direct addition of 10 U of exonuclease I (USB) and 200 U of Exonuclease III (Epicentre) to the reaction. This mix was incubated for 1 h at 37°C, followed by heat inactivation for 20 min at 95°C, and was held at 4°C.
The reactions were then treated with sodium bisulfite to convert unmethylated cytosines to uracil. This was achieved by using the EZ DNA Methylation Gold Bisulfite Treatment Kit (Zymo Research) following the manufacturer's instructions, with one exception. Since the sample volume after the exonuclease treatment is 27 μL, the CT Conversion Reagent from the kit is made by adding 830 μL of dH2O instead of 900 μL of dH2O. The DNA is eluted from the columns in the final step with 10 μL of M-Elution buffer.
For the Universal PCR, we added reagents to the 10 μL of column elution to result in these final concentrations in 50 μL: 0.5 μM 454A:Sample Specific Barcode:U1, 0.5 μM 454B:Sample Specific Bar code:U2′, 10 U of Platinum Taq polymerase (Invitrogen), 0.5 mM each dNTP, 2 mM MgCl2, 0.5 M Betaine, 20 mM Tris-HCl (pH 8.4), and 50 mM KCl. This reaction was incubated for 2 min at 93°C, followed by 30 sec at 93°C and 6 min at 57°C for 35 cycles, and was held at 4°C. The PCR product smear between the expected sizes was confirmed by running 20 μL of the PCR product from each sample on a 3% Metaphor agarose gel (Lonza). We then pooled 5 μL from each sample into a single tube and purified this pool on a Qiaquick Spin Column (Qiagen). The eluted DNA was quantified on the NanoDrop (http://www.nanodrop.com) as well as on a plate reader (BioTek Synergy HT) using PicoGreen (Invitrogen) following the manufacturer's instructions. This pooled sample was then prepared and sequenced on the 454 Life Sciences (Roche) FLX machine following the manufacturer's instructions.
Sequencing data analysis
We obtained 97,115 sequencing reads, with an average read length of 228 bp. The sequence reads and quality scores are available in a gzipped fastq format file (Supplemental Data 2). To determine which sequences matched our targets, we aligned the reads against a database of reference sequences for each target using WU-BLASTN (http://blast.wustl.edu). Since the sequences are sodium bisulfite–treated, we substituted a T in place of C in the genomic sequence at the non-CpG positions in the reference sequences. We then determined how many reads matched significantly to each promoter (BLAST smallest sum P < 0.001) and put all reads from each promoter in a separate file. We computed the correlation between the number of reads and the amplicon length for each promoter using linear regression. We identified which sample each read came from by matching the first five bases of the read to the list of sample-specific barcode and corresponding patients. To determine the reproducibility of the method, we computed the number of reads for each locus in each sample, and calculated the correlation coefficient between two samples for all possible pairs of samples; the mean of these correlation coefficients represents the average correlation between the number of reads per locus across samples. For each promoter, we used ClustalW to generate multiple sequence alignments of all of the reads and the reference sequence (Larkin et al. 2007). We identified germline SNPs in the sequences by looking for variants in the reads and comparing these to known SNPs reported on the UCSC Genome Browser (http://www.genome.ucsc.edu). To visualize these multiple sequence alignments, we create one matrix per promoter, where the first column identifies the sample from which the read originated (1–48), and the remaining columns are coded for the base in the read, where Cs are replaced with 8, the two alleles at SNP positions are replaced with 5 and 12, and the remaining bases are converted to 0. This matrix was then visualized as an image using the Matlab software package (The Mathworks, Inc.). The matrix was sorted by sample type (the first column) and further calculations regarding the amount of methylation per read, and per sample were computed using Matlab (The MathWorks, Inc.).
To quantify the sensitivity and specificity of each locus exhibiting tumor-specific methylation, we used a threshold to classify a locus as methylated or unmethylated in each sample. We queried many CpGs for each locus with the bisulfite sequencing. We used this information to find the optimal classifier of DNA methylation to distinguish tumor and normal samples. We searched across all possible values for two parameters: percentage of CpGs per molecule and percentage of reads per sample. We found that the optimal classifier between tumor and normal was to classify a sample as “methylated” if more than 20% of CpG positions per molecule were methylated in more than 35% of molecules. The fraction of samples that were classified as methylated is listed in Table 2 for each locus.
Acknowledgments
We thank Jeffrey Gordon, Brian Muegge, and Jill Manchester for generously sequencing the samples on their 454 Life Sciences (Roche) FLX machine. We thank Kay Tweedy for assistance in preparing the samples. We thank Jason Gertz for suggestions, discussion, and critical reading of the manuscript. We thank Maximiliaan Schillebeeckx, Lee Tessler, Todd Druley, Adam Joyce, David Mayhew, Francesco Vallania, Michael Brooks, and Yue Yun for helpful discussions. This work was supported by a Siteman Cancer Center Endometrial Cancer Working Group Research Development Award, the Genome Analysis Training Program (T32 HG000045), a Center for Excellence in Genome Sciences grant from the National Human Genome Research Institute (5P50HG003170-03), and a Technology Development in Epigenetics grant from the National Institutes of Health (1R01DA025744-01).
Footnotes
[Supplemental material is available online at http://www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.101212.109.
References
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM 2009. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol 27: 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP 1998. Alterations in DNA methylation: A fundamental aspect of neoplasia. Adv Cancer Res 72: 141–196 [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TA, Glockner S, Yi JM, Chen W, Van Neste L, Cope L, Herman JG, Velculescu V, Schuebel KE, Ahuja N, et al. 2008. Convergence of mutation and epigenetic alterations identifies common genes in cancer that predict for poor prognosis. PLoS Med 5: e114 doi: 10.1371/journal.pmed.0050114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE 2008. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452: 215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, et al. 2009. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol 27: 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW 2000. MethyLight: A high-throughput assay to measure DNA methylation. Nucleic Acids Res 28: e32 http://nar.oxfordjournals.org/cgi/content/abstract/28/8/e32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den Boom D 2005. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci 102: 15785–15790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG 2000. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 343: 1350–1354 [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci 89: 1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, et al. 2007. Genome-wide in situ exon capture for selective resequencing. Nat Genet 39: 1522–1527 [DOI] [PubMed] [Google Scholar]
- Hodges E, Smith A, Kendall J, Xuan Z, Ravi K, Rooks M, Zhang M, Ye K, Battacharjee A, Brizuela L et al. 2009. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Research 19: 1593–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun HJ, Woolfenden S, Coven S, Lane K, Bronson R, Housman D, Charest A 2009. Epigenetic regulation of c-ROS receptor tyrosine kinase expression in malignant gliomas. Cancer Res 69: 2180–2184 [DOI] [PubMed] [Google Scholar]
- Kim JY, Tavare S, Shibata D 2006. Human hair genealogies and stem cell latency. BMC Biol 4: 2 doi: 10.1186/1741-7007-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarmann GJ, Decker A, Farrar WL 2008. Epigenetic gene silencing in the Wnt pathway in breast cancer. Epigenetics 3: 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova Y, Maloney RK, Lakey N, Citek RW, Bacher B, Budiman A, Ordway JM, McCombie WR, Leon J, Jeddeloh JA, et al. 2008. Massively parallel bisulphite pyrosequencing reveals the molecular complexity of breast cancer-associated cytosine-methylation patterns obtained from tissue and serum DNA. Genome Res 18: 19–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW 2005. Cancer epigenetics. Hum Mol Genet 14: R65–R76 [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. ClustalW and ClustalX version 2. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F, Brown R 2005. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst 97: 1498–1506 [DOI] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454: 766–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson K, Clark J, Lamparska-Kupsik K, Smith SS 2007. Recovery of bisulfite-converted genomic sequences in the methylation-sensitive QPCR. Nucleic Acids Res 35: 2893–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabilsi NH, Broaddus RR, Loose DS 2009. DNA methylation inhibits p53-mediated survivin repression. Oncogene 28: 2046–2050 [DOI] [PubMed] [Google Scholar]
- Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, et al. 2007. Multiplex amplification of large sets of human exons. Nat Methods 4: 931–936 [DOI] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Toyota M, Carraway H, Gabrielson E, Ohmura T, Fujikane T, Nishikawa N, Sogabe Y, Nojima M, Sonoda T, et al. 2008. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer 98: 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KH, Kramer RS, Davis JW, Guo J, Duff DJ, Xu D, Caldwell CW, Shi H 2007. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res 67: 8511–8518 [DOI] [PubMed] [Google Scholar]
- Tomii K, Tsukuda K, Toyooka S, Dote H, Hanafusa T, Asano H, Naitou M, Doihara H, Kisimoto T, Katayama H, et al. 2007. Aberrant promoter methylation of insulin-like growth factor binding protein-3 gene in human cancers. Int J Cancer 120: 566–573 [DOI] [PubMed] [Google Scholar]
- Ushijima T 2005. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer 5: 223–231 [DOI] [PubMed] [Google Scholar]
- Varley KE, Mitra RD 2008. Nested Patch PCR enables highly multiplexed mutation discovery in candidate genes. Genome Res 18: 1844–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley KE, Mutch DG, Edmonston TB, Goodfellow PJ, Mitra RD 2009. Intra-tumor heterogeneity of MLH1 promoter methylation revealed by deep single molecule bisulfite sequencing. Nucleic Acids Res 37: 4603–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veigl ML, Kasturi L, Olechnowicz J, Ma AH, Lutterbaugh JD, Periyasamy S, Li GM, Drummond J, Modrich PL, Sedwick WD, et al. 1998. Biallelic inactivation of hMLH1 by epigenetic gene silencing: A novel mechanism causing human MSI cancers. Proc Natl Acad Sci 95: 8698–8702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Siegmund KD, Muller HM, Fiegl H, Marth C, Muller-Holzner E, Jones PA, Laird PW 2004. Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res 64: 3807–3813 [DOI] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. 2007. The genomic landscapes of human breast and colorectal cancers. Science 318: 1108–1113 [DOI] [PubMed] [Google Scholar]