Abstract
Objective
Although twin and family studies have shown attention deficit/hyperactivity disorder (ADHD) to be highly heritable, genetic variants influencing the trait at a genome-wide significant level have yet to be identified. Thus, additional genomewide association studies (GWAS) are needed.
Method
We used case-control analyses of 896 cases with DSM-IV ADHD genotyped using the Affymetrix 5.0 array and 2,455 repository controls screened for psychotic and bipolar symptoms genotyped using Affymetrix 6.0 arrays. A consensus SNP set was imputed using BEAGLE 3.0, resulting in an analysis dataset of 1,033,244 SNPs. The data were analyzed using a generalized linear model.
Results
No genome-wide significant associations were found. The most significant results implicated the following genes: PRKG1, FLNC, TCERG1L, PPM1H, NXPH1, PPM1H, CDH13, HK1 and HKDC1.
Conclusions
The current analyses are a useful addition to the present literature and will make a valuable contribution to future meta-analyses. The candidate gene findings are consistent with a prior meta-analysis in suggesting that the effects of ADHD risk variants must, individually, be very small and/or include multiple rare alleles.
Keywords: ADHD, genetics, genome-wide association, imputation
Attention deficit/hyperactivity disorder (ADHD) is among the most common childhood onset psychiatric disorders. The world-wide prevalence of ADHD in children is eight to twelve percent 1 and the prevalence of ADHD in adults in the United States is about four percent 2, 3. Early studies found the risk of ADHD among parents of children with ADHD to be increased by between two and eight-fold with similarly elevated risk among the siblings of ADHD subjects (for a review of this literature see 4). Faraone et al 5 extended these findings to families ascertained via adult probands meeting criteria for either full DSM-IV ADHD or late-onset ADHD.
Adoption and twin studies are necessary to disentangle genetic from environmental sources of transmission observed in family studies. Three studies found that biological relatives of ADHD 6 or hyperactive children 7, 8 were more likely to have hyperactivity than adoptive relatives. A more direct method of examining the heritability of ADHD is to study twins: the extent to which monozygotic twins are more concordant for ADHD than dizygotic twins can be used to compute the degree to which variability in ADHD in the population can be accounted for by genes (i.e., heritability). Reviews of twin studies from the United States, Australia, Scandinavia and the European Union show ADHD's heritability to be around 75%, which places it among the most heritable of psychiatric disorders 9-11.
In an attempt to find regions of chromosomes which might harbor genes for ADHD, several groups have conducted genome-wide linkage scans. This approach examines many DNA markers across the genome to determine if any chromosomal regions are shared more often than expected among ADHD family members. These have produce mixed results, with some reporting evidence of linkage e.g.12, 13 and others not e.g.14. To determine if there were any significant linkage signals among these studies, Zhou et al. 15 conducted a Genome Scan Meta-Analysis of these data. They reported genome-wide significant linkage (PSR =0.00034, POR=0.04) for a region on chromosome 16 between 64Mb and 83Mb. Although this finding is intriguing and worthy of follow-up, the lack of significant findings for other loci suggests that many genes of moderately large effect are unlikely to exist and that the method of association will be more fruitful in the search for ADHD susceptibility genes.
Candidate gene association studies have focused heavily on catecholaminergic pathways16-20; the major target of most pharmacotherapies for ADHD 21. However, genes within the serotonergic and neuro-developmental pathways have also been examined. While a meta-analysis found nominally significant (p < .05) association at SLC6A3/DAT1 (3′UTR VNTR & rs27072); DRD4 (exon 3 VNTR & rs1800955); DRD5 (148-bp allele); SLC6A4/5HTT (5HTTLPR); HTR1B (rs6296) and SNAP-25 (rs3746544) 16, these effects, if present, are likely to be small and have not been unequivocally confirmed by prior genome-wide association scans of ADHD 22-25.
The present work continues the search for ADHD susceptibility genes by completing a new, independent multi-site case-control GWAS of DSM-IV ADHD, using the Affymetrix 5.0 and 6.0 arrays.
Method
Participants
The 1,150 cases used in the present analysis consist of a) samples collected by a subset of the International Multicenter ADHD Genetics (IMAGE) Project sites but not included in the IMAGE GWAS 23; and b) samples collected at additional sites (Frankfurt/Homburg, Trier, Wuerzburg, Germany, Scotland and Cardiff, UK) that were assessed in a manner similar to IMAGE samples. Cases were identified mainly through outpatient clinics at the data collection sites. They were predominantly of European origin from the U.K., Ireland, Germany, The Netherlands and the USA. Eighty-one percent of the cases met criteria for DSM-IV ADHD. Children had been referred for assessment of hyperactive, disruptive or disorganized behavior and had been clinically diagnosed as ADHD (or hyperkinetic disorder, the most closely equivalent category in the ICD-10 nomenclature used at some of the clinics). Clinical and demographic features of the case sample stratified by site are provided in Table 1. All case data were collected with informed consent of parents and with the approval of the site's Institutional Review Board (IRB) or Ethical Committee.
Table 1.
| ADHD Subtype | Comorbidity | ||||||
|---|---|---|---|---|---|---|---|
| Site | Sex (Male) N (%) |
Age (yr) mean±SD |
IA N (%) |
HI N (%) |
C N (%) |
ODD N (%) |
CD N (%) |
| Cardiff | 34 (97) | 8.9±2.0 | 3 (9) | 6 (18) | 25 (75) | 22 (63) | 4 (11) |
| Homburg | 156 (85) | 9.7±1.8 | 45 (24) | 17 (9) | 121 (66) | 64 (47)a | 40 (23) |
| IMAGE I | 40 (77) | 11.5±3.0 | 0 (0) | 0 (0) | 52 (100) | 34 (65) | 20 (39) |
| Ireland | 123 (88) | 10.9±4.2 | 10 (8) | 13 (11) | 99 (81) | 75 (55) | 27 (20) |
| MGH | 61 (69) | 9.3±2.5 | 26 (30) | 10 (11) | 52 (59) | 53 (61) | 13 (15) |
| Netherlands | 66 (92) | 11.2±2.8 | 6 (11) | 1 (2) | 49 (88) | 19 (33) | 3 (5) |
| St. Andrews | 47 (96) | 10.5±3.0 | 2 (4) | 4 (8) | 43 (88) | 16 (34) | 8 (17) |
| Wurzburg | 203 (82) | 11.0±2.7 | 18 (7) | 0 (0) | 229 (93) | 90 (36) | 34 (14) |
|
| |||||||
| Total | 730 (84) | 10.5±2.9 | 110 (13) | 51 (6) | 670 (81) | 373 (47) | 149 (18) |
Note: Additional clinical information was missing for 29 subjects (MGH N=1, Ireland N=2, St. Andrews N=1, Wurzburg N=20, and Homburg N=5). ADHD = Attention-Deficit / Hyperactivity Disorder; C = Combined type; HI = hyperactive-impulsive; IA = Inattentive; IMAGE = International Multicenter ADHD Genetics Project; MGH = Massachusetts General Hospital.
Oppositional Defiant Disorder (ODD) criteria were not assessed in subjects meeting criteria for conduct disorder (CD) at this site and were therefore not available in 40 subjects.
At the IMAGE sites, parents of children were interviewed with the Parental Account of Childhood Symptom (PACS), a semi-structured, standardized, investigator-based interview developed as an instrument to provide an objective measure of child behavior. Both parents and teachers completed the respective versions of the Conners ADHD rating scales and the Strengths and Difficulties Questionnaire. Exclusion criteria were autism, epilepsy, IQ < 70, brain disorders and any genetic or medical disorder associated with externalizing behaviors that might mimic ADHD.
In Germany, families were recruited in order of clinical referral in the outpatient clinics in Wuerzburg, Homburg, and Trier. Families were of German, Caucasian ancestry. All cases met DSM-IV criteria for ADHD. The index child was aged 6 years or above, further affected siblings were included when aged at least 6 years. All children were assessed by full semi-structured interview (Kiddie-Sads-PL-German Version or Kinder-DIPS) and parent and teacher ADHD DSM-IV based rating scales to ensure pervasiveness of symptoms. Exclusion criteria were IQ below 80, comorbid autistic disorders or somatic disorders (hyperthyroidism, epilepsy, neurological diseases, severe head trauma etc.), primary affective disorders, Tourette-syndrome, psychotic disorders or other severe primary psychiatric disorders, and birth weight below 2000 grams.
At the Cardiff site, children ages 6 to 16, of British, Caucasian ancestry, were assessed by interviewing parents with the Parent Child and Adolescent Psychiatric Assessment (CAPA)-a semi-structured research diagnostic interview and a telephone interview with the teacher using the Child ADHD Teacher Telephone Interview. All cases met diagnostic criteria for DSM-IV ADHD or ICD-10 hyperkinetic disorder or DSM-III-R ADHD and had IQ test scores above 70. Exclusion criteria were pervasive developmental disorder, Tourette syndrome, psychosis or any neurological conditions.
At the Scottish site, children ages 6 to 16, of British, Caucasian ancestry, were assessed by interviewing parents with the Parent Child and Adolescent Psychiatric Assessment (CAPA)-a semi-structured research diagnostic interview. To confirm pervasiveness, teachers completed the Conners Teacher Rating Scale. All cases met diagnostic criteria for DSM-IV ADHD. Children with an IQ below 70, autistic spectrum disorder, head injury, known chromosomal abnormality, encephalitis or significant medical conditions such as epilepsy were excluded.
The control sample (2,653 population controls of European ancestry) was collected for an IRB approved GWAS of schizophrenia and have been described elsewhere 26. Briefly, the control participants were drawn from a US Nationally representative survey panel (of approximately 60,000 adult individuals at any one time, with constant turnover) ascertained via random digit dialing. Participants were screened for psychosis and bipolar disorder. Control participants were not screened for ADHD. A blood sample was collected via a US national phelbotomy service. Control participants gave written consent for their DNA to be used for medical research at the discretion of NIMH.
Genotyping
Cases were genotyped using the Affymetrix 5.0 array at the State University of New York Upstate Medical University, Syracuse using the standard protocol issued by Affymetrix. The genotypes were called using both BRLMM-P and BIRDSUITE 27, with any calling discrepancies coded as missing. Controls were genotyped using the Affymetrix 6.0 array, at the Broad Institute National Center for Genotyping and Analysis. Genotype calls were made with the BIRDSEED program, a module of the BIRDSUITE package.
The control genotype data were initially quality controlled (QC) by The National Center for Biotechnology Information (NCBI). The QC of the control data has been described in detail elsewhere 26. Briefly, the 2,653 control samples used in the present analyses had call rates >97%, genders consistent with site reports, 26-28.5% heterozygous genotypes, and were of European ancestry (as evaluated by EIGENSTRAT). The prior data cleaning efforts for this set of genotypes include SNP call rate < 95%, HWE p-value < 10−6, MAF < 1%, plate effects, and removal of SNPs showing more than 2 Mendelian errors (from a set of trios that are not included in these analyses) or discordant genotypes for duplicate samples.
Quality Control and Statistical Analyses
As the cases and controls were genotyped using different platforms, we undertook additional quality control checks prior to conducting imputation. To ensure imputation quality, we applied more stringent quality control exclusion thresholds and carefully examined differences between cases and controls. Our key criterion for quality control consideration was call-rate at the sample- and SNP- level, as well as call-rate-differences between cases and controls. These sample and SNP exclusion criteria are found in Table 2.
Table 2.
Summary of Case and Control Quality Control (QC) filtering and exclusion criteria (in order of operation)
| Quality control metric | Cases - Affymetrix 5.0 | Controls - Affymetrix 6.0 |
|---|---|---|
| Quality Control of Individuals | ||
| N Individuals prior to QC | 1,150 | 2,653 |
| N Individuals Call Rate < 98% | 126 | 0 |
| N Related individuals removed | 52 | 0 |
| N Ancestry outliers removed | 76 | 198 |
| N Individuals following QC stage 2 | 896 | 2,455 |
| Quality Control of SNPs | ||
| N SNPs on the Array | 500,568 | 906,600 |
| SNPs dropped due to genotyping failure/lack of annotation/previous NCBI QC |
116,295 | 251,240 |
| N SNPs prior to QC stage 1 | 384,273 | 671,422 |
| Merge SNPs | 340,536 | 340,536 |
| SNP Call Rate 98% (N SNPs) | 9,349 | 611 |
| SNP missing difference < .5% | 61,383 | a |
| Allele frequency difference >15% cf HapMap3 | 12 | a |
| Hardy-Weinberg Disequilibrium (P < 0.000001) | 4,822 | a |
| 1% MAF | 768 | a |
| QC Stage 3 - following imputation | ||
| Final SNP Count for imputation | 263,591 | |
| N SNPs following imputation | 1,253,831 | |
| Quality Score >.6 | 1,033,244 | |
Note: MAF = minor allele frequency; SNP = single-nucleotide polymorphism.
The control data were screened for individual call rates > 98%, MAF > 1% prior to National Center for Biotechnology Information (NCBI) release. The slight changes occurred when ancestral outliers were excluded.
In the first stage of QC, the case and control samples were screened for low call rate, low MAF, allele frequency differences relative to the European-American and Toscani Italian HapMap samples (CEU and TSI). The two datasets were then merged, resulting in a dataset of 896 cases and 2,455 controls, genotyped on 340,536 SNPs common to both samples. To define the set of SNPs which were included in the analysis, we conducted a first pass case/control analysis in PLINK 28 and examined the distribution of association test statistics, with a particular focus on the λ statistic (defined as the observed median χ2 divided by the expected median χ2). We observed a relatively high λ of 1.20. A strong correlation was also observed between significance-values and call-rate. This relationship was more pronounced for SNPs where call-rate-differences were observed between cases and controls.
Based on these results we undertook a further round of quality control excluding SNPs with a call rate < 99%, call rate-difference case-control > .5 %, MAF < 1. This led to the exclusion of an additional 76,334 SNPs. We also increased the stringency of call rate at the individual level to 98%, which excluded an additional 6 cases. We chose these exclusion thresholds because SNPs in the excluded categories had a much higher rate of missingness than other SNPs, which could lead to spurious evidence for association. The resulting λ was 1.16 with only little evidence for technical inflation of test-statistics as indicated by further meta-data such as call rate and HWE P-value.
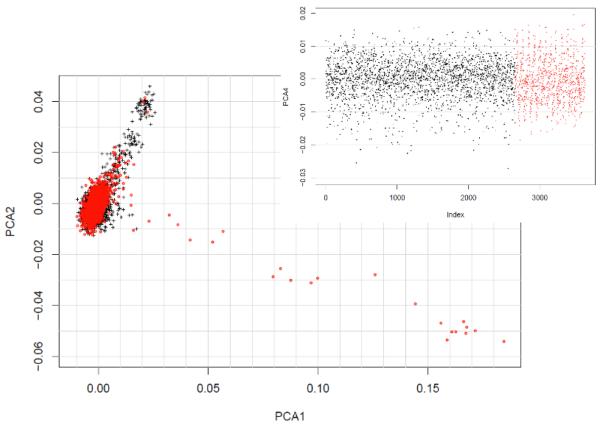
With the merged dataset, before QC, we estimated genome-wide identity-by-descent using PLINK which derives this information from the pair-wise identity-by-state patterns within the data. Based on these results, we excluded an additional 52 cases (7 avuncular/halfsibs, 35 siblings, 10 identical). To further examine population substructure within our sample we applied the multi-dimensional scaling algorithm (MDS) in PLINK to a linkage disequilibrium pruned sample, after cleaning. This MDS algorithm is numerically equivalent to the principal component analysis implemented in Eigenstrat. These units represent the contribution of many hundreds to thousands of SNPs, which share covariance induced by technical artifacts or, more commonly, population stratification. The main plot in Figure 1 shows the first two multidimensional scaling dimensions (PCA1 & PCA2). As shown in Figure 1, the majority of subjects were tightly clustered indicating most of the subjects were of European ancestry. The following exclusion criteria were applied based on visual inspection of the MDS plots:
MDS dimension 1 > 0.01
MDS dimension 2 > 0.01 & < −0.01
MDS dimension 4 < −0.02
Figure 1.
Population substructure within the case (red dots) and control (black dots) as assessed by multidimensional scaling. Note: PCA1 = first multidimensional scaling dimension
Based on these thresholds, an additional 274 (198 controls, 76 cases) samples were excluded from the analysis. Thus, the final sample consisted of 3,357 individuals (896 cases, 2,455 controls), genotyped on 226,110 SNPs. The λ arising from a PLINK case-control analysis was calculated as 1.08 suggesting that the methodological and technical confounds have mostly been accounted for. These data were then imputed using BEAGLE 3.0 29-31, using the HapMap 3 phased CEU and TSI samples as a reference (410 haplotypes) 32, 33. The imputation procedure was conducted jointly on cases and controls.
In order to restrict the analysis to well imputed SNPs we applied a threshold of 0.6 to the quality score resulting in an analysis dataset of 1,033,244 SNPs. The quality score was defined as the ratio of the variance of the dosages, as compared to the variance that is predicted from the allele frequency. This can be interpreted as the proportion of information generated by imputing versus genotyping the variant.
Statistical Analyses
Following the QC, we conducted association analysis using a generalized linear model (GLM) in R, using the 10 principal components from the MDS procedure as covariates. This model removes most of the effects of population stratification and any residual technical bias, which helps to control for false positives. The p-value reported is the Wald statistic from the GLM which is asymptotically distributed as a chi-square with 1 degree of freedom under the null hypothesis.
Results
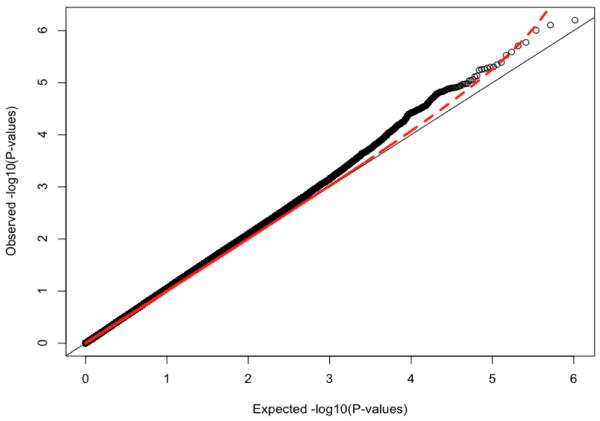
Because genotyping artifacts and ethnic differences between populations can confound GWAS analyses, it is essential to show the absence of artifacts. We do this with the quantile-quantile (QQ) plot in Figure 2. It plots, for each SNP, the observed versus expected p-value. In the presence of spurious associations, evidence for association would be seen across all SNPs, i.e., the plotted line would be above the diagonal line throughout the range plotted. We see some inflation above the diagonal, but this is small as measured by a lambda statistic (λ) of 1.08, which is not much greater than the value of 1.0 expected when there is no inflation. Thus, the QQ plot suggests that our quality control procedures removed most association signals that could be attributed to either technical artifacts or population stratification. We further examined the distribution of test statistics and λ by imputation status (genotyped vs. imputed) and by minor allele frequency (using 10 equally spaced bins). There was no obvious correlation of inflation of test-statistics with either MAF or imputation status. Similarly there was no relationship between quality score and the distribution of the test statistic.
Figure 2.
Quartile-Quartile Plot of Genome-wide Association of Attention-Deficit / Hyperactivity Disorder.
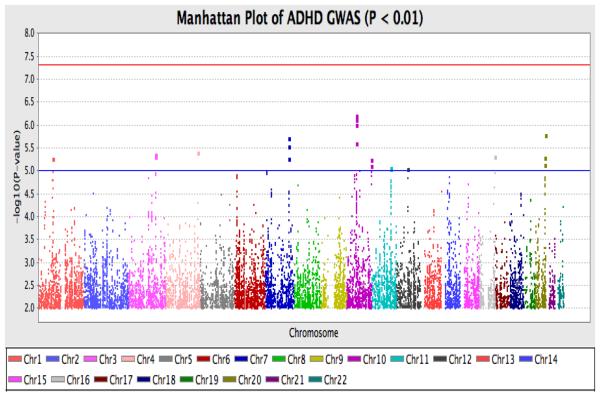
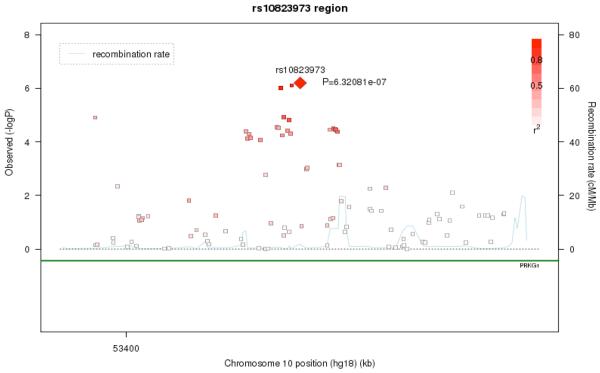
As shown in Figure 3, the strongest signal was observed on Chromosome 10 (10q21.1; p = 6.32e-07) driven by rs10823973 (C/G). The association signal from this SNP is supported by association from SNPs with which it is in strong LD. In this sample, the C allele was more common in cases versus controls with a frequency of 57.8% in cases and 51.2% in controls. The SNP is located in PRKG1. As Figure 4 shows, the association signal is also seen for nearby SNPs in the region, which suggests that the association is true rather than being caused by technical artifact or population stratification. This result lies in a well-established chromosome 10 inversion 34, spanning 10q11.2 to 10q21.
Figure 3.
Manhattan plot from the genome wide generalized linear model (GLM) analysis. Note: ADHD = Attention-deficit / Hyperactivity Disorder; Chr = Chromosome; GWAS = Genome-wide association.
Figure 4.
Regional association and Linkage Disequilibrium (LD) plot for the 10q21.1 region. Note: The most associated genotyped single-nucleotide polymorphism (SNP) is shown in the diamond in bright red and the color of the remaining markers reflects the linkage disequilibrium (r2) with the top SNP in each panel (increasing red hue associated with increasing r2). The recombination rate (right-hand y axis) is plotted in light blue and is based on the European-American (CEU) HapMap Phase III population.
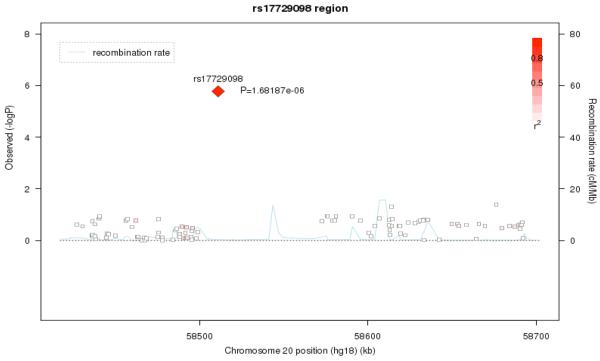
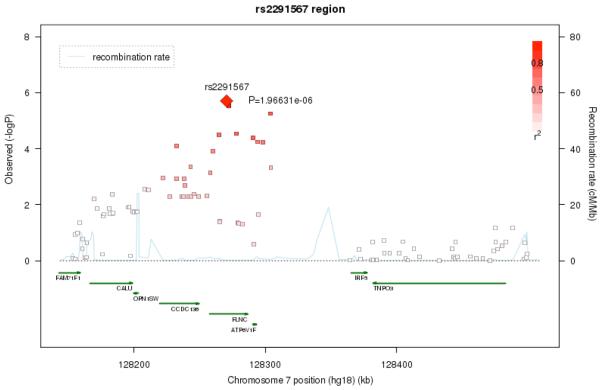
The next strongest signals were located on chromosomes 20q13.33 (rs17729098, p= 1.68e-06) and 7q31.1 (rs2291567, p=1.97e-06). The association signal on chromosome 20 is likely artifactual, as there are no other SNPs in the region which show similar levels of association (Figure 5a), although this conclusion is limited by the poor coverage of SNPs in the region. The chromosome 7 finding, on the other hand, does show a broad region of association suggesting that this is not due to a technical artifact or population stratification (Figure 5b). We also examined the results for SNPs in the 18 genes considered in a recent meta-analysis 16. However, none of these SNPs were significant at a p<= .001 level.
Figure 5a.
Regional association and Linkage Disequilibrium (LD) plot for the 20q13.33 region
Figure 5b.
Regional association and Linkage Disequilibrium (LD) plot for the 7q32.1 region.
Table 3 presents the top 100 hits based on p-value significance in our imputed dataset. The table indicates the gene name if the SNP fall within a gene. We and colleagues from three other GWAS Consortia have completed a meta-analysis which includes the data presented in this paper (see ADHD GWAS Consortium, this issue). That meta-analysis comprised 2,064 trios, 896 cases and 2,455 controls. In Table 3, we also give the p-value for each SNP from the meta-analysis.
Table 3.
Top 100 Single Nucleotide Polymorphisms from imputed International Multicenter ADHD Genetics Project (IMAGE) II dataset
| CHR | BP | SNP | A1 | A2 | IMAGE 2 P- value |
Meta- Analysis p- value |
Case Freq |
Control Freq |
GLM Estimate |
Gene |
|---|---|---|---|---|---|---|---|---|---|---|
| 10 | 53446292 | rs1903977 | A | T | 1.50E-05 | .1749 | 0.348 | 0.403 | 0.56072 | PRKG1 |
| 10 | 53449486 | rs10823973 | C | G | 6.32E-07 | .3663 | 0.578 | 0.512 | 0.563132 | PRKG1 |
| 10 | 53447049 | rs10823964 | T | G | 7.84E-07 | .2744 | 0.565 | 0.501 | 0.553325 | PRKG1 |
| 10 | 53443918 | rs10762524 | T | G | 9.89E-07 | .3336 | 0.577 | 0.512 | 1.6774 | PRKG1 |
| 20 | 58511043 | rs17729098 | T | C | 1.68E-06 | 1.68e-06 | 0.978 | 0.951 | 0.883769 | |
| 7 | 128270463 | rs2291567 | T | C | 1.97E-06 | .04204 | 0.856 | 0.816 | 0.644948 | FLNC |
| 10 | 53377755 | rs7092969 | A | T | 2.57E-06 | .05565 | 0.792 | 0.739 | 0.870689 | PRKG1 |
| 7 | 128272052 | rs3734973 | A | G | 2.97E-06 | .0661 | 0.861 | 0.822 | −0.565169 | FLNC |
| 4 | 177360844 | rs11731185 | A | C | 4.09E-06 | .03243 | 0.602 | 0.662 | 0.894289 | |
| 3 | 145754148 | rs10490827 | T | C | 4.48E-06 | .01823 | 0.106 | 0.071 | −0.53528 | |
| 16 | 86126636 | rs4488454 | C | G | 4.97E-06 | .003164 | 0.32 | 0.381 | 0.88796 | |
| 3 | 145768265 | rs12633370 | A | G | 5.03E-06 | .01686 | 0.104 | 0.069 | −0.635448 | |
| 20 | 54266838 | rs6014657 | A | C | 5.25E-06 | .2581 | 0.191 | 0.243 | −1.07922 | |
| 1 | 79972153 | rs6672282 | A | G | 5.49E-06 | .01273 | 0.929 | 0.957 | 0.906555 | |
| 7 | 128303706 | rs13238831 | T | C | 5.57E-06 | .0005903 | 0.87 | 0.835 | −0.71242 | |
| 10 | 132876636 | rs11593241 | T | C | 5.73E-06 | .06239 | 0.833 | 0.876 | −0.609587 | TCERG1L |
| 20 | 54282384 | rs6014664 | T | C | 7.38E-06 | .2728 | 0.211 | 0.263 | 0.700502 | |
| 10 | 132883393 | rs2033791 | A | G | 7.66E-06 | .1028 | 0.165 | 0.122 | −0.506491 | TCERG1L |
| 11 | 102923368 | rs716274 | A | G | 8.67E-06 | .01452 | 0.378 | 0.437 | −0.507027 | |
| 11 | 102923078 | rs716273 | T | C | 8.98E-06 | .01426 | 0.378 | 0.437 | 1.31019 | |
| 12 | 61326877 | rs12317552 | T | C | 9.14E-06 | .005663 | 0.065 | 0.044 | 0.693778 | PPM1H |
| 10 | 70699308 | rs2394534 | T | G | 1.04E-05 | .02563 | 0.804 | 0.761 | −1.8826 | |
| 7 | 8595016 | rs17151821 | C | G | 1.05E-05 | .014 | 0.967 | 0.982 | 1.22331 | NXPH1 |
| 1 | 79991162 | rs7543390 | A | C | 1.06E-05 | .1833 | 0.052 | 0.028 | −0.663373 | |
| 10 | 132874346 | rs1370868 | A | G | 1.07E-05 | .2392 | 0.815 | 0.858 | −0.862547 | TCERG1L |
| 3 | 145781004 | rs12631326 | T | C | 1.14E-05 | .01534 | 0.9 | 0.933 | 0.520709 | |
| 16 | 86128662 | rs11117283 | T | C | 1.15E-05 | .005478 | 0.668 | 0.61 | 2.28425 | |
| 7 | 8603077 | rs2107281 | T | C | 1.19E-05 | .05902 | 0.023 | 0.012 | −0.508054 | NXPH1 |
| 10 | 53444832 | rs1903979 | T | C | 1.21E-05 | .1637 | 0.346 | 0.401 | 0.591023 | PRKG1 |
| 10 | 53391182 | rs1903960 | T | G | 1.24E-05 | .03183 | 0.784 | 0.735 | 1.34502 | PRKG1 |
| 6 | 18588998 | rs567621 | T | C | 1.25E-05 | .3004 | 0.97 | 0.945 | −0.49569 | |
| 10 | 44401400 | rs1999774 | A | T | 1.25E-05 | .1014 | 0.438 | 0.5 | −1.35758 | |
| 6 | 18584639 | rs532347 | A | G | 1.26E-05 | .2679 | 0.031 | 0.055 | −0.495269 | |
| 10 | 44393217 | rs1021055 | A | G | 1.27E-05 | .1103 | 0.438 | 0.5 | 0.857156 | |
| 3 | 145784349 | rs16855659 | T | C | 1.28E-05 | .01951 | 0.1 | 0.067 | −0.495811 | |
| 10 | 44392825 | rs899889 | A | G | 1.32E-05 | .1055 | 0.44 | 0.501 | 1.33796 | |
| 6 | 18594466 | rs490618 | A | T | 1.32E-05 | .2535 | 0.97 | 0.945 | 0.688136 | |
| 10 | 132873811 | rs2397752 | T | C | 1.33E-05 | .4843 | 0.298 | 0.256 | −0.62526 | TCERG1L |
| 20 | 54274113 | rs1980818 | C | G | 1.37E-05 | .2628 | 0.203 | 0.25 | −0.579146 | |
| 14 | 43155518 | rs8011986 | T | C | 1.40E-05 | .2581 | 0.692 | 0.739 | 1.3277 | |
| 6 | 18594397 | rs536700 | C | G | 1.42E-05 | .2551 | 0.97 | 0.945 | −1.25684 | |
| 12 | 61331522 | rs10506445 | A | G | 1.44E-05 | .0199 | 0.938 | 0.959 | 0.528822 | PPM1H |
| 3 | 107394229 | rs7633167 | A | C | 1.49E-05 | .04074 | 0.603 | 0.548 | 0.491204 | |
| 10 | 44393389 | rs10900075 | T | C | 1.50E-05 | .1204 | 0.561 | 0.5 | −0.50273 | |
| 10 | 44424421 | rs7069381 | T | G | 1.50E-05 | .1185 | 0.439 | 0.5 | −0.491285 | |
| 10 | 44398176 | rs11239157 | T | C | 1.52E-05 | .1195 | 0.439 | 0.5 | −0.490879 | |
| 20 | 54277553 | rs1326021 | T | C | 1.55E-05 | .1755 | 0.736 | 0.682 | 0.545545 | |
| 1 | 79963454 | rs6424669 | C | G | 1.60E-05 | .05057 | 0.948 | 0.971 | −1.20443 | |
| 11 | 102909121 | rs7117196 | T | C | 1.64E-05 | .3623 | 0.672 | 0.616 | 0.509248 | |
| 20 | 54277432 | rs1326022 | A | G | 1.64E-05 | .1636 | 0.266 | 0.32 | −0.546283 | |
| 1 | 74879329 | rs17095690 | A | G | 1.65E-05 | .256 | 0.163 | 0.126 | 0.740204 | C1orf173 |
| 11 | 102899137 | rs1481963 | A | G | 1.78E-05 | .8727 | 0.749 | 0.698 | 0.557108 | |
| 10 | 44391896 | rs7083920 | T | C | 1.80E-05 | .1134 | 0.439 | 0.5 | −0.486633 | |
| 14 | 43160678 | rs7146388 | A | G | 1.85E-05 | .2889 | 0.694 | 0.74 | −0.574504 | |
| 10 | 132874204 | rs1583670 | T | C | 1.94E-05 | .1733 | 0.189 | 0.145 | 0.635465 | TCERG1L |
| 15 | 44499663 | rs11853869 | T | C | 1.97E-05 | .34 | 0.975 | 0.988 | −1.7472 | |
| 20 | 54279203 | rs6064362 | A | G | 2.00E-05 | .1074 | 0.688 | 0.632 | 0.519797 | |
| 11 | 102909072 | rs7105630 | T | C | 2.04E-05 | .9765 | 0.252 | 0.303 | −0.548703 | |
| 8 | 20988666 | rs6986709 | T | C | 2.12E-05 | .02959 | 0.104 | 0.071 | 0.795838 | |
| 20 | 54274415 | rs6064361 | T | C | 2.20E-05 | .1339 | 0.311 | 0.366 | −0.521177 | |
| 11 | 102863301 | rs10895437 | A | G | 2.22E-05 | .712 | 0.256 | 0.307 | −0.542526 | |
| 16 | 81813774 | rs8045006 | A | C | 2.28E-05 | .000158 | 0.057 | 0.035 | 1.1611 | CDH13 |
| 10 | 44409059 | rs2061971 | A | G | 2.42E-05 | .06829 | 0.552 | 0.493 | 0.478701 | |
| 1 | 79931809 | rs1524174 | A | C | 2.47E-05 | .009319 | 0.071 | 0.044 | 0.998981 | |
| 11 | 102899049 | rs1481964 | A | G | 2.49E-05 | .1488 | 0.578 | 0.521 | 0.478905 | |
| 7 | 34663070 | rs1023556 | T | C | 2.61E-05 | .4215 | 0.271 | 0.225 | 0.564398 | |
| 14 | 33797982 | rs11625217 | A | G | 2.71E-05 | .05322 | 0.53 | 0.474 | 0.472106 | |
| 10 | 70703703 | rs2394538 | T | C | 2.77E-05 | .03182 | 0.627 | 0.676 | −0.527049 | HK1 |
| 6 | 18606326 | rs10498690 | A | G | 2.80E-05 | .3496 | 0.028 | 0.051 | −1.34235 | |
| 11 | 102923294 | rs10895458 | A | C | 2.81E-05 | .009655 | 0.449 | 0.506 | −0.473362 | |
| 13 | 112568363 | rs1278776 | A | G | 2.85E-05 | .0000285 | 0.048 | 0.027 | 1.19599 | ATP11A |
| 10 | 53442805 | rs7893146 | T | C | 2.87E-05 | .02307 | 0.227 | 0.275 | −0.549052 | PRKG1 |
| 7 | 128277789 | rs13227216 | T | C | 2.90E-05 | .1392 | 0.837 | 0.798 | 0.668785 | FLNC |
| 3 | 145751545 | rs12631899 | T | C | 2.98E-05 | .03321 | 0.889 | 0.922 | −0.782344 | |
| 14 | 33801343 | rs12432593 | A | C | 3.00E-05 | .07092 | 0.53 | 0.474 | 0.468952 | |
| 10 | 53443371 | rs10823954 | A | G | 3.01E-05 | .02442 | 0.225 | 0.273 | −0.551318 | PRKG1 |
| 7 | 34595629 | rs2893482 | A | G | 3.01E-05 | .2577 | 0.722 | 0.768 | −0.538738 | |
| 14 | 33814177 | rs4624074 | T | C | 3.10E-05 | .04751 | 0.5 | 0.555 | −0.471712 | |
| 2 | 50122120 | rs17495366 | T | C | 3.12E-05 | .01472 | 0.762 | 0.712 | 0.53582 | NRXN1 |
| 7 | 128264708 | rs2291560 | T | C | 3.21E-05 | .002541 | 0.898 | 0.866 | 0.858583 | FLNC |
| 20 | 54285153 | rs932792 | A | C | 3.22E-05 | .1638 | 0.678 | 0.623 | 0.502806 | |
| 14 | 43080539 | rs8005625 | T | G | 3.24E-05 | .6327 | 0.2 | 0.163 | 0.667015 | |
| 18 | 62468199 | rs545245 | T | C | 3.24E-05 | .0188 | 0.606 | 0.661 | −0.479264 | |
| 10 | 53458887 | rs10762539 | T | G | 3.24E-05 | .1289 | 0.58 | 0.534 | 0.556207 | PRKG1 |
| 14 | 43175448 | rs7140723 | A | C | 3.27E-05 | .7938 | 0.184 | 0.147 | 0.668814 | |
| 20 | 54269761 | rs913000 | T | C | 3.34E-05 | .1422 | 0.269 | 0.321 | −0.523468 | |
| 11 | 102900172 | rs1481961 | A | C | 3.36E-05 | .1996 | 0.578 | 0.522 | 0.470946 | |
| 5 | 113707103 | rs36936 | T | C | 3.36E-05 | .0002923 | 0.234 | 0.19 | 0.566459 | |
| 7 | 34604539 | rs6462569 | T | C | 3.40E-05 | .2659 | 0.278 | 0.232 | 0.535921 | |
| 10 | 53459394 | rs7901487 | T | C | 3.42E-05 | .1268 | 0.419 | 0.464 | −0.556738 | PRKG1 |
| 10 | 53457963 | rs10823978 | A | G | 3.51E-05 | .02782 | 0.221 | 0.265 | −0.593313 | PRKG1 |
| 3 | 127402754 | rs4374483 | A | G | 3.57E-05 | .07212 | 0.475 | 0.415 | 0.471741 | |
| 7 | 34566246 | rs10258145 | A | G | 3.58E-05 | .3281 | 0.28 | 0.234 | 0.53732 | |
| 18 | 62467515 | rs556372 | T | C | 3.59E-05 | .01625 | 0.606 | 0.66 | −0.477123 | |
| 11 | 102904902 | rs10895454 | A | C | 3.61E-05 | .1831 | 0.578 | 0.522 | 0.467765 | |
| 10 | 53459673 | rs10762540 | A | C | 3.62E-05 | .1287 | 0.581 | 0.535 | 0.555974 | PRKG1 |
| 10 | 29605989 | rs2796304 | A | C | 3.65E-05 | .09506 | 0.577 | 0.524 | 0.484793 | |
| 7 | 34572861 | rs12690808 | T | G | 3.72E-05 | .3395 | 0.278 | 0.233 | 0.533947 | |
| 10 | 70696516 | rs906219 | A | C | 3.73E-05 | .0368 | 0.377 | 0.33 | 0.525993 | HKDC1 |
| 6 | 18614743 | rs10456831 | T | C | 3.74E-05 | .5029 | 0.027 | 0.049 | −1.34042 |
Note: A = allele; BP = base pair; CHR = chromosome; SNP = single-nucleotide polymorphism; GLM = Generalized linear model; IMAGE = International Multicenter ADHD Genetics Project.
Discussion
We have reported an independent genome-wide association scan of ADHD. None of the SNPs achieved genome-wide significance (p<5.0e-08) in either the sample reported here or in a meta-analysis or our results with other samples (see ADHD GWAS Consortium, this issue). Given the extent to which ADHD is genetic, it is highly likely that within the set of SNPs with P-value < 10−3 there are true associations for which we do not yet have sufficient power to unequivocally detect.
Although no finding achieved genome-wide significance, several of our top findings deserve further comment. The PRKG1 gene regulates neuronal migration, signal transduction, dendrite development, long term potentiation and forebrain development 35-38. Thus, it is a reasonable candidate for a gene that might lead to brain abnormalities and ADHD.
One of our top findings was in the CDH13 gene (p=2.28E-05). CDH13 was implicated by a GWAS of 343 ADHD adults and 250 controls 22 and in the IMAGE GWAS of ADHD symptom counts 24, 25. This gene lies under a linkage peak implicated in a meta-analysis of ADHD linkage studies 15 and has been implicated in substance use disorders 39, which co-occur with ADHD 40, 41.
Column seven of Table 3 helps with the interpretation of our findings in the context of other ADHD GWAS studies. This column gives the p-value from a meta-analysis (see ADHD GWAS Consortium, this issue) that comprises the data in this paper and data from three other consortia: IMAGE 23, PUWMa (See Mick et al., this issue) and CHOP (unpublished data). The Table shows that most SNPs show a dramatically higher p-value on the combined sample. No SNP increases in significance and a few retain significance levels less than .0001 . This does not create confidence in the idea that many of our top 100 SNPs are true associations.
Our negative results indicating the existence of very small genetic effects when individual variants are considered alone is not surprising. GWAS findings are now emerging for other psychiatric disorders. There have been replicated copy number variation associations for schizophrenia 42, 43 and for autism 44-46, a genome-wide significant association for bipolar disorder 47, and a significant association from a schizophrenia-bipolar dataset 48. These early GWAS results suggest that, due to the many statistical comparisons required to scan the genome, large samples are needed to detect some genes and extremely large samples are needed to detect many.
For example, the successful bipolar disorder GWAS, which detected two loci at genome-wide levels of significance, required 4387 cases and 6209 controls 47. Studies of this size and larger implicated several genes for diabetes 49 but a pooled sample of 60,000 subjects was required to definitively implicate a large set of genes 50. For Type II Diabetes and Crohn's disease mapping of one or a small number of disease-associated variants was successful in studies with similar sample sizes to the present study, but the vast majority of findings have emerged with the incorporation of multiple scans involving sample sizes many times larger than the one presented here 50, 51 and in most cases consisted of genetic loci conferring odds ratio in the regions of 1.1 – 1.4. The statistical requirement for large sample size for GWAS studies should not be interpreted as meaning that the effects of individual genetic variants are very much smaller than the effects of individual environmental variants. In fact, the latter are small as well 52.
The general expectation from GWAS of complex disorders is for multiple genes of very small effect 53. Backward power calculations on some of the initial true results from these diseases indicate that many of the identified candidates were extremely unlikely to be detected from the initial study 53. Thus, these initial studies were either fortunate or many such effects (potentially one hundred or more) with a similar effect size must exist. In this study we have not been fortunate, insofar as we did not identify a variant above genome-wide significance, which we define as 5×10-8 54, 55. Concerning the existing candidate genes for ADHD, the genome-wide association data do not provide genome-wide significant support for any of the previously postulated candidates. That is not to say that these genes should be rejected from consideration, but rather that the effect sizes for each of these variants must be small if they are real effects, which is consistent with the meta-analyses of candidate gene studies 10, 11, 16.
We have considered the pathophysiological and clinical implications of genetic effects so small that they cannot be detected with our current sample size. Such small effects can arise for several potential reasons. First it may be correct that genetic risks for ADHD are due to numerous small additive effects of common risk variants. However, it is also possible that multiple rare variants of small to moderately large effect size could account for these findings 56. Alternative explanations include sample heterogeneity, the possible interaction of genetic variants either within or between genes; and their interaction with environmental risk factors. Although the heritability of ADHD is high, this does not give an indication of the underlying genetic architecture, although it does imply that genetic influences are important for the etiology of the disorder. Recent modeling of complex behavioral and biological traits in the mouse suggests that as heritability increases the number of genetic variants involved increases, though effect sizes of individual variants remain small 57. For ADHD, our expectation is that novel genes for ADHD will be identified from GWAS once sufficient whole genome association data has been accumulated from the analysis of 5,000 – 10,000 cases.
Given the expense of GWAS, it is reasonable to ask if genes of very small effect are worth discovering. Theoretical considerations suggest that the smallness of a gene effect should not be confused with the potential importance of its discovery. For example, should we someday discover a rare variant or a common variant of small effect that implicates a new biological pathway in ADHD, that pathway could then be searched for biological targets that might yield treatments which are more efficacious than standard therapies for the disorder. The discovery of such a variant would also focus research on the implicated gene and pathway, which could lead the discovery of similar variants.
The need to search for DNA variants that lead to ADHD cannot be understood without placing the disorder in the context of current knowledge. ADHD is a common disorder affecting up to 10% of children 1. In the majority of cases, the disorder persists into adulthood 58 and is associated with serious impairments including traffic accidents 58, increased health care utilization 58, substance abuse 58, unemployment 58, divorce 58, and risk behaviors for acquired immunodeficiency syndrome (AIDS) 59. About 25 percent of ADHD patients do not respond well to currently available therapies 60, 61. Moreover, the currently preferred treatment for ADHD is stimulant medication. Although medications for ADHD are effective in controlling symptoms for many patients, they do not “cure” the disorder. Even those receiving treatment are at risk for adverse outcomes 62. Currently available treatments improve outcome, but leave patients with much residual disability and do not markedly improve the executive dysfunction seen in many ADHD patients. These treatments also have adverse effects, including delays in growth 63.
The outcome of the present study may have been influenced by a number of limitations. Most notably is the issue of power. We had 80% power for an odds ratio of 1.65 assuming a multiplicative model and a 10% minor allele frequency. Thus, we did not have sufficient power to detect smaller effects or the same effect at lower allele frequencies. Much larger samples or meta-analyses of the current samples will provide a stronger strategy for advancing knowledge regarding the molecular genetics of ADHD along with paradigms designed to search for rare genetic variants. Another limitation of the current study was the differences in genotyping platforms used to analyze case and control samples. This design limitation lead to the exclusion of numerous SNPs through additional QC steps prior to imputation. While it is possible that this might lead to artificial inflation of the test statistics it is unlikely that this had much influence on the outcome of the study given the relatively low genomic control value and the lack of significant results.
Imputation analysis, while extremely useful at generating estimates of association evidence at genetic loci that have not been typed is not perfect. The uncertainty inherent in these analyses reduces the effective sample size, thus limiting power. Also, imputation, like genome-wide association, has limited capacity for the analysis of rarer variation. These analyses used population-based controls not screened for ADHD. Although this will have reduced power somewhat, given the prevalence of the disorder, we do not expect that this had much impact on the results. Finally, ADHD very likely is genetically heterogeneous, such that many different genetic architectures give rise to similar clinical presentations. This includes the possibility that rare variants account for part of the disorder's heritability. Such genetic heterogeneity and complexity reduces power to detect significant association.
In summary, although the current analyses have not identified any convincing results the sample is a useful addition to the present literature and has made a valuable contribution to the current meta-analysis of ADHD GWAS studies (see ADHD GWAS Consortium, this issue), which combines data from four ADHD GWAS studies.
Acknowledgments
This work was supported by the following grants: US NIH Grants R13MH059126, R01MH62873, U01MH085518 and R01MH081803 to S.V. Faraone; Affymetrix Power Award, 2007 to B. Franke; NHMRC (Australia), a Sidney Sax Public Health Fellowship (443036) to S.E. Medland; Wellcome Trust, UK to L Kent, an Internal Grant of Radboud University Nijmegen Medical Centre to J. Buitelaar, Wellcome Trust, UK to A Thapar, M O'Donovan and M Owen, the Deutsche Forschungsgemeinschaft (KFO 125 to K.P. Lesch and A. Reif, SFB 581, GRK 1156 to K.P. Lesch, RE 1632/5-1 to A. Reif, SFB TRR58 to K.P. Lesch and A. Reif, ME 1923/5-1, ME 1923/5-3 to C. Freitag and J. Meyer, SCHA 542/10-3 to H. Schäfer) and the Bundesministerium für Bildung und Forschung (BMBF 01GV0605 to K.P. Lesch).
We thank the patients and the family members who provided data and the research coworkers who helped collect, manage and discuss data, especially Mick O'Donovan, Kate Langley, Michael Owen , Peter Holmans, and Nigel Williams, all of Cardiff University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This article represents one of several articles published in the August and September issues of the Journal of the American Academy of Child and Adolescent Psychiatry that explores the intersection of genetics and mental health disorders in children and adolescents. The editors invite the reader to investigate the additional articles on this burgeoning area of developmental psychopathology.
Disclosure: Dr. Buitelaar has, in the past three years, served as a consultant to, member of the advisory board, and/or served on the speakers' bureau for Janssen Cilag BV, Eli Lilly and Co., Bristol-Myers Squibb, Organon/Shering Plough, UCB, Shire, Medice, Servier, and Servier. Dr. Faraone has, in the past year, received consulting fees and has served on advisory boards for Eli Lilly and Co., Ortho-McNeil, and Shire Development, and has received research support from Eli Lilly and Co., Pfizer, Shire, and the National Institutes of Health. In previous years, Dr. Faraone has received consulting fees, served on advisory boards, or been a speaker for Shire, McNeil, Janssen, Novartis, Pfizer, and Eli Lilly and Co. In previous years he has received research support from Eli Lilly and Co., Shire, Pfizer and the National Institutes of Health. Dr. M. Romanos, in the past three years, has served on the speakers' bureau for Janssen-Cilag. In previous years he has served on the speakers' bureau for MEDICE. Dr. Freitag, in the past three years, has served on the speakers' bureau for Eli-Lilly and Co., Shire, Novartis, and Janssen-Cilag. Dr. Mick receives research support from Ortho-McNeil Janssen Scientific Affairs, Pfizer, Shire Pharmaceuticals, and has been an advisory board member for Shire Pharmaceuticals. Dr. Biederman receives research support from Alza, AstraZeneca, Bristol Myers Squibb, Eli Lilly and Co., Janssen Pharmaceuticals Inc., McNeil, Merck, Organon, Otsuka, Shire, the National Institute of Mental Health, and the National Institute of Child Health and Human Development. In 2009, Dr. Biederman served on the speakers' bureau for Fundacion Areces, Medice Pharmaceuticals, and the Spanish Child Psychiatry Association. In previous years, Dr. Biederman has received research support, consultation fees, or speaker's fees for/from Abbott, AstraZeneca, Celltech, Cephalon, Eli Lilly and Co., Esai, Forest, Glaxo, Gliatech, Janssen, McNeil, the National Alliance for Research on Schizophrenia and Depression, the National Institute on Drug Abuse, New River, Novartis, Noven, Neurosearch, Pfizer, Pharmacia, The Prechter Foundation, Shire, The Stanley Foundation, UCB Pharma, Inc. and Wyeth. Dr. Sergeant has served on the advisory boards for Eli Lilly and Co. and Shire. He has received research grant support from Eli Lilly and Co. and served on the speakers' bureau for Shire, Eli Lilly and Co., Janssen-Cilag, and Novartis. Dr. Schäfer has served as a consultant for Daiichi Sankyo, served on the speakers' bureau for the European School of Oncology, and receives research grants from the German Federal Government, and the German Research Foundation. Drs. Neale, Medland, Ripke, Anney, Asherson, Franke, Gill, Kent, Holmans, Middleton, Thapar, Lesch, Daly, Nguyen, Steinhausen, Reif, Renner, J. Romanos, Warnke, Walitza, Meyer, Palmason, Rothenberger, Hawi, and Roeyers report no biomedical financial interests or potential conflicts of interest.
References
- 1.Faraone SV, Sergeant J, Gillberg C, Biederman J. The Worldwide Prevalence of ADHD: Is it an American Condition? World Psychiatry. 2003;2(2):104–113. [PMC free article] [PubMed] [Google Scholar]
- 2.Kessler RC, Adler L, Barkley R, et al. The prevalence and correlates of adult ADHD in the United States: Results from the national comorbidity survey replication. American Journal of Psychiatry. 2006;163(4):716–723. doi: 10.1176/appi.ajp.163.4.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faraone SV, Biederman J. What is the prevalence of adult ADHD? Results of a population screen of 966 adults. Journal of Attention Disorders. 2005;9(2):384–391. doi: 10.1177/1087054705281478. [DOI] [PubMed] [Google Scholar]
- 4.Faraone SV, Biederman J. Nature, nurture, and attention deficit hyperactivity disorder. Developmental Review. 2000;20:568–581. [Google Scholar]
- 5.Faraone SV, Biederman J, Spencer TJ, et al. Diagnosing Adult attention deficit hyperactivity disorder: Are Late Onset and Subthreshold Diagnoses Valid? American Journal of Psychiatry. 2006;163(10):1720–1729. doi: 10.1176/ajp.2006.163.10.1720. [DOI] [PubMed] [Google Scholar]
- 6.Sprich S, Biederman J, Crawford MH, Mundy E, Faraone SV. Adoptive and biological families of children and adolescents with ADHD. Journal of the American Academy of Child and Adolescent Psychiatry. 2000;39(11):1432–1437. doi: 10.1097/00004583-200011000-00018. [DOI] [PubMed] [Google Scholar]
- 7.Cantwell DP. Genetics of hyperactivity. Journal of Child Psychology and Psychiatry. 1975;16:261–264. doi: 10.1111/j.1469-7610.1975.tb01275.x. [DOI] [PubMed] [Google Scholar]
- 8.Morrison JR, Stewart MA. The psychiatric status of the legal families of adopted hyperactive children. Archives of General Psychiatry. 1973;28(June):888–891. doi: 10.1001/archpsyc.1973.01750360098015. [DOI] [PubMed] [Google Scholar]
- 9.Freitag CM, Rohde LA, Lempp T, Romanos M. Phenotypic and measurement influences on heritability estimates in childhood ADHD. Eur Child Adolesc Psychiatry. 2010 Mar;19(3):311–323. doi: 10.1007/s00787-010-0097-5. [DOI] [PubMed] [Google Scholar]
- 10.Faraone SV, Perlis RH, Doyle AE, et al. Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2005 Jun 1;57(11):1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 11.Faraone SV, Mick E. Molecular Genetics of Attention Deficit Hyperactivity Disorder. Psychiatric Clinics of North America. 2010;33(1):159–180. doi: 10.1016/j.psc.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asherson P, Zhou K, Anney RJ, et al. A high-density SNP linkage scan with 142 combined subtype ADHD sib pairs identifies linkage regions on chromosomes 9 and 16. Mol Psychiatry. 2008 May;13(5):514–521. doi: 10.1038/sj.mp.4002140. [DOI] [PubMed] [Google Scholar]
- 13.Zhou K, Asherson P, Sham P, et al. Linkage to Chromosome 1p36 for Attention-Deficit/Hyperactivity Disorder Traits in School and Home Settings. Biol Psychiatry. 2008 Apr 23; doi: 10.1016/j.biopsych.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faraone SV, Doyle AE, Lasky-Su J, et al. Linkage analysis of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2007 Dec 14;147B(8):1387–1391. doi: 10.1002/ajmg.b.30631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou K, Dempfle A, Arcos-Burgos M, et al. Meta-analysis of genome-wide linkage scans of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008 Nov 5;147B(8):1392–1398. doi: 10.1002/ajmg.b.30878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gizer IR, Ficks C, Waldman ID. Candidate gene studies of ADHD: a meta-analytic review. Hum Genet. 2009 Jul;126(1):51–90. doi: 10.1007/s00439-009-0694-x. [DOI] [PubMed] [Google Scholar]
- 17.Kebir O, Tabbane K, Sengupta S, Joober R. Candidate genes and neuropsychological phenotypes in children with ADHD: review of association studies. J Psychiatry Neurosci. 2009 Mar;34(2):88–101. [PMC free article] [PubMed] [Google Scholar]
- 18.Brookes K, Xu X, Chen W, et al. The analysis of 51 genes in DSM-IV combined type attention deficit hyperactivity disorder: association signals in DRD4, DAT1 and 16 other genes. Mol Psychiatry. 2006 Aug 8; doi: 10.1038/sj.mp.4001869. [DOI] [PubMed] [Google Scholar]
- 19.Lasky-Su J, Banaschewski T, Buitelaar J, et al. Partial replication of a DRD4 association in ADHD individuals using a statistically derived quantitative trait for ADHD in a family-based association test. Biological Psychiatry. 2007 doi: 10.1016/j.biopsych.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Guan L, Wang B, Chen Y, et al. A high-density single-nucleotide polymorphism screen of 23 candidate genes in attention deficit hyperactivity disorder: suggesting multiple susceptibility genes among Chinese Han population. Mol Psychiatry. 2008 Jan 8; doi: 10.1038/sj.mp.4002139. [DOI] [PubMed] [Google Scholar]
- 21.Kratochvil CJ, Vaughan BS, Barker A, Corr L, Wheeler A, Madaan V. Review of pediatric attention deficit/hyperactivity disorder for the general psychiatrist. Psychiatr Clin North Am. 2009 Mar;32(1):39–56. doi: 10.1016/j.psc.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Lesch KP, Timmesfeld N, Renner TJ, et al. Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm. 2008 Oct 7;115:1573–1585. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- 23.Neale BM, Lasky-Su J, Anney R, et al. Genome-wide association scan of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008 Nov 3;147B(8):1337–1344. doi: 10.1002/ajmg.b.30866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lasky-Su J, Anney RJ, Neale BM, et al. Genome-wide association scan of the time to onset of attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2008 Oct 20;147B(8):1355–1358. doi: 10.1002/ajmg.b.30869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lasky-Su J, Neale BM, Franke B, et al. Genome-wide association scan of quantitative traits for attention deficit hyperactivity disorder identifies novel associations and confirms candidate gene associations. Am J Med Genet B Neuropsychiatr Genet. 2008 Sep 26;147B(8):1345–1354. doi: 10.1002/ajmg.b.30867. [DOI] [PubMed] [Google Scholar]
- 26.Shi J, Levinson DF, Duan J, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009 Aug 6;460(7256):753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korn JM, Kuruvilla FG, McCarroll SA, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008 Oct;40(10):1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007 Sep;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009 Feb;84(2):210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Browning BL, Browning SR. Haplotypic analysis of Wellcome Trust Case Control Consortium data. Hum Genet. 2008 Apr;123(3):273–280. doi: 10.1007/s00439-008-0472-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007 Nov;81(5):1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project Web site. Genome Res. 2005 Nov;15(11):1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibbs RA, Belmont J, Hardenbol P, et al. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 34.Pierotti MA, Santoro M, Jenkins RB, et al. Characterization of an inversion on the long arm of chromosome 10 juxtaposing D10S170 and RET and creating the oncogenic sequence RET/PTC. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1616–1620. doi: 10.1073/pnas.89.5.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Demyanenko GP, Halberstadt AI, Pryzwansky KB, Werner C, Hofmann F, Maness PF. Abnormal neocortical development in mice lacking cGMP-dependent protein kinase I. Brain Res Dev Brain Res. 2005 Nov;160(1):1–8. doi: 10.1016/j.devbrainres.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 36.Calabresi P, Gubellini P, Centonze D, et al. Dopamine and cAMP-regulated phosphoprotein 32 kDa controls both striatal long-term depression and long-term potentiation, opposing forms of synaptic plasticity. J Neurosci. 2000 Nov;20(22):8443–8451. doi: 10.1523/JNEUROSCI.20-22-08443.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleppisch T, Wolfsgruber W, Feil S, et al. Hippocampal cGMP-dependent protein kinase I supports an age- and protein synthesis-dependent component of long-term potentiation but is not essential for spatial reference and contextual memory. J Neurosci. 2003 Jul;23(14):6005–6012. doi: 10.1523/JNEUROSCI.23-14-06005.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Werner CG, Godfrey V, Arnold RR, et al. Neutrophil dysfunction in guanosine 3′,5′-cyclic monophosphate-dependent protein kinase I-deficient mice. J Immunol. 2005 Aug;175(3):1919–1929. doi: 10.4049/jimmunol.175.3.1919. [DOI] [PubMed] [Google Scholar]
- 39.Uhl GR, Drgon T, Liu QR, et al. Genome-wide association for methamphetamine dependence: convergent results from 2 samples. Arch Gen Psychiatry. 2008 Mar;65(3):345–355. doi: 10.1001/archpsyc.65.3.345. [DOI] [PubMed] [Google Scholar]
- 40.Faraone SV, Adamson JJ, Wilens TE, Monuteaux MC, Biederman J. Familial transmission of derived phenotypes for molecular genetic studies of substance use disorders. Drug Alcohol Depend. 2008 Jan 1;92(1-3):100–107. doi: 10.1016/j.drugalcdep.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faraone SV, Adamson JJ, Wilens T, Monuteaux MC, Biederman J. Deriving phenotypes for molecular genetic studies of substance use disorders: A family approach. Drug and Alcohol Dependence. 2007;88(2-3):244–250. doi: 10.1016/j.drugalcdep.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 42.Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008 Jul 30; doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stone JL, O'Donovan MC, Gurling H, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008 Jul 30; doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar RA, KaraMohamed S, Sudi J, et al. Recurrent 16p11.2 microdeletions in autism. Hum Mol Genet. 2008 Feb 15;17(4):628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 45.Marshall CR, Noor A, Vincent JB, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008 Feb;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss LA, Shen Y, Korn JM, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008 Feb 14;358(7):667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 47.Ferreira MA, O'Donovan MC, Meng YA, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008 Aug 17; doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Donovan MC, Craddock N, Norton N, et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008 Jul 30; doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 49.Frayling TM. Genome-wide association sudies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007 Sep;8(9):657–662. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- 50.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008 May;40(5):638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007 May;39(5):596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Banerjee TD, Middleton F, Faraone SV. Environmental risk factors for attention-deficit hyperactivity disorder. Acta Pediatrica. 2007 Sep;96(9):1269–1274. doi: 10.1111/j.1651-2227.2007.00430.x. [DOI] [PubMed] [Google Scholar]
- 53.Altshuler D, Daly M. Guilt beyond a reasonable doubt. Nat Genet. 2007 Jul;39(7):813–815. doi: 10.1038/ng0707-813. [DOI] [PubMed] [Google Scholar]
- 54.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008 Apr;32(3):227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pe'er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008 May;32(4):381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 56.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8(1):e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flint J, Munafo MR. The endophenotype concept in psychiatric genetics. Psychol Med. 2007 Feb;37(2):163–180. doi: 10.1017/S0033291706008750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biederman J, Faraone SV. Attention Deficit Hyperactivity Disorder. Lancet. 2005;366(9481):237–248. doi: 10.1016/S0140-6736(05)66915-2. [DOI] [PubMed] [Google Scholar]
- 59.Barkley RA, Murphy KR, Fischer M. ADHD in Adults: What the Science Says. Guilford Press; New York, NY: 2008. [Google Scholar]
- 60.Biederman J, Mick E, Prince J, et al. Systematic chart review of the pharmacologic treatment of comorbid attention deficit hyperactivity disorder in youth with bipolar disorder. Journal of Child and Adolescent Psychopharmacology. 1999;9(4):247–256. doi: 10.1089/cap.1999.9.247. [DOI] [PubMed] [Google Scholar]
- 61.Biederman J. Treatment of ADHD with an emphasis on stimulant medication; Paper presented at: The evaluation and treatment of ADHD in pediatrics; Boston, MA. October 28, 1997; 1997. [Google Scholar]
- 62.Biederman J, Faraone SV, Milberger S, et al. Predictors of persistence and remission of ADHD: results from a four-year prospective follow-up study of ADHD children. Journal of the American Academy of Child and Adolescent Psychiatry. 1996;35(3):343–351. doi: 10.1097/00004583-199603000-00016. [DOI] [PubMed] [Google Scholar]
- 63.Faraone SV, Biederman J, Morley CP, Spencer TJ. Effect of stimulants on height and weight: a review of the literature. J Am Acad Child Adolesc Psychiatry. 2008 Sep;47(9):994–1009. doi: 10.1097/CHI.ObO13e31817eOea7. [DOI] [PubMed] [Google Scholar]