Summary
The Cre-loxP system is widely used for making conditional alterations to the mouse genome. Cre-mediated recombination is frequently monitored using reporter lines in which Cre expression activates a reporter gene driven by a ubiquitous promoter. Given the distinct advantages of fluorescent reporters, we developed a transgenic reporter line, termed IRG, in which DsRed-Express, a red fluorescent protein (RFP) is expressed ubiquitously prior to Cre-mediated recombination and an enhanced green fluorescent protein (EGFP) following recombination. Besides their utility for monitoring Cre-mediated recombination, we show that in IRG mice red and green native fluorescence can be imaged simultaneously in thick tissue sections by confocal microscopy allowing for complex reconstructions to be created that are suitable for analysis of neuronal morphologies as well as neurovascular interactions in brain. IRG mice should provide a versatile tool for analyzing complex cellular relationships in both neural and nonneural tissues.†
Keywords: Cre recombinase, loxP, conditional gene activation, DsRed-express, red fluorescent protein, enhanced green fluorescent protein, transgenic mice
INTRODUCTION
Many transgenic lines exist that express Cre recombinase either broadly or in cell type or tissue-specific manners (http://nagy.mshri.on.ca/cre/) making the system widely applicable for inducing cell type- or tissue-specific gene deletions as well as lineage tracing during development. When used for lineage tracing, Cre-expressing lines are crossed to reporter lines in which Cre-mediated excision activates a reporter gene. The efficacy and specificity of Cre excision in gene ablation studies is also frequently monitored with crosses to Cre reporter lines.
Several reporter lines have been developed (Kawamoto et al., 2000; Lobe et al., 1999; Luche et al., 2007; Mao et al., 1999, 2001; Muzumdar et al., 2007; Novak et al., 2000; Soriano, 1999; Srinivas et al., 2001; Vintersten et al., 2004; Yamauchi et al., 1999; Zinyk et al., 1998). A number of single reporters have, for example, targeted the ROSA26 locus with lacZ (Mao et al., 1999; Soriano, 1999) or a variety of fluorescent proteins (Luche et al., 2007; Mao et al., 2001; Srinivas et al., 2001). Single reporters however suffer from the limitation that there is no reporter expression until Cre excision and thus no internal control for expression in cell types or tissues where reporter expression may be weak or the “ubiquitous” promoter may in fact not be active. As an alternative, double reporter lines have been developed in which one reporter is ubiquitously expressed prior to Cre-mediated recombination while a second is activated following Cre excision (Lobe et al., 1999; Muzumdar et al., 2007; Novak et al., 2000; Vintersten et al., 2004). These mice offer the advantage that one reporter must always be “on” in any individual cell, thus creating an internal control.
Most dual reporter systems described to date have combined an enzymatic reporter (lacZ or alkaline phosphatase) with either green or red fluorescent proteins (Lobe et al., 1999; Novak et al., 2000; Vintersten et al., 2004). Although histochemical or immunocytochemical detection of enzymes such as β-galactosidase is simple, the sensitivity of this method may be problematic and vary depending on the amount of protein synthesized or other factors that can affect its subcellular distribution leading to an underestimation of the extent of staining, particularly in the nervous system (Friedrich et al., 1993). By contrast, fluorescent proteins offer the advantage that they can be visualized in tissues without fixation and cells can be isolated using fluorescence-activated cell sorting (FACS) without exogenous substrates. However, to date, only one dual reporter using fluorescent proteins has been developed (Muzumdar et al., 2007). These authors targeted a red fluorescent protein (RFP) from the family of reef coral fluorescent proteins known as tandem dimer Tomato along with an enhanced green fluorescent protein (EGFP) to the ROSA26 locus.
We developed a two-color fluorescent protein reporter by modifying the plasmid used to generate Z/EG mice (Novak et al., 2000). This vector utilizes the chicken β-actin promoter (CCAG) and first intron coupled to the CMV immediate early enhancer with a downstream polyadenylation signal derived from the rabbit β-globin gene (Niwa et al., 1991). Downstream of the CCAG promoter is a loxP flanked lacZ cDNA coupled to an EGFP reporter. We replaced the lacZ cDNA with a cDNA for DsRed-Express, an RFP derived from the Dicosoma sp. of reef coral fluorescent proteins. These proteins offer the advantage that due to their low homology with the Aequorea fluorescent proteins (Matz et al., 1999), specific monoclonal and polyclonal antibodies against them exist that do not cross-react with EGFP. Thus, in addition to spontaneous fluorescence, they can be distinguished from Aequorea GFPs by immunohistochemical means. To minimize integration site effects and possible promoter interference, we flanked the construct at each end with two paired copies of the 1.2-kb HS4 insulator sequence, found near the 5′-end of the chicken β-globin locus (Chung et al., 1993; Hebbes et al., 1994). This insulator has been previously shown to largely protect integrated transgenes from position site effects (Ciana et al., 2001; Guglielmi et al., 2003; Potts et al., 2000). A diagram of the final construct, which we termed IRG, for “insulator/red/green” is shown in Figure 1a.
FIG. 1.
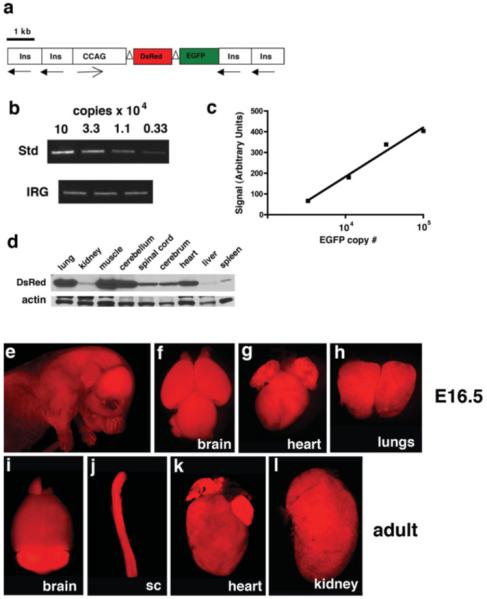
RFP expression in transgenic IRG mice. Panel a shows the design of the IRG transgene with insulator sequences (Ins), the chicken-CMV β-actin promoter (CCAG), as well as the DsRed express and EGFP cDNAs indicated. loxP sites are indicated as open triangles around the DsRed express cDNA and the directional orientation of the insulator sequences and CCAG promoter are indicated by arrows. Transgene copy number was estimated by semiquantitative PCR amplification of the EGFP cDNA (b and c). A standard curve was established by amplifying different amounts of the pIRG plasmid. In parallel, genomic DNA was amplified from IRG mice. In (b), a standard curve from a representative experiment is shown in the upper panel and amplification from three IRG transgenic mice is shown in the lower panel. Transgene copy number was extrapolated from the standard curve (c) and the copy number was calculated from the amount of starting genomic template DNA (12 ng) assuming a DNA content of 6 pg/cell. Western blotting for DsRed expression in the indicated tissues from an adult IRG transgenic animal is shown in (d). The lower panel shows the blot reprobed for β-actin. In e–h, spontaneous RFP fluorescence is viewed in an E16.5 embryo (e) as well as embryonic brain (f), heart (g) and lungs (h), whereas i–l show expression in brain (i), spinal cord (j), heart (k), and kidney (l) from an adult IRG transgenic mouse. In (e–l), all panels were photographed using a ×1 stereoscopic lens except panel i which was photographed with an ×0.75 lens.
Transgenic mice were generated by pronuclear injection of linearized IRG plasmid DNA and analysis of mice from a line established from founder five is described. By semiquantitative PCR, this line was estimated to contain seven copies of the transgene (Fig. 1b,c). Multiple generations of IRG mice from Line 5 have not exhibited any signs of toxicity due to RFP expression up to 15 months of age and both males as well as females are fertile.
Transgenic IRG mice demonstrated red fluorescence under the appropriate fluorescent excitation filter for RFP without any green fluorescence (Figs. 1 and 2a,b). All internal organs examined showed red fluorescence without detectable green fluorescence above that seen in nontransgenic embryos. Examples of tissues from both embryonic and adult IRG mice are shown in Figure 1e–l. Western blotting of tissues from adult IRG mice showed that although the level of DsRed-Express protein varied from tissue to tissue, expression was detectable in all organs examined (Fig. 1d).
FIG. 2.
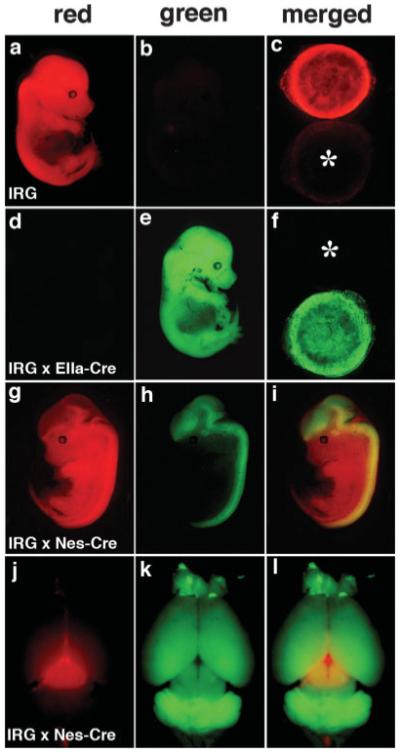
Cre-mediated recombination in IRG reporter mice. In (a, b), an E14.5 single transgenic IRG embryo is shown viewed under red (a) or green (b) filters. In (d, e), an IRG/EIIa-Cre double transgenic embryo is shown viewed under red (d) or green (e) filters. In (c) and (f), a merged image of the placentas from the two embryos sequentially photographed under red and green filters is shown. Asterisks in each panel indicate the location of the nonfluorescent placenta. An IRG/Nestin-Cre double transgenic embryo (E13.5) is shown in g–i, photographed under red (g) or green filters (h) or presented as a merged image (i). The brain from an adult IRG/Nestin-Cre double transgenic animal is shown in (j–l), photographed under red (j)/green (k) filters or shown as a merged image (l).
To determine the extent of Cre-mediated recombination that could be induced, we crossed IRG mice with either EIIa-Cre (Lakso et al., 1996) or CMV-Cre mice (Schwenk et al., 1995), two general Cre deletor lines. EIIa-Cre mice express Cre under the control of the adenovirus EIIa promoter, which drives Cre expression as early as the preimplantation embryo (Lakso et al., 1996), whereas in CMV-Cre mice, Cre expression is driven by the human cytomegalovirus minimal promoter (Schwenk et al., 1995). Both lines have been shown to be efficient inducers of widespread Cre-mediated recombination. As shown in Figure 2d–f, the embryo as well as the placenta from double transgenic IRG/EIIa-Cre mice showed green fluorescence. Widespread EGFP activation was also seen in IRG/CMV-Cre mice (Supplementary Fig. S1). Therefore widespread Cre-mediated activation of EGFP is possible in IRG mice, thus demonstrating their suitability as a general Cre reporter.
We next examined Cre-mediated excision in the central nervous system by crossing IRG mice to Nestin-Cre mice. Nestin is expressed in neural stem/progenitor cells and regarded as a general marker of this cell population. Nestin expression in the nervous system is controlled by an intron 2 enhancer that has been widely used to express heterologous genes in the neural stem/progenitor cell population (Zimmerman et al., 1994). In double transgenic IRG/Nestin-Cre embryos, EGFP expression was activated in brain and spinal cord (Fig. 2g–i). Internal organs exhibited only spotty green fluorescence in heart and kidneys (data not shown). Double transgenic mice showed EGFP activation throughout the brain (Fig. 2j–l). The only grossly nonrecombined region was an area of the dorsal midbrain (Fig. 2j), which based on immunohistochemical staining corresponds to prominent RFP expression within the superior colliculus (Supplementary Fig. S2).
Immunohistochemical staining of single transgenic IRG mice showed widespread RFP expression throughout the adult brain without any EGFP expression. Examples of RFP staining in the hippocampus and cerebellum of IRG single transgenic mice are shown in Figure 3. In double transgenic IRG/Nestin-Cre mice, immunohistochemical staining documented widespread EGFP activation in both embryonic and adult brain as expected for a Nestin-Cre transgene. Although EGFP activation did not occur in all cells, the pattern of activation was very similar to that observed in Nestin-Cre crosses to Z/EG reporter mice (see Fig. 4). IRG mice should thus be suitable for two-color fluorescent protein imaging in the brain as well as other organs.
FIG. 3.
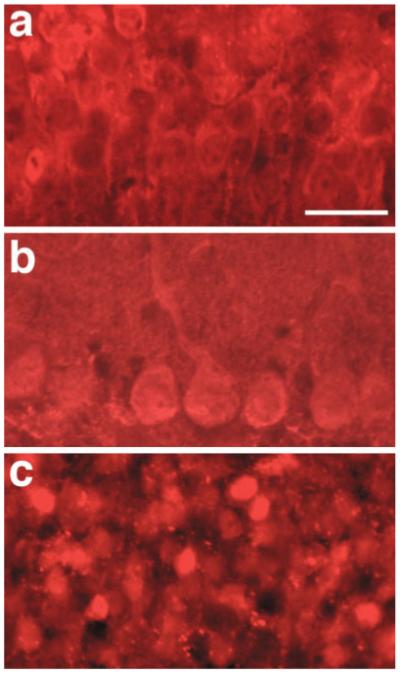
DsRed expression in adult brain of single transgenic IRG mice. Shown is immunohistochemical staining for DsRed in CA1 pyramidal neurons of the hippocampus (a), Purkinje cells of the cerebellum (b), and the hippocampal granule cell layer (c). Scale bar: 20 μm.
FIG. 4.
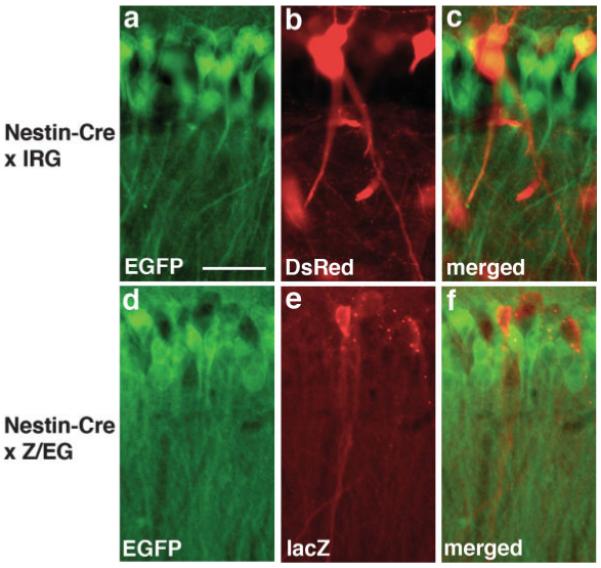
EGFP activation in the adult brain of IRG/Nestin-Cre double transgenic mice. Pyramidal cells in the CA1 region of the hippocampus from Nestin-Cre mice crossed to ether IRG (a–c) or Z/EG (d–f) reporter mice. In (a–c) sections were double immunostained for EGFP (a) and DsRed (b), whereas in (d–f) immunostaining was performed for EGFP (d) and LacZ (e). Merged images are shown in Panels c and f. A cluster of nonrecombined DsRed or lacZ expressing cells is apparent in both animals. Scale bar: 20 μm.
Having two fluorescent proteins offers the opportunity of using FACS to separate recombined from nonrecombined cell populations without the need for exogenous substrates. To determine whether RFP and EGFP expressing cells could be separated using IRG mice, we collected E14.5 embryonic brain from IRG as well as IRG crossed to Nestin-Cre mice. The tissues were dissociated into single cell suspensions and analyzed by FACS using cells from nontransgenic littermates to set levels of background fluorescence (Fig. 5b). Using a 488-nm excitation laser, 54.6% of the cells sorted from the IRG brains were RFP positive. We suspect that the relatively low percentage of RFP positive cells reflects the fact that optimal excitation for DsRed-Express is 557 nm (Biosciences, 2003) and thus using the available 488 nm laser, we likely failed to detect weakly positive cells. Supporting this notion when we immunostained single cell suspensions from brains of the same-age embryos, we found that 96% of the cells were DsRed-immunolabeled (Fig. 5a). Muzumdar et al. (2007) have reported a similar discrepancy between detection of tandem dimer tomato positive cells by FACS and visual inspection when using a 488-nm laser for FACS detection. No EGFP positive cells were detected either by FACS and or by immunostaining in single transgenic IRG mice.
FIG. 5.
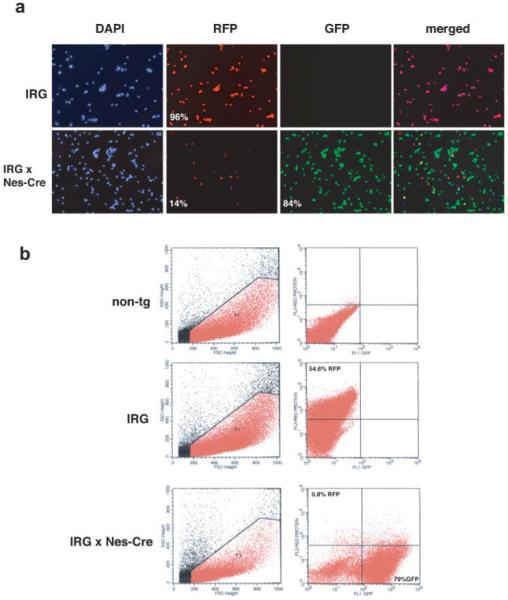
Analysis of single cell suspensions from embryonic brain of IRG and IRG/Nestin-Cre transgenic mice by immunostaining and FACS. In (a) the telencephalon from brains of E14.5 IRG single transgenic or IRG crossed to Nestin-Cre double transgenic mice were dissociated into single cell suspensions and plated on coverslips. Immunostaining was performed with anti-DsRed (RFP) and anti-EGFP (GFP), and the nuclei were visualized with DAPI. A merged image is shown in the far right panel. Numbers in the panels indicate the percentage of DAPI stained cell that were immunolabeled for DsRed or EGFP. One hundred cells per coverslip were counted from three coverslips for each genotype. In (b), FACS was performed on cell suspensions from E14.5 embryonic telencephalon from IRG, IRG/Nes-Cre, or nontransgenic littermate mice. Excitation was performed using a 488-nm blue argon laser. Gates were established to exclude 100% of the cells from the nontransgenic littermates.
When Nestin-Cre crossed to IRG, mice were examined by FACS (Fig. 5b), the number of EGFP positive cells was very similar between FACS (79%) and visual inspection of immunostained single cell suspensions (84%) likely reflecting that the optimal excitation for EGFP is 484 nm (Biosciences, 2003), which is much closer to that of the laser available for FACS detection. In these crosses, 0.8% of cells were RFP positive (i.e., nonrecombined) by FACS analysis and 14% RFP positive by immunostaining (see Fig. 5), again likely representing an under estimation of RFP expressing cells by FACS.
Structural changes in the brain occur in normal aging and with a variety of pathological states. Increasingly it is being realized that detailed reconstructions of brain structure at various levels of resolution can contribute to understanding disease states. Such imaging requires imaging in thick materials, whole brains, or thick tissue slabs. Immunolabeling within thick sections is often compromised because of the poor tissue penetration of most immunostaining methods. Fluorescent proteins offer the possibility of overcoming the limitations of immunostaining by allowing native fluorescence to be imaged in situ. In addition, having two fluorescent proteins in the IRG mouse and the ability to activate them selectively based on the choice of Cre driver creates the possibility of imaging distinct structures or cell types within the same section. Nestin-Cre, for example, activates widespread EGFP expression in neurons but because the nestin intron 2 enhancer is not active in vascular elements (Wen et al., 2005), cerebral blood vessels which are mesenchymally derived remain nonrecombined and red. Thus, neurons and vascular structures can in principle be imaged simultaneously.
Two of us (PRH and SLW) recently described algorithms for reconstructing 3D images from original grayscale data obtained by laser-scanning confocal microscopy (Rodriguez et al., 2006; Wearne et al., 2005). Using double transgenic IRG/Nestin-Cre mice, we determined whether these algorithms could be used to image native fluorescence in thick sections of brain tissue to visualize simultaneously neuronal and vascular structures. Using 200-μm thick sections of brain tissue, native RFP and EGFP fluorescence was imaged by laser-scanning confocal microscopy (see Materials and Methods). Figure 6 shows high-resolution confocal imaging of the CA1 pyramidal layer of the hippocampus. Recombined pyramidal neurons appear green and nonrecombined blood vessels are red. An additional example of simultaneous vascular and neuronal imaging in the cerebellar cortex is shown in Supplementary Figure S3.
FIG. 6.
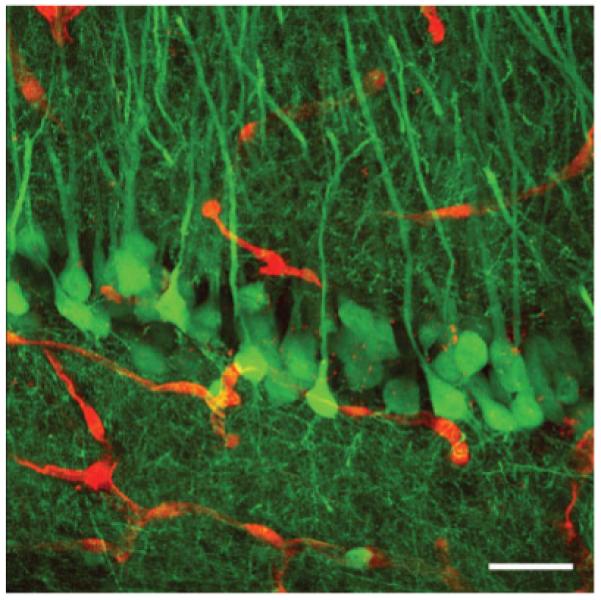
Maximal two-dimensional projection of a three-dimensional reconstruction of deconvolved and merged z-stacks of native red and green fluorescence from 200-μm thick tissue sections using confocal laser-scanning microscopy. The CA1 pyramidal cell layer in the hippocampus from an adult IRG/Nestin-Cre double transgenic mouse is shown. Recombined pyramidal cells are green and nonrecombined blood vessels are red. Scale bar: 20 μm.
Native fluorescence offers the advantage that it can be detected in thick tissue sections at depths where immunostaining is difficult. To determine whether native expression and immunostaining have comparable sensitivities at tissue thicknesses where immunostaining is possible, we took sections of brain from EIIa-Cre mice crossed to IRG mice where recombination is essentially complete and performed immunostaining for EGFP using an AlexaFluor568-conjugated secondary. We then imaged native EGFP fluorescence (green) and EGFP immunostaining (red) in the same sections. The patterns of EGFP expression detected by native fluorescence and immunostaining appeared essentially identical. An example of imaging from the hippocampal granule cell layer is shown in Supplementary Figure S4. To address whether RFP imaging had similar sensitivities, sections of brain from single transgenic IRG mice were immuno-stained for DsRed-Express using an AlexaFluor488-conjugated secondary antibody and then native RFP fluorescence (red) and DsRed-Express immunostaining (green) were compared. The patterns of RFP expression detected by native fluorescence and immunostaining were essentially identical (Supplementary Fig. 4). Thus detection of EGFP and RFP by native fluorescence and immunostaining appear equally sensitive under the conditions tested.
These studies show that in addition to serving as a general reporter for Cre-mediated recombination, IRG mice can be used to image complex cellular relationships in thick tissue sections, which cannot currently be reliably achieved by standard immunostaining approaches. By choice of Cre driver it should be possible to image a variety of complex cellular relationships in situ. IRG mice should thus provide a versatile tool for a variety of applications utilizing the Cre/loxP recombination system. Future modifications of this system could also be envisioned whereby modulation of the temporal onset of recombination could be regulated to study processes such as adult neurogenesis.
Finally it is of note that to our knowledge this is the first reporter line created using pronuclear injection as opposed to targeting an endogenous gene locus by homologous recombination success that we suspect may be attributed to the inclusion of the insulator sequences. If so, insulator sequences may provide a mechanism for expressing other reporter constructs without the need for gene targeting.
MATERIALS AND METHODS
Plasmid Construction
A 1.8-kb SalI fragment containing the CMV-chicken β-actin promoter (CCAG) and one loxP site was obtained by digestion of the plasmid pZ/EG (Novak et al., 2000) (gift of Dr. Corrine Lobe, University of Toronto, Ontario, Canada) and cloned into the SalI site of pSP72 (Promega Corporation, Madison, WI) to generate the plasmid pCCAG. The DsRed express fragment was obtained from the pDS Red-Express vector (Clontech, Mountain View, CA) and was cloned into a unique BamHI site down-stream of the loxP site of pCCAG. The 3xpA cassette containing three SV40 polyadenylation sequences was obtained by PCR from pZ/EG and cloned downstream of the DsRed express cDNA into a unique KpnI site to give the plasmid pCCAG-R. A fragment containing a loxP site, the coding sequence for the EGFP and rabbit β-globin polyadenylation sequence (loxP-EGFP-globin poly A) was excised from pZ/EG by digestion with SmaI/Hin-dIII, end repaired, ClaI linkered and cloned into a unique ClaI site downstream of the 3xpA cassette to give the plasmid pCCAG-R/G. A 2.4-kb fragment containing tandem repeats of the 5′-HS4 insulator sequences from the chicken β-globin gene was excised form pJC13 (Chung et al., 1993) (gift of Dr. Gary Felsenfeld National Institutes of Health, Bethesda MD) by BamHI/SalI digestion, SphI linkered, and cloned upstream of the CCAG promoter to give pInsCCAG-R/G. To insert a second insulator cassette at the 3′-end of the loxP-EGFP-globin poly A sequence, the EcoRV site downstream of EGFP was converted into a PacI site while the 2.4-kb insulator fragment was subcloned from pJC-13 into the EcoRI/BamHI sites of pSP72 and the upstream SalI site was converted into a PacI site. The Ins-CCAG-DsRed-EGFP cassette was excised from pInsCCAG-R/G by digestion with XhoI/PacI and cloned upstream of the insulator cassette, resulting in the final construct pIRG in which the CCAG-DsRed-EGFP (CCAG-R/G) fragment is flanked both 5′ and 3′ by two tandem repeats of the 5′-HS4 insulator sequence both oriented antisense with respect to the CCAG promoter.
Transgenic Mice
The 9.4-kb insert from the plasmid pIRG was excised from the plasmid backbone by XhoI/EcoRV digestion, gel purified and injected into fertilized oocytes of C6B3 F1 hybrid mice. Founders were identified by PCR analysis of DNA isolated from tail clips using primers specific for EGFP (5′-CGTAAACGGCCAGTTCAG and 5′-ATGCGCTTCTTCTGCTTGTCG) and for DsRed express (5′-GCCGACATCCCCGACTAC-3′ and 5′-GCTTCAGGGCCTTGTGGATCT-3′). Founders were bred to C57BL/6 wild type mice. Transgene copy number was estimated in the established transgenic line by semiquantitative PCR. A portion of the EGFP cDNA was amplified using the primers described earlier from different amounts of the pIRG plasmid to establish a standard curve and from genomic DNA isolated from IRG mice. After 28 cycles, the amplified DNAs were visualized by UV-ethidium-bromide-induced fluorescence and quantitated on an Alpha-Imager 2200 (Alpha Innotech, San Leandro, CA). Transgene copy number was extrapolated from the standard curve, and the copy number in IRG line 5 was calculated from the amount of starting genomic template DNA (12 ng) assuming a DNA content of 6 pg/cell. Nestin-Cre (Strain Name: B6.Cg-Tg (Nes-cre) 1Kln/J; Stock Number: 003771), CMV-Cre (Strain name: B6.C-Tg (CMV-cre) 1Cgn/J; Stock Number: 006054) and Z/EG transgenic mice (Stain name: lacZ/EGFP; stock name: Tg (ACTB-Bgeo/GFP); stock #:003920) were purchased from the Jackson Laboratories (Bar Harbor, MA). EIIa-Cre mice (Lakso et al., 1996) were kindly provided by Drs. Erwin Bottinger and Sandra Merscher-Gomez (Mount Sinai School of Medicine, New York, NY). Cre recombinase transgenic mice were identified with Cre-specific primers as previously described (Wen et al., 2005). Animals were housed on 12-h light/dark cycles and received food and water ad libitum. All protocols were approved by the Institutional Animal Care and Use Committees of the James J. Peters Department of Veterans Affairs Medical Center and the Mount Sinai School of Medicine and were conducted in conformance with the National Institutes of Health “Guide for the Care and Use of Laboratory Animals” (NIH publication 80–23).
Western Blotting
Telencephalon, cerebellum, spinal cord, liver, lung, muscle, spleen, and kidney were dissected from a 5-month-old IRG transgenic animal and homogenized in 10 volumes of 50 mM Tri HCl pH 7.6, 0.15 M NaCl, 1 mM EDTA, 1% Triton X100, 1% sodium deoxycholate, 0.1% SDS with the addition of the HALT protease and phosphatase inhibitor cocktails (Pierce Biotechnologies, Rockford, IL). The samples were centrifuged at 15,000 rpm for 20 min, and then the supernatant was collected. Protein concentration was determined using the BCA reagent assay (Pierce) as described by the manufacturer. DsRed expression in the tissue extracts was analyzed by Western blotting. Twenty micrograms of protein was separated on SDS-PAGE gels and blotted onto polyvinylidene difluoride (PVDF) membranes (Millipore Corporation, Billerica MA). The blots were blocked with 50 mM Tris HCl, pH 7.6, 0.15 M NaCl, 0.1% Tween-20 (TBST), 5% non-fat dry milk and probed with a rabbit polyclonal anti-DsRed (Clontech; 1:10,000 dilution in blocking solution) followed by secondary detection with horseradish peroxidase goat anti-rabbit IgG (GE Healthcare Bio-Sciences Corporation, Piscataway, NJ). As a loading control the blot was stripped and reprobed with mouse monoclonal anti-β-actin antibody (Sigma-Aldrich, St. Louis, MO; 1:3,000 dilution). The bands were visualized with the ECL+ reagent (GE Healthcare Bio-Sciences Corporation) and exposure to CL-Xposure film (Pierce).
Imaging of Spontaneous Fluorescence in Whole Embryos
Embryos were isolated from timed-pregnant female mice with the day a vaginal plug was observed considered as E0.5. Following euthanasia of the mother with CO2, embryos were removed and provisionally genotyped by observation under a Nikon SMZ 1500 stereoscope (Nikon Corporation, Melville, NY) equipped with a P-FLA epifluorescence attachment equipped with GFP (excitation 450–490 nm, emission 525–550 nm) and RPF (excitation 530–560 nm, emission 590–650 nm) filters. Images of embryos were photographed with a Nikon Digital Sight DS-5M camera and processed with Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA).
Analysis of RFP and EGFP Expression in Single Cell Suspensions
Brains isolated from E14 embryos were dissociated in 0.006% trypsin (Invitrogen, Carlsbad, CA) in phosphate-buffered saline (PBS) by trituration with a fire polished pipette. The reaction was stopped by addition of Dulbecco’s Modified Eagles Medium (DMEM, Invitrogen)/10% fetal calf serum (FCS, Summit Biotechnology, Fort Collins, CO) and the cell suspension was centrifuged at 1,500 rpm for 5 min. To lyse the red blood cells, samples were treated with 1 ml of 0.15 M ammonium chloride, overlayed onto 4 ml of DMEM medium and centrifuged at 1,500 rpm for 5 min. The cell pellet was resuspended in medium containing 60 units/ml DNAse I (Sigma-Aldrich) and filtered through a 75-μm nylon mesh. For plating, the cell suspension was centrifuged at 1,500 rpm for 5 min, and the cell pellet was resuspended in Eagle’s minimal essential medium (ATCC, Manassas, VA) containing 10% horse serum (Biowhittaker, Walkersville, MD) (106 cell/ml), plated onto poly-d-lysine- (Sigma-Aldrich) coated glass slides (Nunc, Naperville, IL) and allowed to adhere for 2 h. The cells were then fixed 15 min with 4% paraformaldehyde and rinsed with PBS. Immunostaining was performed using the antibodies described later. Nuclei were counterstained with 1 μg/ml 4′, 6-diamidino-2-phenylindole (DAPI) and the coverslips imaged with the Zeiss Axioimager (Zeiss, Thorn-wood, NY). One hundred cells per coverslip (3 coverslips/genotype) in randomly chosen fields were examined and the percentage of DAPI stained cells expressing DsRed express and EGFP were determined.
Fluorescence-Activated Cell Sorting
FACS analysis was performed on cell suspensions prepared as described earlier in the Mount Sinai School of Medicine Flow Cytometry Shared Research Facility with a Becton Dickinson FACS Vantage cell sorter equipped with a 488-nm blue argon laser and FACS DIVA software (BD Biosciences, San Jose, CA). Cells from nontransgenic littermates were used to set background fluorescence levels and a size threshold was used to gate out cell debris. For each sample, 100,000 events were collected.
Immunohistochemistry
Adult mice were anesthetized with 150 mg/kg ketamine and 30 mg/kg xylazine and then sacrificed by transcardial perfusion with cold 1% paraformaldehyde in 0.1 M PBS pH 7.4 for 1 min, followed by cold 4% paraformaldehyde in PBS for 10 min. After perfusion, brains were removed and postfixed in the 4% paraformaldehyde for 48 h and then transferred to 0.1 M PBS, and stored at 4°C until sectioning. Fifty-micrometer thick coronal sections were cut using a Vibratome.
Immunohistochemistry was performed on free-floating sections. The primary antibodies used were anti-EGFP (rabbit polyclonal 1:1,000 dilution or mouse monoclonal clone 3E6, 1:500 dilution, both from Molecular Probes/Invitrogen, Carlsbad, CA), rabbit polyclonal anti-DsRed (1:5,000 dilution; Clontech), and anti-β-galactosidase (lacZ; mouse monoclonal, 1:200 dilution; Promega, Madison WI). Sections were blocked with Tris-buffered saline (TBS; 50 mM Tris-HCl, 0.15 M NaCl, pH 7.6, 0.15 M NaCl)/0.1% Triton X-100/5% goat serum (TBS-TGS) for 1.5 h and the primary antibody was applied overnight in TBS-TGS at room temperature. Following washing in PBS for 1 h, immunofluorescence staining was detected by incubation with species-specific Alexa Fluor secondary antibody conjugates (1:400, Molecular Probes/Invitrogen) for 2 h in TBS-TGS. After washing in PBS, sections were mounted on slides using Gel/Mount (Biomeda, Foster City, CA). Sections were photographed on a Zeiss Axioimager microscope using the AxioVision Release 4.3 program. Digital images were color balanced using Adobe Photoshop 7.0 (Adobe Systems, San Jose, CA).
Confocal Imaging of Native EGFP and DsRed Fluorescence
The fluorescence emitted by EGFP and DsRed-Express (emission maxima at 509 and 579 nm, respectively), was simultaneously detected using confocal laser scanning microscopy with a Radiance 2000 MP system (Biorad, Hercules, CA) with an ArKr laser. This system feeds into an upright Olympus BX50 WIF microscope equipped with UPLApo 60x (1.2 NA) objective lens using adapted band pass filters. Each series of images was acquired with a voxel resolution of 0.1 μm × 0.1 μm × 0.1 μm, respectively, for the x, y, z axes. Each stack of images was deconvolved (Autodeblur, Media Cybernetics, Bethesda, MD) and merged with LaserSharp imaging software.
Supplementary Material
ACKNOWLEDGMENTS
Transgenic mice were generated by the Mount Sinai School of Medicine Mouse Genetics Shared Research Facility and Flow cytometry was performed through the Mount Sinai School of Medicine Flow Cytometry Shared Research Facility. We thank Drs. Corrine Lobe and Gary Felsenfeld for providing plasmids and Drs. Erwin Bottinger and Sandra Merscher-Gomez for providing EIIa-Cre mice.
Grant Support: AG021305, AG023599, AG02219, AG05138; Contract grant sponsor: National Institute of Mental Health; Contract grant numbers: MH070603, MH58911; Contract grant sponsor: National Cancer Institute; Contract grant number: 5R24 CA095823-04; Contract grant sponsor: National Science Foundation; Contract grant number: DBI-9724504; Contract grant sponsors: United States Department of Veterans Affairs, Merit Award, National Alliance for Research in Schizophrenia and Affective Disorders (NARSAD), Young Investigator Award.
Footnotes
This article is a US Government work and, as such, is in the public domain in the United States of America.
This article contains supplementary material available via the Internet at http://www.interscience.wiley.com/jpages/1526-954X/suppmat
LITERATURE CITED
- Biosciences B BD Living Colors User Manual. 2003;II www.bdbiosciences.com.
- Chung JH, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- Ciana P, Di Luccio G, Belcredito S, Pollio G, Vegeto E, Tatangelo L, Tiveron C, Maggi A. Engineering of a mouse for the in vivo profiling of estrogen receptor activity. Mol Endocrinol. 2001;15:1104–1113. doi: 10.1210/mend.15.7.0658. [DOI] [PubMed] [Google Scholar]
- Friedrich VL, Jr, Holstein GR, Li X, Gow A, Kelley KA, Lazzarini RA. Intracellular distribution of transgenic bacterial β-galactosidase in central nervous system neurons and neuroglia. J Neurosci Res. 1993;36:88–98. doi: 10.1002/jnr.490360110. [DOI] [PubMed] [Google Scholar]
- Guglielmi L, Le Bert M, Truffinet V, Cogne M, Denizot Y. Insulators to improve expression of a 3′IgH LCR-driven reporter gene in transgenic mouse models. Biochem Biophys Res Commun. 2003;307:466–471. doi: 10.1016/s0006-291x(03)01185-9. [DOI] [PubMed] [Google Scholar]
- Hebbes TR, Clayton AL, Thorne AW, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Niwa H, Tashiro F, Sano S, Kondoh G, Takeda J, Tabayashi K, Miyazaki J. A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett. 2000;470:263–268. doi: 10.1016/s0014-5793(00)01338-7. [DOI] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobe CG, Koop KE, Kreppner W, Lomeli H, Gertsenstein M, Nagy A. Z/AP, a double reporter for cre-mediated recombination. Dev Biol. 1999;208:281–292. doi: 10.1006/dbio.1999.9209. [DOI] [PubMed] [Google Scholar]
- Luche H, Weber O, Nageswara Rao T, Blum C, Fehling HJ. Faithful activation of an extra-bright red fluorescent protein in “knock-in”. Cre-reporter mice ideally suited for lineage tracing studies. Eur J Immunol. 2007;37:43–53. doi: 10.1002/eji.200636745. [DOI] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97:324–326. doi: 10.1182/blood.v97.1.324. [DOI] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Orkin SH. Improved reporter strain for monitoring cre recombinase-mediated DNA excisions in mice. Proc Natl Acad Sci USA. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for highexpression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Potts W, Tucker D, Wood H, Martin C. Chicken β-globin 5′HS4 insulators function to reduce variability in transgenic founder mice. Biochem Biophys Res Commun. 2000;273:1015–1018. doi: 10.1006/bbrc.2000.3013. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Ehlenberger DB, Hof PR, Wearne SL. Rayburst sampling, an algorithm for automated three-dimensional shape analysis from laser scanning microscopy images. Nat Protoc. 2006;1:2152–2161. doi: 10.1038/nprot.2006.313. [DOI] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vintersten K, Monetti C, Gertsenstein M, Zhang P, Laszlo L, Biechele S, Nagy A. Mouse in red: Red fluorescent protein expression in mouse ES cells, embryos, and adult animals. Genesis. 2004;40:241–246. doi: 10.1002/gene.20095. [DOI] [PubMed] [Google Scholar]
- Wearne SL, Rodriguez A, Ehlenberger DB, Rocher AB, Henderson SC, Hof PR. New techniques for imaging, digitization and analysis of three-dimensional neural morphology on multiple scales. Neuroscience. 2005;136:661–680. doi: 10.1016/j.neuroscience.2005.05.053. [DOI] [PubMed] [Google Scholar]
- Wen PH, De Gasperi R, Sosa MA, Rocher AB, Friedrich VL, Jr, Hof PR, Elder GA. Selective expression of presenilin 1 in neural progenitor cells rescues the cerebral hemorrhages and cortical lamination defects in presenilin 1-null mutant mice. Development. 2005;132:3873–3883. doi: 10.1242/dev.01946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Abe K, Mantani A, Hitoshi Y, Suzuki M, Osuzu F, Kuratani S, Yamamura K. A novel transgenic technique that allows specific marking of the neural crest cell lineage in mice. Dev Biol. 1999;212:191–203. doi: 10.1006/dbio.1999.9323. [DOI] [PubMed] [Google Scholar]
- Zimmerman L, Parr B, Lendahl U, Cunningham M, McKay R, Gavin B, Mann J, Vassileva G, McMahon A. Independent regulatory elements in the nestin gene direct transgene expression to neural stem cells or muscle precursors. Neuron. 1994;12:11–24. doi: 10.1016/0896-6273(94)90148-1. [DOI] [PubMed] [Google Scholar]
- Zinyk DL, Mercer EH, Harris E, Anderson DJ, Joyner AL. Fate mapping of the mouse midbrain-hindbrain constriction using a site-specific recombination system. Curr Biol. 1998;8:665–668. doi: 10.1016/s0960-9822(98)70255-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


