Abstract
The gel mobility shift assay is routinely used to visualize protein-RNA interactions. Its power resides in the ability to resolve free from bound RNA with high resolution in a gel matrix. In this chapter, we review the quantitative application of this approach to elucidate thermodynamic properties of protein-RNA complexes. Assay designs for titration, competition, and stoichiometry experiments are presented for two unrelated model complexes.
Keywords: gel mobility shift, RNA-binding protein, free energy, equilibrium dissociation constant, titration, competition binding
1. Introduction
1.1 Gel mobility shift assay
The gel mobility shift assay is a simple yet powerful tool that is commonly used to visualize the interaction between proteins and nucleic acid. This method, also referred to as electrophoretic mobility shift assay or EMSA, relies on the property that nucleic acid will migrate through an agarose or polyacrylamide gel matrix towards an anode upon application of an electric field. In a somewhat simplified view, the migration of RNA through a gel is governed by three primary factors, the molecular weight (and hence the charge) of the RNA, its three-dimensional shape, and the physical properties of the gel substrate. Interaction of a protein that modulates the RNA conformation or substantially increases the molecular weight of the ribonucleoprotein particle can lead to differential mobility in the gel. The choice of the gel matrix can amplify or dampen this effect depending upon the size and shape of the RNA-protein complex. By this approach, formation of most RNA-protein complexes can be conveniently monitored by comparing the mobility of the nucleic acid in the presence and absence of the protein.
This method is generalizable and has been used to study a wide variety of protein-RNA complexes. Some of the first examples involved characterization of polyribosomes on bacterial mRNA and the interaction of small ribosomal subunit proteins with 16S rRNA (1, 2). Since then, the approach has been used to analyze the interaction of structured and linear RNA with protein and small molecule co-factors in a variety of systems (3–8). In this overview, we will compare and contrast the gel mobility shifts of two systems with distinct binding properties in order to highlight the useful quantitative analyses that can be performed with this method (Fig. 1). In the first example, we review the ability of the translation repressor GLD-1 from Caenorhabditis elegans to shift the mobility of its cognate 28-nucleotide linear RNA target (termed TGE) from the 3’-untranslated region of tra-2 mRNA (9). In the second, we review binding of the small ribosomal subunit protein S15 from the hyperthermophilic bacterium Aquifex aeolicus to a fragment of 16S rRNA termed Afr1 (10). The accompanying protocol is relevant to both systems, even though the mechanism of the shift is different. In an accompanying chapter in this volume, the binding properties of two related systems will be compared using isothermal titration calorimetry.
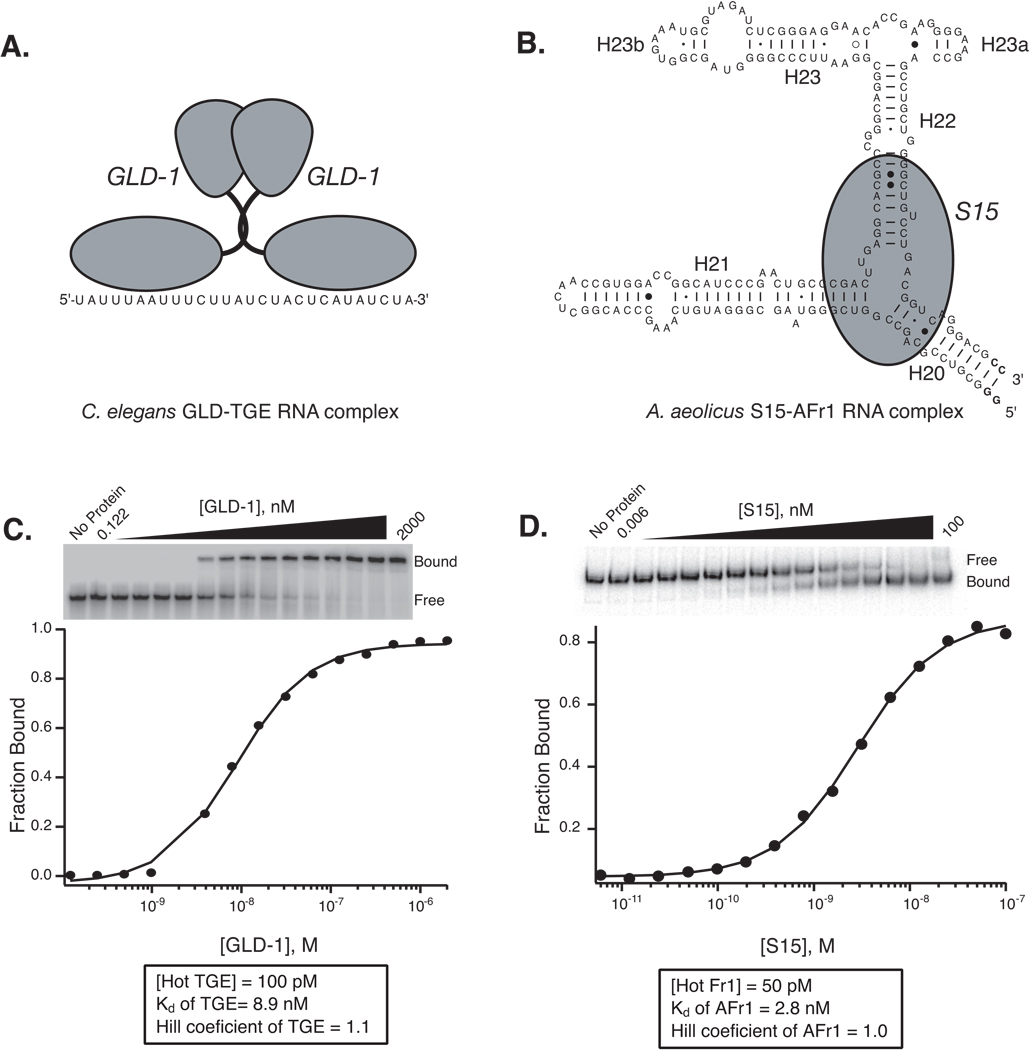
Fig 1.
Measurement of the equilibrium dissociation constant by gel mobility shift assay for two divergent systems A. Model of the interaction between C. elegans GLD-1 and TGE RNA. GLD-1 binds to RNA as a pre-formed stable homodimer (9, 20). B. Model of the interaction between A. aeolicus S15 with Afr1 RNA, derived from the central domain of 16S rRNA. C. The binding of recombinant GLD-1 induces a mobility shift in TGE RNA. GLD-1 RNA-binding domain was expressed and purified as a fusion protein with maltose binding protein (MBP) (9). In this experiment, the mobility of the TGE RNA (MW = 8.7 kDa) is retarded by the interaction with homodimeric GLD-1 (MW = 125 kDa). Bound and free RNA populations are labeled. The fraction of bound RNA is plotted as a function of the monomeric concentration of GLD-1. A fit to the Hill equation (1) is shown. The Kd and Hill coefficient for this particular experiment is given. The binding is not cooperative (n = 1), suggesting the GLD-1 dimers bind as a stable, pre-formed unit. D. Gel mobility shift of Afr1 RNA due to S15 binding. In this case, the RNA species is much larger than the protein. Also, the mobility of the RNA is accelerated, likely indicating that a protein dependent conformational change in the RNA structure mediates the shift. As in panel C, the plot of the data and a fit are shown.
1.2 Determination of the equilibrium dissociation constant
A frequent application of the gel mobility shift assay is to probe the competency of a protein to bind to a specific RNA sequence. In this case, labeled RNA is incubated in the presence or absence of the protein and the mobility of bound versus free RNA is compared side by side on a native gel. Differential mobility in the presence of protein is indicative of an interaction between the protein and the RNA (see Fig. 1C for an example of gel retardation; Fig 1D for an example of gel acceleration). The fraction of bound RNA (f) can be determined by measuring the counts present in the bound species and dividing by the total counts present in the lane.
This type of experiment is often performed with a single concentration of protein or cell extract, and is sufficient to provide a quick yes or no answer or to corroborate binding observed by a separate method. However, a single concentration is not suitable to make definitive statements concerning the ability of a protein to discriminate between RNA sequences or to compare the binding properties of different RNA-binding proteins. This is because the fraction of bound RNA is sensitive to changes in protein concentration for only a narrow range surrounding the equilibrium dissociation constant (Kd). For experiments that utilize only one protein concentration, this insensitivity can lead to serious misinterpretations of binding specificity.
The equilibrium dissociation constant for the binding reaction in scheme (1) is defined by equation (2):
| (1) |
| (2) |
where [R], [P], and [RP] are the molar concentrations of free RNA, free protein, and bound RNA-protein complex at equilibrium, respectively. Theoretically, if one could monitor the equilibrium concentrations of all three species, then the Kd could be determined from a single equilibration. However, the gel shift experiment monitors the fraction of bound RNA rather than the free protein concentration. The fraction of bound RNA is related to the Kd by equation (3)
| (3) |
where Pt is the total protein concentration used in the equilibration. This equation assumes that the RNA is in trace, such that [RP] is negligible and [Pt] approximates the free protein concentration at equilibrium. By determining f for multiple values of Pt, the equilibrium dissociation constant can be derived from a non-linear least squares fit. The equilibrium dissociation constant has units of concentration and is a meter of the binding affinity; weak interactions have relatively large values of Kd while strong interactions have smaller values.
1.3 The relationship between the equilibrium dissociation constant and free energy
The equilibrium dissociation constant is directly related to the Gibbs standard free energy change (DDG°) for a dissociation reaction via the following equation (4):
| (4) |
where R is the Gas constant and T is the temperature. Therefore, measurement of the equilibrium dissociation constant defines a thermodynamic parameter necessary to describe the energetics of an interaction. Of particular value, the ability of an RNA-binding protein to discriminate between two sequences can be quantified in energetic terms by calculating the change in the standard free energy change (DDG°) using equation (5):
| (5) |
where Kd1 and Kd2 represent the equilibrium dissociation constants for each sequence. For example, if a protein binds to sequence two five-fold weaker than sequence one, then the corresponding DDG° is approximately equivalent to one kcal/mol at 37°C, the thermodynamic equivalent of a single hydrogen bond in an RNA-protein complex (11).
Determination of the equilibrium dissociation constant of an RNA-protein interaction eliminates the risk that specificity will be misjudged due to unresponsive assay conditions, defines a useful thermodynamic parameter, and provides a quantitative platform to assess the energetic penalty of defined mutations. Below, we present protocols to derive this parameter from direct titration and competition-binding experiments by gel mobility shift. In addition, we include a stoichiometric-binding protocol to assess the active fraction of protein or the ratio of protein to RNA in the shifted complex.
1.4 Direct titration experiments
As mentioned above, the radiolabeled RNA concentration used in the equilibration reactions must be trace in order to satisfy the assumptions in equation (3). Under ideal conditions, the final molar concentration of RNA in each reaction is 10- to 100-fold less than the Kd. However, the acheivable lower limit of the RNA concentration is defined by the specific activity of the radiolabel. In practice, it is unfeasible to utilize less than 10 pM RNA in the binding reactions due to the sensitivity of the phosphorimaging screens. Therefore, interactions with Kd values tighter than 0.1 nM are difficult to analyze quantitatively by this method.
Conversely, weak interactions are difficult to measure because significant dissociation of the ribonucleoprotein complex can occur during the time that it takes to load and run the native gel. Dissociation can lead to smearing of the bound species and difficulty in quantitation. A good rule of thumb is that equilibrium dissociation constants greater than 1–3 µM typically cannot be accurately determined by standard gel mobility shift. Weaker interactions can be monitored using the competition experiments described below or by using polyacrylamide coelectrophoresis (PACE), a variant of the gel shift where the ligand is polymerized into the gel matrix. PACE has been reviewed elsewhere, and as such will not be described further here (12).
For an initial experiment, it is usually safe to design the experiment assuming that the binding constant will fall between 1 and 100 nM. However, the assay conditions must eventually be optimized once an initial determination of the Kd has been made. The concentration of protein used in the titration should span at least two orders of magnitude above and below the binding constant. The protein concentration stocks should be prepared by serial dilution in order to minimize variation and pipet error that can skew the results. The final concentration series used for the measurement should be chosen to maximize the number of data points within the binding transition. Usually, a 1:1 or 2:1 protein to buffer dilution series works well. The concentrations used in the protocols below are optimized for the interaction of GLD-1 with TGE RNA. A comparison of the gel shifts observed for GLD-1 binding to TGE and S15 binding to Afr1 can be found in figure 1.
1.5 Competition binding experiments
Two common goals in the analysis of an RNA-binding protein are to find the minimal RNA fragment that binds with high affinity and to define the sequence specificity of that interaction by probing a series of RNA mutants. These goals can be met using direct titration if every RNA fragment or mutation to be tested is individually radiolabeled and the gel shift protocol for each species is optimized. This can be an expensive, arduous, and somewhat time-consuming task.
As an alternative, quantitative competition-binding experiments can be performed. These experiments utilize a well-defined shift as the signal to probe the binding competency of unlabeled RNA variants. One advantage of this approach is that several mutants can be characterized using only one labeled species. A second is that weak interactions can be measured since there is no theoretical limit to the amount of competitor that can be added in the equilibration. Competitor RNA is titrated into the labeled complex and the ability of the RNA to disrupt the complex is determined by native gel electrophoresis. The efficiency of unlabeled RNA binding is determined by plotting the fraction of bound radiolabeled RNA as a function of competitor concentration.
If the binding conditions are chosen carefully, the equilibrium dissociation constant of the competitor (Kc) can be determined. The amount of protein included in the equilibration must be below the saturation limit for the Kd of the labeled species. The amount of protein used in the competion experiment should give 60–90% maximal binding of the labeled RNA in the absence of competitor. This is to ensure that competition occurs during the equilibration. Also, calculation of the Kc presumes that the competitor species binds with identical stoichiometry to the same binding site as the labeled species. Before the binding of any mutation or fragment series is measured by competition experiments, self-competion using unlabeled wild-type RNA should be performed to ensure that the Kc compares favorably to the Kd determined by direct titration. The protocol below details experimental conditions for setting up a self-competition binding experiment using the GLD-1/TGE complex and unlabeled TGE RNA as a competitor. Self-competition plots are compared for GLD-1 and S15 in figure 2. Detailed fitting procedures are presented in section 3.5 below.
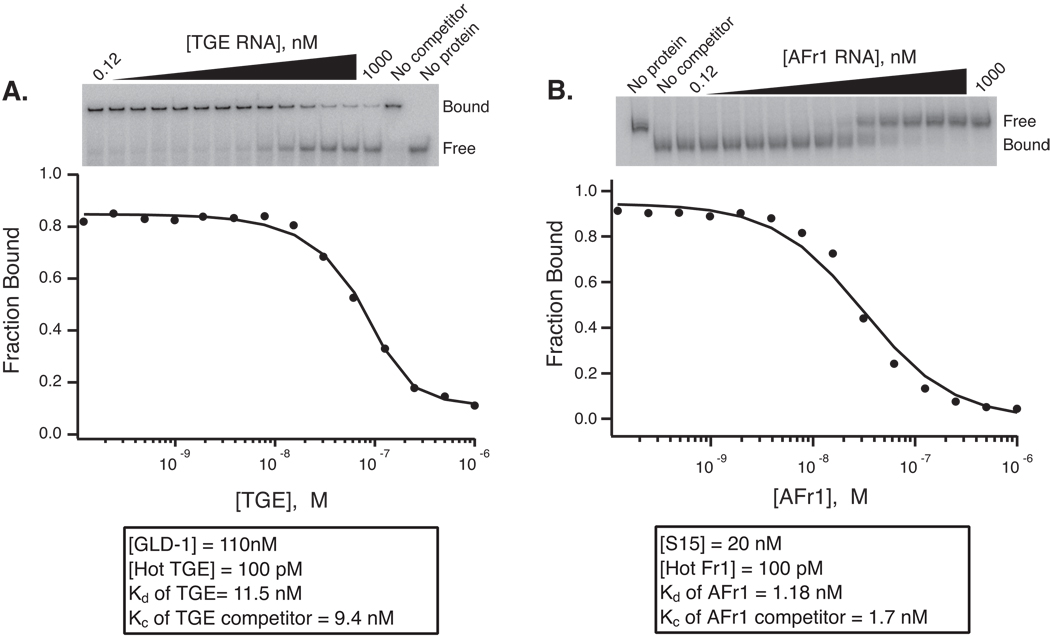
Fig 2.
Competition mobility shift experiments. A. Titration of cold TGE RNA into a GLD-1-TGE complex. A fit of the fraction bound versus competitor RNA concentration is shown. The concentration of GLD-1, labeled TGE RNA, and the Kd and Kc of GLD-1 for TGE RNA are shown. The Kc was determined of a fit to the data using the Weeks and Crothers solution of the Lin and Riggs equation (18, 19). B. Competition of unlabeled AFr1 RNA into an S15-AFr1 complex. The data is plotted and fit as in panel A. In both cases, the Kc is equivalent within error to the Kd determined by direct titration.
1.6 Stoichiometric binding experiments
If the active concentration of protein is known, then the stoichiometry of an RNA-protein interaction can be inferred from gel mobility shift experiments using conditions where the RNA concentration is not in trace. In this experiment, the protein is titrated into RNA of known concentration that has been spiked with a trace amount of radiolabeled RNA. The molar equivalent of protein necessary to saturate binding is determined by measuring the fraction of bound radiolabeled RNA and plotting the data as a function of molar equivalents of protein to RNA. A comparison of the results with theoretical saturation curves defines the stoichiometry of binding. For an accurate determination, the concentration of RNA used in the experiment should be 50 to 100 fold greater than the Kd.
If the active protein concentration is not known, then the interpretation of the above experiment can be ambiguous. If the stoichiometry of binding is known to be 1:1 by another method, then same approach gives an approximation of the active protein concentration. For example, stoichiometric binding of GLD-1 to TGE RNA best matches the 2:1 theoretical curve (Fig. 3). This can indicate that two molecules of GLD-1 bind to this RNA, or it can mean that the protein preparation is only 50% active. In this case, GLD-1 is known to form stable homodimers, consistent with the 2:1 model. In addition, the stoichiometry was validated with ITC (see accompanying paper) using this and a shorter RNA construct in order to distinguish between these two possibilities. In either case, stoichiometric binding experiments serve as a valuable control in defining a model for any system. A protocol for determining binding stoichiometry is given below. The concentrations given are relevant to GLD-1 binding to TGE RNA.
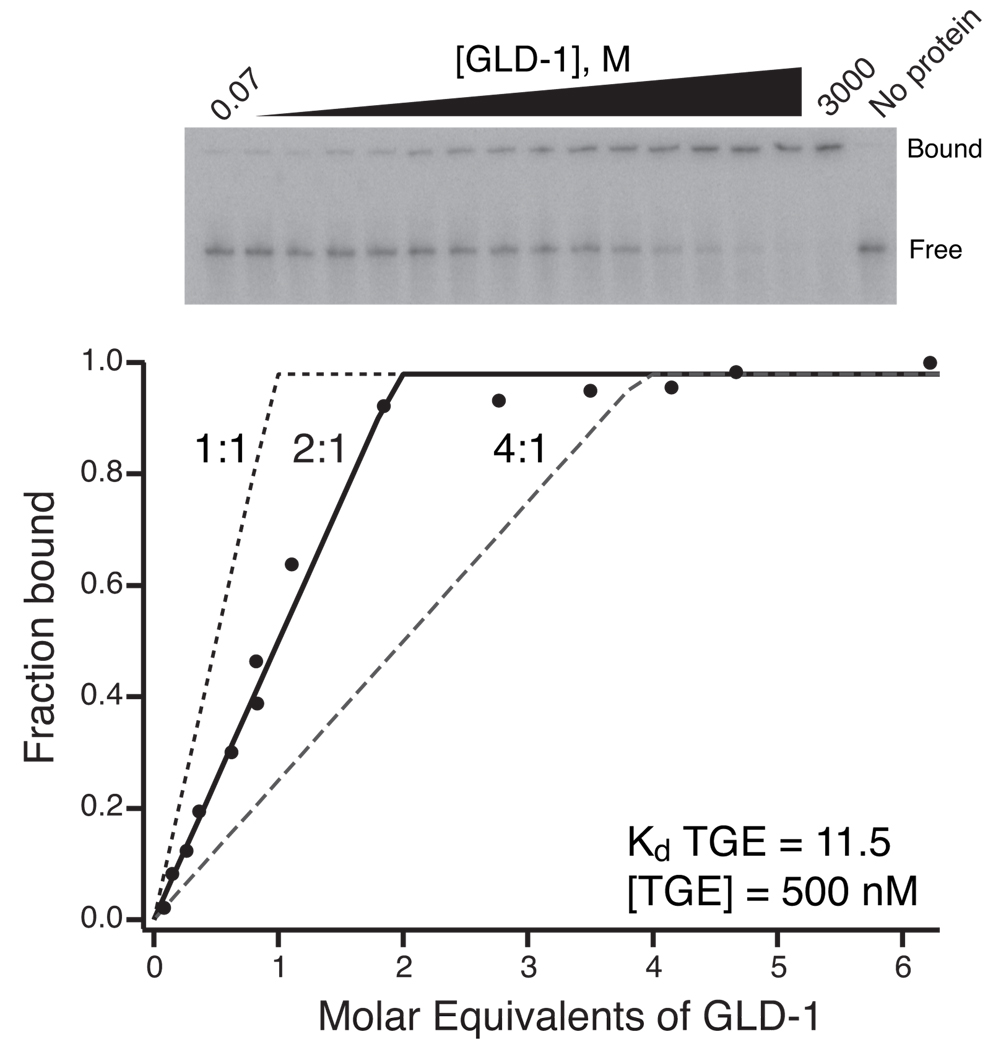
Fig 3.
Stoichiometry shift of GLD-1 binding to TGE RNA. The data are plotted as the fraction of bound labeled RNA versus molar equivalents of GLD-1 to TGE RNA. The total RNA concentration used in the equilibrations and the Kd of GLD-1 for TGE RNA determined by direct titration is given. The data are compared to theoretical saturation curves for 1:1, 2:1, and 4:1 protein to RNA stoichiometry. The 2:1 curve most closely approximates the data, indicating that two copies of GLD-1 interact with a single TGE RNA molecule. This is consistent with literature indicating that this protein forms a stable homodimer (9, 20). This figure is adapted with permission from Ryder et al., 2004 (9).
2. Materials
RNA suitable for quantitative measurements, prepared by chemical synthesis or by in vitro transcription. The molar concentration of the RNA should be measured as accurately as possible with UV spectrophotometry using base-hydrolized RNA (see note 1).
32P-labeled RNA prepared by end- or body-labeling. The molar concentration of labeled RNA should be estimated from the specific activity of the label using liquid scintillation or Cerenkov counting.
RNA-binding protein. The protein should be purified as much as possible and concentrated to at least 20 µM. The concentration of protein should be determined by UV spectroscopy. An online calculator to estimate the extinction coefficient of any peptide sequence based on the method of Gill and von Hippel is available at http://us.expasy.org/tools/protparam.html (13). An estimate of the active protein concentration can be determined by stoichiometric-binding experiments outlined below.
- Buffers and reagents. All buffers should be prepared with distilled, deionized H2O (ddH2O) and filter sterilized prior to use. These buffers may be prepared in advance and stored at −20°C. To minimize ribonuclease contamination, it is safest to work with gloves and certified RNase-free tubes. The contents of the binding buffer, including buffer and pH, ion concentration, detergent, and non-specific competitors should be optimized for each system (see note 2)
- 2X GLD-1 binding buffer: 20 mM Tris-HCl pH 8.0, 50 mM NaCl, 0.2 mM EDTA
- 2X S15 binding buffer: 40 mM K-HEPES, pH 7.6, 660 mM KCl, 20 mM MgCl2, 0.2 mM EDTA
- 1 mg/mL tRNA, prepared from baker’s yeast (Sigma R9001)
- 50 µg/mL heparin, sodium salt (USB 16920)
- 1% (v/v) IGEPAL CA-630 (Sigma I8896)
- 6X loading buffer: 30% (v/v) Glycerol, 0.25% (w/v) xylene cyanol, 0.25% (w/v) bromophenol blue
- Electrophoresis stock solutions. These solutions should be prepared with ddH2O and stored at room temperature. The gel mix is stable for at least one month if stored in an amber bottle, but we usually prepare it fresh immediately before use.
- 10X TBE: 108 g Tris base, 55 g boric acid, and 9.3 g EDTA, tetra-sodium salt dissolved into ddH2O to a final volume of 1L. Dilute a sufficient amount to 0.5X for use as running buffer in a standard vertical electrophoresis apparatus.
- Gel mix (GLD-1): 6% (v/v) 29:1 acrylamide/bis acrylamide solution (available as a 40% stock from Fisher BP1408-1), 0.5X TBE
- Gel mix (S15): 15% (v/v) 29:1 acrylamide/bis acrylamide solution, 0.5X TBE
- 10% (w/v) ammonium persulfate
- N,N,N’,N’ tetramethylethylenediamine (TEMED, Fisher BP150-20)
Electrophoresis equipment, including a power supply that can be used at 4°C.
PhosphorImager and imaging screens or equivalent.
3. Methods
3.1 Binding reactions for direct titration
The final equilibration reactions contain 1X binding buffer, 0.01% IGEPAL CA-630, 0.1 mg/mL of tRNA, 5 µg/mL of heparin, 100 pM radiolabeled RNA, and protein concentrations ranging from 2 µM to 120 pM (see Table 1 for an example, see Note 2 for an explanation of additives used in the equilibration reaction). Typical reaction volumes are 20 µL.
Prepare a master mix (MM) that contains 1.11X concentration of all of the reagents excluding protein and heparin. Enough mix should be prepared for at least sixteen equilibrations. In practice it is wise to prepare excess master mix. The master mix should be heated and then allowed to cool to the equilibration temperature in order to disrupt RNA aggregates and to promote proper folding (see Note 3).
Prepare 30 µL of 20 µM protein in 50 µg/mL heparin to make the master protein (MP) stock. Generate a 1:1 serial dilution series by transfering 15 µL of the master protein stock into 15 µL of 50 µg/mL heparin and mixing by gently pipeting up and down. Repeat in order to generate a series of 10X protein stocks ranging from 20 µM to 1.22 nM.
Aliquot 18 µL of master mix into sixteen RNase-free tubes. To 15 of these, add 2 µL of one of the 10X protein stocks and mix by pipeting up and down. To the last tube, add 2 µL of H2O. This reaction is the no protein control.
Incubate the reactions at a constant temperature until equilibrium is reached. The time necessary to achieve equilibrium depends on the association and dissociation rate constants. Since the apparent association rate depends upon the protein concentration, it takes more time for the reactions with low protein concentrations to reach equilibrium.
Add 4 µL of 6X loading buffer to each tube. Working as quickly as possible, load 5 µL of each reaction onto a pre-run, pre-cleaned native polyacrylamide gel (see section 3.4) at 4°C. Run the gel at 600 V for as long as necessary to achieve separation. The time necessary is highly variable and system dependent. It must be optimized for each RNA or protein construct used. For example, adequate separation of the GLD-1/TGE complex from free TGE RNA is achieved in 30 minutes on a 6% native gel, while separation of S15 bound Afr1 RNA requires at least 15 hours on a 15% gel.
Take down the gel, dry it, and expose the gel to a phosphorimager screen. Quantitate and fit the data as detailed in section 3.5.
Table 1.
Equilibration reactions for a direct titration experiment Lane
| Lane | MM (µL) | Protein Stock (µL) | Dilution | Final [Protein] (nM) | Buffer (µL) | Total Vol (µL) |
|---|---|---|---|---|---|---|
| 1 | 18 | 0 | - | - | 2 | 20 |
| 2 | 18 | 2 | 1:0 | 2000 | 0 | 20 |
| 3 | 18 | 2 | 1:2 | 1000 | 0 | 20 |
| 4 | 18 | 2 | 1:4 | 500 | 0 | 20 |
| 5 | 18 | 2 | 1:8 | 250 | 0 | 20 |
| 6 | 18 | 2 | 1:16 | 125 | 0 | 20 |
| 7 | 18 | 2 | 1:32 | 62.5 | 0 | 20 |
| 8 | 18 | 2 | 1:64 | 31.3 | 0 | 20 |
| 9 | 18 | 2 | 1:128 | 15.6 | 0 | 20 |
| 10 | 18 | 2 | 1:256 | 7.8 | 0 | 20 |
| 11 | 18 | 2 | 1:512 | 3.9 | 0 | 20 |
| 12 | 18 | 2 | 1:1024 | 2.0 | 0 | 20 |
| 13 | 18 | 2 | 1:2048 | 1.0 | 0 | 20 |
| 14 | 18 | 2 | 1:4096 | 0.50 | 0 | 20 |
| 15 | 18 | 2 | 1:8192 | 0.25 | 0 | 20 |
| 16 | 18 | 2 | 1:16,384 | 0.12 | 0 | 20 |
3.2 Competition binding reactions
The final equilibration reactions are identical to the direct titration shift above, except a constant concentration of protein is included and variable concentrations of competitor RNA are used (1X binding buffer, 0.01% IGEPAL CA-630, 0.1 mg/mL of tRNA, 5 µg/mL of heparin, 100 pM radiolabeled RNA, 100 nM GLD-1, and cold RNA concentrations ranging from 1 µM to 120 pM). Again, the usual reaction volume is 20 µL.
Prepare a master mix (MM) that contains 1.25X concentration of all of the reagents excluding protein and heparin. Heat and cool the mix to fold the labeled RNA as necessary. Be sure to prepare enough mix for at least 16 reactions.
Prepare a master protein (MP) mix that contains a 10X concentration of the protein dissolved in 50 µg/mL of heparin. Enough MP solution should be prepared for at least sixteen equilibrations.
Prepare 30 µL of 10 µM competitor RNA to make the master competitor (MC) stock. Heat and cool this mix in order to fold the competitor RNA as necessary. Prepare a series of 10X stocks by serial dilution of 15 µL of the MC stock into an equal volume of H2O to generate a range from 10 µM to 1.22 nM.
Add 0.125 volumes of the MP solution to one volume of the MM solution to generate the master mix plus protein (MMP) stock. Be sure to retain at least 16 µL of the MM solution to use in the no protein control equilibration
Aliquot 18 µL of MMP solution into 15 individual tubes. To a separate tube, add 16 µL of MM solution. Add 2 µL of one of the 10X competitor stocks to 14 of the 15 MMP tubes and mix by pipeting up and down. To the remaining MMP tube, add 2 µL of H2O and mix. This is the no competitor control. To the MM tube, add 2 µL of 50 µg/mL heparin stock and 2 µL of H2O. This is the no protein control.
Follow steps five through seven for the direct titration protocol in section 3.1 above.
3.3 Stoichiometric binding reactions
Stoichiometric binding reactions are set up exactly like the direct titration binding reactions except a constant concentration of unlabeled RNA is included in each equilibration. The final concentrations for GLD-1 binding to TGE RNA are 1X binding buffer, 0.01% IGEPAL CA-630, 0.1 mg/mL of tRNA, 5 µg/mL of heparin, 100 pM radiolabeled RNA, 500 nM cold RNA, and protein concentrations ranging from 4 µM to 0.07 µM. Once again, typical binding reaction volumes are 20 µL.
Prepare a master mix (MM) at 1.33X concentration including every reagent except for protein and heparin. Fold the RNA by heating and cooling as necessary.
Prepare 60 µL of 16µM (4X) master protein (MP) stock by diluting the protein into 50 µg/mL heparin. Generate a 3:1 protein to buffer serial dilution series by pipeting 45 µL of the MP stock into 15 µL of 50 µg/mL heparin and mixing carefully. This gives a series of 4X stocks that will span the range of 0.14 to 8 molar equivalents of protein to RNA in the final equilibrations
Aliquot 15 µL of MM into 16 tubes and add 5 µL of one of the 4X mP stocks to all but one of the tubes. To the remaining tube, add 5 µL of 50 µg/mL heparin to serve as the no protein control.
Follow steps 5–7 from the direct titration protocol in section 3.1.
3.4 Native gel electrophoresis
Central to the gel mobility shift assay is the native gel. Native gels are differentiated by the lack of denaturant, such as urea, formamide, or sodium dodecyl sulfate (SDS). The primary assumption of the gel mobility shift assay is that the binding reaction is quenched upon entrance into the gel. In other words, the concentration of bound and free RNA should not change after loading. The veracity of this assumption depends on the dissociation rate constant of the interaction and the “caging effect”, which has been reviewed extensively elsewhere and will not be considered further here (14).
For the purpose of this protocol, we will describe the preparation of a 29:1 acrylamide to bis-acrylamide, 0.5X TBE native polyacrylamide gel for use in a vertical gel apparatus. This gel substrate is relevant to many binding interactions including both GLD-1 and S15, and the separation efficiency can be easily tuned by the modulating the percentage of acrylamide. We typically pour gels between glass plates that are 35 cm wide by 25 cm tall (inner plate, outer plate is 27 cm) using 0.05 cm teflon spacers and run the gel with an Owl Scientific ADJ3 or equivalent apparatus. The gel can be poured in advance or while the reaction is coming to equilibrium.
Clean and assemble the glass plates and the spacers. Use binder clips to hold the plates together.
Prepare 50 mL of gel mix using the recipe in the materials section above.
Add 1/1000th volume of TEMED, 1/100th volume of 10% APS to the gel mix and mix thoroughly.
Working quickly, pour the mix into the space between the plates being careful to avoid air bubbles. If the plates are placed on a flat surface, the gel can be poured without tape by applying the gel mix as a bead across the top of the plate and displacing air from the bottom. Alternatively, the sides and bottom can be taped and the gel poured while at an angle.
Prior to polymerization, insert a comb into the top of the gel. We use combs with 36 teeth that are 0.5 cm wide by 1.0 cm tall and are spaced 0.3 cm apart. These combs enable two simultaneous 16-point titrations on a single gel. Wait until polymerization is complete, approximately 15 minutes.
Remove the comb and assemble the gel into the apparatus in a 4°C room. Using a syringe, clear out bubbles and unpolymerized acrylamide from the wells. Add chilled 0.5X TBE running buffer to the top and bottom buffer chamber, and pre-run the gel at 600 volts until the temperature has equilibrated (at least one hour).
Immediately before loading, turn down the power supply to 100 volts and re-clean the wells. Be careful to avoid coming into contact with both buffer chambers as current is flowing through the gel. Once all of the lanes have been loaded, turn the power supply back up to 600 volts. Run the gel as long as necessary in order to achieve separation, which unfortunately must be determined empirically.
Remove the gel from the apparatus and empty the buffer chambers. Make sure that the lower buffer chamber is not radioactive. If it is, then carefully remove the bottom buffer and dispose of in an approved container. Separate the plates, transfer the gel to Whatman filter paper, cover with saran wrap and dry with a gel drying apparatus.
3.5 Data analysis
Each of the fitting methods below requires determination of the fraction of bound RNA. This is most easily achieved using phosphorimaging plates. Other methods do exist but are less accurate, and have been reviewed previously (15). The dried gel should be exposed to the phosphorimaging plate for at least one overnight period, however, low concentrations of labeled RNA or a low specific activity may necessitate longer exposure times. The fraction of bound RNA is determined by measuring the volume of counts in both the bound and free species using the software associated with the phosphorimager. The fraction bound equals the counts in the bound population divided by the sum of the bound plus free counts.
Direct titration reactions should be plotted as fraction of bound RNA versus the protein concentration. Displaying the protein concentration axis on the log scale facilitates visual inspection of the data. For bimolecular interactions, the midpoint of the transition is the apparent Kd. The transition should span two orders of magnitude in protein concentration. If this is not the case, then binding may be proceeding by a more complex mechanism or equilibrium may not have been reached during the incubations.
The data can be fit to equation (3) using non-linear least squares methods. In practice, it is usually necessary to fit the data to a modified form of the Hill equation (6) that can compensate for deviations from ideal conditions, including incomplete binding due to an inactive population of RNA, loss of protein sample at low concentrations, cooperative binding, oligomerization, or other more complex mechanisms of binding (16). The modified Hill equation is shown below
| (6) |
where m and b are normalization factors that represent the fraction of bound RNA at the upper and lower asymptotes of the titration and n is the Hill coefficient. The Hill coefficient is a measure of the cooperativity of binding, and for bimolecular association of protein and RNA its value is one. Deviations from unity may indicate cooperative binding of multiple proteins or it may indicate that the binding reactions have not reached equilibrium. Small deviations from integer values are common and are usually caused by some protein or RNA sticking to the equilibration vessel.
We favor IGOR (Wavemetrics) software, which incorporates the Levenberg-Marquadt algorithm and statistical analysis tools into a convenient graphical rendering program (17). Using this software, the fit error can be estimated by weighting the data such that the c2 value is equivalent to n-1 the number of data points used in the fit. The final reported value of the equilibrium dissociation constant should be an average of three independent determinations. If the data are clean, then the standard deviation of three determinations should be greater than the estimated error of each individual fit.
Competition reactions should be plotted as the fraction of bound labeled RNA versus the concentration of unlabeled competitor. The concentration of RNA that disrupts 50% of the bound labeled complex (IC50) can be derived from a fit of the data to a sigmoidal dose-response curve (7)
| (7) |
where C is the concentration of unlabeled competitor. If the protein concentration, labeled RNA concentration, and equilibrium dissociation constant of the labeled species are known to a high degree of accuracy, then the IC50 can be converted into the equilibrium dissociation constant for the competitor (Kc) using the Lin and Riggs equation (8) (18)
| (8) |
where P is the protein concentration, R is the labeled RNA concentration, and Kd is the equilibrium dissociation constant of the labeled RNA. In practice, it is more efficient to fit the data to a quadratic solution of the Lin and Riggs equation (9) determined by Weeks and Crothers (19)
| (9) |
where each of the fitted parameters can be held constant, constrained, or allowed to float during the fit as is appropriate. The Lin and Riggs expression assumes that the competitor binds with the same stoichiometry to the same site as the labeled species. If this is not the case, then this correction should not be used.
Stoichiometric binding data should be plotted as the fraction of bound RNA versus the molar equivalent of protein to RNA in each reaction. On a linear scale, the data should appear as two lines that intersect at a defined point. The value of the x-axis at this point is the apparent stoichiometry of protein to RNA in the complex. For bimolecular association, this value should be one molar equivalent. A value of two may indicate that two copies of the protein are bound to the RNA in the shifted species, as is the case with homodimeric GLD-1 binding to TGE RNA, or it may indicate that 50% of the protein is not active. If the data appears to form a curved line rather that two distinct lines, then it is likely that insufficient cold RNA was used in the equilibration. The intersection of the lines can be derived from linear fits of each phase of the data or by comparison to theoretical saturation curves as in figure 3.
Acknowledgements
The authors thank Ivan Baxter for the alkaline hydrolysis protocol and Dana Abramovitz for generation of the GLD-1 expression construct. S.P.R. is supported by a Damon Runyon Cancer Research Foundation Fellowship (DRG-1723). M.I.R was supported by a Research Scholar grant from the American Cancer Society (#PF-01-087-01-GMC). This research was supported by grants from the National Institutes of Health (NIH, GM53320 and GM53757).
Footnotes
- Aliquot 2 µL of the RNA sample into three 0.5 ml microcentrifuge tubes.
- Aliquot 2 µL of ddH2O each into three more tubes.
- Add 8 µL of 1 M NaOH into each tube.
- Incubate the tubes at 37° C for at least one hour.
- Add 8 µL of 1 M HCl to each tube.
- Add 282 µL of ddH2O.
- Using the water tubes as blanks, take the absorbance of each solution at 260 nm.
- Average the three readings and use that value to determine the concentration.
Several additives may be used in equilibration reactions in order to improve the quality of the shift. It is often useful to include tRNA, heparin, or salmon sperm DNA to inhibit non-specific binding and serve as a carrier nucleic acid to inhibit sample sticking to tubes, pipet tips, and the wells of the gel. Detergent can help to solubilize the protein at high concentration and may help the ribonucleoprotein particles enter the gel matrix. If the protein has a tendency to stick to surfaces, then bovine serum albumin (BSA) can be included in the titration to minimize this effect. The identity and concentration of the additives used in the equilibration should be optimized for each system. The additives used in this protocol are generally useful and work well for both S15 and GLD-1.
Storage of RNA in a −20°C freezer increases its usable life span. Unfortunately, it also promotes denaturation of RNA structure and aggregation. Therefore, it is usually necessary to refold RNA prior to use in gel mobility shift experiments. There are several protocols for folding RNA, including heating the RNA to 65°C or warmer followed by slow-cooling to assay temperature. Other techniques include heating the RNA and snap cooling on ice and prolonged incubation of RNA at an elevated temperature. The folding of some highly structured RNA species is promoted by monovalent and divalent cations, which may be added to the folding reaction. The state of the folded RNA can be assessed on a native gel. Typically, the unbound RNA should migrate through the gel as a single species. Multiple bands may indicate that the RNA is not properly folded or that it has begun to degrade.
References
- 1.Dahlberg AE, Dingman CW, Peacock AC. Electrophoretic characterization of bacterial polyribosomes in agarose-acrylamide composite gels. J Mol Biol. 1969;41:139–147. doi: 10.1016/0022-2836(69)90131-4. [DOI] [PubMed] [Google Scholar]
- 2.Schaup HW, Green M, Kurland CG. Molecular interactions of ribosomal components. I. Identification of RNA binding sites for individual 30S ribosomal proteins. Mol Gen Genet. 1970;109:193–205. doi: 10.1007/BF00267007. [DOI] [PubMed] [Google Scholar]
- 3.Murphy FL, Wang YH, Griffith JD, Cech TR. Coaxially stacked RNA helices in the catalytic center of the Tetrahymena ribozyme. Science. 1994;265:1709–1712. doi: 10.1126/science.8085157. [DOI] [PubMed] [Google Scholar]
- 4.Samuels ME, Bopp D, Colvin RA, Roscigno RF, Garcia-Blanco MA, Schedl P. RNA binding by Sxl proteins in vitro and in vivo. Mol Cell Biol. 1994;14:4975–4990. doi: 10.1128/mcb.14.7.4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yakhnin AV, Trimble JJ, Chiaro CR, Babitzke P. Effects of mutations in the L-tryptophan binding pocket of the Trp RNA-binding attenuation protein of Bacillus subtilis. J Biol Chem. 2000;275:4519–4524. doi: 10.1074/jbc.275.6.4519. [DOI] [PubMed] [Google Scholar]
- 6.Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 7.Batey RT, Williamson JR. Interaction of the Bacillus stearothermophilus ribosomal protein S15 with 16 S rRNA: I. Defining the minimal RNA site. J Mol Biol. 1996;261:536–549. doi: 10.1006/jmbi.1996.0481. [DOI] [PubMed] [Google Scholar]
- 8.Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- 9.Ryder SP, Frater L, Abramovitz DL, Goodwin EB, Williamson JR. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat Struct Mol Biol. 2004;11:20–28. doi: 10.1038/nsmb706. [DOI] [PubMed] [Google Scholar]
- 10.Recht MI, Williamson JR. Central Domain Assembly: Thermodynamics and Kinetics of S6 and S18 Binding to an S15-RNA Complex. J Mol Biol. 2001;313:35–48. doi: 10.1006/jmbi.2001.5018. [DOI] [PubMed] [Google Scholar]
- 11.delCardayre SB, Raines RT. A residue to residue hydrogen bond mediates the nucleotide specificity of ribonuclease A. J Mol Biol. 1995;252:328–336. doi: 10.1006/jmbi.1995.0500. [DOI] [PubMed] [Google Scholar]
- 12.Cilley CD, Williamson JR. PACE analysis of RNA-peptide interactions. Methods Mol Biol. 1999;118:129–141. doi: 10.1385/1-59259-676-2:129. [DOI] [PubMed] [Google Scholar]
- 13.Gill S, von Hippel P. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 14.Cann JR. Phenomenological theory of gel electrophoresis of protein-nucleic acid complexes. J Biol Chem. 1989;264:17032–17040. [PubMed] [Google Scholar]
- 15.Setzer DR. Measuring equilibrium and kinetic constants using gel retardation assays. Methods Mol Biol. 1999;118:115–128. doi: 10.1385/1-59259-676-2:115. [DOI] [PubMed] [Google Scholar]
- 16.Hill AV. The possible effects of the aggregation of the molecules of haemoglobin on its oxygen dissociation curve. J Physiol (London) 1910;40:4–7. [Google Scholar]
- 17.Marquardt D. An algorithm for least-squares estimation of nonlinear paramaters. J Soc Ind Appl Math. 1963;11:431–441. [Google Scholar]
- 18.Lin SY, Riggs AD. Lac repressor binding to non-operator DNA: detailed studies and a comparison of equilibrium and rate competition methods. J Mol Biol. 1972;72:671–690. doi: 10.1016/0022-2836(72)90184-2. [DOI] [PubMed] [Google Scholar]
- 19.Weeks KM, Crothers DM. RNA binding assays for Tat-derived peptides: implications for specificity. Biochemistry. 1992;31:10281–10287. doi: 10.1021/bi00157a015. [DOI] [PubMed] [Google Scholar]
- 20.Chen T, Damaj BB, Herrera C, Lasko P, Richard S. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: role of the KH domain. Mol Cell Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]