Abstract
Aging is a multifaceted process characterized by genetic and epigenetic changes in the genome. The genetic component of aging received initially all of the attention. Telomere attrition and accumulation of mutations due to a progressive deficiency in the repair of DNA damage with age remain leading causes of genomic instability. However, epigenetic mechanisms have now emerged as key contributors to the alterations of genome structure and function that accompany aging. The three pillars of epigenetic regulation are DNA methylation, histone modifications, and noncoding RNA species. Alterations of these epigenetic mechanisms affect the vast majority of nuclear processes, including gene transcription and silencing, DNA replication and repair, cell cycle progression, and telomere and centromere structure and function. Here, we summarize the lines of evidence indicating that these epigenetic defects might represent a major factor in the pathophysiology of aging and aging-related diseases, especially cancer.
Keywords: cancer, epigenetic changes
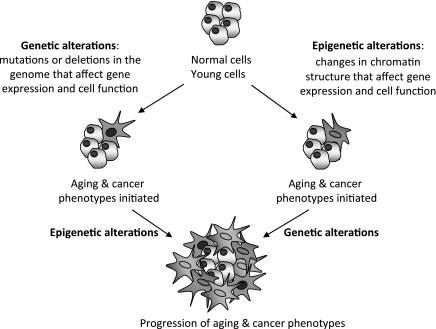
a new consensus for the definition of epigenetics has recently been reached: “An epigenetic trait is a stably heritable phenotype resulting from changes in a chromosome without alterations in the DNA sequence” (8). A cascade of signaling events, activated by an environmental cue referred to as “epigenator”, is required to effect these chromosomal changes. Next, an “epigenetic initiator” establishes the region of the chromosome that will undergo epigenetic regulation, and an “epigenetic maintainer” executes the epigenetic modifications that change chromatin structure (8). The main players that orchestrate chromatin regulation are DNA methylation, histone modifications, and RNA species. Extensive cross talk among these players regulates gene expression and maintains facultative and constitutive heterochromatin. Establishment of different combinations of chromatin marks sets codes that can be read by chromatin-associating complexes (58, 59, 66, 118). These complexes, in turn, execute additional chromatin modifications and/or recruit effectors that regulate the specific biological process: transcription, DNA replication and repair, cell division, and centromere and telomere function, among others (7, 9, 15, 46, 103). Compelling evidence indicates that the combination of genetic and epigenetic alterations contributes to human disease, including neurological and cardiovascular diseases, autoimmune disorders, cancer, and aging. Genetic alterations, such as mutations, deletions, and translocations or telomere loss, are among the leading causes of genomic instability in aging and cancer. A whole body of evidence indicates that epigenetic alterations also contribute to aging-associated pathologies, especially cancer. Epigenetic changes may initiate aging and cancer phenotypes, or prime cells, in such a way as to make them more susceptible to subsequent genetic or epigenetic alterations. The accumulation of further genetic or epigenetic changes over time would promote the progression of aging and cancer phenotypes (Fig. 1).
Fig. 1.
Model of how genetic and epigenetic alterations contribute to aging and cancer. Genetic and epigenetic alterations contribute toward the pathogenesis of aging and aging-related diseases, especially cancer. Genetic alterations, such as chromosomal deletions, DNA rearrangements, gene amplifications, and mutations, can initiate aging and cancer phenotypes. Epigenetic alterations, including changes in DNA methylation, histone modifications, and levels of expression of noncoding RNAs, also contribute to these phenotypes. Epigenetic changes may directly trigger aging and cancer phenotypes, or prime cells to make them more susceptible to subsequent genetic or epigenetic alterations.
EPIGENETIC MECHANISMS: DNA METHYLATION, HISTONE MODIFICATIONS, AND NONCODING RNAs
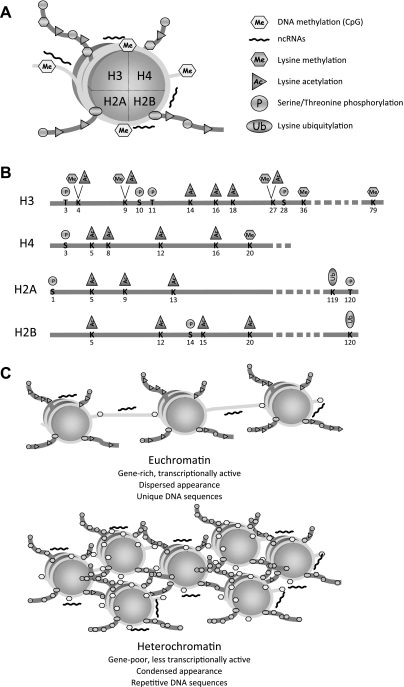
The basic unit of chromatin, the nucleosome, consists of DNA that wraps around an octamer of histones (78) (Fig. 2A). Many different cellular activities target the nucleosome as to modulate chromatin structure, either at specific genomic loci, or at large chromosomal domains (115). The histone tails protrude from the nucleosome core and are subjected to different posttranslational modifications (59). An extremely complicated picture has emerged about the types of posttranslational modifications of histones and the enzymatic activities responsible for these modifications (64, 66, 83). Histone modifications include serine and threonine phosphorylation, lysine acetylation, lysine and arginine methylation, ADP-ribosylation of glutamic acids, lysine sumoylation and ubiquitylation, and proline isomerization (Fig. 2B). The main purpose of these histone modifications is to present a platform for binding of a variety of chromatin associating/remodeling activities that, in turn, allow or restrict the access of regulatory proteins to DNA.
Fig. 2.
The nucleosome, chromatin, and specific marks. A: diagram of a nucleosome, formed by an octamer of histones wrapped around by DNA. Histone tails protruding from the nucleosome core are shown with a variety of posttranslational modifications. DNA methylation and binding of noncoding RNAs (ncRNAs) are also shown. B: the four histone tails are shown with the corresponding posttranslational modifications at specific residues. C: diagram illustrating the differences between euchromatin and heterochromatin domains. Euchromatin is characterized by hyperacetylation of histones and hypomethylation of histones and DNA. In contrast, heterochromatin shows hypoacetylation of histones and hypermethylation of histones and DNA.
Methylation of nucleosomal DNA by DNA methyltransferase activities (DNMTs) is established as a means to modulate the transcriptional activation or repression of many genes. DNA methylation consists of the addition of a methyl group to the cytosine of the CpG dinucleotide. CpGs are found at low frequency in the genome, but accumulate at specific gene promoters, CpG islands (11, 65). The bulk of DNA methylation is found at repetitive sequences, such as transposons and pericentric and subtelomeric chromatin. Methylation of repetitive sequences prevents recombination or translocation throughout the genome (20).
The types of posttranslational modifications of histones and the degree of DNA methylation determine a specific chromatin structure (71, 135). In eukaryotes, two morphologically distinct types of chromatin can be distinguished, namely euchromatin and heterochromatin (28, 39) (Fig. 2C). Euchromatin is associated with gene-rich and transcriptionally active domains of dispersed appearance, characterized biochemically by hyperacetylation of histones and hypomethylation of both histones and DNA. In contrast, highly condensed heterochromatin is linked to gene-poor and transcriptionally inactive domains, such as centromeres and telomeres. Biochemically, heterochromatin is characterized by hypoacetylation of histones and hypermethylation of histones and DNA (44, 82). However, these two morphologically distinct chromatin domains can also have common features. For example, euchromatic promoters assemble a battery of chromatin-modifying activities that induce a local closing of chromatin around the promoter sufficient to silence transcription. This closed chromatin state around the promoter shares many similarities with the mechanism of global heterochromatic silencing (42).
While histone modifications and DNA methylation have been recognized as canonical epigenetic modifications, noncoding RNAs (ncRNAs) have been recently recognized as players in chromatin remodeling, primarily involved in heterochromatin formation and transcriptional or posttranscriptional gene silencing. Three major species of ncRNAs have been identified in mammalian cells: 1) small interfering RNAs (siRNAs), molecules of 21–25 nucleotides originated from double-stranded RNA that are targeted to chromatin and participate in gene silencing and in the assembly of heterochromatic domains (85); 2) micro-RNAs (miRNAs), generated from individual RNA molecules that form hairpin structures, induce posttranscriptional silencing by inhibiting translation (50); 3) Piwi-associated RNAs, molecules of 28–33 nucleotides function in preventing the mobility of transposons (3). The role of ncRNAs in genome function and integrity is only beginning to be unraveled.
DNA METHYLATION
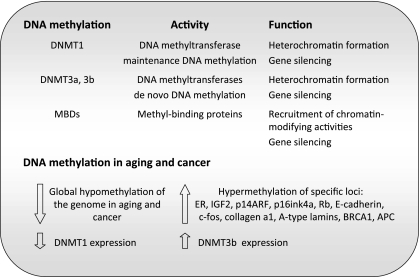
DNA methylation is one of the best understood modifications of chromatin, with crucial roles in gene expression and imprinting, defense against viral sequences, inhibition of recombination, as well as assembly of heterochromatin (11). Three active DNMT activities (DNMT1, DNMT3a, and DNMT3b) have been identified in humans and mice (93, 94) (Fig. 3). DNMT1 functions primarily as maintenance DNMT, responsible for copying the parental-strand DNA methylation pattern onto the daughter strand after each round of DNA replication. DNMT3a and DNMT3b function as de novo DNMTs, although they can also maintain methylation patterns. DNA methylation is associated with the induction of a closed chromatin state by recruitment of protein complexes that bear repressive chromatin modifying activities, including MBD (methyl-CpG-binding domain) proteins and histone deacetylase (HDAC)-containing complexes (4, 65). Cytosine methylation at euchromatic promoters participates in switching off the corresponding genes. At heterochromatic domains, it participates in the assembly of the highly compacted chromatin characteristic of these domains. Aberrant DNA methylation patterns have been linked to genomic instability and increased mutation rates, hallmarks of disease, especially cancer (23, 74).
Fig. 3.
DNA methylation in aging and cancer. The different proteins playing a role in DNA methylation are shown with their reported molecular activity and cellular function. A summary of the changes observed in DNA methylation patterns during cancer and aging by different studies are also shown. Both changes at a global level and at specific gene promoters are indicated. ER, estrogen receptor; BRCA1, breast cancer 1; APC, adenomatosis polyposis coli.
Ground-breaking studies performed in rat brain and heart showed a global loss of DNA methylation during aging (127), which was later confirmed in different tissues from rat, mouse, and cow (102), in primary fibroblasts from mice, hamsters, and humans grown in culture (134), and in human lymphocytes (29) and peripheral blood cells (14, 38). Paradoxically, a variety of specific loci become hypermethylated in normal tissues during aging (35). A study using T-lymphocytes isolated from individuals of different ages, ranging from newborns to elderly people, showed hypermethylation of ∼1% of the loci examined with increasing age. Comparable changes in DNA methylation at these loci were observed in other tissues (124). Similarly, a study of changes in DNA methylation in the human cerebral cortex identified a marked increase in cytosine methylation at 8 out of 50 loci examined (114). Specific examples of genes that are hypermethylated during aging include the estrogen receptor (ER) (55), insulin-like growth factor-II (IGF-II) (56); p14ARF (110); p16ink4a (116), E-cadherin (17), c-fos (24), and collagen-α1 (121). Interestingly, many of these promoters are also hypermethylated during tumorigenesis, suggesting a role in the increased cancer susceptibility associated with aging.
These different lines of evidence indicate that a global hypomethylation of the genome accompanies the aging process, concomitant with increased methylation of specific gene promoters (19, 32) (Fig. 3). The molecular mechanisms behind these changes in DNA methylation patterns during aging remain unknown. Transcriptional control of DNMTs was shown to be altered in aged and transformed human fibroblasts (21). In fact, it was proposed that the global decrease in DNA methylation could stem from a progressive decrease in the efficacy of DNMT1 activity on heterochromatic domains during aging (21, 35). In contrast, the hypermethylation of specific promoter CpG islands could be brought about by overexpression of the de novo DNMTs. In particular, upregulation of DNMT3b was reported in cultured fibroblasts (21, 35). Future studies need to determine whether transcriptional changes are indeed responsible for the accumulation of DNA methylation alterations during aging, and whether these changes increase cancer susceptibility with age.
HISTONE ACETYLATION/DEACETYLATION
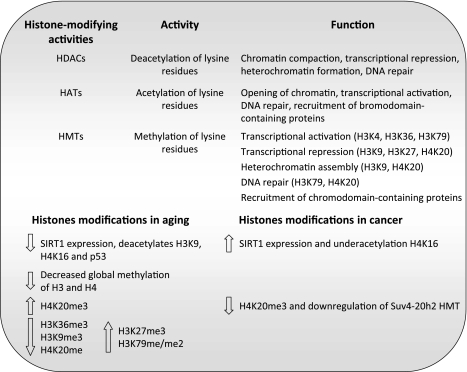
Histone acetyltransferases (HAT) and HDAC modulate the addition or removal of acetyl groups on histones, as means to regulate transcription (7) (Fig. 4). Acetylated lysine residues serve as binding platforms for proteins that contain the bromodomain protein module, found in some transcription factors, as well as in subunits of chromatin-remodeling complexes (27, 49, 57). HAT activities are usually found in large multisubunit complexes. Overall, acetylation of lysine residues on histones is associated with transcriptional activation and DNA repair (47, 69). Conversely, histone deacetylation allows the acquisition of a closed chromatin state. HDAC enzymes deacetylate all four histones and can act on nonhistone substrates as well. Overall, histone deacetylation is associated with chromatin recondensation, transcriptional repression, and assembly of heterochromatin.
Fig. 4.
Histone-modifying activities in aging and cancer. Listing of the major classes of histone-modifying activities indicate their specific activities and reported cellular function. HDACs, histone deacetylases; HATs, histone acetyltransferases; HMTs, histone methyltransferases. In the case of HMTs, the specific lysine residues that are methylated and their function are indicated. A summary of the changes observed in histones modifications during aging and cancer is also shown. Note that the changes in SIRT1 expression and H4K20me3 levels during aging and cancer go in the opposite direction. See text for definition of histone acronyms.
HDAC activities, such as the yeast HDAC Sir2 (silent information regulator 2), and its functional mammalian orthologs, the sirtuins, are the best example of an epigenetic change that is linked to aging. Pioneer studies by Brian Kennedy and colleagues (62) demonstrated a clear correlation between expression of proteins of the Sir complex, Sir2/Sir3/Sir4, and longevity in yeast. Deletion of Sir4 abolished silencing of yeast-mating loci and telomeres and caused a decrease in lifespan. In contrast, a gain-of-function mutation in Sir4 induced the relocation of the Sir complex from mating loci and telomeres to the nucleolus and extended lifespan by 30% (63). Furthermore, increased Sir2 activity has anti-aging effects in yeast (61), C. elegans (123), and D. melanogaster (101). Interestingly, the closest mammalian ortholog of Sir2, SIRT1, has also been correlated with aging. SIRT1 deacetylates histone H4 at lysine 16 (H4K16) and histone H3 at lysine 9 (H3K9) positions (99, 129) and deacetylates also p53, decreasing its activity. A decline in the levels of SIRT1 protein was observed concomitant with decreased mitotic activity in mouse and human cells. Marked decreases were especially found during aging or in premature aging mice (Fig. 4). In contrast, SIRT1 increases when cells are stimulated to divide in vitro, and upregulation of SIRT1 is a hallmark of a whole spectrum of human tumors. Accordingly, loss of H4K16 acetylation was identified as a common event in human cancer (34). These data suggest that the“ downregulation of SIRT1 during aging might represent a tumor-suppressor mechanism that stabilizes p53 at the expense of limiting the cellular lifespan. A variety of molecular mechanisms could be behind SIRT1 function in longevity and tumor suppression. SIRT1 can regulate gene expression (92, 98, 99), formation of facultative and constitutive heterochromatin (128, 129), signaling through the DNA damage response pathway (80, 130), and DNA methylation patterns (90). Thus it is tempting to speculate that a profound decrease in the levels of SIRT1 during aging could increase genomic instability. This can lead to decreased cell viability, but also an increase in cancer susceptibility. The upregulation of SIRT1 during the transformation of normal cells to malignant derivatives could promote the longevity of the transformed cells. The fact that increased SIRT1 expression promoted survival in a mouse model of genomic instability (91) supports this model.
SIRT6, another member of the sirtuins family, has been shown to be required for DNA repair and genomic integrity. Accordingly, SIRT6 deficiency leads to a degenerative aging-like phenotype in mice, including growth retardation, lymphopenia, loss of subcutaneous fat, lordokyphosis, and severe metabolic defects, dying at around 4 wk of age (87). Overall, these studies provide evidence of sirtuins playing a role in the pathophysiology of aging and cancer, making these proteins ideal targets for cancer therapeutics. In fact, recent studies have shown that inhibitors of sirtuins exhibit strong cancer-specific proapoptotic effects (72). It is important to note that, despite all that we have learned about sirtuin function in cancer and aging, the role that most HATs and HDACs play in these processes remains unknown.
HISTONE METHYLATION/DEMETHYLATION
Histone methylation plays an important role in transcriptional regulation and in the assembly of facultative and constitutive heterochromatin (70, 84, 105, 112, 115). Histone methyltransferases (HMTs) are the enzymes responsible for the transfer of methyl groups to lysine or arginine residues on histones H3 and H4 (Fig. 4). Lysines 4, 9, 27, 36, and 79 on H3 and lysine 20 on H4 are frequently methylated. Methylation of lysine residues can have transcriptional repressive or activating effects on chromatin, depending on the particular lysine that undergoes methylation. Thus methylation of H3K9, H3K27, and H4K20 is generally associated with transcriptional repression, while methylation of H3K4 (H3K4me3), H3K36 (H3K36me3), and H3K79 is associated with active chromatin (67). Genomewide chromatin-state maps of stem, progenitor, or differentiated cells have shown that H3K4me3 and H3K27me3 discriminate genes poised for transcriptional activation or repression, respectively. While H3K36me3 marks sites of transcription of coding and ncRNAs, H3K9me3, and H4K20me3 mark heterochromatic domains, such as pericentric and telomeric chromatin (86).
Another level of complexity in the histone code is the fact that lysine residues can undergo mono-, di-, or trimethylation, each providing a specific degree of chromatin compaction and a different binding platform (137). As for lysine acetylation, methylation of lysine residues in histones creates binding sites for chromodomain-containing proteins; such is the case of H3K9 methylation, binding site for HP1 (heterochromatin protein 1), and H3K27 methylation, binding site for polycomb proteins (137). Recruitment of HP1 is important for the maintenance of heterochromatin domains, and occupancy of gene promoters by polycomb complexes favors an environment for transcriptional repression. Histone demethylation has also been described and is believed to antagonize the function of lysine methylation in diverse biological processes (65, 111, 126, 133, 136).
Changes in histone methylation patterns have been observed during aging (Fig. 4). Early studies in rats showed that methylation of histones H3 and H4 declines gradually with increasing age (122). A systemic investigation of posttranslational modifications of histones in the brain of senescence-accelerated prone mouse 8 (SAMP8) model has revealed a number of changes during aging (132). Seven methylation sites were detected in histones H3 and H4 of aged mice. Of these, H4K20me (H3 monomethylated on lysine 20) and H3K36me3 (H3 trimethylated on lysine 36) decreased significantly in the brain of 12-mo-old SAMP8 mice compared with 3-mo-old mice. In contrast, the abundance of H3K27me3, H3K79me, and H3K79me2 increased in old-aged mouse brains. In another study performed in kidneys and liver of aged rats, the levels of mono- and dimethylated H4K20 did not significantly vary with age. Conversely, H4K20me3 was greatly increased during aging in these tissues (106). Similarly, the levels of H4K20me3 increase in senescent cells (35). This histone mark is stabilized by the coordinated action of HMTs Suv4–20h1 and Suv4–20h2 and the retinoblastoma family of proteins (pRb, p107, and p130) (6, 43). Thus alterations in the expression of these proteins or in their activities during aging could be behind H4K20me3 changes.
Changes in histone modifications at specific genomic loci have also been reported during senescence. For example, polycomb proteins and associated repressive epigenetic marks regulate replication timing and silencing of the ink4a/ARF locus (1). In young cells, the polycomb repressive complex (PRC2) member EZH2 (enhancer of zeste homolog 2) and members of the PRC1 complex Bmi1 (B-cell-specific Moloney murine leukemia virus integration site 1) and M33 are highly expressed and targeted to the ink4a/ARF locus. This results in silencing of the locus and replication during late S phase. Upon entry into senescence, PRC1 and PRC2 complexes are lost at this locus, leading to decreased H3K27me3 levels. Concomitantly, upregulation of the histone demethylase jumonji domain containing 3 and recruitment of the mixed lineage leukemia gene 1 protein to the locus results in expression of the ink4a/ARF genes and replication of the locus during early S phase. These studies show that changes in the epigenetic status of specific loci can play a key role during senescence.
Overall, changes in the methylation status of histone residues during aging are beginning to be reported. However, the significance of these findings for the pathophysiology of aging is far from being understood. Given that histone methylation/demethylation can regulate transcriptional activation or repression, one can anticipate that changes in this mark can switch on/off many genes that collectively promote aging or aging-associated diseases, such as cancer. Furthermore, the role of methylation of certain histone residues on DNA repair and maintenance of heterochromatin domains, such as telomeres and centromeres, suggests that a proper set of histone methylation marks could help prevent genomic instability.
EXPRESSION OF miRNAs
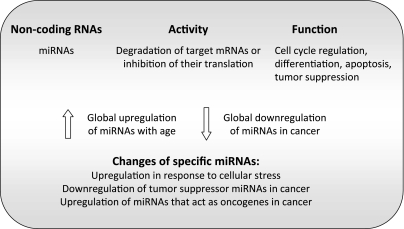
The roles of ncRNA species in genome function and integrity are only beginning to be unraveled. The best characterized ncRNAs are miRNAs. A whole variety of functions have been attributed to miRNAs, including cell cycle regulation, differentiation, apoptosis, and tumor suppression (45) (Fig. 5). It has been estimated that a single miRNA can have hundreds of targets, illustrating the complexity of miRNA transcription for genome function. These miRNAs negatively regulate gene expression by either degrading target messenger RNAs or inhibiting their translation (50, 75). It has been proposed that ∼60% of human genes are controlled by an ever growing list of miRNAs (5, 37). Recent genomewide analyses have shown that 95% of the human genome is transcribed, and that the vast majority of transcripts are noncoding (13, 131). Interestingly, epigenetic mechanisms can modulate the expression of miRNA (104), and miRNAs can impact on the levels of chromatin-modifying activities, such as DNMTs and HMTs (33, 36).
Fig. 5.
Noncoding RNAs [micro-RNAs (miRNAs)] in aging and cancer. Summary is given of some of the functions reported for miRNAs and changes that could have an impact in cancer and aging phenotypes.
The first evidence about a miRNA-regulating life span came from studies in C. elegans (73). The miRNA lin-4 was shown to regulate the protein lin-14, an essential transcription factor that affects a variety of signaling pathways controlling developmental timing and life span (16, 54). The increase in longevity on upregulation of lin-4 appears to be mediated by the insulin/IGF signaling pathway. Since then, a variety of miRNAs that modulate different players in this pathway in mammalian cells have been identified (45). Future studies need to determine whether these miRNAs have a conserved role in organismal aging. Only a few reports have shown a correlation between changes in miRNA expression and aging (75). In liver, for example, array profiling of global miRNA expression showed upregulation of miRNAs during aging (81).
The accumulation of genomic instability is considered a key factor in the aging process. Interestingly, a number of miRNAs are induced in response to cellular stress, leading to decreased levels of proteins involved in DNA repair (22). In particular, the hypoxia-inducible factor-1α (HIF-1α) activates miRNAs (100, 120). Importantly, hypoxia-induced miRNAs are overexpressed in some human tumors, suggesting a role for these ncRNAs in the increased susceptibility to cancer during aging (68). Furthermore, p53 is upregulated in response to a number of genomic insults. Intriguingly, p53 regulates the expression of miRNAs while being an indirect target of miRNA action (51, 95). The regulation of p53 by miRNAs places these ncRNAs at a central position in the control of genomic stability.
EPIGENETIC ALTERATIONS IN CANCER
The evidence to date suggests that aging is associated with alterations in epigenetic control. However, how this contributes to the pathophysiology of aging remains unclear. One exception is the clear connection that has been established between epigenetic deregulation and tumor progression. Given that aging represents the highest risk factor for cancer, it is considered that cancer susceptibility increases as cells accumulate mutations and epigenetic alterations during aging, which, in turn, provide proliferative advantages and increased genomic instability (Fig. 1).
Cancer is characterized by global DNA hypomethylation and site-specific promoter hypermethylation (32, 60) (Fig. 3). DNA hypomethylation initiates chromosome instability and activates protooncogenes, increasing tumor frequency in mouse models (30, 53). CpG islands located in gene promoters are unmethylated in normal tissues (12). De novo methylation of these islands occurs in a variety of tumors, leading to transcriptional repression of tumor suppressor genes. One of the first links between CpG island methylation, gene silencing, and tumorigenesis was provided by studies with the ER gene. Methylation of the ER gene CpG island was shown to increase in normal colonic mucosa as a function of age and be present in colonic tumors (55). Interestingly, in cultured colon cancer cells, methylation-induced silencing of the ER gene leads to increased growth, and expression of exogenous ER results in marked growth inhibition. Other genes modulated by hypermethylation in this tissue are the N33 and MYOD genes (2). The same group of investigators also showed alterations in DNA methylation patterns in the imprinted gene for IGF-II in human tumors, including colon, breast, lung, and leukemias (56). Classical tumor suppressor genes such as retinoblastoma, p16ink4a, breast cancer 1, adenomatosis polyposis coli, von Hippel-Lindau, and E-cadherin are also silenced by methylation during tumorigenesis (32). Thus increased gene silencing by methylation has emerged as a leading cause of tumorigenesis.
Alterations of histone modifications are also a hallmark of cancer cells (109) (Fig. 4). In particular, global decreases in H4K16ac (34) and increased levels of HDACs are often found in cancer cells (48, 117). These changes in histone acetylation could promote the silencing of tumor suppressor genes. In contrast, the decreased levels of H4K20me3 (34) and H3K9me3 (88) observed in human tumors could induce genomic instability by impacting on the structure of heterochromatin or provide a proliferative advantage by altering telomere metabolism (42, 43). Importantly, a comparative study of cancer cells representing different stages of human breast cancer revealed that the more malignant cells exhibited the highest decrease in H4K20me3 levels (125). The decrease in H4K20me3 was concomitant with lower expression of Suv4–20h2, suggesting that this enzyme might regulate this histone modification in tumors. Interestingly, the more malignant cells also have a more prominent loss of DNA methylation, which is accompanied by altered expression of DNMT1 and methyl-binding proteins MeCP2 and MBD2. These results correlate malignant progression with more extensive epigenetic alterations. More studies are needed to clearly determine the functional interplay between specific alterations of DNA methylation and histone modifications and the cancer phenotype.
Several reports indicate that expression of miRNAs is altered during malignancy (Fig. 5). In fact, miRNA expression profiles classify human cancers (77). The authors found that miRNA expression distinguishes tumors of different origins. In addition, a general downregulation of miRNAs was observed in tumors compared with the corresponding normal tissues. Furthermore, these profiling experiments revealed that miRNAs are induced during differentiation. Overall, these studies support the notion that changes in miRNA expression are linked to tumorigenesis. In particular, miRNAs can act as tumor suppressors, being downregulated during malignancy. Examples of miRNAs with tumor-suppressive functions are miR15 and miR16, which are located in a region that is lost in the majority of chronic lymphocytic leukemias (18). Interestingly, a recently identified molecular target of miR15 and miR16 is Bmi1, a gene that is upregulated in a variety of epithelial malignancies. Downregulation of miR15 and miR16 in ovarian cancer is correlated with Bmi1 protein expression (10). However, miRNAs can also act as oncogenes. The miR-17–92 cluster of miRNAs cooperates with c-myc to promote tumor development. In particular, an upregulation of miRNAs derived from this locus is substantially increased in B-cell lymphoma. Furthermore, enforced expression of miR-17–92 cluster and c-myc accelerated tumor progression in a mouse model of B-cell lymphoma (52). Another study showed that myc binds directly to the miR-17–92 locus and activates expression of the miRNA cluster (89). At the same time, miRNAs from this cluster were shown to inhibit the expression of the transcription factor E2F1. In recent years, many more miRNAs have been identified whose expression is associated with cancer (25, 79). Some of these miRNAs act as tumor suppressors and are downregulated in tumors, while others act as oncogenes and are upregulated in cancer.
In summary, these studies revealed that a variety of processes contributing to tumorigenesis, activation of oncogenes, silencing of tumor suppressor genes, cell cycle defects, and increased genomic instability, are subjected to epigenetic deregulation. Understanding how epigenetic alterations contribute to human cancer is an active area of research, with high potential for therapeutics.
EPIGENETIC ALTERATIONS IN PREMATURE AGING
Premature aging syndromes have been instrumental in the characterization of molecular mechanisms that contribute to physiological aging. One of the most devastating syndromes of premature aging in humans is Hutchinson Gilford Progeria Syndrome (HGPS). Patients with HGPS appear normal at birth, but develop severe growth abnormalities by 2 yr of age. Children with progeria continue to exhibit characteristics associated with aging and generally die due to atherosclerosis in their teens. HGPS is caused by a mutation in the gene encoding A-type lamins, lamins A and C, structural components of the nuclear lamina and the inner nuclear matrix (26, 31). The mutation in the LMNA gene associated with progeria leads to the expression of a truncated form of lamin A known as progerin, which is toxic to the cells. Interestingly, normal human fibroblasts from aged individuals also express progerin, linking alterations of A-type lamins to physiological aging (107, 108). At the cellular level, common defects in HGPS include ultrastructural defects of the nuclear envelope, loss of heterochromatin from the nuclear periphery (97, 119), increased genomic instability (76), and alterations in epigenetic marks characteristic of constitutive heterochromatin (113). In particular, decreased levels of H3K9me3 at pericentric heterochromatin and decreased binding of HP1 to these domains were observed in fibroblasts from HGPS patients. In addition, these authors showed that progeric fibroblasts exhibit decreased levels of H3K27me3, concomitant with decreased levels of EZH2, the HMT responsible for establishing this chromatin mark. Furthermore, an increase in H4K20me3 levels was reported in these cells. A recent study has provided a mechanism responsible for the decrease in H3K9me3 levels and HP1 binding. These defects were linked to a reduction of NURD (nucleosome remodeling and histone deacetylation) complex subunits, specifically RBBP4 (retinoblastoma binding protein 4), RBBP7 (retinoblastoma binding protein 7), and HDAC1, which interact with lamin A (96). The authors found that the loss of NURD subunits in HGPS cells was dependent on the presence of progerin. All together, these studies support the notion that defects in the structural nuclear proteins A-type lamins that occur in aging impact on epigenetic mechanisms.
Further support for a role of A-type lamins in epigenetic regulation is the fact that abrogation of A-type lamins in the Lmna−/− mouse model induces yet a different pattern of changes in chromatin structure (40, 41). Fibroblasts from these mice exhibit a marked decrease in the levels of H4K20me3, while maintaining the levels of H3K9me3, both at pericentric and telomeric heterochromatin. The concomitant decrease of retinoblastoma family members observed in these cells is likely to be responsible for this epigenetic defect. Thus A-type lamins have emerged as novel players in epigenetic regulation. Expression of the mutant progerin protein leads to defects in histones marks characteristic of physiological aging, while loss of A-type lamins recapitulates phenotypes of cancer cells.
SUMMARY
A whole body of evidence indicates that alterations in the three major epigenetic mechanisms, DNA methylation, histone modifications, and ncRNAs, are a hallmark of aging and cancer. These alterations can impact large chromosomal domains or specific genomic loci within the chromosome. The main cellular consequences of epigenetic deregulation are changes in the transcriptional activation and repression of many genes, an increase in genomic instability, and structural defects in heterochromatic domains. Given the variety of cellular processes that are affected by epigenetic mechanisms, it has been proposed that epigenetic alterations could represent an important factor in pathophysiology of aging and aging-related diseases, especially cancer. Interestingly, some epigenetic alterations are common in aging and cancer, suggesting that the accumulation of epigenetic defects with age might promote cellular transformation and increase cancer susceptibility. This is the case of global DNA hypomethylation and promoter hypermethylation. In contrast, other epigenetic alterations are modulated in an opposite way in aging and cancer. The best examples are H4K20me3 levels and expression of SIRT1. The levels of H4K20me3 increase during normal aging and in progeria, while decreasing during tumorigenesis. Similarly, SIRT1 is downregulated during aging and upregulated in cancer. These data suggest that the transformation of normal cells to malignant derivatives might be accompanied by an epigenetic switch involving a variety of mechanisms. A big challenge in the field is to identify the molecular mechanisms responsible for this switch, which could putatively act as master epigenetic regulators. This will require establishing the epigenetic signature of young and old cells, as well as normal and tumor cells. High-throughput methods need to be conducted for the concomitant analysis of an elevated number of epigenetic modifications, including DNA methylation, histones modifications, and expression of miRNAs. These types of studies, in combination with genomewide expression profiles, will be fundamental for determining how epigenetic changes contribute to aging and cancer phenotypes. The findings have the potential to impact cellular diagnosis for cancer screening and accelerate drug discovery.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
REFERENCES
- 1. Agherbi H, Gaussmann-Wenger A, Verthuy C, Chasson L, Serrano M, Djabali M. Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence. PLoS One 4: e5622, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res 58: 5489–5494, 1998 [PubMed] [Google Scholar]
- 3. Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747, 2007 [DOI] [PubMed] [Google Scholar]
- 4. Ballestar E, Esteller M. Methyl-CpG-binding proteins in cancer: blaming the DNA methylation messenger. Biochem Cell Biol 83: 374–384, 2005 [DOI] [PubMed] [Google Scholar]
- 5. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T, Blasco MA. Suv4–20h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol 178: 925–936, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Berger SL. The complex language of chromatin regulation during transcription. Nature 447: 407–412, 2007 [DOI] [PubMed] [Google Scholar]
- 8. Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev 23: 781–783, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell 128: 669–681, 2007 [DOI] [PubMed] [Google Scholar]
- 10. Bhattacharya R, Nicoloso M, Arvizo R, Wang E, Cortez A, Rossi S, Calin GA, Mukherjee P. MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res 69: 9090–9095, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21, 2002 [DOI] [PubMed] [Google Scholar]
- 12. Bird AP. CpG-rich islands and the function of DNA methylation. Nature 321: 209–213, 1986 [DOI] [PubMed] [Google Scholar]
- 13. Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, Drenkow J, Bell I, Zhao X, Srinivasan KG, Sung WK, Ooi HS, Chiu KP, Foissac S, Alioto T, Brent M, Pachter L, Tress ML, Valencia A, Choo SW, Choo CY, Ucla C, Manzano C, Wyss C, Cheung E, Clark TG, Brown JB, Ganesh M, Patel S, Tammana H, Chrast J, Henrichsen CN, Kai C, Kawai J, Nagalakshmi U, Wu J, Lian Z, Lian J, Newburger P, Zhang X, Bickel P, Mattick JS, Carninci P, Hayashizaki Y, Weissman S, Hubbard T, Myers RM, Rogers J, Stadler PF, Lowe TM, Wei CL, Ruan Y, Struhl K, Gerstein M, Antonarakis SE, Fu Y, Green ED, Karaoz U, Siepel A, Taylor J, Liefer LA, Wetterstrand KA, Good PJ, Feingold EA, Guyer MS, Cooper GM, Asimenos G, Dewey CN, Hou M, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Huang H, Zhang NR, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Seringhaus M, Church D, Rosenbloom K, Kent WJ, Stone EA, Batzoglou S, Goldman N, Hardison RC, Haussler D, Miller W, Sidow A, Trinklein ND, Zhang ZD, Barrera L, Stuart R, King DC, Ameur A, Enroth S, Bieda MC, Kim J, Bhinge AA, Jiang N, Liu J, Yao F, Vega VB, Lee CW, Ng P, Shahab A, Yang A, Moqtaderi Z, Zhu Z, Xu X, Squazzo S, Oberley MJ, Inman D, Singer MA, Richmond TA, Munn KJ, Rada-Iglesias A, Wallerman O, Komorowski J, Fowler JC, Couttet P, Bruce AW, Dovey OM, Ellis PD, Langford CF, Nix DA, Euskirchen G, Hartman S, Urban AE, Kraus P, Van Calcar S, Heintzman N, Kim TH, Wang K, Qu C, Hon G, Luna R, Glass CK, Rosenfeld MG, Aldred SF, Cooper SJ, Halees A, Lin JM, Shulha HP, Zhang X, Xu M, Haidar JN, Yu Y, Ruan Y, Iyer VR, Green RD, Wadelius C, Farnham PJ, Ren B, Harte RA, Hinrichs AS, Trumbower H, Clawson H, Hillman-Jackson J, Zweig AS, Smith K, Thakkapallayil A, Barber G, Kuhn RM, Karolchik D, Armengol L, Bird CP, de Bakker PI, Kern AD, Lopez-Bigas N, Martin JD, Stranger BE, Woodroffe A, Davydov E, Dimas A, Eyras E, Hallgrimsdottir IB, Huppert J, Zody MC, Abecasis GR, Estivill X, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Koriabine M, Nefedov M, Osoegawa K, Yoshinaga Y, Zhu B, de Jong PJ. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekstrom TJ, Harris TB, Launer LJ, Eiriksdottir G, Leppert MF, Sapienza C, Gudnason V, Feinberg AP. Intra-individual change over time in DNA methylation with familial clustering. JAMA 299: 2877–2883, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet 8: 299–309, 2007 [DOI] [PubMed] [Google Scholar]
- 16. Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science 310: 1954–1957, 2005 [DOI] [PubMed] [Google Scholar]
- 17. Bornman DM, Mathew S, Alsruhe J, Herman JG, Gabrielson E. Methylation of the E-cadherin gene in bladder neoplasia and in normal urothelial epithelium from elderly individuals. Am J Pathol 159: 831–835, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 99: 15524–15529, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Calvanese V, Lara E, Kahn A, Fraga MF. The role of epigenetics in aging and age-related diseases. Ageing Res Rev 8: 268–276, 2009 [DOI] [PubMed] [Google Scholar]
- 20. Callinan PA, Feinberg AP. The emerging science of epigenomics. Hum Mol Genet 15, Spec No 1: R95–R101, 2006 [DOI] [PubMed] [Google Scholar]
- 21. Casillas MA, Jr, Lopatina N, Andrews LG, Tollefsbol TO. Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem 252: 33–43, 2003 [DOI] [PubMed] [Google Scholar]
- 22. Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res 69: 1221–1229, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature 395: 89–93, 1998 [DOI] [PubMed] [Google Scholar]
- 24. Choi EK, Uyeno S, Nishida N, Okumoto T, Fujimura S, Aoki Y, Nata M, Sagisaka K, Fukuda Y, Nakao K, Yoshimoto T, Kim YS, Ono T. Alterations of c-fos gene methylation in the processes of aging and tumorigenesis in human liver. Mutat Res 354: 123–128, 1996 [DOI] [PubMed] [Google Scholar]
- 25. Davalos V, Esteller M. MicroRNAs and cancer epigenetics: a macrorevolution. Current opinion in oncology 22: 35–45, 2010 [DOI] [PubMed] [Google Scholar]
- 26. De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I, Lyonnet S, Stewart CL, Munnich A, Le Merrer M, Levy N. Lamin a truncation in Hutchinson-Gilford progeria. Science 300: 2055, 2003 [DOI] [PubMed] [Google Scholar]
- 27. Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399: 491–496, 1999 [DOI] [PubMed] [Google Scholar]
- 28. Dillon N, Festenstein R. Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet 18: 252–258, 2002 [DOI] [PubMed] [Google Scholar]
- 29. Drinkwater RD, Blake TJ, Morley AA, Turner DR. Human lymphocytes aged in vivo have reduced levels of methylation in transcriptionally active and inactive DNA. Mutat Res 219: 29–37, 1989 [DOI] [PubMed] [Google Scholar]
- 30. Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300: 455, 2003 [DOI] [PubMed] [Google Scholar]
- 31. Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 423: 293–298, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Esteller M. Epigenetics in cancer. N Engl J Med 358: 1148–1159, 2008 [DOI] [PubMed] [Google Scholar]
- 33. Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A 104: 15805–15810, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Perez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 37: 391–400, 2005 [DOI] [PubMed] [Google Scholar]
- 35. Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet 23: 413–418, 2007 [DOI] [PubMed] [Google Scholar]
- 36. Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, Zhou X, Jones PA. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res 69: 2623–2629, 2009 [DOI] [PubMed] [Google Scholar]
- 37. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, Miyazaki T, Ogura C, Okazaki Y, Jinno Y. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann Hum Genet 68: 196–204, 2004 [DOI] [PubMed] [Google Scholar]
- 39. Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118: 555–566, 2004 [DOI] [PubMed] [Google Scholar]
- 40. Gonzalez-Suarez I, Redwood AB, Gonzalo S. Loss of A-type lamins and genomic instability. Cell Cycle 8: 3860–3865, 2009 [DOI] [PubMed] [Google Scholar]
- 41. Gonzalez-Suarez I, Redwood AB, Perkins SM, Vermolen B, Lichtensztejin D, Grotsky DA, Morgado-Palacin L, Gapud EJ, Sleckman BP, Sullivan T, Sage J, Stewart CL, Mai S, Gonzalo S. Novel roles for A-type lamins in telomere biology and the DNA damage response pathway. EMBO J 28: 2414–2427, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gonzalo S, Blasco MA. Role of Rb family in the epigenetic definition of chromatin. Cell Cycle 4: 752–755, 2005 [DOI] [PubMed] [Google Scholar]
- 43. Gonzalo S, Garcia-Cao M, Fraga MF, Schotta G, Peters AH, Cotter SE, Eguia R, Dean DC, Esteller M, Jenuwein T, Blasco MA. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat Cell Biol 7: 420–428, 2005 [DOI] [PubMed] [Google Scholar]
- 44. Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet 8: 35–46, 2007 [DOI] [PubMed] [Google Scholar]
- 45. Grillari J, Grillari-Voglauer R. Novel modulators of senescence, aging, and longevity: small non-coding RNAs enter the stage. Exp Gerontol 45F: 302–311, 2010 [DOI] [PubMed] [Google Scholar]
- 46. Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell 128: 721–733, 2007 [DOI] [PubMed] [Google Scholar]
- 47. Grunstein M. Histone acetylation in chromatin structure and transcription. Nature 389: 349–352, 1997 [DOI] [PubMed] [Google Scholar]
- 48. Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate 59: 177–189, 2004 [DOI] [PubMed] [Google Scholar]
- 49. Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111: 369–379, 2002 [DOI] [PubMed] [Google Scholar]
- 50. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531, 2004 [DOI] [PubMed] [Google Scholar]
- 51. He L, He X, Lowe SW, Hannon GJ. microRNAs join the p53 network–another piece in the tumour-suppression puzzle. Nat Rev Cancer 7: 819–822, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature 435: 828–833, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Howard G, Eiges R, Gaudet F, Jaenisch R, Eden A. Activation and transposition of endogenous retroviral elements in hypomethylation induced tumors in mice. Oncogene 27: 404–408, 2008 [DOI] [PubMed] [Google Scholar]
- 54. Hristova M, Birse D, Hong Y, Ambros V. The Caenorhabditis elegans heterochronic regulator LIN-14 is a novel transcription factor that controls the developmental timing of transcription from the insulin/insulin-like growth factor gene ins-33 by direct DNA binding. Mol Cell Biol 25: 11059–11072, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet 7: 536–540, 1994 [DOI] [PubMed] [Google Scholar]
- 56. Issa JP, Vertino PM, Boehm CD, Newsham IF, Baylin SB. Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proc Natl Acad Sci U S A 93: 11757–11762, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science 288: 1422–1425, 2000 [DOI] [PubMed] [Google Scholar]
- 58. Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33, Suppl: 245–254, 2003 [DOI] [PubMed] [Google Scholar]
- 59. Jenuwein T, Allis CD. Translating the histone code. Science 293: 1074–1080, 2001 [DOI] [PubMed] [Google Scholar]
- 60. Jones PA, Baylin SB. The epigenomics of cancer. Cell 128: 683–692, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13: 2570–2580, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kennedy BK, Austriaco NR, Jr, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell 80: 485–496, 1995 [DOI] [PubMed] [Google Scholar]
- 63. Kennedy BK, Gotta M, Sinclair DA, Mills K, McNabb DS, Murthy M, Pak SM, Laroche T, Gasser SM, Guarente L. Redistribution of silencing proteins from telomeres to the nucleolus is associated with extension of life span in S. cerevisiae. Cell 89: 381–391, 1997 [DOI] [PubMed] [Google Scholar]
- 64. Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell 116: 259–272, 2004 [DOI] [PubMed] [Google Scholar]
- 65. Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31: 89–97, 2006 [DOI] [PubMed] [Google Scholar]
- 66. Kouzarides T. Chromatin modifications and their function. Cell 128: 693–705, 2007 [DOI] [PubMed] [Google Scholar]
- 67. Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev 12: 198–209, 2002 [DOI] [PubMed] [Google Scholar]
- 68. Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M. A microRNA signature of hypoxia. Mol Cell Biol 27: 1859–1867, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20: 615–626, 1998 [DOI] [PubMed] [Google Scholar]
- 70. Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol 14: 286–298, 2002 [DOI] [PubMed] [Google Scholar]
- 71. Lachner M, O'Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J Cell Sci 116: 2117–2124, 2003 [DOI] [PubMed] [Google Scholar]
- 72. Lara E, Mai A, Calvanese V, Altucci L, Lopez-Nieva P, Martinez-Chantar ML, Varela-Rey M, Rotili D, Nebbioso A, Ropero S, Montoya G, Oyarzabal J, Velasco S, Serrano M, Witt M, Villar-Garea A, Imhof A, Mato JM, Esteller M, Fraga MF. Salermide, a Sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene 28: 781–791, 2009 [DOI] [PubMed] [Google Scholar]
- 73. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854, 1993 [DOI] [PubMed] [Google Scholar]
- 74. Lengauer C, Kinzler KW, Vogelstein B. DNA methylation and genetic instability in colorectal cancer cells. Proc Natl Acad Sci U S A 94: 2545–2550, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liang R, Bates DJ, Wang E. Epigenetic control of MicroRNA expression and aging. Curr Genomics 10: 184–193, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liu B, Wang J, Chan KM, Tjia WM, Deng W, Guan X, Huang JD, Li KM, Chau PY, Chen DJ, Pei D, Pendas AM, Cadinanos J, Lopez-Otin C, Tse HF, Hutchison C, Chen J, Cao Y, Cheah KS, Tryggvason K, Zhou Z. Genomic instability in laminopathy-based premature aging. Nat Med 11: 780–785, 2005 [DOI] [PubMed] [Google Scholar]
- 77. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature 435: 834–838, 2005 [DOI] [PubMed] [Google Scholar]
- 78. Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8. A resolution. Nature 389: 251–260, 1997 [DOI] [PubMed] [Google Scholar]
- 79. Lujambio A, Esteller M. How epigenetics can explain human metastasis: a new role for microRNAs. Cell Cycle 8: 377–382, 2009 [DOI] [PubMed] [Google Scholar]
- 80. Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107: 137–148, 2001 [DOI] [PubMed] [Google Scholar]
- 81. Maes OC, An J, Sarojini H, Wang E. Murine microRNAs implicated in liver functions and aging process. Mech Ageing Dev 129: 534–541, 2008 [DOI] [PubMed] [Google Scholar]
- 82. Maison C, Bailly D, Peters AH, Quivy JP, Roche D, Taddei A, Lachner M, Jenuwein T, Almouzni G. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet 30: 329–334, 2002 [DOI] [PubMed] [Google Scholar]
- 83. Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev 15: 163–176, 2005 [DOI] [PubMed] [Google Scholar]
- 84. Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6: 838–849, 2005 [DOI] [PubMed] [Google Scholar]
- 85. Mello CC, Conte D., Jr Revealing the world of RNA interference. Nature 431: 338–342, 2004 [DOI] [PubMed] [Google Scholar]
- 86. Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124: 315–329, 2006 [DOI] [PubMed] [Google Scholar]
- 88. Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JC, Liang G, Jones PA. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res 62: 6456–6461, 2002 [PubMed] [Google Scholar]
- 89. O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843, 2005 [DOI] [PubMed] [Google Scholar]
- 90. O'Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet 4: e1000155, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, Wright SM, Mills KD, Bonni A, Yankner BA, Scully R, Prolla TA, Alt FW, Sinclair DA. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135: 907–918, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Oberdoerffer P, Sinclair DA. The role of nuclear architecture in genomic instability and ageing. Nat Rev Mol Cell Biol 8: 692–702, 2007 [DOI] [PubMed] [Google Scholar]
- 93. Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257, 1999 [DOI] [PubMed] [Google Scholar]
- 94. Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 19: 219–220, 1998 [DOI] [PubMed] [Google Scholar]
- 95. Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol 16: 23–29, 2009 [DOI] [PubMed] [Google Scholar]
- 96. Pegoraro G, Kubben N, Wickert U, Gohler H, Hoffmann K, Misteli T. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol 11: 1261–1267, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Pendas AM, Zhou Z, Cadinanos J, Freije JM, Wang J, Hultenby K, Astudillo A, Wernerson A, Rodriguez F, Tryggvason K, Lopez-Otin C. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat Genet 31: 94–99, 2002 [DOI] [PubMed] [Google Scholar]
- 98. Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 429: 771–776, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet 2: e40, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Pulkkinen K, Malm T, Turunen M, Koistinaho J, Yla-Herttuala S. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett 582: 2397–2401, 2008 [DOI] [PubMed] [Google Scholar]
- 101. Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A 101: 15998–16003, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Romanov GA, Vanyushin BF. Methylation of reiterated sequences in mammalian DNAs. Effects of the tissue type, age, malignancy and hormonal induction. Biochim Biophys Acta 653: 204–218, 1981 [DOI] [PubMed] [Google Scholar]
- 103. Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat Rev Mol Cell Biol 7: 437–447, 2006 [DOI] [PubMed] [Google Scholar]
- 104. Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle 5: 2220–2222, 2006 [DOI] [PubMed] [Google Scholar]
- 105. Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411, 2002 [DOI] [PubMed] [Google Scholar]
- 106. Sarg B, Koutzamani E, Helliger W, Rundquist I, Lindner HH. Postsynthetic trimethylation of histone H4 at lysine 20 in mammalian tissues is associated with aging. J Biol Chem 277: 39195–39201, 2002 [DOI] [PubMed] [Google Scholar]
- 107. Scaffidi P, Gordon L, Misteli T. The cell nucleus and aging: tantalizing clues and hopeful promises. PLoS Biol 3: e395, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science 312: 1059–1063, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis 31: 27–36, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Shen L, Kondo Y, Hamilton SR, Rashid A, Issa JP. P14 methylation in human colon cancer is associated with microsatellite instability and wild-type p53. Gastroenterology 124: 626–633, 2003 [DOI] [PubMed] [Google Scholar]
- 111. Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953, 2004 [DOI] [PubMed] [Google Scholar]
- 112. Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem 75: 243–269, 2006 [DOI] [PubMed] [Google Scholar]
- 113. Shumaker DK, Dechat T, Kohlmaier A, Adam SA, Bozovsky MR, Erdos MR, Eriksson M, Goldman AE, Khuon S, Collins FS, Jenuwein T, Goldman RD. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci U S A 103: 8703–8708, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS One 2: e895, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Sims RJ, 3rd, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet 19: 629–639, 2003 [DOI] [PubMed] [Google Scholar]
- 116. So K, Tamura G, Honda T, Homma N, Waki T, Togawa N, Nishizuka S, Motoyama T. Multiple tumor suppressor genes are increasingly methylated with age in non-neoplastic gastric epithelia. Cancer Sci 97: 1155–1158, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Song J, Noh JH, Lee JH, Eun JW, Ahn YM, Kim SY, Lee SH, Park WS, Yoo NJ, Lee JY, Nam SW. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS 113: 264–268, 2005 [DOI] [PubMed] [Google Scholar]
- 118. Strahl BD, Allis CD. The language of covalent histone modifications. Nature 403: 41–45, 2000 [DOI] [PubMed] [Google Scholar]
- 119. Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol 147: 913–920, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Taguchi A, Yanagisawa K, Tanaka M, Cao K, Matsuyama Y, Goto H, Takahashi T. Identification of hypoxia-inducible factor-1 alpha as a novel target for miR-17–92 microRNA cluster. Cancer Res 68: 5540–5545, 2008 [DOI] [PubMed] [Google Scholar]
- 121. Takatsu M, Uyeno S, Komura J, Watanabe M, Ono T. Age-dependent alterations in mRNA level and promoter methylation of collagen alpha1(I) gene in human periodontal ligament. Mech Ageing Dev 110: 37–48, 1999 [DOI] [PubMed] [Google Scholar]
- 122. Thakur MK, Kanungo MS. Methylation of chromosomal proteins and DNA of rat brain and its modulation by estradiol and calcium during aging. Exp Gerontol 16: 331–336, 1981 [DOI] [PubMed] [Google Scholar]
- 123. Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410: 227–230, 2001 [DOI] [PubMed] [Google Scholar]
- 124. Tra J, Kondo T, Lu Q, Kuick R, Hanash S, Richardson B. Infrequent occurrence of age-dependent changes in CpG island methylation as detected by restriction landmark genome scanning. Mech Ageing Dev 123: 1487–1503, 2002 [DOI] [PubMed] [Google Scholar]
- 125. Tryndyak VP, Kovalchuk O, Pogribny IP. Loss of DNA methylation and histone H4 lysine 20 trimethylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferase 1, Suv4–20h2 histone methyltransferase and methyl-binding proteins. Cancer Biol Ther 5: 65–70, 2006 [DOI] [PubMed] [Google Scholar]
- 126. Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439: 811–816, 2006 [DOI] [PubMed] [Google Scholar]
- 127. Vanyushin BF, Nemirovsky LE, Klimenko VV, Vasiliev VK, Belozersky AN. The 5-methylcytosine in DNA of rats. Tissue and age specificity and the changes induced by hydrocortisone and other agents. Gerontologia 19: 138–152, 1973 [PubMed] [Google Scholar]
- 128. Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature 450: 440–444, 2007 [DOI] [PubMed] [Google Scholar]
- 129. Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell 16: 93–105, 2004 [DOI] [PubMed] [Google Scholar]
- 130. Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107: 149–159, 2001 [DOI] [PubMed] [Google Scholar]
- 131. Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science 291: 1304–1351, 2001 [DOI] [PubMed] [Google Scholar]
- 132. Wang CM, Tsai SN, Yew TW, Kwan YW, Ngai SM. Identification of histone methylation multiplicities patterns in the brain of senescence-accelerated prone mouse 8. Biogerontology 11: 87–102, 2010 [DOI] [PubMed] [Google Scholar]
- 133. Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125: 467–481, 2006 [DOI] [PubMed] [Google Scholar]
- 134. Wilson VL, Jones PA. DNA methylation decreases in aging but not in immortal cells. Science 220: 1055–1057, 1983 [DOI] [PubMed] [Google Scholar]
- 135. Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem 67: 545–579, 1998 [DOI] [PubMed] [Google Scholar]
- 136. Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125: 483–495, 2006 [DOI] [PubMed] [Google Scholar]
- 137. Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15: 2343–2360, 2001 [DOI] [PubMed] [Google Scholar]