Abstract
Background
Warfarin demonstrates wide interindividual variability that is partly mediated by variants in CYP2C9 and VKORC1. Whether variants in CALU (vitamin K reductase regulator) influence warfarin dose is unknown.
Methods and Results
We resequenced CALU regions in a discovery cohort of dose-outliers: patients with high(>90th percentile, n=55) or low(<10th percentile, n=53) dose requirements(after accounting for known genetic and nongenetic variables). One CALU variant, rs339097, was associated with high-doses(p=0.01). We validated this variant as a predictor of higher warfarin doses in two replication cohorts: 1)496 patients of mixed ethnicity, 2)194 African-American patients. The G allele of rs339097(African-American and Caucasian allele frequency 0.14 and 0.002, respectively), was associated with a 14.5%(SD±7%) greater therapeutic dose(p=0.03) in the first replication cohort and a higher than predicted dose in the second replication cohort(allele frequency=0.14, one-sided p=0.03).
Conclusions
CALU rs339097 A>G is associated with higher warfarin dose requirements independent of known genetic and nongenetic predictors of warfarin dose in African-Americans.
Keywords: anticoagulants, calumenin, genetic polymorphism, pharmacogenetics, warfarin/administration & dosage
Introduction
Warfarin sodium is a widely used anticoagulant for the prevention and treatment of venous thromboembolism, myocardial infarction, and stroke. Warfarin is characterized by a wide interindividual variability in dose response. In addition, warfarin has a narrow therapeutic index: International Normalized Ratio (INR) values greater than 3 or 4 increase the risk of bleeding while those significantly less than 2 are less effective.(1) Several single nucleotide polymorphisms (SNPs) in Cytochrome P450 2C9 and 4F2 (CYP2C9 and CYP4F2, respectively) and vitamin K 2,3-epoxide reductase complex subunit 1 (VKORC1) are associated with dose-response, but together explain only 35% of the variation in warfarin dose requirements in Caucasians (2, 3) and 10% in African-Americans(4)
Although recent whole genome association studies failed to identify novel variants outside of these three loci, they were limited to Caucasian patients.(5–7) Thus, additional genes may help predict warfarin dose, especially in African-American populations, where VKORC1 and CYP2C9 SNPs are not as predictive of the dose-response(8, 9) and where warfarin resistance may be more common(10).
Studies of warfarin resistant rats and in vitro mRNA silencing experiments have identified calumenin (CALU) as a regulator of vitamin K 2,3-epoxide reductase (VKOR) and warfarin sensitivity.(11–13) In humans, limited studies of genetic variation in CALU in unselected warfarin treated patients have yielded unconvincing results. For example, an exonic CALU SNP, rs2290228, was found in one patient with an exceptionally high warfarin dose(14) but CALU SNPs have not been significantly associated with dose in other studies.(15, 16)
To systematically investigate if genetic variation in CALU is associated with warfarin sensitivity or resistance, we chose a two-stage study design (Figure 1). In the first stage we selected patients from a well-characterized parent cohort who were at the extremes of warfarin dose requirements after accounting for known genetic and nongenetic factors from a parent cohort of patients taking warfarin chronically (Figure 1, left hand panel). In the second stage, we used two replication cohorts: 1) a larger, mixed ethnicity sample of the parent cohort (Figure 1, right hand panel) and 2) an external cohort of African-American patients. We used these replication cohorts to validate how CALU SNPs correlated with higher warfarin doses while accounting for known genetic and clinical factors. This two-stage strategy has been shown to be powerful(17) and successful at identifying novel variants using candidate genes (ABCA1 for HDL cholesterol(18) and PCSK9 for LDL cholesterol(19)) or whole genome (NOS1AP for QT interval(20)) approaches. To identify novel variants in CALU we directly sequenced exonic regions of CALU as well as evolutionary conserved putative regulatory regions. To identify these regulatory elements we used a comparative genomics approach to identify noncoding elements that were conserved in the rat and mouse orthologs. We hypothesized that patients who required unusually high or low doses of warfarin (compared to their predicted dose based on a validated pharmacogenetic algorithm that incorporates genetic and nongenetic factors(9)) would be more likely to have clinically relevant SNPs in CALU. We then validated these associations in two larger replication cohorts.
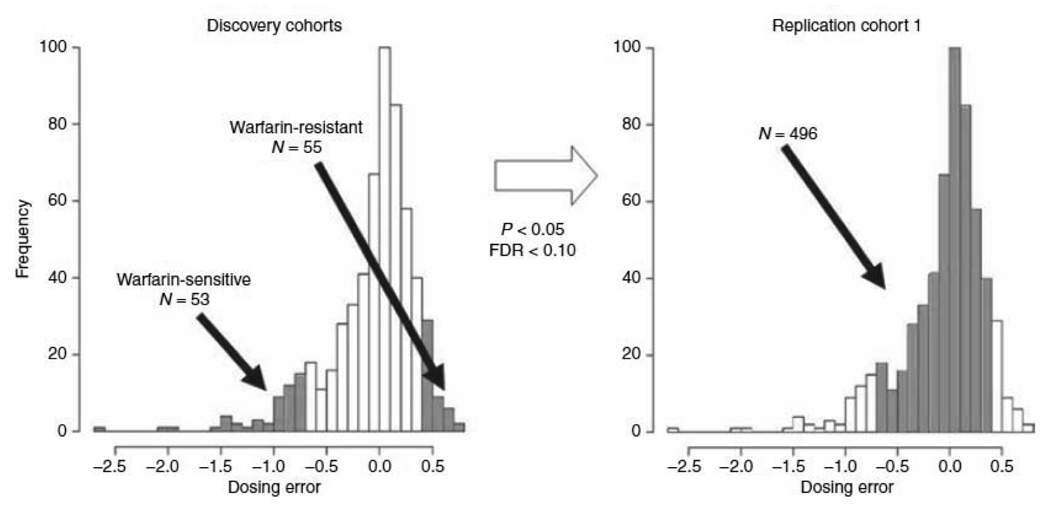
Figure 1.
Schematic of two-stage study design using a parent cohort of patients on stable warfarin therapy. In the discovery cohort (left panel, we sequenced CALU in patients who had the largest or smallest residual dosing errors (shaded) within the parent cohort. In the second stage, significant (P < 0.05, FDR < 0.010) SNPs from the discovery stage were then genotyped in the remainder of the parent cohort (shaded, replication cohort 1, right panel). Predicted doses were based on a validated pharmacogenetics algorithm.9 CALU, calumenin; FDR, false-discovery rate; SNP; single-nucleotide polymorphism.
Results
Discovery Cohort
From the parent cohort we selected 108 outliers: 53 patients formed the low-dose group and 55 patients formed the high-dose group (Figure 1, left panel). Despite having similar predicted doses (4.7±1.9 vs. 5.2±1.9) based on the validated pharmac algorithm (9), the average dose in the high-dose group was five times greater than in the low-dose group (Table 1). As designed, the dose ratio (therapeutic/predicted doses) was higher in the high-dose group compared to the low-dose group: 2.4 ± 1.4 vs. 0.55 ± 0.07, p < 0.0001. The two groups were well matched with respect to other important determinants of warfarin responsiveness. (Table 1)
Table 1.
Baseline Characteristics of Discovery and Replication Cohorts
| Discovery | Replication Cohort #1 (n = 496) |
Replication Cohort #2 (n = 194) |
||
|---|---|---|---|---|
| Low-Dose (n=53) |
High-Dose (n=55) |
|||
| Therapeutic dose, mean±SD (mg/day) † |
2.6±1.1 | 12.7±8.3 | 4.8 ± 1.6 | 5.9 ± 2.3 |
| Age, mean±SD (years) | 62±15 | 59±15 | 61 ± 15 | 57 ± 16 |
| BSA, mean±SD (m2) | 2.01±0.41 | 2.04±0.31 | 2.0 ± 0.3 | 2.07 ± 0.33 |
| Current Smoker, N (%) | 7 (13) | 11 (20) | 60 (12) | 35 (18) |
| Medications | ||||
| Amiodarone, N (%) | 4 (8) | 1 (2) | 9 (2) | 4 (2) |
| Simvastatin, N (%)* | 4 (8) | 0 (0) | 56 (11) | 64 (33) |
| Male | 20 (38%) | 29 (53%) | 255 (51%) | 47 (24%) |
| Race | ||||
| Black, N (%) | 18 (34) | 21 (38) | 70 (14) | 194 (100) |
| White, N (%) | 34 (64) | 33 (60) | 407 (82) | 0 |
| Other, N (%) | 1 (2) | 1 (2) | 20 (4) | 0 |
| CYP2C9*2 (% carriers) | 30 | 23 | 11 | 5 |
| CYP2C9*3 (% carriers) | 13 | 15 | 6 | 2 |
| VKORC1-1639 A (% carriers) | 51 | 49 | 33 | 18 |
BSA = Body surface area; CYP2C9 = Cytochrome P450; VKORC1 = Vitamin K epoxide reductase, complex subunit 1; SD = standard deviation; High and low dose groups based on residual dosing error = (therapeutic dose – predicted dose)/therapeutic dose; Predicted doses based on a validated pharmacogenetic algorithm.(9)
p = 0.05 between discovery groups;
p<0.01
The comparative genomics approach identified 10 highly conserved regions for resequencing that included all known CALU exons, portions of the 5’ and 3’ UTRs and promoter region, and several conserved transcription factor binding sites. (Supplementary Methods) Our resequencing strategy identified eight polymorphisms in CALU: seven previously identified in public databases (dbSNP Build ID: 130) and one novel SNP. (Table 2) All eight SNPs were in Hardy-Weinberg Equilibrium (data not shown). Among these eight SNPs, the proportion of patients carrying the minor allele of one SNP, rs339097, was significantly different between groups. (Table 2) This A>G SNP was a significant (p=0.01, false discovery rate 9%) predictor of the high-dose group among African-American outliers: it was present in 7 of 17 (41%) high-dose outliers (minor allele frequency [MAF] 20%) vs. 1 of 19 (5%) low-dose outliers (MAF = 3%). In contrast, the allele was invariant in Caucasian outliers.
Table 2.
Variants discovered in CALU and association with high dose in outlier group (n = 108)
| SNP | Region | Position | Alleles | MAF by Race in low dose group (AA/white/O,%) |
MAF by Race in high dose group (AA/white/O,%) |
P-value in white pts |
P-value in AA pts |
|---|---|---|---|---|---|---|---|
| rs12538139 | 5' utr | 128175777 | G/A | 10/23/0 | 8/21/50 | 0.7 | 0.9 |
| rs2290228 | exon | 128175884 | G/A | 12/16/50 | 10/23/0 | 0.09 | 0.5 |
| rs2290227 | intron | 128176217 | G/A | 0/1/0 | 5/6/0 | 0.1 | 0.9 |
| rs2307040 | intron | 128181842 | C/T | 12/37/0 | 13/26/50 | 0.3 | 0.8 |
| rs41274227 | intron | 128182044 | C/T | 0/3/0 | 5/6/0 | 0.1 | 0.9 |
| rs339097 | intron | 128186460 | A/G | 3/0/0 | 20/0/0 | 1.0 | 0.01 |
| 38412 | intron | 128195082 | C/T | 6/0/0 | 3/0/0 | 1.0 | 0.6 |
| rs1043550 | 3' utr | 128196461 | A/G | 16/36/0 | 15/32/50 | 0.8 | 0.9 |
AA = African–American; MAF = minor allele frequency; O = Other; pts = patients; SNP = Single Nucleotide Polymorphism labeled as dbSNP reference number or internal identification number; utr = untranslated region.
Replication Cohorts
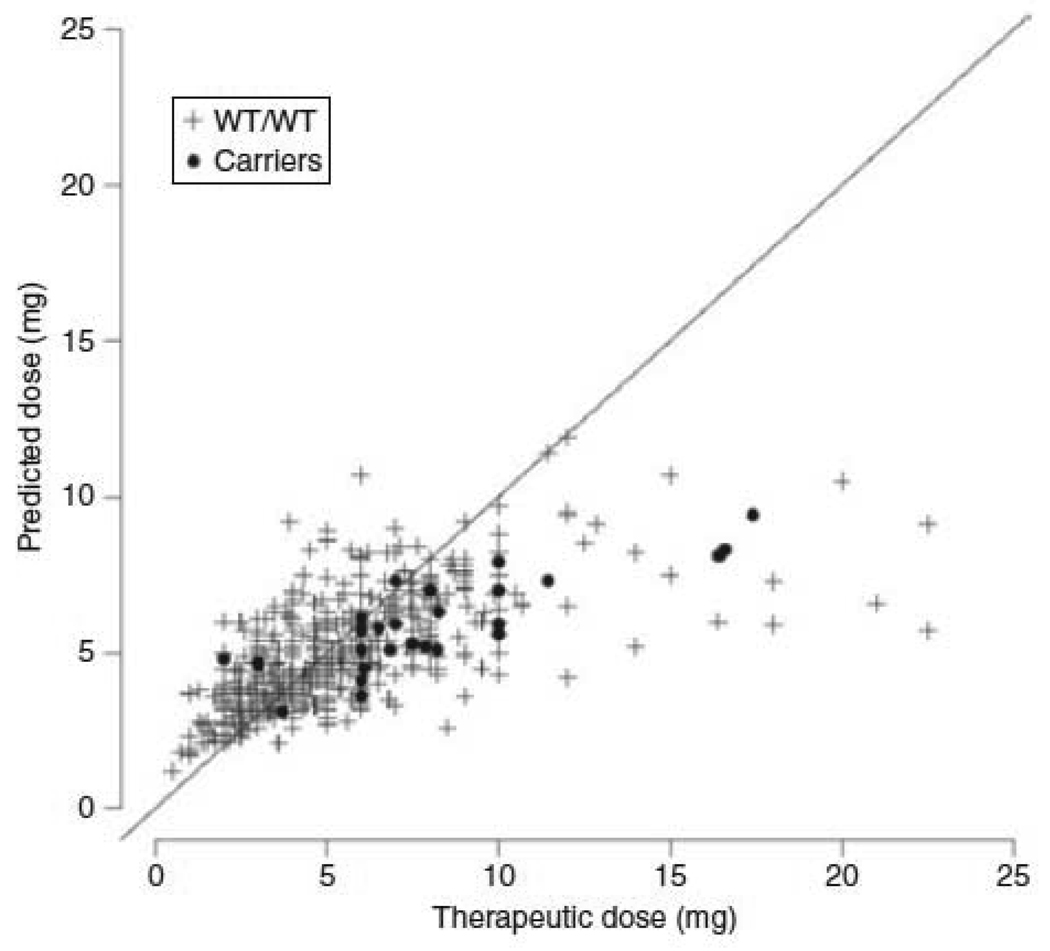
To validate these findings we first identified 496 patients from the parent cohort who had DNA and complete clinical data (replication cohort #1, Figure 1 right hand panel). We genotyped these participants for CALU rs339097 and three CALU SNPs identified in the literature (rs2290228, rs2290227, and rs1043550)(14, 15, 21). In a univariate analysis, only the CALU SNP rs339097 was significantly (p = 0.003) associated with therapeutic dose. Carriers of the minor (G) allele of rs339097 were more likely African-American (Table 3). In a multivariable regression model that accounted for CYP2C9 and VKORC1 genotypes and clinical factors including race, carriers of the G allele of CALU SNP rs339097 averaged (± SD) 14.5% (± 7%) higher therapeutic doses compared to noncarriers (p=0.03). In the entire parent cohort (discovery plus replication cohort #1), 25 out of the 31 carriers of the CALU rs339097 minor allele (83%) had larger therapeutic doses than predicted by the validated pharmacogenetic algorithm(9) (Figure 2). In African-Americans, rs339097 individually explained 5.7% (R2 = 0.057) of the residual variation in warfarin. However, the allele was too rare in Caucasians to evaluate its influence on the R2 in that group. There was no significant two-way interaction between the CALU SNPs and either VKORC1 (VKORC1 T-1639A) or CYP2C9 SNPs (*2, and *3).
Table 3.
Characteristics of replication cohorts, stratified by CALU SNP rs339097
| Replication Cohort #1 | Replication Cohort #2 | |||
|---|---|---|---|---|
| Characteristic | rs339097 GG or AG (n = 21) |
rs339097 AA (n= 475) |
rs339097 GG or AG (n = 52) |
rs339097 AA (n= 142) |
| Therapeutic warfarin dose (mg/day, mean§ ± SD ) |
6.5 ± 1.4 | 4.8 ± 1.6** | 6.3 ± 2.3 | 5.7 ± 2.3 |
| Age (years, mean ± SD) |
57 ± 19 | 62 ± 15 | 58 ± 15 | 56 ± 16 |
| Race (%) | ||||
| African-American | 86% | 11%** | 100% | 100% |
| Caucasian | 10% | 85% | 0% | 0% |
| Other | 5% | 4% | 0% | 0% |
| BSA (m2, mean ± SD) |
2.0 ± 0.2 | 2.0 ± 0.3 | 2.0 ± 0.4 | 2.1 ± 0.3 |
| VKORC1 -1639 A† | 10% | 37% | 12% | 20% |
| CYP2C9*2† | 2% | 12% | 2% | 6% |
| CYP2C9*3† | 2% | 6% | 2% | 1% |
| Current smoker | 29% | 14%* | 10% | 21% |
| Current amiodarone use |
16% | 2%* | 2% | 2% |
BSA = Body surface area; CYP2C9*2 = Cytochrome P450 2C9*2; CYP2C9*3 = Cytochrome P450*3; SD = standard deviation; DVT = deep venous thrombosis; PE = pulmonary embolus; Comparisons to heterozygotes/homozygotes for rs339097:
p<0.05;
p<0.01;
represented as percentage with minor allele;
geometric mean
Figure 2.
Carriers of CALU SNP rs339097 require higher-than-predicted therapeutic warfarin doses in the combined discovery and replication cohort 1. Therapeutic dose vs. predicated dose (based on a validated pharmacogenetic algorithm9) in carriers (•) and noncarriers (+) of the G allele for CALU rs339097. In this combined cohort, 25 of 31 (83%) carriers of the G allele required a higher-than-predicated warfarin dose to achieve a therapeutic INR. CALU, calumenin; INR, international normalized ratio; SNP, single-nucleotide polymorphism; WT, wild type.
Because the prevalence of the G allele for rs339097 varied significantly by race, we sought additional replication in 194 African-American patients on warfarin (replication cohort #2). The baseline characteristics of this group were similar, except for higher average dose requirements and a lower percentage of men (Table 1). In this second cohort, carriers of the G allele (MAF 14%) had a higher residual dosing error (therapeutic dose − predicted dose(9)) compared to noncarriers: mean residual (± SD) in carriers vs. noncarriers: 0.20 (± 2.0) vs. − 0.41 ± 1.9, one-sided p-value = 0.03, two-sided p-value = 0.06), confirming that despite known genetic/nongenetic factors, carriers of the G for CALU rs339097 allele required higher warfarin doses.
In a pooled analysis of all African-American patients who participated in this study, the minor allele of CALU rs339097 was associated with an 11% increase in warfarin dose requirements after accounting for other factors (Table 4).
Table 4.
Influence of pharmacogenetic factors on therapeutic warfarin dose in pooled African-American patients (n = 241)
| Factor | Effect on warfarin dose | |
|---|---|---|
| Age in years, mean (SD) | 58 (16) | −6% per decade |
| BSA, m2, mean (SD) | 2.1 (0.4) | 24% per 0.5m2 |
| Percent Smokers | 20% | 13% |
| Percent Using Amiodarone | 3% | −16% |
| CALU Allele frequency | 0.14 | 11% per allele |
| CYP2C9 *2 Allele Frequency | 0.04 | −20% per allele |
| CYP2C9 *3 Allele Frequency | 0.01 | −34% per allele |
| VKORC1 3673A Frequency | 0.11 | −24% per allele |
| Percent with indication of DVT or PE | 43% | 2% |
| Target INR | 2.5 (0.13) | 4% per 0.25 increase in target INR |
SD = standard deviation; INR = international normalized ratio; BSA = body surface area; m = meter; DVT = deep venous thrombosis; PE = pulmonary embolus
In silico validation
Because hepatic CALU is overexpressed in rodents with warfarin resistance(11) and rs339097 is intronic, we used publicly available gene expression data to determine whether genetic variation at rs339097 altered CALU gene expression in humans. In immortalized lymphoblastoid cell lines of 30 trios of African descent (parents, n = 60 and their adult children, n = 30), the number of G alleles was associated with higher CALU gene expression (mean normalized CALU expression ± SE in those with 0, 1, or 2 alleles: 10.01 ± 0.02, 10.17 ± 0.69, and 10.33 ± 0.69; respectively; p-value: 0.02).
Discussion
Warfarin is the most widely prescribed oral anticoagulant and is characterized by wide interindividual variability. Although recent studies have identified SNPs in VKORC1 and CYP2C9 in conjunction with nongenetic factors (9, 22, 23) as significant predictors of dose requirements, more than half of the variability in warfarin dose has remained unexplained. Calumenin, a recently identified regulator of VKOR(13), is overexpressed in warfarin resistant rats(11) and has been suggested to underlie warfarin sensitivity in humans(14), but had not been evaluated thoroughly.
In this pharmacogenetic study we found that carriers of the minor allele of the third intronic CALU SNP rs339097 required a dose that was 14.5% greater than predicted by VKORC1, CYP2C9 and clinical factors. The A>G allele was rare in Caucasian (allele frequency 0.2%), but common in African-Americans (allele frequency 11%–14%).
This genetic association study had several methodological strengths: 1) use of a candidate gene approach, 2) resequencing of conserved regions to identify influential regions within this gene, and 3) employing a two-stage approach(18–20) that first examines extreme phenotypes in a population and then carries forward significant SNPs to a larger, more general samples of patients. The two–stage approach allowed us to discover a CALU SNP associated with a high-dose warfarin group and then demonstrate that carriers of the minor, G allele, of rs339097 had higher than predicted doses in two general populations. The decision to study CALU was backed by evidence of its role in rodent warfarin resistance(11) and in vitro evidence of calumenin inhibiting VKOR(13). We resequenced highly conserved regions of CALU to maximize our ability to identify influential regions outside of the coding regions, and used the false discovery rate to account for multiple comparisons.
Calumenin is a member of the CREC family of proteins and localizes in the endoplasmic reticulum,(24) where γ – carboxylase(25) and VKOR(26) also localize. In humans, two calumenin isoforms are expressed that use two different second exons (exon 2-1 and exon 2-2) and both isoforms were sequenced. In the rat, calumenin interacts with and inhibits VKOR, thereby inhibiting γ - carboxylase. Furthermore, certain warfarin-resistant rats have increased hepatic levels of calumenin that protect VKOR from inhibition by warfarin. The VKOR and γ - carboxylase isolated from these rats have normal warfarin sensitivity.(11, 12) In cells genetically engineered cells to produce Factor IX or VII, in vitro silencing of CALU mRNA increases VKOR and γ - carboxylase activity and production of these VKOR dependent clotting factors.(13, 27) In humans, a SNP in the 3’ UTR of CALU was associated with increased activity of one other vitamin-K dependent protein (protein S).(15) Therefore, the available animal and in vitro evidence suggest that calumenin regulates VKOR and γ – carboxylase.
Although the molecular implications of rs3390907 are not known, using publicly available gene expression data from lymphoblastoid cell lines generated from trios of the YRI cohort(28) we observed that the G allele of rs3390907 is associated with higher expression of CALU. Interestingly, this association with higher gene expression in carriers of the G allele for rs339097 is consistent with the findings in warfarin resistant rats where CALU was found to be overexpressed(11). Based on HapMap linkage disequilibrium data of the YRI population, rs339097 is in the center of an 18-Kb haplotype block (Haploview(29)). This large block contains several conserved regions, particularly in the second intron, that were not examined as part of the current study. Therefore it is unclear if this SNP directly leads to alterations in calumenin expression or mRNA stability or is in linkage disequilibrium with a causative SNP within this haplotype block.
The CALU SNP identified in our populations, rs339097, had minor allele frequencies of 11% to 14% in African-Americans and 0.2% in Caucasians. Although we recruited subjects with a wide variety of clinical conditions, these allele frequencies are similar to those found in the YRI and CEU (Centre d'Etude du Polymorphisme Humain from Utah) populations studied by the HapMap consortium (19.5% and 0%, respectively). Thus, among patients starting warfarin, this SNP will have greater potential to benefit patients of African descent.
In the multivariable regression model that accounted for race, rs339097 remained a significant predictor of higher warfarin dose in the first replication cohort. Though the overall contribution of this SNP to the residual variance in African-Americans was modest, patients who carried this SNP had a 14.5% increase in their warfarin requirement per allele. The dose-effect is similar to the CYP2C9*2 allele, which is associated with a 19% decrease, and to the CYP4F2 rs2108622 allele, which is associated with a 4% to 12% increase in warfarin requirements. Identification of novel SNPs in the African-American population is important because lower allele frequencies of VKORC1 and CYP2C9 SNPs in this population make current pharmacogenetic models of warfarin dosing less accurate in that population.(8, 9, 30) Therefore, future studies of pharmacogenetic warfarin therapy, particularly those with larger numbers of patients of African descent, could use additional SNPs and perhaps initial INR response(31) to improve predictive accuracy. To facilitate these studies, we have incorporated the present findings into an online dosing algorithm: www.warfarindosing.org.
In summary, we found that carriers of the minor allele of the CALU SNP rs339097 require higher doses of warfarin. Because the incremental cost of adding a single SNP into commercial genotyping platforms is low, CALU rs339097 could be incorporated into these platforms and into clinical trials that include populations with African ancestry.
Methods
Patient Selection
Our parent cohort is comprised of approximately 700 patients on therapeutic doses of warfarin who attended an outpatient anticoagulation clinic affiliated with Barnes–Jewish Hospital at Washington University Medical Center (St. Louis), who participated in the PREVENT (PREvention of VENous Thromboembolism) study, or who were initiated on warfarin during a hospitalization for orthopedic surgery.(2, 9, 32, 33) These patients provided written informed consent, DNA, demographic variables, and medication histories as previously described.(9) The parent cohort is representative of most ambulatory patients on warfarin therapy (Table 1) except that it also included four patients who were referred for unusual warfarin doses (> 20 mg/day or < 1 mg/day). Because of missing data or unavailable DNA, only 604 (86%) patients were available to construct the discovery and replication cohorts. The research protocol was approved by the institutional review board at Washington University in St. Louis.
Calumenin SNP discovery cohort
For all patients in the parent cohort, the therapeutic daily warfarin dose (in mg) was compared to the dose predicted by a validated pharmacogenetic algorithm (9)). We then chose the 10% of this cohort with the largest residual dosing error (defined as therapeutic dose - predicted dose/therapeutic dose) to identify 108 outliers with DNA who were either warfarin sensitive (N = 53) or resistant (N = 55) (Figure 1, left panel).
Calumenin SNP replication cohorts
From the remaining patients in the parent cohort we selected the 496 patients with DNA and complete clinical data recruited at Washington University Medical Center for further genotyping (Replication cohort #1, Figure 1, right panel). Of these, three did not reach a stable dose and their therapeutic dose was estimated by two independent clinical pharmacists (PEM and GRG). The pharmacists were blinded to genotype and agreed in their estimation of therapeutic dose.
To validate our findings in an African-American population we (SRP, LHC) genotyped 194 African-American patients at the University of Illinois at Chicago (replication cohort #2)(34). We excluded those (n = 7) with missing VKORC1 and CYP2C9 genotypes or those on known inducers of Cytochrome P450 enzymes.
CALU in silico gene expression replication
Using software developed by Ge et al.,(35) we merged publicly available gene expression data(36) from lymphoblastoid cell lines with genotype data from the HapMap project to test for an association between the minor (G) allele of rs339097 and CALU gene expression in trios of Yoruba people of Ibadan, Nigeria (YRI population, HapMap project).
CALU Target Region Selection
CALU is a 32Kb gene located on human 7q32 and contains six exons. In humans two CALU isoforms are expressed that use two different exons 2 (exon 2-1 and exon 2-2).
We used two complementary methods to target region selection for direct sequencing of CALU. We used two comparative genomics approaches to find 10 highly conserved (>90% across rat and mouse orthologs) regions of CALU and 5Kb of its 5' and 3' flanking regions using the VISTA Browser (http://genome.lbl.gov/vista/index.shtml) and the University of California at Santa Cruz (UCSC) Genome Browser's "knownGene" database file and mRNAs from GenBank (http://genome.ucsc.edu/cgi-bin/hgTrackUi?hgsid=66289578&c=chr7&g=knownGene, March 2004). These 10 regions contained all six exons and the alternatively spliced exon 2 (as expected) and a 69-bp segment of the 5’UTR. In addition, we identified two regions in the 3’UTR and one in the 5’UTR that were highly conserved. We designed primers and sequenced these 10 regions as previously described (correction to original description (37): 100 µM each of the 4 dNTPs was used). (Supplementary Methods).
Genotyping Protocols
To validate the SNPs identified by sequencing we first genotyped the 108 outliers for the SNPs identified by resequencing using PCR and Pyrosequencing® (38, 39). PCR primers were designed using the UCSC Golden Path Human Genome Browser May 2004 Build (http://genome.ucsc.edu/cgi-bin/hgGateway) and CALU Refseq NM_001219. This confirmatory step revealed eight polymorphic CALU SNPs for analysis in the discovery cohort: c38412t, rs41274227, rs1043550, rs12538139, rs2290227, rs2290228, rs2307040, rs339097. For the replication cohort, we used Pyrosequencing® (40) to genotype selected SNPs.
Statistical Analyses
In the SNP discovery analysis, we used a chi-squared analysis to compare the prevalence of SNPs between the high-dose and low-dose groups and the Fisher’s exact test when cell frequencies were low. As recommended,(41) we calculated and report false discovery rates.
In replication cohort #1, we tested CALU SNPs (rs339097, rs2290228, rs2290227, and rs1043550) in all subjects and in the subsets of Caucasians and African-Americans. We tested for Hardy-Weinberg Equilibrium using Haploview 3.32; if the SNP was in HWE (all were), we used simple linear regression to regress the therapeutic daily warfarin dose (log transformed) onto the number of minor alleles in univariate and multivariate models that included CYP2C9 and VKORC1 genotypes, age, smoking, body surface area, concomitant medications, target INR, venous thromboembolism indication, and race. We used an unpaired t-test to compare therapeutic doses (log transformed) of carriers and non carriers of the CALU minor allele. We tested for two-way interactions between SNPs in CALU, VKORC1, and CYP2C9.
In replication cohort #2, we tested for association of CALU SNP rs339097 A>G with residual dosing error (ln(therapeutic) − ln(predicted warfarin dose)(9)) using a one-sided (unpaired) t-test. The normality of residuals was confirmed using the Kolmogorov-Smirnov test.
For the in silico replication we use a generalized estimating equation to regress the normalized CALU gene expression onto the number of minor alleles of rs339097 (coded as 0, 1, 2) in YRI children and parents using SAS procedure GENMOD to account for the family structure of the trios. Normality of residuals was confirmed Kolmogorov-Smirnov test.
We performed all statistical analyses in SAS (Version 9.1; SAS Institute, Inc; Cary, NC) except that replication cohort #2 was analyzed in SPSS (Release 15.0; SPSS Inc., Chicago, IL). Statistical significance refers to a p-value < 0.05.
Supplementary Material
Acknowledgements
This study was funded by the National Institutes of Health, R01 HL074724
Footnotes
Authorship and conflicts of interest
Voora D: responsible for study design, analytical approach, analysis and interpretation of data, and drafting manuscript; no conflict of interest
Koboldt DC: responsible for study design, interpretation of sequence analysis, and critical revision of manuscript; no conflict of interest
King CR: responsible for study design, interpretation and performing genotype analysis, and critical revision of manuscript; no conflict of interest
Lenzini P: responsible for study design, interpretation and performing statistical analysis, and critical revision of manuscript; no conflict of interest
Eby C: responsible for study design, performing genotype and sequence analyses, and critical revision of manuscript; no conflict of interest
Porche-Sorbet R: responsible for performing genotype analysis, and critical revision of manuscript; no conflict of interest
Deych E: responsible for study design, interpretation and performing statistical analysis, and critical revision of manuscript; no conflict of interest
Crankshaw M: responsible for interpretation and performing sequence analysis and critical revision of manuscript; no conflict of interest
Milligan PE: responsible for participant recruitment and warfarin dose management, and critical revision of manuscript; no conflict of interest
McLeod HL: responsible for study design and critical revision of manuscript; no conflict of interest
Patel SR: responsible for genotyping and genotype analysis for replication cohort #2 and critical revision of the manuscript; no conflict of interest
Cavallari LH: responsible for participant recruitment and data analysis for replication cohort #2 and critical revision of the manuscript; no conflict of interest
Ridker P: responsible for providing PREVENT study samples and critical revision of manuscript; no conflict of interest
Grice GR: responsible for participant recruitment and, warfarin dose management, and critical revision of manuscript; no conflict of interest
Miller RD: responsible for study design, interpretation of sequence analysis, and critical revision of manuscript; no conflict of interest
Gage BF: responsible for study design, analytical approach, analysis and interpretation of data, obtaining funding, and critical revision of manuscript; no conflict of interest
Washington University in St. Louis may file for a patent for CALU rs339097.
References
- 1.Oden A, Fahlen M, Hart RG. Optimal INR for prevention of stroke and death in atrial fibrillation: a critical appraisal. Thromb Res. 2006;117:493–499. doi: 10.1016/j.thromres.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 2.Rieder MJ, Reiner AP, Gage BF, Nickerson DA, Eby CS, McLeod HL, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 3.Caldwell MD, Awad T, Johnson JA, Gage BF, Falkowski M, Gardina P, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008 doi: 10.1182/blood-2007-11-122010. blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Limdi NA, Arnett DK, Goldstein JA, Beasley TM, McGwin G, Adler BK, et al. Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European-Americans and African-Americans. Pharmacogenomics. 2008;9:511–526. doi: 10.2217/14622416.9.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008 doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teichert M, Eijgelsheim M, Rivadeneira F, Uitterlinden AG, van Schaik RHN, Hofman A, et al. A genome-wide association study of acenocoumarol maintenance dosage. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp309. ddp309. [DOI] [PubMed] [Google Scholar]
- 8.Schelleman H, Chen Z, Kealey C, Whitehead AS, Christie J, Price M, et al. Warfarin response and vitamin K epoxide reductase complex 1 in African Americans and Caucasians. Clin Pharmacol Ther. 2007;81:742–747. doi: 10.1038/sj.clpt.6100144. [DOI] [PubMed] [Google Scholar]
- 9.Gage B, Eby C, Johnson J, Deych E, Rieder M, Ridker P, et al. Use of Pharmacogenetic and Clinical Factors to Predict the Therapeutic Dose of Warfarin. Clin Pharmacol Ther. 2008 doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diab F, Feffer S. Hereditary warfarin resistance. South Med J. 1994;87:407–409. doi: 10.1097/00007611-199403000-00023. [DOI] [PubMed] [Google Scholar]
- 11.Wallin R, Hutson SM, Cain D, Sweatt A, Sane DC. A molecular mechanism for genetic warfarin resistance in the rat. Faseb J. 2001;15:2542–2544. doi: 10.1096/fj.01-0337fje. [DOI] [PubMed] [Google Scholar]
- 12.Wajih N, Sane DC, Hutson SM, Wallin R. The inhibitory effect of calumenin on the vitamin K-dependent gamma-carboxylation system. Characterization of the system in normal and warfarin-resistant rats. J Biol Chem. 2004;279:25276–25283. doi: 10.1074/jbc.M401645200. [DOI] [PubMed] [Google Scholar]
- 13.Wajih N, Hutson SM, Wallin R. siRNA silencing of calumenin enhances functional factor IX production. Blood. 2006;108:3757–3760. doi: 10.1182/blood-2006-02-004671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vecsler M, Loebstein R, Almog S, Kurnik D, Goldman B, Halkin H, et al. Combined genetic profiles of components and regulators of the vitamin K-dependent gamma-carboxylation system affect individual sensitivity to warfarin. Thromb Haemost. 2006;95:205–211. doi: 10.1160/TH05-06-0446. [DOI] [PubMed] [Google Scholar]
- 15.Kimura R, Kokubo Y, Miyashita K, Otsubo R, Nagatsuka K, Otsuki T, et al. Polymorphisms in vitamin K-dependent gamma-carboxylation-related genes influence interindividual variability in plasma protein C and protein S activities in the general population. Int J Hematol. 2006;84:387–397. doi: 10.1532/IJH97.06082. [DOI] [PubMed] [Google Scholar]
- 16.Wadelius M, Chen LY, Eriksson N, Bumpstead S, Ghori J, Wadelius C, et al. Association of warfarin dose with genes involved in its action and metabolism. Hum Genet. 2007;121:23–34. doi: 10.1007/s00439-006-0260-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satagopan JM, Venkatraman ES, Begg CB. Two-stage designs for gene-disease association studies with sample size constraints. Biometrics. 2004;60:589–597. doi: 10.1111/j.0006-341X.2004.00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 19.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 20.Arking DE, Pfeufer A, Post W, Kao WH, Newton-Cheh C, Ikeda M, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez-Conejero R, Corral J, Roldan V, Ferrer F, Sanchez-Serrano I, Sanchez-Blanco JJ, et al. The genetic interaction between VKORC1 c1173t and calumenin a29809g modulates the anticoagulant response of acenocoumarol. J Thromb Haemost. 2007;5:1701–1706. doi: 10.1111/j.1538-7836.2007.02630.x. [DOI] [PubMed] [Google Scholar]
- 22.Wadelius M, Chen LY, Lindh JD, Eriksson N, Ghori MJR, Bumpstead S, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2008 doi: 10.1182/blood-2008-04-149070. blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thacker SM, Grice GR, Milligan PE, Gage BF. Dosing anticoagulant therapy with coumarin drugs: is genotyping clinically useful? Yes. J Thromb Haemost. 2008;6:1445–1449. doi: 10.1111/j.1538-7836.2008.03074.x. [DOI] [PubMed] [Google Scholar]
- 24.Vorum H, Liu X, Madsen P, Rasmussen HH, Honore B. Molecular cloning of a cDNA encoding human calumenin, expression in Escherichia coli and analysis of its Ca2+-binding activity. Biochim Biophys Acta. 1998;1386:121–131. doi: 10.1016/s0167-4838(98)00089-2. [DOI] [PubMed] [Google Scholar]
- 25.Presnell SR, Stafford DW. The vitamin K-dependent carboxylase. Thromb Haemost. 2002;87:937–946. [PubMed] [Google Scholar]
- 26.Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hortnagel K, Pelz HJ, et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 27.Wajih N, Owen J, Wallin R. Enhanced functional recombinant factor VII production by HEK 293 cells stably transfected with VKORC1 where the gamma-carboxylase inhibitor calumenin is stably suppressed by shRNA transfection. Thromb Res. 2008;122:405–410. doi: 10.1016/j.thromres.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative Impact of Nucleotide and Copy Number Variation on Gene Expression Phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 30.Limdi NA, Beasley TM, Crowley MR, Goldstein JA, Rieder MJ, Flockhart DA, et al. VKORC1 polymorphisms, haplotypes and haplotype groups on warfarin dose among African-Americans and European-Americans. Pharmacogenomics. 2008;9:1445–1458. doi: 10.2217/14622416.9.10.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenzini PA, Grice GR, Milligan PE, Dowd MB, Subherwal S, Deych E, et al. Laboratory and clinical outcomes of pharmacogenetic vs. clinical protocols for warfarin initiation in orthopedic patients. J Thromb Haemost. 2008 doi: 10.1111/j.1538-7836.2008.03095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voora D, Eby C, Linder MW, Milligan PE, Bukaveckas BL, McLeod HL, et al. Prospective dosing of warfarin based on cytochrome P-450 2C9 genotype. Thromb Haemost. 2005;93:700–705. doi: 10.1160/TH04-08-0542. [DOI] [PubMed] [Google Scholar]
- 33.Ridker PM, Goldhaber SZ, Danielson E, Rosenberg Y, Eby CS, Deitcher SR, et al. Long-term, low-intensity warfarin therapy for the prevention of recurrent venous thromboembolism. N Engl J Med. 2003;348:1425–1434. doi: 10.1056/NEJMoa035029. [DOI] [PubMed] [Google Scholar]
- 34.Momary KM, Shapiro NL, Viana MA, Nutescu EA, Helgason CM, Cavallari LH. Factors influencing warfarin dose requirements in African-Americans. Pharmacogenomics. 2007;8:1535–1544. doi: 10.2217/14622416.8.11.1535. [DOI] [PubMed] [Google Scholar]
- 35.Ge D, Zhang K, Need AC, Martin O, Fellay J, Urban TJ, et al. WGAViewer: Software for genomic annotation of whole genome association studies. Genome Res. 2008 doi: 10.1101/gr.071571.107. gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller RD, Duan S, Lovins EG, Kloss EF, Kwok PY. Efficient high-throughput resequencing of genomic DNA. Genome Res. 2003;13:717–720. doi: 10.1101/gr.886203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marsh S, King CR, Garsa AA, McLeod HL. Pyrosequencing of clinically relevant polymorphisms. Methods Mol Biol. 2005;311:97–114. doi: 10.1385/1-59259-957-5:097. [DOI] [PubMed] [Google Scholar]
- 39.King CR, Scott-Horton T. Pyrosequencing: a simple method for accurate genotyping. Methods Mol Biol. 2007;373:39–56. doi: 10.1385/1-59745-377-3:39. [DOI] [PubMed] [Google Scholar]
- 40.Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363. doi: 10.1126/science.281.5375.363. 5. [DOI] [PubMed] [Google Scholar]
- 41.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.