Abstract
An important facet of transcriptional repression by Polycomb repressive complex 1 (PRC1) is the mono-ubiquitination of histone H2A by the combined action of the Posterior sex combs (Psc) and Sex combs extra (Sce) proteins. Here, we report that two ubiquitin-specific proteases, USP7 and USP11, co-purify with human PRC1-type complexes through direct interactions with the Psc orthologues MEL18 and BMI1, and with other PRC1 components. Ablation of either USP7 or USP11 in primary human fibroblasts results in de-repression of the INK4a tumour suppressor accompanied by loss of PRC1 binding at the locus and a senescence-like proliferative arrest. Mechanistically, USP7 and USP11 regulate the ubiquitination status of the Psc and Sce proteins themselves, thereby affecting their turnover and abundance. Our results point to a novel function for USPs in the regulation and function of Polycomb complexes.
Keywords: chromatin, INK4a, Polycomb, transcription, ubiquitination
Introduction
Ubiquitination of chromosome-associated proteins is important for many aspects of DNA repair and transcriptional regulation (Vissers et al, 2008; Weake and Workman, 2008). In yeast, for example, dynamic cycles of ubiquitination and de-ubiquitination of H2B appear to be a pre-requisite for active transcription, whereas in higher eukaryotes, mono-ubiquitination of H2A on lysine 119 is associated with transcriptional repression. There is, therefore, a great deal of interest in the enzymes that catalyse the addition and removal of these histone marks. The first H2A ubiquitin ligases to be identified belonged to the Polycomb group (PcG) of transcriptional repressors, which are important for developmental patterning and maintenance of pluripotency (Sparmann and van Lohuizen, 2006; Schwartz and Pirrotta, 2007). Originally characterised in Drosophila, PcG proteins participate in two distinct multi-protein complexes termed Polycomb repressive complex 1 (PRC1) and PRC2. PRC2 functions as a histone methyl transferase that trimethylates histone H3 on lysine 27 (H3K27me3) (Cao and Zhang, 2004). This mark acts as a recruiting platform for the PRC1 or maintenance complex, which catalyses the ubiquitination of histone H2A on Lys119 (de Napoles et al, 2004; Wang et al, 2004; Sparmann and van Lohuizen, 2006; Schwartz and Pirrotta, 2007).
In Drosophila, the PRC1 complex is composed of stoichiometric amounts of the Polycomb (Pc), Posterior sex comb (Psc), Polyhomeotic (Ph) and Sex combs extra (Sce) proteins (Shao et al, 1999; Saurin et al, 2001). Expansion of the PcG genes has led to a more complicated picture in mammalian cells in which there are five Pc (CBX2, CBX4, CBX6, CBX7 and CBX8), six Psc (MEL18, BMI1, MBLR, RNF159, NSPC1 and RNF3), three Ph (HPH1, HPH2 and HPH3) and two Sce (RING1 and RING2) orthologues (Gil and Peters, 2006; Whitcomb et al, 2007). However, experiments in mammalian cells have established that the ubiquitin ligase activity of the complex is attributable to the combined action of the Psc and Sce proteins (Cao et al, 2005; Ben-Saadon et al, 2006; Buchwald et al, 2006; Li et al, 2006; Elderkin et al, 2007; Wu et al, 2008).
As well as homeotic and lineage-specific transcription factors, one of the critical targets of PcG-mediated repression in mammals is the INK4a/ARF locus (Sharpless, 2005; Gil and Peters, 2006; Sparmann and van Lohuizen, 2006). This unusual locus encodes two unrelated tumour suppressor proteins: p16INK4a, which activates the pRb pathway by inhibiting the cyclin D-dependent kinases CDK4 and CDK6, and p14ARF (p19Arf in mice), which activates the p53 pathway by inhibiting the MDM2 ubiquitin ligase. The INK4a/ARF locus has an important function in cellular senescence, the state of irreversible growth arrest engaged by various forms of stress, including telomere erosion, DNA damage, cytotoxic drugs and oncogenic signalling (Campisi and d'Adda di Fagagna, 2007; Collado et al, 2007). Importantly, cellular senescence is now recognised as a major facet of tumour suppression in vivo (Collado and Serrano, 2006) and has recently been shown to impair the reprogramming of somatic cells into pluripotent stem cells (Banito et al, 2009; Hong et al, 2009; Kawamura et al, 2009; Li et al, 2009; Marion et al, 2009; Utikal et al, 2009).
There are several lines of evidence linking PcG proteins to the regulation of INK4a/ARF. For example, mice lacking particular PcG genes display defects in stem and progenitor cell renewal that is attributable to de-repression of Ink4a/Arf (Jacobs et al, 1999; Lessard and Sauvageau, 2003; Molofsky et al, 2003; Park et al, 2003; Isono et al, 2005; Cales et al, 2008). In human fibroblasts, on the other hand, ectopic expression and shRNA-mediated knockdown approaches have implicated a number of PRC1 components, notably CBX7, CBX8, BMI1, MEL18 and RING2, as regulators of INK4a (Jacobs et al, 1999; Itahana et al, 2003; Gil et al, 2004; Dietrich et al, 2007; Kotake et al, 2007; Maertens et al, 2009). Indeed, we recently reported that several distinct PRC1 complexes bind simultaneously and interdependently to the same region of the locus (Maertens et al, 2009).
To try to understand the reasons for this complexity, we affinity purified the PRC1 complexes formed by the Psc proteins MEL18 and BMI1. In addition to known PcG proteins, we found two de-ubiquitinating enzymes, USP7 and USP11. Here, we show that USP7 and USP11 are associated with chromatin and bind to several PRC1 components in high-molecular-weight complexes. Importantly, shRNA-mediated knockdown of either USP results in de-repression of INK4a and displacement of PRC1 proteins from the locus. Knockdown of USP7 or USP11 causes increased turnover of chromatin-bound MEL18 and BMI1, whereas over-expression of the USPs reduces the levels of mono- and poly-ubiquitination of these proteins. Our findings underscore the importance of ubiquitination in the regulation and function of Polycomb complexes and provide new insights into the function of de-ubiquitinases in these events.
Results
Two de-ubiquitinating enzymes co-purify with MEL18 and BMI1
To gain a better insight into the mechanisms underlying PRC1-mediated repression, we isolated human PRC1 complexes by tandem affinity purification of the Psc homologues, MEL18 and BMI1, and identified co-purifying proteins by mass spectrometry. A full list of the identified proteins can be found in Supplementary Table S1 and images of the Coomassie-stained gels have appeared elsewhere (Elderkin et al, 2007; Maertens et al, 2009). As previously reported, the MEL18- and BMI1-based PRC1 complexes contained several known PcG proteins, the most prominent of which were CBX8, the short isoform of HPH2, and RING1 and RING2 (Elderkin et al, 2007). Importantly, MEL18 and BMI1 did not co-purify with one another and in several independent preparations as well as reconstruction experiments, we have never observed more than one representative of the Pc, Psc, Ph or Sce families within the same soluble complex (Maertens et al, 2009). As anticipated for PRC1 function, the purified MEL18 and BMI1 complexes were capable of ubiquitinating H2A on K119 (Elderkin et al, 2007). It was, therefore, intriguing to find that two ubiquitin-specific proteases, USP7 and USP11, co-purified in the MEL18 complex and that USP11 was also present in the BMI1 complex.
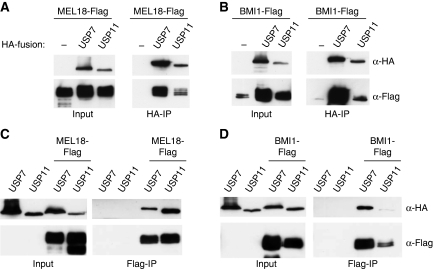
Although neither of these proteins was detected in the negative control (parallel purification from cells expressing the TAP-only vector), it was important to confirm their association with PcG proteins. To this end, HA-tagged versions of USP7 and USP11 were transiently co-expressed with Flag-tagged versions of MEL18 and BMI1 in HEK293T cells. Cell lysates were then analysed directly, to establish the expression levels of the input proteins, or immunoprecipitated with monoclonal antibodies against the HA or Flag epitopes (Figure 1). As indicated, Flag-tagged MEL18 (Figure 1A) and BMI1 (Figure 1B) were precipitated with anti-HA beads only when HA-tagged USP7 or USP11 were co-expressed. In the reciprocal precipitation of Flag-tagged MEL18 (Figure 1C) or BMI1 (Figure 1D) with anti-Flag beads, HA-tagged USP7 and USP11 were co-precipitated only when either MEL18 or BMI1 were present. Thus, although no USP7-derived peptides were detected by mass spectrometry of the gel bands that co-purified with TAP-tagged BMI1, we could show that these two proteins do associate when ectopically expressed.
Figure 1.
USP7 and USP11 interact with MEL18 and BMI1. (A, B) 293T cells were transiently transfected with either MEL18-Flag (A) or BMI-Flag (B) alone or together with HA-USP7 or HA-USP11. Cell lysates were analysed directly (Input) or after immunoprecipitation with HA antibody and immunoblotted for HA and Flag. (C, D) After co-expression of either MEL18-Flag (C) or BMI1-Flag (D) with HA-USP7 or HA-USP11, the cell lysates were analysed directly (Input) or after immunoprecipitation with Flag antibody and immunoblotted for HA and Flag.
USP7 and USP11 associate with other PRC1 components
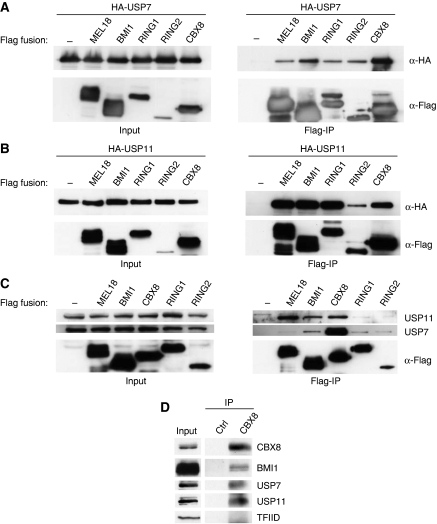
Although the co-precipitation data implied that both USP7 and USP11 can interact with MEL18 and BMI1, they did not address whether this occurred within the context of a canonical PRC1 complex. We, therefore, investigated whether the HA-tagged USPs were able to interact with Flag-tagged versions of CBX8, RING1 and RING2; BMI1 and MEL18 served as positive controls with Flag alone as a negative control. HA-USP7 (Figure 2A) and HA-USP11 (Figure 2B) were efficiently co-precipitated with all PRC1 proteins tested. In the reciprocal immunoprecipitations, we confirmed that CBX8 and RING1 co-precipitated with HA-tagged USP7 and USP11, but only detected RING2 in the HA-USP7 precipitate (Supplementary Figure S1). Note that Flag-RING2 was expressed at significantly lower levels compared with Flag-RING1 or Flag-CBX8 (Figure 2B), which might explain the poor recovery in the HA immunoprecipitates. Importantly, by using a commercial USP7 antibody and a rabbit antiserum that we generated against USP11, it was possible to confirm that the endogenous USP7 and USP11 in 293T cells co-precipitated with Flag-tagged PRC1 proteins (Figure 2C).
Figure 2.
USP7 and USP11 bind to PRC1 components. (A, B) Co-precipitation of HA-USP7 (A) and HA-USP11 (B) with Flag-tagged MEL18, BMI1, CBX8, RING1 and RING2 from transiently transfected 293T cells. Input samples are shown in the left panels, and the Flag IPs in the right panels. (C) Co-precipitation of endogenous USP7 and USP11 with transiently expressed Flag-tagged MEL18, BMI1, CBX8, RING1 and RING2. (D) Co-precipitation of endogenous CBX8, BMI1, USP7 and USP11 from nuclear extracts of 293T cells using an affinity-purified CBX8 antiserum immobilised on Sepharose beads.
In these co-precipitation analyses, the interactions with CBX8 seemed particularly robust, and in a direct comparison between Pc family members, USP7 displayed a clear preference for CBX8, whereas USP11 showed the strongest binding to CBX6 and CBX8 (Supplementary Figure S2). Taking a cue from these observations, we used an affinity-purified antiserum (Maertens et al, 2009) to precipitate the endogenous CBX8 from nuclear extracts of 293T cells. As shown in Figure 2D, immunoblotting revealed that endogenous USP7 and USP11 co-precipitated with CBX8 under these conditions. BMI1 and TFIID were used as positive and negative controls, respectively.
USP7 and USP11 are bound to chromatin and form part of the PRC1 complex
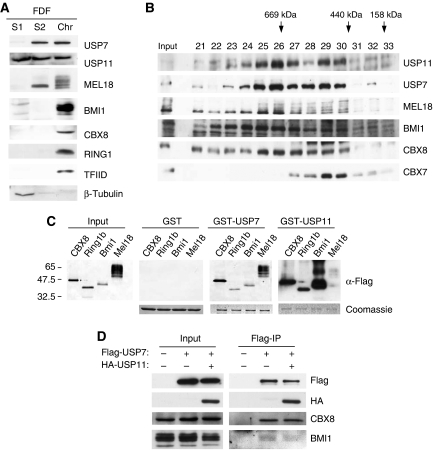
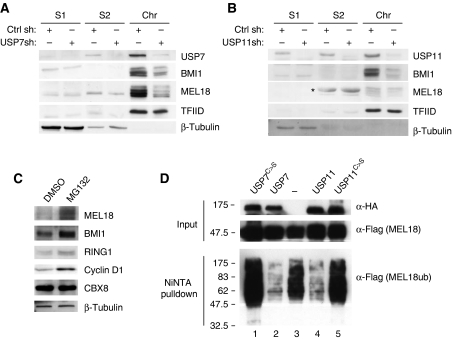
To consolidate the idea that these interactions take place in the context of PRC1 complexes, we next asked whether endogenous USP7 and USP11 have an appropriate intracellular localisation. These experiments were conducted in foetal dermal fibroblasts (FDFs), in which the functions of PRC1 complexes are less likely to have been perturbed (Figure 3A) and in 293T cells (not shown). A standard cell fractionation protocol was used to recover cytosolic (S1), nucleoplasmic (S2) and chromatin fractions (Chr), and the proteins were analysed by sodium dodecyl sulphate (SDS)–PAGE and immunoblotting. TFIID (TBP) was used as a marker for the chromatin fraction, whereas β-tubulin was used as a control for the soluble fractions, S1 and S2. As anticipated, the bulk of the MEL18, BMI1 and other PRC1 components, such as CBX8 and RING1, were bound to chromatin (Figure 3A). In FDFs, virtually all of the USP7 was found in the nuclear fraction with almost 50% bound to chromatin. In contrast, USP11 was equally distributed in all three fractions. Interestingly, 293T cells showed a somewhat different pattern with a higher proportion of each USP in the soluble fractions. The reasons for these differences remain unclear, but it was previously reported that USP7 and USP11 are nuclear proteins (Ideguchi et al, 2002; Fernandez-Montalvan et al, 2007) and that Drosophila USP7 is bound to polytene chromosomes (van der Knaap et al, 2005). Our data suggested that a substantial proportion but not all of the USP7 and USP11 are in the same nuclear compartment as PRC1 proteins.
Figure 3.
Association of USP7 and USP11 with chromatin and high-molecular weight PRC1-like complexes. (A) Primary human fibroblasts (FDF) were used to prepare cytosolic (S1), nucleoplasmic (S2) and chromatin fractions (Chr) after standard procedures (Wysocka et al, 2001). Equivalent amounts of each fraction were separated by SDS–PAGE and immunoblotted for the indicated proteins. TFIID was used as a control for chromatin and β-tubulin as a non-chromatin protein. (B) Nuclear extracts (∼2 mg) from FDF cells were subjected to gel filtration on a Superose 6 column. Fractions (0.5 ml) were TCA precipitated, separated by SDS–PAGE and immunoblotted for the indicated proteins. Input refers to a sample of the cell extract before gel filtration and numbers above the gel specify relevant fraction numbers. Molecular weight standards (as indicated) were analysed separately on the same column and under the same conditions. (C) GST-pull-down assays using GST alone, GST-USP7 or GST-USP11 as indicated incubated with FlagHis12-tagged mouse Ring1b, Bmi1 and Mel18 produced from baculovirus vectors or human CBX8 expressed in E. coli. The left-hand panel shows the relative input levels of the different target proteins. The Coomassie-stained images below the panels confirm that equal amounts of recombinant protein were present in each pull-down assay. The PRC1 proteins were detected using a rabbit anti-Flag antibody and either alkaline phosphatase-conjugated anti-rabbit IgG (Input, GST- and GST-USP7-bound proteins) or horse radish peroxidase-conjugated anti-rabbit IgG (GST-USP11-bound proteins) as described in Materials and methods. (D) Co-precipitation of ectopically expressed Flag-USP7 and HA-USP11 from nuclear extracts of 293T cells. Endogenous CBX8 and BMI1 served as positive controls.
To assess whether USP7 and USP11 associate with bona fide PRC1 complexes, nuclear extracts from FDFs (Figure 3B) and 293T cells (not shown) were subjected to size exclusion chromatography on a Superose 6 10/300 column, and individual fractions were analysed by SDS–PAGE and immunoblotting. Most of the USP7 and USP11 eluted in high-molecular weight complexes of between 400 and 700 kDa (Figure 3B). The majority of the MEL18, BMI1 and CBX8 were also present in these fractions, whereas CBX7 peaked in complexes of around 400–500 kDa.
USP7 and USP11 bind directly to multiple PcG proteins
In view of the co-precipitation and size fractionation data, it was of interest to know whether USP7 and USP11 interact directly with MEL18 and BMI1 or other PRC1 components. To address this issue, we purified recombinant GST-USP7- and GST-USP11-fusion proteins from bacteria and asked whether they interact with purified FlagHis12-tagged Bmi1, Mel18 and Ring1b produced in insect cells using baculovirus vectors (Cao et al, 2005; Buchwald et al, 2006; Li et al, 2006; Elderkin et al, 2007) or with CBX8-Flag purified from bacteria. Note that our baculovirus constructs encode the mouse homologues of the Psc and Sce proteins, but the findings were confirmed using 35S-labelled versions of human CBX8, BMI1, MEL18, RING1 and RING2 produced by coupled in vitro transcription and translation (Supplementary Figure S3A). Strikingly, both GST-USP7 and GST-USP11 were able to pull down all of the PRC1 components tested, suggesting that they interact directly with multiple proteins (Figure 3C; Supplementary Figure S3A).
Using a series of deletion mutants of MEL18, we further showed that both USP7 and USP11 associate with the amino terminal region of the protein, which includes the RING domain (Supplementary Figure S3B and C). Importantly, USP7 and USP11 have also been shown to associate with one another (Sowa et al, 2009), and we have confirmed this both in transfected cells and with the recombinant proteins (Figure 3D and data not shown). It is, therefore, conceivable that they bind simultaneously within the same complex.
USP7 and USP11 contribute to the regulation of p16INK4a
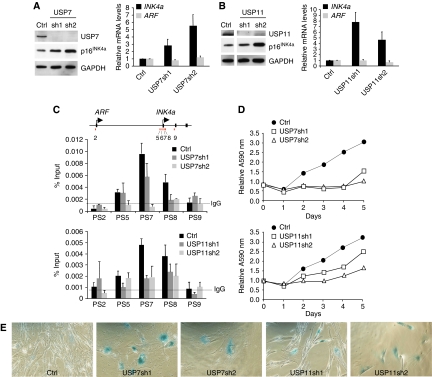
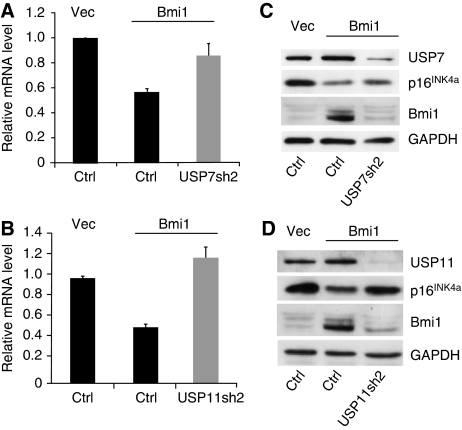
We previously showed that BMI1 and MEL18 associate directly with and are required for the repression of the INK4a tumour suppressor gene in human fibroblasts (Maertens et al, 2009). The presence of USP7 and USP11 in the MEL18 and BMI1 complexes prompted us to ask whether these USPs have a function in the regulation of INK4a. To this end, we ablated the expression of USP7 and USP11 in FDFs by lentiviral delivery of shRNAs targeting their respective transcripts; an shRNA that has no known target in the human genome was used as a negative control. Knockdown of USP7 (Figure 4A) or USP11 (Figure 4B) with two independent shRNAs caused a demonstrable increase in the p16INK4a transcript and protein levels, with no corresponding changes in ARF mRNA; the low levels of p14ARF in FDFs preclude detection with the available antibodies. Importantly, knockdown of either USP7 or USP11 resulted in decreased binding of MEL18 to the INK4a locus, as judged by chromatin immunoprecipitation (Figure 4C), consistent with loss of PcG-mediated repression (Bracken et al, 2007; Dietrich et al, 2007; Maertens et al, 2009). Similar effects were observed for the association of BMI1, CBX8, RING1 and EZH2 with the INK4a locus, as well as the extent of H3K27 trimethylation (Supplementary Figure S4). Cells transduced with the USP shRNAs also displayed characteristics of senescence, including proliferation arrest, an enlarged flattened appearance, single prominent nucleoli and SA-β-Gal activity (Figures 4D, E and 5E).
Figure 4.
Knockdown of USP7 or USP11 increases expression of INK4a. (A, B) Knockdown of USP7 (A) or USP11 (B) in FDF cells using two different shRNAs (sh1 and sh2) leads to up-regulation of p16INK4a protein (left panels) and INK4a RNA (right panels) with no equivalent effect on ARF RNA. (C) Chromatin immunoprecipitation showing that knockdown of USP7 or USP11 results in decreased binding of MEL18 to the INK4a locus. The map of the INK4a-ARF locus and PS numbers show the location of previously described primer sets used to interrogate the precipitated DNA by qPCR (Bracken et al, 2007). IgG shows the maximum background signal obtained using an irrelevant rabbit antibody. (D) FDFs transduced with independent shRNAs against USP7 and USP11 show impaired proliferation relative to cells expressing control shRNA, as judged by crystal violet staining. Each time point represents an average of six replicates and the A595 nm values were normalised to the absorbance at day 0 (first day after plating). (E) SA-β-Gal staining of FDFs expressing control shRNA (Ctrl) or two independent shRNAs against USP7 or USP11.
Figure 5.
Effects of USP7 or USP11 knockdown in INK4a-deficient cells. FDFs or the Leiden strain of INK4a-deficient human fibroblasts (Brookes et al, 2002) were infected with lentiviruses encoding two independent USP7 or USP11 shRNAs (sh1 and sh2) or a control shRNA (Ctrl). The effects on INK4a and the respective target genes were assessed by qRT–PCR (A, B) and immunoblotting (C, D) using an antibody that detects the mutant form of p16INK4a present in Leiden cells. (E) Phase contrast images showing that FDFs transduced with either USP7 or USP11 shRNAs display a senescence-like phenotype with single prominent nucleoli (arrowheads). Whereas Leiden cells transduced with USP7 shRNAs also display this phenotype, Leiden cells expressing USP11 shRNAs continued to proliferate and resembled cells expressing the control shRNA. (F) Semi-quantitative assessment of the senescence phenotype in FDFs and Leiden cells based on the percentage of cells with a single nucleolus.
To assess whether the senescence-like arrest was wholly dependent on the de-repression of INK4a, a similar experiment was conducted in the Leiden strain of human fibroblasts. As we have previously reported, these cells were derived from an individual who is homozygous for a germline mutation that inactivates p16INK4a, but not p14ARF (Brookes et al, 2002). As Leiden cells stain weakly for SA-β-Gal activity (Jacobs et al, 1999; Itahana et al, 2003; Brookes et al, 2004), we used the appearance of single prominent nucleoli as a simple, non-invasive marker of senescence. USP11 knockdown did not cause senescence in these cells, despite the up-regulation of mutant p16INK4a, implying that the effects of USP11 knockdown are p16INK4a dependent (Figure 5). In contrast, knockdown of USP7 with two independent shRNAs caused senescence in both the Leiden and control fibroblasts. The effects of USP7 knockdown would be consistent with its predicted impact on the MDM2-p53 pathway (Li et al, 2002, 2004; Cummins and Vogelstein, 2004; Cummins et al, 2004; Brooks et al, 2007). However, in these experiments, neither the USP7 nor the USP11 shRNAs had any discernible effects on the steady state levels of p53 or p21CIP1 (Supplementary Figure S5).
USP7 and USP11 affect the ability of Bmi1 to repress INK4a
We next asked whether the previously documented ability of mouse Bmi1 to down-regulate INK4a (Jacobs et al, 1999; Itahana et al, 2003; Brookes et al, 2004) is dependent on USP7 or USP11. FDF cells were first infected with a vesicular stomatitis virus G (VSV-G) pseudotyped retroviral vector encoding mouse Bmi1, or an empty vector control (pBABE-Hygro). After selection in hygromycin, the two cell populations were infected with lentiviral vectors encoding shRNAs against either USP7 (USP7sh2) or USP11 (USP11sh2) and selected in puromycin. Non-specific shRNA was included as a negative control. As expected, ectopic expression of Bmi1 in FDFs resulted in down-regulation of p16INK4a at both the protein and RNA level. However, this effect was negated after knockdown of either USP7 (Figure 6A) or USP11 (Figure 6B) and these findings were reproduced with independent shRNAs against each USP (data not shown).
Figure 6.
Knockdown of USP7 or USP11 impairs Bmi1-mediated repression of INK4a. Ectopic expression of mouse Bmi1 in FDFs results in decreased expression of p16INK4a RNA (A, B, middle lanes) and protein (C, D, middle lanes). Knockdown of USP7 (A, C) or USP11 (B, D) reverses the effects of Bmi1 (right lane in each panel).
Effects of USP7 and USP11 on ubiquitination of histones H2A and H2B
The effects of USP7 and USP11 shRNAs on p16INK4a invite different interpretations. One scenario would be that PRC1 proteins recruit the USPs to modulate the ubiquitination of a specific target protein, such as H2A or H2B. Indeed, when tested in vitro, Flag-USP7 was capable of de-ubiquitinating both H2Aub and H2Bub (Supplementary Figure S6A and B). However, Flag-USP11 had no effect on H2Aub or H2Bub under these conditions. We also performed de-ubiquitination assays using nucleosomes purified from cells stably expressing Flag-H2A. The purification was carried out in the presence of iodoacetamide to protect H2Aub from de-ubiquitination during purification. Again, USP7 was capable of de-ubiquitinating H2Aub, but USP11 was not (Supplementary Figure S6C). However, we were unable to establish an equivalent assay for H2B de-ubiquitination because of the relatively low levels of H2Bub in mammalian cells (West and Bonner, 1980). As an alternative approach, we transfected the cells with a plasmid encoding His6-tagged ubiquitin and recovered ubiquitinated proteins on Ni+-agarose beads (Supplementary Figure S6D). Immunoblotting for H2A showed that over-expression of either active or catalytically inactive USP7 or USP11 did not result in a significant change in the levels of H2Aub. There did not appear to be any discernible reduction in the low levels of H2Bub, although there was an obvious increase in the cells expressing USP7C223S, suggesting that it might be having a dominant negative effect. Conversely, we could not observe an increase in H2Aub or H2Bub on knockdown of either USP7 or USP11 in human fibroblasts (data not shown). Taken together, we found no evidence that USP7 or USP11 directly affects the steady state levels of H2Aub or H2Bub.
Knockdown of USP7 or USP11 affects the levels of PRC1 components
An alternative scenario would be that the USPs influence the functionality of the PRC1 complex. A striking feature of the data in Figure 6 was that knockdown of either USP7 or USP11 had a dramatic effect on the total levels of Bmi1, detected with an antibody that recognises both the mouse and human homologues of the protein. This suggested that USP7 and USP11 might directly or indirectly affect the stability of Bmi1.
To focus on the effects of USP knockdown on the endogenous PRC1 proteins, FDFs transduced with USP7 or USP11 shRNAs were used to prepare different subcellular fractions as described above. Knockdown of either USP7 or USP11 caused a substantial decrease in the chromatin-associated pool of BMI1 and MEL18 (Figure 7A and B). This effect was specific for the PRC1 proteins as the association of TFIID with chromatin was unaffected.
Figure 7.
USP7 and USP11 affect the ubiquitination status of MEL18 and BMI1. (A, B) Cellular fractionation was performed as in Figure 3 using FDF cells transduced with control shRNA (Ctrl) and either USP7shRNA (A) or USP11 shRNA (B) as indicated. Ablation of USP7 or USP11 significantly reduced the amount of chromatin-associated BMI1 and MEL18, but had little effect on the distribution of TFIID or β-tubulin. ‘*' Indicates a non-specific band. (C) FDFs were treated with MG132 or the solvent control (DMSO) for 4 h and the levels of the indicated proteins were analysed by immunoblotting. (D) 293T cells that stably express MEL18-Flag were transfected with plasmids encoding His6-ubiquitin alone or in combination with HA-USP7WT, HA-USP7C223S, HA-USP11WT or HA-USP11C275S. Ubiquitinated proteins were recovered on NiNTA beads under denaturing conditions and analysed by SDS–PAGE and immunoblotting. In the absence of USPs, MEL18 is clearly ubiquitinated (lane 3), whereas co-expression of wild-type USP7 or USP11 removes the ubiquitin marks (lanes 2 and 4). The catalytically inactive USP7 and USP11 mutants (USPC>S) were unable to remove the ubiquitin mark from MEL18.
As there did not appear to be a corresponding increase in the soluble pool of BMI1 or MEL18, the most likely scenario would be that knockdown of USP7 or USP11 caused a reduction in the total levels of endogenous BMI1 and MEL18, presumably through increased turnover. To explore this possibility, FDFs were transduced with two independent shRNAs against USP7 and USP11 and the levels of BMI1 and MEL18 protein, and RNA were monitored by immunoblotting and qRT–PCR, respectively. Knockdown of USP7 and USP11 resulted in a reduction in the total BMI1 and MEL18 protein levels with no equivalent change in the transcript levels (Supplementary Figure S7).
This implied that USP7 and USP11 were influencing the stability of MEL18 and BMI1. Treating the FDFs with the proteasome inhibitor MG132 significantly increased the total levels of MEL18 and BMI1 indicating that they are subject to proteasomal degradation (Figure 7C). Although the control for these experiments, cyclin D1, was clearly stabilised by MG132, it was not affected by knockdown of USP7 or USP11 (Supplementary Figure S5), suggesting that the effects were specific.
USP7 and USP11 regulate the ubiquitination status of MEL18 and BMI1
To determine whether USP7 and USP11 were directly affecting the ubiquitination status of Psc proteins, 293T cells that stably express MEL18-Flag were transfected with a plasmid encoding His6-ubiquitin, and the ubiquitinated proteins were purified on NiNTA beads under denaturing conditions (Figure 7D, lane 3). Under these conditions, MEL18 is both mono- and poly-ubiquitinated. Importantly, over-expression of HA-tagged USP7 or USP11 reduced the overall ubiquitination of MEL18 (Figure 7D, lanes 2 and 4). This seemed to be a direct effect because the catalytically inactive point mutants, USP7C223S and USP11C275S, respectively, had little impact on the ubiquitination status of MEL18 (Figure 7D, lanes 1 and 5). Analogous results were obtained for de-ubiquitination of BMI1 and RING1 (Supplementary Figure S8).
Discussion
In this study, we identify two ubiquitin-specific proteases that are physically and functionally associated with human PRC1 complexes and provide novel insights into their modes of action. At first sight, it seemed odd to find two distinct USPs associated with a complex whose principal function is to act as an E3 ubiquitin ligase for histone H2A (Cao et al, 2005; Buchwald et al, 2006; Li et al, 2006; Elderkin et al, 2007). USP7 and USP11 show no obvious similarities in their primary sequence apart from the catalytic domains common to all USPs (Nijman et al, 2005), and although they have been classified within the same group of USPs based on the number and functional diversity of their binding partners (Sowa et al, 2009), there are few overlaps among the interacting proteins.
Curiously, PcG proteins were not identified in these screens, although there have been previous reports that USP7 co-purifies with PRC1 components in both human and insect cells (van der Knaap et al, 2005; Sanchez et al, 2007). Our data strongly suggest that USP7 and USP11 bind directly to several components of the PRC1 complex, as confirmed using purified recombinant proteins. It will be interesting to determine whether the USPs associate simultaneously with representatives of the Pc, Psc and Sce families and if so through which domains, but there is currently very little information about the interactions between PRC1 components. As a first step, we showed that USP7 and USP11 both associate with the amino terminal region of MEL18, which contains the RING domain. This region of MEL18 (residues 1–121) shares 77% similarity with equivalent regions in BMI1, RING1 and RING2, and it is, therefore, possible that the RING domain represents a common binding site for the two USPs. However, it is not clear whether they bind simultaneously within the same complex and the situation is complicated by the fact that USP7 and USP11 also interact with one another (Sowa et al, 2009; Figure 3D). Interestingly, knockdown of USP7 caused a concomitant decrease in the levels of chromatin-bound USP11, underscoring the close association between these two proteins (Supplementary Figure S7C).
The physiological relevance of this interaction remains unclear as USP7 and USP11 can function independently and have different target specificities. Although USP11 has been shown to interact with a diverse array of cellular proteins (Bouwmeester et al, 2004; Yamaguchi et al, 2007), its de-ubiquitination activity has only been shown for a few substrates (Ideguchi et al, 2002; Lin et al, 2008). Here, we show that USP11 is capable of removing ubiquitin from several components of the PRC1 complex, including BMI1, MEL18 and RING1, but had no apparent effect on the ubiquitination status of H2A and H2B under the assay conditions used.
In contrast, recombinant USP7 was able to de-ubiquitinate both H2A and H2B in vitro. Given the precedents in Drosophila (van der Knaap et al, 2005; Sanchez et al, 2007), we initially thought that human USP7 might be contributing to PcG-mediated silencing by reversing the mono-ubiquitination of histone H2B. However, in the in vitro assay, we did not observe specificity for H2Bub over H2Aub. As the purified USP7 used in these experiments was extracted under high salt conditions, it is possible that an essential co-factor, such as GMP synthetase (van der Knaap et al, 2005), was missing. To try to circumvent the possible limitations of in vitro assays, we also asked whether knockdown of USP7 in FDFs would change the global levels of H2Aub and H2Bub. However, we saw no discernible increase in the extent of ubiquitination on these histones (data not shown). Conversely, over-expression of USP7 did not result in a significant decrease in H2Aub and H2Bub levels. As the human genome encodes at least four times as many deubiquitinating enzymes as Drosophila, it is possible that a different USP has assumed the specialised function of regulating the ubiquitination of H2B in human cells.
Similar to USP11, ectopically expressed USP7 was able to reverse the ubiquitination of the PRC1 proteins themselves. Although complexes formed between recombinant BMI1 or MEL18 with RING1 or RING2 become auto-ubiquitinated (Cao et al, 2005; Buchwald et al, 2006; Li et al, 2006; Elderkin et al, 2007; Supplementary Figure S6), it is not clear whether this accounts for the mono- and poly-ubiquitination of these proteins observed in vivo. Other E3 ligases might be involved, such as the CULLIN3/SPOP complex, which has been shown to ubiquitinate BMI1 (Hernandez-Munoz et al, 2005) or the putative E3 ligase HECD1, which was among the proteins identified in our mass spectrometry analyses of the BMI1 complex (Supplementary Table S1).
It is conceivable that mono- and poly-ubiquitination have different effects in this setting, but the simplest explanation of our findings would be that USP7 and USP11 control the ubiquitin-mediated turnover of the PRC1 proteins. Importantly, knockdown of either USP7 or USP11 reduces the total and chromatin-bound pool of BMI1 and MEL18, and the effects can be mitigated by inhibiting proteasome function. From a functional standpoint, this reduces the availability of PRC1 components resulting in de-repression of target genes such as INK4a. Thus, knockdown of either USP7 or USP11 in primary human fibroblasts resulted in loss of PRC1 and PRC2 occupancy at the INK4a promoter and up-regulation of p16INK4a. The effects were consistently observed with independent shRNAs and the obvious phenotypic consequence was a senescence-like proliferative arrest.
In the case of USP11, the arrest appeared to be entirely explained by de-repression of p16INK4a because the Leiden strain of INK4a-deficient fibroblasts were unaffected by USP11 shRNAs. USP11 might participate in DNA damage repair within the BRCA2 pathway (Schoenfeld et al, 2004), but had no apparent effect on p53 (Li et al, 2002; Supplementary Figure S5). In contrast, USP7 (also known as HAUSP), which was originally identified through an interaction with a Herpes simplex virus immediate-early gene (Everett et al, 1997), is credited with a pivotal, but complex function in the interplay between p53 and MDM2 (Li et al, 2002, 2004; Cummins and Vogelstein, 2004; Cummins et al, 2004; Brooks et al, 2007). Although these effects are generally interpreted in terms of the auto-ubiquitination of MDM2 and the ubiquitin-mediated turnover of p53, MDM2 has also been reported to ubiquitinate H2B (Minsky and Oren, 2004), albeit on residues that may not be relevant for transcriptional activation. Other studies suggest that USP7 influences the nuclear accumulation of PTEN (Song et al, 2008), an effect that is opposed by the PML–DAXX complex. These links have provided an attractive rationale for the over-expression of USP7 commonly seen in human cancers (reviewed in Hussain et al, 2009). On the basis of the findings we report here, the function of USP7 in PcG-mediated repression of INK4a offers an alternative explanation. However, the senescence-like arrest caused by knockdown of USP7 did not appear to be INK4a dependent.
The initial motivation for this study was to better understand the transcriptional regulation of the INK4a tumour suppressor, and in this regard, we have added two ubiquitin-specific proteases, USP7 and USP11, to the extensive list of proteins that are directly or indirectly involved. A future priority will be to explore the physiological contexts in which USP7 and USP11 might be deployed to regulate PRC1 function. The central function of ubiquitination in the regulation of INK4a and the possibility that USPs can promote the bypass of oncogene-induced senescence by stabilising the PRC1 complex provide opportunities for using specific USP inhibitors in a variety of therapeutic contexts.
Materials and methods
Tissue culture
Human dermal fibroblasts (FDF and Leiden strains), 293T- and 293T-derived cell lines were cultured in Dulbecco modified Eagle's medium supplemented with 10% foetal calf serum, 100 IU/ml penicillin and 100 μg/ml streptomycin. Lentiviral vector production and infections were performed as described (Maertens et al, 2009). Stably expressing cell lines were made by infecting 293T cells with VSV-G—pseudotyped retrovirus carrying the pQMEL18-Flag-puro, pQBMI1-Flag-puro, pQFlag-USP7-puro, pQFlag-USP11-puro or pQFlag-H2A-puro vectors. The cells were selected with 2.5 μg/ml puromycin. In some experiments, cells were treated with 20 μM MG132 for 4 h to inhibit proteasome function. The control cells were treated with DMSO. Cell proliferation assays were performed by staining with crystal violet, as previously described (McConnell et al, 1998). Five days post-selection, 5 × 103 cells were plated out into each well of a 24-well plate and 6 wells were used per time point.
Plasmid construction
Primer sequences used for PCR are listed in Supplementary Table S2. To make pGM-Flag-RING1 and pGM-Flag-RING2, the open-reading frames (ORFs) were PCR amplified from cDNA, obtained from 293T cells, using primer sets GM38/107 and GM36/109, respectively. The PCR products were sub-cloned between the BamHI and EcoRI sites of pGM-Flag vector (Maertens et al, 2006). pQFlag-puro was engineered by ligating annealed primers GM66/GM67 in NotI- and BamHI-digested pQCXIP (Clontech), giving the following MCS: NotI-Kozak-MfeI-Flag-AgeI-XhoI-BamHI. To clone pQHA-puro, the Flag encoding sequence was replaced by the HA encoding sequence by ligating the annealed oligonucleotides GM68 and GM69 with MfeI- and AgeI-digested pQFlag-puro. To express HA- or Flag-tagged USP7, the USP7 ORF was PCR amplified from 293T-derived cDNA using primers GM131 and GM132, digested with AgeI and BamHI and ligated with similarly digested pQHA-puro or pQFlag-puro. A similar approach was taken for USP11 ORF using primers GM135 and GM136 and ligation into with AgeI- and XhoI-digested vectors. Catalytic site mutations were introduced into pQFlag-USP7-puro and pQFlag-USP11-puro using primer sets GM142/143 and GM148/149, respectively. To express USP7 and USP11 as GST-fusion proteins in Escherichia coli, the ORFs of wild-type and mutant cDNAs were sub-cloned in pGEX-6P-3 in frame with GST giving pGM-GST-USP7, pGM-GST-USP7C223S, pGM-GST-USP11 and pGM-GST-USP11C275S.
MEL18 ORF was amplified from pGM-MEL18-cTAP (Elderkin et al, 2007) using primers GM153 and GM169. The PCR product was digested with BamHI and MfeI and ligated with BamHI- and EcoRI-digested pGM-Ram2-Flag, giving pGM-MEL18-Flag. To create pQMEL18-Flag-puro, the insert generated from HindIII and XbaI digestion of pGM-MEL18-Flag was treated with Pfu Ultra in the presence of dNTPs to fill in the overhangs, and ligated into filled in NotI-EcoRI-cut pQHA puro. PGM-BMI1-Flag was generated by amplifying the BMI1 ORF from pQBT (Elderkin et al, 2007) using primers GM230 and GM238. The PCR fragment digested with MfeI and BamHI was ligated with BamHI- and EcoRI-digested pGM-Ram2-Flag (Maertens et al, 2006). For the generation of pQBMI1-Flag puro, pGM-BMI1-Flag was digested with BamHI and XhoI and ligated with BamHI- and XhoI-cut pQMEL18-Flag-puro. The constructs expressing CBX2-Flag/HA, CBX4-Flag/HA, CBX6-Flag/HA, CBX7-Flag/HA and CBX8-Flag/HA are described elsewhere (El Messaoudi et al, 2010). Flag-histone H2A was expressed from pQFlag-H2A-puro, which was generated by amplifying H2A with primers GM191 and GM192 using cDNA made from 293T cells. The amplicon was digested with AgeI and BamHI and ligated into AgeI- and BamHI-cut pQFlag-puro. To purify CBX8-Flag from bacteria, the CBX8-Flag ORF was amplified from pGM-CBX8-Flag (Maertens et al, 2009) with primers GM300 and GM301. The amplicon was digested with BglII and ligated into SmaI- and BamHI-cut pCPH6P-BIV IN (Cherepanov, 2007), replacing the BIV IN sequence, giving pGMH6P-CBX8-Flag. The constructs used for baculovirus expression of Bmi1, Mel18 and Ring1b were described elsewhere (Elderkin et al, 2007). All constructs were sequence verified. Lentiviral vectors targeting the USP7 or USP11 transcript were purchased from Sigma-Aldrich; USP7sh1: NM_003470 1249; USP7sh2: NM_003470 1648; USP11sh1: NM_0046512 1800; USP11sh2: NM_0046512 1695.
Immunoblotting
Cells were harvested by trypsinisation and washed extensively in ice-cold phosphate-buffered saline (PBS). Proteins were extracted in five volumes of RIPAS (high salt containing RIPA buffer: 20 mM Hepes-NaOH, pH 7.6, 0.3 M NaCl, 0.01% NP-40, 1% sodium deoxycholate, 0.1% SDS and 2 mM EDTA in the presence of protease inhibitors (Roche) and 1 mM PMSF). Protein concentration was determined using BCA assay (Pierce) and samples (10–40 μg) of total protein were separated on a 12% SDS–PAGE gel. Proteins were transferred onto nitrocellulose membrane and immunoblotted using the following dilutions of antibody: rabbit anti-MEL18 1:500 (Santa Cruz), goat anti-MEL18 1:1000 (Abcam), mouse anti-Flag M2 1:3000 (Sigma-Aldrich), mouse anti-Flag conjugated to horse radish peroxidase (HRP) 1:3000 (Sigma-Aldrich), rabbit anti-RING1 (Cell Signaling), mouse anti-BMI1 (1:500) (Abcam), rabbit anti-β-tubulin 1:1000 (Santa Cruz), rabbit anti-TFIID (TBP) 1:1000 (Santa Cruz), mouse anti-HA 16B12 1:2000 (Covance), rabbit anti-HA 1:1000 (Santa Cruz), mouse anti-p16 JC8 1:5, mouse anti-p53 1:1000 (DO-1, Santa Cruz), mouse anti-cyclin D1 1:1000 (BD Pharmingen), mouse anti-GAPDH HRP-conjugated 1:10 000 (Abcam), rabbit anti-USP7 1:500 (Abcam), rabbit anti-CBX8 (Bethyl Laboratories), rabbit anti-CBX7 (Abcam), rabbit anti-histone H2A (Upstate) and rabbit anti-histone H2B (Upstate). For the generation of polyclonal anti-USP11 antiserum, three synthetic peptides corresponding to amino acids 587–605, 716–734 and 909–920 were fused to Keyhole Limpet Hemocyanin, and a mixture of the three peptides was used to immunise rabbits. The IgG fraction was purified from the isolated serum and used at a 1:500 dilution. HRP-conjugated secondary antibodies were purchased from GE Healthcare and used at 1:2000 dilution. The blots were developed using ECL plus (Pierce). In some experiments, recombinant proteins were detected with rabbit anti-Flag antibody 1:1000 (Cell Signaling) followed by 1:2000 dilution of alkaline phosphatase-conjugated anti-rabbit IgG (Sigma). The blots were stained by washing in 100 mM Tris–HCl pH 9.5 followed by incubation in staining solution (100 mM Tris–HCl pH 9.5, 5 mM MgCl2, 100 mM NaCl, 0.33 mg/ml nitro blue tetrazolium chloride (Sigma) and 0.165 mg/ml 5-bromo-4 chloro-3 indolyl phosphate (Sigma)). The staining was stopped by rinsing the blots with water followed by air drying.
Immunoprecipitation
Cells were harvested by trypsinisation and washed extensively in ice-cold PBS. All further procedures were performed on ice or at 4°C. The proteins were extracted for 10 min in five volumes of lysis buffer (1% NP-40, 10 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 30 mM sodium pyrophosphate, 50 mM sodium fluoride, 10% glycerol and Roche protease inhibitors) and insoluble debris was removed by centrifugation at 15 000 g for 30 min. In some experiments, the analyses were conducted with nuclear extracts prepared as described (Wysocka et al, 2001). Flag- and HA-tagged proteins were precipitated with 30 μl of anti-Flag-agarose beads (Sigma-Aldrich) or anti-HA affinity resin (Covance). Protein complexes were allowed to precipitate by end-over-end rocking for 3 h. After extensive washing, bound proteins were eluted in Laemmli sample buffer or by rocking for 30 min in lysis buffer supplemented with 0.1 mg/ml Flag-peptide (Sigma-Aldrich). Chromatin immunoprecipitations were performed as described in Maertens et al (2009).
Quantitative real-time PCR
RNA was isolated using the RNA purification kit (Roche) and reverse transcribed using random hexamers and MultiScribe RT (Applied Biosystems). Quantitative PCR was performed using SYBR GREEN master mix (ABI) as advised by the manufacturer. Primer pairs USP7Q2S/AS and USP11Q2S/AS were used to detect USP7 and USP11, respectively (Supplementary Table S3). The primers used to detect the p16INK4a, MEL18, BMI1, ARF and GAPDH transcripts were described previously (Maertens et al, 2009). Annealing/extension was performed at 60°C apart from the detection of p16INK4a mRNA, which was at 66°C.
Cellular fractionation and size exclusion chromatography
Cellular fractionation was as described (Wysocka et al, 2001). For gel filtration of nuclear extracts, nuclei were isolated as before (Wysocka et al, 2001) and proteins were extracted in the buffer used for gel filtration: 50 mM Tris–HCl pH 8.0, 1 mM EDTA, 150 mM NaCl, 0.5% CHAPS supplemented with complete, EDTA-free protease inhibitors (Roche) and 1 mM PMSF. Samples (200 μl corresponding to ∼2 mg of protein) were loaded on a Superose 6 10/300 GL column (GE Healthcare); 0.5-ml fractions were collected and precipitated overnight in 10% trichloroacetic acid. After centrifugation at 16 000 g for 30 min at 4°C, the pellets were washed twice with ice-cold acetone, air dried and dissolved in 50 μl Laemmli buffer. Samples (10 μl) of the relevant fractions were separated on 10% SDS–PAGE gels and the proteins were detected by immunoblotting.
Purification of recombinant proteins and glutathione- S-transferase pull downs
To produce GST, GST-USP7 and GST-USP11, the Rosetta (DE3) strain (Novagen) was transformed with the corresponding recombinant-expression vectors. Expression was induced with 1 mM IPTG at 18°C overnight. The cells were harvested and lysed in ice-cold sonication buffer (50 mM Tris–HCl pH 7.4, 2 mM EDTA, 0.25% Triton-X-100, 1 mM PMSF) supplemented with lysozyme. All further procedures were performed at 4°C. The sonicated sample was cleared by centrifugation at 16 000 g for 30 min and the supernatant was run through a column of 1 ml pre-equilibrated glutathione sepharose resin (GE Healthcare). The column was washed extensively in 50 mM Tris–HCl pH 7.4, 2 mM EDTA, 500 mM NaCl, 1 mM PMSF and the proteins were eluted in 50 mM Tris–HCl pH 8.0, 10 mM glutathione. Positive fractions were pooled, dialysed overnight in 25 mM Tris–HCl pH 7.4, 150 mM NaCl, 5 mM DTT, 10% glycerol and concentrated on Microcon spin columns with a MW cutoff of 10 kDa (Millipore). Glycerol was added to final concentration of 20%, and the proteins were snap frozen in liquid nitrogen and stored at −80°C.
Full-length mouse Mel18, Bmi1 and Ring1B were fused to a His12FLAG tag and cloned into pDEST8 using the Gateway cloning system (Invitrogen). Recombinant baculovirus was generated using the Bac-to-Bac baculovirus system (Invitrogen). Wild-type virus, vMel18-FlagHis, vBmi1-FlagHis and vRing1B-FlagHis were used to infect Sf9 insect cells for 60 h. Cells were harvested by centrifugation at 1500 g and resuspended in lysis buffer (20 mM Tris–HCl pH 7.5, 250 mM NaCl, 10% glycerol, 1 mM EDTA, 0.1% triton, 2 mM 2-mecaptoethanol and protease inhibitors). The lysate was then sonicated for five 30 s cycles, followed by centrifugation at 15 000 r.p.m. for 2 h. The cell lysate was loaded onto a 5 ml Hi-Trap Ni2+ chelating column (GE Healthcare) equilibrated with Buffer A (20 mM Tris–HCl pH 7.5, 500 mM NaCl and 10% glycerol) and washed with 10 column volumes of Buffer A containing 40 mM imidazole. Proteins were eluted with a gradient of 40–1000 mM imidazole in buffer A over 10 column volumes. Protein samples were then dialysed against storage buffer (25 mM Tris–HCl pH 7.5, 10% glycerol, 150 mM NaCl, 1 mM DTT) for 4 h with two changes. Aliquots were frozen in liquid nitrogen and stored at −80°C.
To produce CBX8-Flag, the Rosetta2 (DE3) strain (Novagen) was transformed with pGMH6P-CBX8-Flag. His6-CBX8-Flag expression was induced during 4 h at 25°C using 1 mM IPTG. The cells were harvested and lysed in ice-cold core buffer (25 mM Tris–HCl pH 7.4, 0.5 M NaCl) supplemented with 0.5% Triton-X-100, 1 mM PMSF and lysozyme. All procedures were performed at 4°C. The sonicated sample was cleared by centrifugation at 16 000 g for 30 min at 4°C. The supernatant was supplemented with 20 mM imidazole and run over a column prepared with 3 ml NiNTA resin (Qiagen). After extensive washing, the protein was eluted with core buffer supplemented with 200 mM imidazole. Positive fractions were pooled and the His6-tag was removed by Prescission protease cleavage overnight at 4°C in the presence of 5 mM DTT. The cleaved protein was diluted five-fold in ice-cold 25 mM Tris–HCl pH 7.4, to reduce the NaCl concentration to 100 mM and run over a cation exchange column (GE Healthcare). After extensive washes, the protein was eluted using a linear gradient of 0.1–1 M NaCl in 25 mM Tris–HCl pH 7.4 (CBX8-Flag eluted at 0.5 M NaCl). The positive fractions were pooled and further purified by gel filtration on a HiLoad 16/60 Superdex-200 column (GE Healthcare) in 25 mM Tris–HCl pH 7.4, 0.5 M NaCl. Purified CBX8-Flag was concentrated to 2.5 mg/ml, supplemented with 2 mM DTT, 10% glycerol, snap frozen in liquid nitrogen and stored at −80°C.
The Flag-tagged full-length MEL18, BMI1, RING1, RING2 and CBX8 proteins were expressed using the TNT T7-coupled reticulocyte lysate system (Promega), according to the manufacturer's instructions. The templates were prepared by linearisation of pGM-MEL18-Flag, pGM-BMI1-Flag, pGM-Flag-RING1, pGM-Flag-RING2 and pGM-Flag-CBX8 with EcoRV. Each 50-μl in vitro transcription/translation reaction contained 1 μg linearised plasmid, 30 μCi of [35S]-methionine (GE Healthcare) and 25 μl reticulocyte lysate. The GST pull-down assays were performed as described in Maertens et al (2006) using 5 μl of in vitro translated product per reaction.
Ubiquitination assays
To enrich for ubiquitinated proteins, 293T- or 293T-derived cells were transfected with pMT107 (Treier et al, 1994) to express His6-ubiquitin. After 36 h, the cells were washed and harvested by scraping in ice-cold PBS. After centrifugation, the cells were resuspended in 0.5 ml PBS and an input sample (10 μl) was mixed with 50 μl 2 × Laemmli sample buffer and boiled for 10 min. The remaining cells were resuspended in 1 ml buffer A (6 M guanidinium hydrochloride, 0.1 M Na2HPO4/NaH2PO4 pH 8.0, 10 mM imidazole) and sonicated. The extract was incubated with 50 μl NiNTA agarose (Qiagen) for 3 h at room temperature. The beads were washed twice in buffer A, twice in buffer A/TI (3 volumes buffer TI and 1 volume buffer A) and once in buffer TI (25 mM Tris–HCl pH 6.8, 20 mM imidazole). The ubiquitinated proteins were eluted by boiling for 5 min in Laemmli sample buffer supplemented with 5 mM EDTA, separated in 10% SDS–PAGE gels and detected by immunoblotting.
De-ubiquitination assays
The 293T-derived cell lines that stably express Flag-tagged USP7, USP11 or their active site mutants were harvested by trypsinisation and washed extensively in PBS. Nuclei were prepared by hypotonic lysis and the proteins were extracted with nuclear extraction buffer (10 mM HEPES pH 6.8, 450 mM NaCl, 0.5% NP-40, 1 mM DTT, 1 mM MgCl2, 0.1 mM PMSF and Complete EDTA-free protease inhibitors (Roche)). All procedures were performed on ice or at 4°C. The extracts were cleared by centrifugation at 16 000 g for 30 min and the supernatant was further pre-cleared by end-over-end rocking in the presence of Protein G Sepharose beads. The pre-cleared extracts were then allowed to bind pre-equilibrated 200 μl anti-Flag resin (Sigma) for 3 h. After extensive washes in the nuclear extraction buffer, the beads were washed once in low salt buffer (10 mM HEPES pH 6.8, 150 mM NaCl, 0.5% NP-40, 1 mM DTT, 1 mM MgCl2), and the proteins were eluted in low salt buffer supplemented with Flag peptide (0.1 mg/ml).
Ubiquitinated H2A and H2B substrates were made as described in Elderkin et al (2007). After a 1 h incubation, the ubiquitination reactions were stopped by addition of EDTA to a final concentration of 2 mM and held at 37°C for an additional 15 min. Approximately 125 ng of purified Flag-USP was then added to each reaction and the de-ubiquitination was allowed to occur for 1 h at 37°C. The reactions were stopped by boiling the samples in Laemmli buffer. The proteins were separated on a 15% SDS–PAGE gel, and detected by staining with Coomassie R-250.
Mono-nucleosomes containing Flag-H2A were purified from 293T cells stably expressing Flag-H2A as described previously (Schnitzler, 2001; Hernandez-Munoz et al, 2005).
Supplementary Material
Acknowledgments
We are grateful to Nicolla O'Reily for the design and synthesis of the USP11 peptides used to generate the polyclonal anti-USP11 antibody and thank Haruhiko Koseki for providing the RING2 antibody. We are also indebted to Peter Cherepanov for providing advice and facilities during the revision of the paper. SE is supported by a Wellcome Trust Career Development Award.
Footnotes
The authors declare that they have no conflict of interest.
References
- Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, Vallier L, Gil J (2009) Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev 23: 2134–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A (2006) The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell 24: 701–711 [DOI] [PubMed] [Google Scholar]
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G et al. (2004) A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6: 97–105 [DOI] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, Hansen KH, Helin K (2007) The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 21: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes S, Rowe J, Gutierrez Del Arroyo A, Bond J, Peters G (2004) Contribution of p16(INK4a) to replicative senescence of human fibroblasts. Exp Cell Res 298: 549–559 [DOI] [PubMed] [Google Scholar]
- Brookes S, Rowe J, Ruas M, Llanos S, Clark PA, Lomax M, James MC, Vatcheva R, Bates S, Vousden KH, Parry D, Gruis N, Smit N, Bergman W, Peters G (2002) INK4a-deficient human diploid fibroblasts are resistant to RAS-induced senescence. EMBO J 21: 2936–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Li M, Hu M, Shi Y, Gu W (2007) The p53—Mdm2—HAUSP complex is involved in p53 stabilization by HAUSP. Oncogene 26: 7262–7266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK (2006) Structure and E3-ligase activity of the ring-ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J 25: 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cales C, Roman-Trufero M, Pavon L, Serrano I, Melgar T, Endoh M, Perez C, Koseki H, Vidal M (2008) Inactivation of the polycomb group protein Ring1B unveils an antiproliferative role in hematopoietic cell expansion and cooperation with tumorigenesis associated with Ink4a deletion. Mol Cell Biol 28: 1018–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J, d'Adda di Fagagna F (2007) Cellular senescence: when bad things happen to good cells. Nat Rev 8: 729–740 [DOI] [PubMed] [Google Scholar]
- Cao R, Tsukada Y, Zhang Y (2005) Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell 20: 845–854 [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y (2004) The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev 14: 155–164 [DOI] [PubMed] [Google Scholar]
- Cherepanov P (2007) LEDGF/p75 interacts with divergent lentiviral integrases and modulates their enzymatic activity in vitro. Nucleic Acids Res 35: 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Blasco MA, Serrano M (2007) Cellular senescence in cancer and aging. Cell 130: 223–233 [DOI] [PubMed] [Google Scholar]
- Collado M, Serrano M (2006) The power and the promise of oncogene-induced senescence markers. Nat Rev Cancer 6: 472–476 [DOI] [PubMed] [Google Scholar]
- Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B (2004) Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature 428: 1 p following 486 [DOI] [PubMed] [Google Scholar]
- Cummins JM, Vogelstein B (2004) HAUSP is required for p53 destabilization. Cell Cycle 3: 689–692 [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676 [DOI] [PubMed] [Google Scholar]
- Dietrich N, Bracken AP, Trinh E, Schjerling CK, Koseki H, Rappsilber J, Helin K, Hansen KH (2007) Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. EMBO J 26: 1637–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elderkin S, Maertens GN, Endoh M, Mallery DL, Morrice N, Koseki H, Peters G, Brockdorff N, Hiom K (2007) A phosphorylated form of Mel-18 targets the Ring1B histone H2A ubiquitin ligase to chromatin. Mol Cell 28: 107–120 [DOI] [PubMed] [Google Scholar]
- El Messaoudi-Aubert S, Nicholls J, Maertens GN, Brookes S, Bernstein E, Peters G (2010) Role for the MOV10 RNA helicase in Polycomb-mediated repression of the INK4a tumor suppressor. Nat Struct Molec Biol (advance online publication 13 June 2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J (1997) A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J 16: 1519–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Montalvan A, Bouwmeester T, Joberty G, Mader R, Mahnke M, Pierrat B, Schlaeppi JM, Worpenberg S, Gerhartz B (2007) Biochemical characterization of USP7 reveals post-translational modification sites and structural requirements for substrate processing and subcellular localization. FEBS J 274: 4256–4270 [DOI] [PubMed] [Google Scholar]
- Gil J, Bernard D, Martinez D, Beach D (2004) Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol 6: 67–72 [DOI] [PubMed] [Google Scholar]
- Gil J, Peters G (2006) Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev 7: 667–677 [DOI] [PubMed] [Google Scholar]
- Hernandez-Munoz I, Lund AH, van der Stoop P, Boutsma E, Muijrers I, Verhoeven E, Nusinow DA, Panning B, Marahrens Y, van Lohuizen M (2005) Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc Natl Acad Sci USA 102: 7635–7640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S (2009) Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature 460: 1132–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S, Zhang Y, Galardy PJ (2009) DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle 8: 1688–1697 [DOI] [PubMed] [Google Scholar]
- Ideguchi H, Ueda A, Tanaka M, Yang J, Tsuji T, Ohno S, Hagiwara E, Aoki A, Ishigatsubo Y (2002) Structural and functional characterization of the USP11 deubiquitinating enzyme, which interacts with the RanGTP-associated protein RanBPM. Biochem J 367 (Part 1): 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono K, Fujimura Y, Shinga J, Yamaki M, O-Wang J, Takihara Y, Murahashi Y, Takada Y, Mizutani-Koseki Y, Koseki H (2005) Mammalian polyhomeotic homologues Phc2 and Phc1 act in synergy to mediate polycomb repression of Hox genes. Mol Cell Biol 25: 6694–6706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itahana K, Zou Y, Itahana Y, Martinez JL, Beausejour C, Jacobs JJ, Van Lohuizen M, Band V, Campisi J, Dimri GP (2003) Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol Cell Biol 23: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M (1999) The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397: 164–168 [DOI] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC (2009) Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460: 1140–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y (2007) pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev 21: 49–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G (2003) Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423: 255–260 [DOI] [PubMed] [Google Scholar]
- Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M (2009) The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 460: 1136–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Brooks CL, Kon N, Gu W (2004) A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell 13: 879–886 [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W (2002) Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416: 648–653 [DOI] [PubMed] [Google Scholar]
- Li Z, Cao R, Wang M, Myers MP, Zhang Y, Xu RM (2006) Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J Biol Chem 281: 20643–20649 [DOI] [PubMed] [Google Scholar]
- Lin CH, Chang HS, Yu WC (2008) USP11 stabilizes HPV-16E7 and further modulates the E7 biological activity. J Biol Chem 283: 15681–15688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens GN, Cherepanov P, Engelman A (2006) Transcriptional co-activator p75 binds and tethers the Myc-interacting protein JPO2 to chromatin. J Cell Sci 119(Part 12): 2563–2571 [DOI] [PubMed] [Google Scholar]
- Maertens GN, El Messaoudi-Aubert S, Racek T, Stock JK, Nicholls J, Rodriguez-Niedenfuhr M, Gil J, Peters G (2009) Several distinct polycomb complexes regulate and co-localize on the INK4a tumor suppressor locus. PLoS One 4: e6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA (2009) A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460: 1149–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Starborg M, Brookes S, Peters G (1998) Inhibitors of cyclin-dependent kinases induce features of replicative senescence in early passage human diploid fibroblasts. Curr Biol 8: 351–354 [DOI] [PubMed] [Google Scholar]
- Minsky N, Oren M (2004) The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell 16: 631–639 [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ (2003) Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature 425: 962–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123: 773–786 [DOI] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF (2003) Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423: 302–305 [DOI] [PubMed] [Google Scholar]
- Sanchez C, Sanchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M (2007) Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics 6: 820–834 [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE (2001) A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412: 655–660 [DOI] [PubMed] [Google Scholar]
- Schnitzler GR (2001) Isolation of histones and nucleosome cores from mammalian cells. Curr Protoc Mol Biol Chapter 21: Unit 21 25 [DOI] [PubMed] [Google Scholar]
- Schoenfeld AR, Apgar S, Dolios G, Wang R, Aaronson SA (2004) BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol Cell Biol 24: 7444–7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V (2007) Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet 8: 9–22 [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE (1999) Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98: 37–46 [DOI] [PubMed] [Google Scholar]
- Sharpless NE (2005) INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res 576: 22–38 [DOI] [PubMed] [Google Scholar]
- Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP (2008) The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature 455: 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138: 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M (2006) Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 6: 846–856 [DOI] [PubMed] [Google Scholar]
- Treier M, Staszewski LM, Bohmann D (1994) Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78: 787–798 [DOI] [PubMed] [Google Scholar]
- Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K (2009) Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 460: 1145–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, Karch F, Verrijzer CP (2005) GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell 17: 695–707 [DOI] [PubMed] [Google Scholar]
- Vissers JH, Nicassio F, van Lohuizen M, Di Fiore PP, Citterio E (2008) The many faces of ubiquitinated histone H2A: insights from the DUBs. Cell Div 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y (2004) Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878 [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL (2008) Histone ubiquitination: triggering gene activity. Mol Cell 29: 653–663 [DOI] [PubMed] [Google Scholar]
- West MH, Bonner WM (1980) Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res 8: 4671–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb SJ, Basu A, Allis CD, Bernstein E (2007) Polycomb Group proteins: an evolutionary perspective. Trends Genet 23: 494–502 [DOI] [PubMed] [Google Scholar]
- Wu X, Gong Y, Yue J, Qiang B, Yuan J, Peng X (2008) Cooperation between EZH2, NSPc1-mediated histone H2A ubiquitination and Dnmt1 in HOX gene silencing. Nucleic Acids Res 36: 3590–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Reilly PT, Herr W (2001) Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol Cell Biol 21: 3820–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Kimura J, Miki Y, Yoshida K (2007) The deubiquitinating enzyme USP11 controls an IkappaB kinase alpha (IKKalpha)-p53 signaling pathway in response to tumor necrosis factor alpha (TNFalpha). J Biol Chem 282: 33943–33948 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.