Abstract
We report the crystal structure of a termination complex containing release factor RF1 bound to the 70S ribosome in response to an amber (UAG) codon at 3.6-Å resolution. The amber codon is recognized in the 30S subunit-decoding centre directly by conserved elements of domain 2 of RF1, including T186 of the PVT motif. Together with earlier structures, the mechanisms of recognition of all three stop codons by release factors RF1 and RF2 can now be described. Our structure confirms that the backbone amide of Q230 of the universally conserved GGQ motif is positioned to contribute directly to the catalysis of the peptidyl-tRNA hydrolysis reaction through stabilization of the leaving group and/or transition state. We also observe synthetic-negative interactions between mutations in the switch loop of RF1 and in helix 69 of 23S rRNA, revealing that these structural features interact functionally in the termination process. These findings are consistent with our proposal that structural rearrangements of RF1 and RF2 are critical to accurate translation termination.
Keywords: 70S ribosome, amber codon, RF1–UAG complex, translation termination complex
Introduction
Nonsense codons, known as amber (UAG), ochre (UAA) and opal (UGA), signal termination of protein synthesis on the ribosome (Brenner et al, 1965, 1967). Unlike sense-codon recognition, no tRNAs are involved in nonsense (stop)-codon recognition. Instead, termination of protein synthesis in bacteria depends on the class I release factors RF1 and RF2 (Capecchi, 1967; Vogel et al, 1969; Caskey et al, 1971). Peptidyl-tRNA hydrolysis is promoted by RF1 in response to a UAG or UAA stop codon, and by RF2 in response to a UGA or UAA codon (Scolnick et al, 1968; Capecchi and Klein, 1969; Scolnick and Caskey, 1969). For four decades after the discovery of the release factors, three fundamental questions concerning the mechanism of translation termination remained unresolved: (1) How are the stop codons recognized? Are they recognized directly by the release factors, or indirectly, such as through base pairing with ribosomal RNA? (2) What is the mechanism of peptidyl-tRNA hydrolysis? Is the reaction catalysed directly by the release factors, or by converting the ribosomal peptidyl transferase to an esterase, as suggested by the inhibition of peptidyl-tRNA hydrolysis by peptidyl-transferase inhibitors (Tompkins et al, 1970; Caskey et al, 1971)? (3) How is premature termination kept to levels on the order of 10−5 in the absence of proofreading (Freistroffer et al, 2000)? The structural basis for approaching these long-standing problems first began to emerge from low-resolution cryo-EM and X-ray studies (Klaholz et al, 2003; Rawat et al, 2003, 2006; Petry et al, 2005). More recently, detailed insights into the mechanism of translation termination have come from higher-resolution crystal structures of 70S ribosome termination complexes bound with RF1 in response to a UAA stop codon (Laurberg et al, 2008), and RF2 in response to UAA (Korostelev et al, 2008) or UGA (Weixlbaumer et al, 2008) codons.
The structures showed that the bases of the stop codons are recognized directly by domain 2 of the release factors. The important elements contributing to recognition are (1) the polypeptide backbone at the tip of helix α5, which makes specific hydrogen bonds with the Watson–Crick edge of U1 of the stop codon; (2) the hydroxyl group of Thr186 of the conserved PVT motif of RF1 or Ser209 of the SPF motif of RF2 (unless otherwise noted, we use the numbering for the Thermus thermophilus factors throughout), which H-bond to the second base and (3) the conserved Gln181 and T194 of RF1 or V203 and T216 of RF2, which recognize the third base in a separate pocket of the complex. The structures have also revealed that the universally conserved GGQ motif of domain 3 contacts the acceptor end of the P-site tRNA, positioning the backbone amide group of the conserved Gln to participate in catalysis of the peptidyl-tRNA hydrolysis reaction. Substitution by proline at this position, which eliminates the ability of the backbone to participate in transition-state or product stabilization by H-bonding, abolishes the peptidyl-tRNA esterase activity of the factor (Korostelev et al, 2008). On the basis of the dramatic conformational differences between the structures of the class I release factors in their free (Vestergaard et al, 2001; Shin et al, 2004; Zoldak et al, 2007) and ribosome-bound states (Korostelev et al, 2008; Laurberg et al, 2008), we proposed that rearrangement of the switch loop, which connects domains 3 and 4 of a release factor, is critical for correct positioning of the GGQ motif in the ribosomal peptidyl-transferase centre (PTC) (Korostelev et al, 2008; Laurberg et al, 2008) and, therefore, may have a function in strict coordination of peptidyl-tRNA hydrolysis with stop-codon recognition.
Here, we present the crystal structure of a termination complex containing RF1 bound to the 70S ribosome in response to a UAG stop codon, revealing how RF1 is able to recognize both the UAG and UAA codons, and completing the solution of the four possible structures representing recognition of all three stop codons by the two type I release factors. In addition, we have performed mutational studies that provide evidence that functional interaction between the switch loop of RF1 and helix 69 of 23S rRNA, both of which have been proposed to be involved in positioning the GGQ-bearing domain 3 in the active centre, but are not directly involved in stop-codon recognition or peptidyl-tRNA hydrolysis, has a critical function in translation termination.
Results and Discussion
Conformation of the L1 stalk and intersubunit rotational state
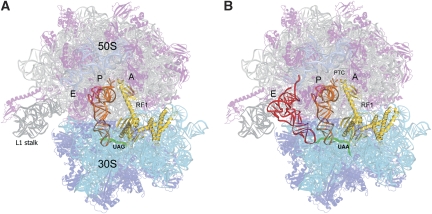
We crystallized a complex containing RF1 bound to the T. thermophilus 70S ribosome in the presence of a UAG-containing mRNA and a deacylated tRNA bound to the P site as described in Materials and methods, and solved its crystal structure at a resolution of 3.6 Å (R/Rfree=0.26/0.29). An important difference between the structure presented here and earlier reported all-atom structures of ribosome functional complexes is that our complex contains a vacant E site (Figure 1). The single notable conformational difference that can be attributed to the absence of an E-site tRNA is opening of the L1 stalk (Supplementary Figure S1), whereas the rest of the structure remains virtually identical to those of the three termination complexes in which an E-tRNA is present (Korostelev et al, 2008; Laurberg et al, 2008; Weixlbaumer et al, 2008). The L1 stalk is rotated by about 5° away from the middle of the ribosome, in a direction consistent with cryo-EM and FRET observations of opening of the L1 stalk in ribosomes containing a vacant E site (Agrawal et al, 1999; Gomez-Lorenzo et al, 2000; Tama et al, 2003; Fei et al, 2008; Cornish et al, 2009). The absence of any other significant differences between the current and E-site bound structures suggests that occupancy of the ribosomal E site is unlikely to influence the catalysis of peptidyl-tRNA hydrolysis.
Figure 1.
Overall structure of 70S translation termination complexes containing RF1 bound in response to (A) UAG (this work) and (B) UAA (Laurberg et al, 2008) stop codons. The positions of P-site tRNA (orange), E-site tRNA (red), mRNA (green) and RF1 (yellow) are shown. In the present structure (A), the ribosomal E site is vacant, resulting in movement of the L1 stalk relative to the structure containing an occupied E site shown in (B).
It was earlier observed, in comparison of the structure of a 70S translation termination complex (Laurberg et al, 2008) with that of a 70S elongation complex (Selmer et al, 2006), that the small ribosomal subunit of the termination complex undergoes a modest clockwise rotation relative to the large subunit (Supplementary Figure S2A) (Zhang et al, 2009). Now that structures for all four translation termination complexes have been solved in the same crystal form as elongation complexes, a statistically significant comparison between them can be made. In all of the RF1- and RF2-bound 70S structures (Korostelev et al, 2008; Laurberg et al, 2008; Weixlbaumer et al, 2008), including the one reported here, the positions of the small subunit relative to the large subunit are very similar (16S rRNA all-atom r.m.s.d.⩽0.8 Å). Comparison of these structures with 70S elongation complexes bound with deacylated tRNAs (Selmer et al, 2006) or acylated tRNAs (Voorhees et al, 2009) shows an ∼1.5° clockwise rotation of the whole 30S subunit and an ∼2° inwards twisting of the head towards domain 1 of the release factor, resulting in an r.m.s.d. change of ∼2.7 Å between the respective 16S rRNAs of the termination and elongation complexes. These changes are accompanied by rearrangement of intersubunit bridge B2a (which is formed by contact between helix 69 of 23S rRNA and front of the mRNA-decoding centre) in a manner that depends on the identity of the A-site substrate. When a tRNA is bound to the A site, the N1 position of the conserved base A1913 at the tip of the helix 69 hairpin loop contacts the 2′-OH of ribose 37 of the tRNA (Supplementary Figure S2B); when a release factor is bound, A1913 stacks on A1493 of 16S rRNA, a change in position of >6 Å (Supplementary Figure S2C). In addition to this local rearrangement, contact between domain 1 of the release factor and the beak of the small subunit head may contribute to the inwards movement of the head.
Recognition of the UAG stop codon
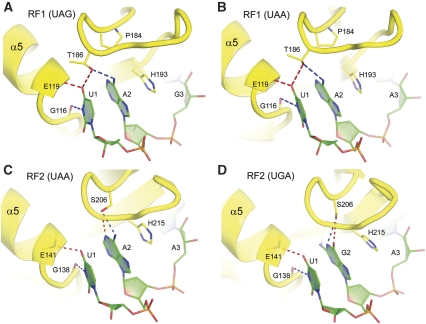
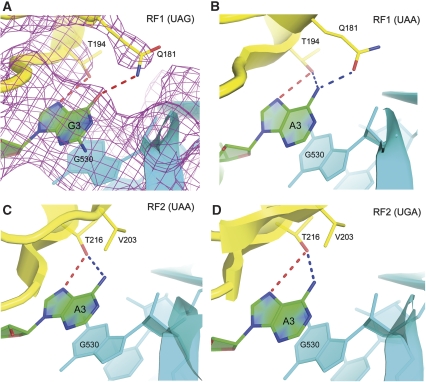
Earlier structures have shown how RF1 and RF2 recognize the UAA and UGA stop codons (Korostelev et al, 2008; Laurberg et al, 2008; Weixlbaumer et al, 2008). The structure described here provides an explanation of how RF1 is able to recognize both UAA and UAG stop codons (Figures 2 and 3). As observed for recognition of a UAA codon, three elements of domain 2 of the factor are involved—(1) The N-terminal end of helix α5: U1 is recognized by specific H-bonds made from the backbone of α5. The α-amide group of Glu119 H-bonds to the O4 oxygen of U1 and the α-carbonyl oxygen of Gly116 accepts an H-bond from the N3 position of U1 (Figure 2A). Identical interactions are made by both RF1 (Figure 2A and B) and RF2 (Figure 2C and D), accounting for the universal occurrence of U in the first position of all three stop codons. (2) The PVT motif: the side chain of Thr186 of the conserved PxT motif hydrogen bonds with the first and second bases of the stop codon (Figure 2A). By donating a hydrogen bond to the 4-keto oxygen of U1, the β-hydroxyl group of Thr186 is thereby constrained to accept a hydrogen bond from the N6-amino group of A2, defining the specificity of RF1 for adenine at the second position. (3) The G530 pocket: the third stop-codon nucleotide is recognized by a separate pocket of domain 2, one face of which is formed by G530 of 16S rRNA, which stacks against the third codon base (Figure 3A; Supplementary Figure S3A), and the other by Ile192 of RF1. The Watson–Crick edge of the third base is packed against phosphate 531 of 16S rRNA, limiting base-specific recognition to its Hoogsteen edge. In both RF1 and RF2, the universally conserved Thr194 donates a hydrogen bond to N7 of the purine ring of the A or G in position 3 (Figure 3; Supplementary Figure S3); Thr194 can also accept a hydrogen bond from the N6 amino group of adenine when a UAA or UGA codon is recognized (Figure 3B–D). Gln181, which is conserved in all RF1 sequences, but replaced by Val in RF2 (Figure 3C and D), H-bonds with the 6-position of the third codon base (Figure 3A and B). By rotation of its amide side chain, Gln181 can either donate a hydrogen bond to the O6-keto group of guanine, or accept a hydrogen bond from the N6 amino group of adenine (Figure 3A and B). Although it is not possible to distinguish directly between the oxygen and nitrogen of the amide group at the resolution of the current crystal structures, rotation of the side-chain amide of Gln181 would explain the ability of RF1 to recognize either G or A in the third position. The importance of Gln181 is emphasized by the fact that in RF2, which is specific for A at the third position, Gln181 is replaced by a hydrophobic side chain, whereas all other side chains proximal to the Hoogsteen edge of A3 are identical to those of RF1.
Figure 2.
Recognition of the first and second nucleotides of stop codons by release factors RF1 and RF2. (A) Recognition of U1 and A2 of the UAG amber codon by RF1 (this work) and (B) the UAA ochre codon by RF1 (Laurberg et al, 2008). (C) Recognition of the UAA ochre codon by RF2 (Korostelev et al, 2008). (D) Recognition of U1 and G2 of the UGA opal codon by RF2 (Weixlbaumer et al, 2008). Steric clash and H-bonds from backbone amide groups at the N-terminal end of helix α5 restrict base 1 of all stop codons to U. The hydroxyl group of T186 of RF1 donates its H-bond to the O4-keto group of U1, restricting its interaction with the second base to accepting an H-bond from the 6-amino group of A2. The position of S206 of RF2 prevents it from H-bonding with U1, allowing its side-chain hydroxyl to H-bond with the Watson–Crick edge of either A or G.
Figure 3.
Recognition of the third nucleotide of the three stop codons takes place in a separate pocket of release factors RF1 and RF2 (this work and Korostelev et al, 2008; Laurberg et al, 2008; Weixlbaumer et al, 2008). In (A), σA-weighted NCS-averaged 3Fo−2Fc electron density (contoured at 1σ) shows well-resolved density for the Gln181 side chain of RF1 (this work); rotation of its amide side chain allows it to H-bond with the Hoogsteen edge of either G or A (B). (C, D) In RF2, Gln181 is replaced by Val, limiting its recognition specificity to A in the third position.
Conformation of the GGQ motif in the PTC suggests a catalytic function for the main-chain amide of Gln230
Upon recognition of a stop codon, release factors RF1 and RF2 promote hydrolysis of peptidyl-tRNA in the peptidyl-transferase center (PTC), located some 75 Å from the position of the stop codon. As in the earlier reported 70S termination complexes (Korostelev et al, 2008; Laurberg et al, 2008; Weixlbaumer et al, 2008), the universally conserved GGQ motif, implicated in the peptidyl-tRNA hydrolysis function by multiple studies (Frolova et al, 1999; Seit-Nebi et al, 2001; Mora et al, 2003; Shaw and Green, 2007; Trobro and Aqvist, 2007), lies next to the ribose of A76 of the deacylated P-site tRNA (Figure 3; Supplementary Figure S4). Methylation of the glutamine of the GGQ motif at the δ-N position has been proposed to be important for the efficiency of termination (Dincbas-Renqvist et al, 2000; Mora et al, 2007). Inactivation of the methyltransferase responsible for Gln230 methylation does not alter the growth rate of bacteria in rich media, but affects their growth in poor media (Mora et al, 2007). The efficiency of termination in the absence of methylation was estimated to be approximately five-fold lower than that observed with methylated release factors in vivo (Mora et al, 2007). The kcat/KM for peptide release reactions catalysed with unmethylated RF2 is at least three times lower than that found for the methylated release factor in vitro (Dincbas-Renqvist et al, 2000), consistent with at least three-fold lower binding affinities of unmethylated release factors for the ribosome (Pavlov et al, 1998). ΔΔG0 values for a hydrophobic cavity-creating mutation of one methylene group are ∼1 kcal mol–1 (Kellis et al, 1988; Serrano et al, 1992; Durr and Jelesarov, 2000), corresponding to an approximately five-fold decrease in binding affinity, further suggesting that methylation of the Gln side chain contributes to the affinity of RF1/RF2 binding to the ribosome rather than to catalysis. In the available release-factor complex structures, the position of the methyl group cannot be observed, because the overexpressed release factors used for crystallization are under-methylated (Dincbas-Renqvist et al, 2000). Therefore, to explore the possible function of methylation, we have modelled a methyl group covalently bound to the δ-N position of Gln230 (Supplementary Figure S5). When the side-chain amide group of Gln230 is oriented with its keto oxygen towards the site of catalysis, the methyl group is positioned in a closely fitting hydrophobic cavity formed by the ribose moieties of U2506 and C2452 (Supplementary Figure S5). The model suggests that the methyl group strengthens the affinity of the release factor for the ribosome through interactions with the hydrophobic pocket, while orienting the side-chain amide group in a way that may support its function in discrimination of attacking nucleophiles (Shaw and Green, 2007).
In three of the four complexes, including the one presented here, the side chain of Gln230 points away from the 3′-OH of ribose 76, the likely position of the scissile bond (Korostelev et al, 2008; Laurberg et al, 2008); in contrast, its backbone amide group is positioned within H-bonding distance of the 3′-OH group of A76 of the P-site tRNA, consistent with its involvement in catalysis, as discussed below. In a fourth termination complex, Weixlbaumer et al (2008) interpreted the conformation of their glutamine side chain to be consistent with two alternative conformations; in one conformation, the side-chain amide pointed away from the 3′-OH group of A76, as observed in the three other termination complexes; in the other conformation, it was oriented towards the position of the scissile bond (Supplementary Figure S6). Although the structures suggested that the side chain of Gln230 might, in principle, contribute to catalysis by orienting the nucleophilic water molecule and/or stabilizing the leaving group (Supplementary Figure S6, REFs), these possibilities have clearly been ruled out by mutational studies, which show that the side chain itself is not important for catalysis (Dincbas-Renqvist et al, 2000; Seit Nebi et al, 2000; Seit-Nebi et al, 2001; Shaw and Green, 2007; Korostelev et al, 2008), but is implicated in discrimination of the nucleophilic water (Shaw and Green, 2007).
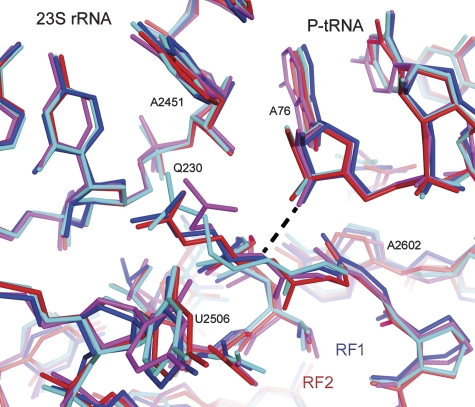
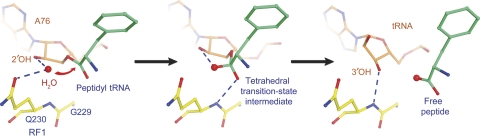
Whereas mutation of Gln230 to several other amino acids has only modest effects on catalysis (Frolova et al, 1999; Shaw and Green, 2007), we have shown that substitution by proline, whose backbone is unable to donate a hydrogen bond, leads to complete inactivation of RF1-mediated peptide release activity (Korostelev et al, 2008). In the structure presented here, the backbone of Gln230 interacts with the 3′-OH of A76 in a way closely resembling that of the other three termination complexes (Figure 4), further supporting a proposed critical function for this moiety in the catalytic reaction (Laurberg et al, 2008). Superposition of the 70S termination complex on the structure of the 70S ribosome bound with peptidyl-tRNA analogues (Voorhees et al, 2009) and that of the 50S subunit containing an analogue of the tetrahedral transition-state intermediate (Schmeing et al, 2005) allow modelling of the substrate and transition-state structures of the termination reaction (Figure 5). In these models, the backbone amide NH of Gln230 is positioned to stabilize the transition state of the hydrolysis reaction by forming a hydrogen bond with the developing oxyanion (Figure 5, middle panel). Thus, the backbone amide of the glutamine is likely involved in catalysis by stabilizing the leaving group (Figure 5, right panel) and/or the transition-state intermediate (Figure 5, middle panel). In addition, the 2′-hydroxyl of A76 of the peptidyl tRNA, shown to be essential for peptide release (Brunelle et al, 2008), participates in catalysis by interactions with the nucleophilic water (Figure 5, left panel) and with the transition state (Figure 5, middle panel), consistent with a proton shuttle model, in which the 2′-hydroxyl facilitates proton movement to the 3′-hydroxyl leaving group of A76 (reviewed in Rodnina et al, 2006; Youngman et al, 2008). Rodnina et al (2006) discuss the proton shuttle model in the context of peptide bond formation.
Figure 4.
Superposition of features of the peptidyl-transferase centres of the RF1–UAA (Laurberg et al, 2008) (light blue), RF1–UAG (dark blue; this work), RF2–UAA (Korostelev et al, 2008) (red) and RF2–UGA(Weixlbaumer et al, 2008) (magenta). The 70S termination complexes show that the backbone amide of Gln230 (T. thermophilus RF1 numbering; Gln235 in E. coli RF1) is positioned to participate in catalysis of peptidyl-tRNA hydrolysis through H-bonding to the leaving group (3′-OH of A76).
Figure 5.
Modelling the reaction mechanism for translation termination. The peptidyl-tRNA (left panel) and transition (middle panel) states were modelled by superimposing the 23S rRNA structure of the RF2 termination complex (Korostelev et al, 2008) with structures of a 70S ribosome complex containing a peptidyl-tRNA analogue (Voorhees et al, 2009) and a 50S subunit containing a transition-state analogue (Schmeing et al, 2005), respectively. (Left panel) Nucleophilic attack. The nucleophilic water molecule is positioned at the Bürgi–Dunitz angle (Bürgi et al, 1974) to the carbonyl centre of the peptidyl-tRNA ester bond, optimal for nucleophilic attack, and at H-bonding distance of the 2′-OH of ribose 76 of the peptidyl tRNA. The position of the side chain of Gln230 interferes with attack by bulkier nucleophiles. (Centre panel) Transition-state stabilization. In the tetrahedral transition state, the developing oxyanion is stabilized by H-bonding with the backbone amide NH group of Gln230. (Right panel) Product stabilization. After hydrolysis, the 3′-hydroxyl leaving group of the deacylated P-site tRNA H-bonds with the backbone amide NH group of Gln230.
Testing the function of the switch loop and h69 in translation termination
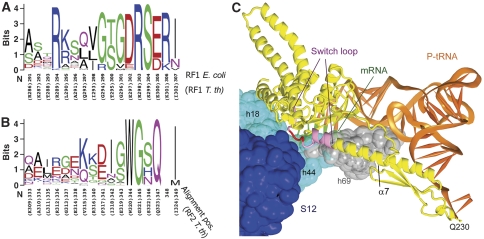
The low error frequency (10−3–10−6) (Freistroffer et al, 2000) of translation termination requires that peptidyl-tRNA hydrolysis is strictly coordinated with stop-codon recognition. In the absence of proofreading (Freistroffer et al, 2000), this might be accomplished by preventing the catalytic site in domain 3 from final docking into the PTC until a stop codon is recognized. On the basis of comparison of the differences between the structures of free release factors (Vestergaard et al, 2001; Shin et al, 2004; Zoldak et al, 2007) and those bound in termination complexes (Supplementary Figure S7), we proposed that the switch loop, which connects domains 3 and 4 of the release factor, regulates the final positioning of domain 3 by sensing changes in the conformation of the ribosomal-decoding centre that occur as a consequence of stop-codon recognition (Korostelev et al, 2008; Laurberg et al, 2008). The compact, or ‘closed', conformation of release factors observed in the free state (Vestergaard et al, 2001; Shin et al, 2004; Zoldak et al, 2007) is incompatible with ribosome binding because of multiple steric clashes with the P-site tRNA and other parts of the 70S–tRNA complex (Supplementary Figure S7), but has been proposed to have functional relevance for methylation of class I release factors (Graille et al, 2005). This suggests that the conformational change between the closed and open forms, observed in solution using small-angle X-ray scattering (Vestergaard et al, 2005; Zoldak et al, 2007), may involve an intermediate conformation, which allows domain 2 to interact with the decoding centre of the ribosome, followed by docking of domain 3 into the PTC upon successful recognition of a stop codon. In termination complexes, the N-terminal end of the switch loop of the release factor is reorganized into a helical extension of α7, which is held in place by interactions with the backbone of h69 of 23S rRNA (Figure 6C; Supplementary Figure S7). The rearranged switch loop packs in a pocket formed by the universally conserved nucleotides A1492 and A1493 of 16S rRNA and A1913 of 23S rRNA, which is repositioned in the termination complex to stack on A1493 (Supplementary Figure S2). This seems to direct helix α7 towards the PTC, docking domain 3 in the PTC. Although the amino-acid sequences of the switch loops of RF1 and RF2 diverge sharply (Figure 6A and B), their overall packing adjacent to the decoding centre (Figure 6C) is similar (Korostelev et al, 2008). Formation of the pocket by A1492, A1493 and A1913 on recognition of a stop codon and packing of the switch loop in this pocket, therefore, seem to be critical for coordinating stop-codon recognition in the decoding centre with peptidyl-tRNA hydrolysis in the PTC.
Figure 6.
The switch loop. Conservation of switch loop sequences in (A) RF1 and (B) RF2. (C) Packing of the switch loop of RF1 (light purple and red) in a pocket formed by conserved elements of 16S rRNA (light blue), ribosomal protein S12 (blue) and helix 69 of 23S rRNA (grey). Residues 302–304 of RF1, which were mutated in this work, are shown in red. The rest of RF1 is shown in yellow.
To test the importance of the switch loop and h69 for signal transduction from the decoding centre to the PTC, we created three mutational variants of RF1 and measured their activities in the peptide release reaction using wild-type ribosomes or ribosomes containing a deletion of helix 69 in 23S rRNA (Δh69) (Ali et al, 2006). Two point mutants, R303A and S304A, contain alanine substitutions of the only conserved side chains in the switch loop of RF1. As the only similarity between the switch loops of RF1 and RF2 is their length, we also tested a mutant version of RF1 carrying a deletion in residues 302–304. This mutation was designed to disrupt interactions of the switch loop with the pocket formed by A1492, A1493 and A1913 and with h69 of 23S rRNA.
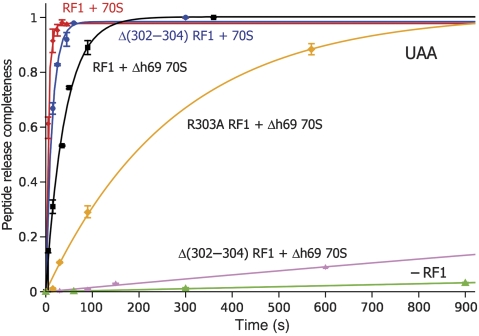
The rates of peptide release by the two point mutants in response to a UAA stop codon were indistinguishable from that of wild-type RF1 (0.2 s−1; Table I) in combination with wild-type 70S ribosomes. The R302A mutant is modestly (six-fold) slower than wild-type RF1 when used with Δh69 mutant 70S ribosomes (Figure 7; Table I). These results show that the identities of the side chains in the switch loop are not critical for RF1 function.
Table 1. Rates of peptide release by mutant versions of RF1 and 70S ribosomes in response to a UAA stop codona.
| Wild-type 70S | Δh69 70S | |
|---|---|---|
| Wild-type RF1 | 0.20±0.01 | 2.5±0.2 × 10−2 |
| RF1 | 0.20±0.01 | 4.0±0.4 × 10−3 |
| S304A RF1 | 0.19±0.01 | ND |
| Δ302–304 RF1 | 7.0±0.4 × 10−2 | 5.1±0.2 × 10−5 |
| No RF1 | 1.6±0.1 × 10−5 | ND |
| aRates are given in units of s−1. | ||
Figure 7.
Time courses for release of [35S]-fMet from [35S]-fMet-tRNA catalysed by wild-type and mutant versions of RF1 with wild-type and Δh69 ribosomes (Ali et al, 2006) in the presence of the UAA stop codon. Each data point represents the average of at least two measurements.
The mutant RF1 containing a deletion in the switch loop showed a modest (three-fold) decrease in peptide release rate with wild-type ribosomes (0.07 s−1). However, the rate of peptide release by Δh69 mutant ribosomes, which is only 10-fold slower than that of wild-type ribosomes when tested with wild-type RF1 (Ali et al, 2006, and this work), is decreased by >103-fold in the presence of the RF1 Δ302–304 deletion mutant. The synthetic-negative behaviour resulting from combining these two mutations indicates that they interact (directly or indirectly) in a common pathway, and suggests that the rate-limiting step(s) for the reactions involving (Δ302–304)RF1 or (Δh69)70S is different from that of the reaction between wild-type RF1 and 70S. We suggest that the pathway in question is a conformational change required for directing domain 3 into the PTC, which becomes rate limiting when the mutant RF1 or ribosomes are used, and that cooperative interaction between an intact h69 and switch loop are critical for docking of the catalytic Gln230 in the active site.
Materials and methods
mRNA, tRNA and RF1
The M1–27 mRNA used for construction of the termination complex was chemically synthesized (IDT) to give the following nucleotide sequence: GGC AAGGAGGU AAAA AUG UAG AAAAAA. The 8 base-pair Shine–Dalgarno sequence places the AUG start codon at the 70S ribosome P site followed immediately by a UAG stop codon. Escherichia coli tRNAfMet was purchased from Sigma-Aldrich. T. thermophilus release factor RF1 was purified as described earlier (Laurberg et al, 2008).
Preparation of 70S ribosomes
The 70S ribosomes were purified from T. thermophilus as earlier described (Laurberg et al, 2008). The ribosomes were dissociated into subunits and reassociated into vacant 70S ribosomes to remove endogenous mRNA and tRNA as described (Laurberg et al, 2008). Ribosomes were adjusted to a concentration of 20 mg ml–1 (8 μM), and aliquots were flash frozen in liquid nitrogen and stored at −80°C until further use.
Complex formation and co-crystallization of RF1–ribosome complexes
Reassociated 70S ribosomes were incubated with M1–27 mRNA and deacylated tRNAfMet at 37°C for 40 min in buffer I (25 mM Tris-OAc (pH 7.0), 50 mM KOAc, 10 mM NH4OAc, 10 mM Mg(OAc)2). RF1 was then added and incubated at 37°C for 30 min. The final concentration of 70S ribosomes was 10 mg ml–1 (4 μM), containing 2.8 mM deoxy-Bigchaps (Hampton Research); the final molar ratios were 70S:M1–27 mRNA:tRNAfMet:RF1=1:2.2:1.4:4.8. The ribosome termination complex was clarified by centrifugation at 16 000 g for 5 min at room temperature before crystallization.
Initial crystallization screening was performed around conditions earlier reported (Selmer et al, 2006; Laurberg et al, 2008) by dispensing 0.2+0.2 μl sitting drops using a Phoenix robotic liquid handling system (Art Robbins) with 96-well plates. After optimization, crystals were grown by the sitting-drop vapour diffusion method dispensed robotically using 1–2 μl ribosome complexes mixed with 1–2 μl reservoir solution (100 mM Tris-OAc, pH 7.0, 200 mM KSCN, 3.4–4.8% PEG20 000, 2.5–11.5% PEG200) at 22.5°C. Crystals emerged after 5–7 days and matured after 2–3 weeks. Crystals were then subjected to cryoprotection by gradually replacing the mother liquor with cryoprotection buffer II (100 mM Tris-OAc, pH 7.0, 200 mM KSCN, 5% PEG20 000, 25% MPD, 14% PEG200 and 10 mM Mg(OAc)2). Crystals were flash frozen by plunging into liquid nitrogen.
Construction and isolation of mutant RF1 and peptide release assay
The RF1 gene from E. coli incorporating a C-terminal 6His-tag (Korostelev et al, 2008) was used to generate mutations at the switch loop of RF1 using Quick-Change mutagenesis (Stratagene, La Jolla, CA). RF1 proteins were expressed and purified on Ni-NTA agarose resin (Qiagen) using standard procedures and stored in 50 mM Tris–HCl (pH 7.0), 60 mM NH4Cl, 10 mM MgCl2 and 5 mM β-mercaptoethanol (βME) at −80°C.
The 70S ribosomes were prepared from salt-washed E. coli MRE600 ribosomes as described (Moazed and Noller, 1986, 1989). The tRNAfMet was aminoacylated and formylated as described (Lancaster and Noller, 2005). Termination complexes were formed by incubating 70S ribosomes (0.4 μM), M0–27 mRNA (1 μM) and [35S]fMet-tRNAfMet (0.2 μM) for 20 min at 37°C in Buffer A (50 mM KHepes (pH 7.6), 75 mM NH4Cl, 20 mM MgCl2, 5 mM βME). The nucleotide sequence of the chemically synthesized M0–27 mRNA (IDT) is GGC AAGGAGGU AAAA AUG UAA AAAAAA, which places the UAA stop codon at the 70S ribosome A site. The [Mg++] was reduced to 10 mM by addition of buffer A lacking MgCl2. For peptide release assays, the complex was added to RF1 and incubated in Buffer A (adjusted to 10 mM MgCl2) at room temperature. Aliquots were removed from the reaction at each time point and quenched in five volumes of 0.1 N HCl; hydrolysed [35S]fMet was extracted with 1 ml of ethyl acetate, 0.8 ml of which was added to scintillation cocktail and counted. Reactions were performed using a 2- to 10-fold molar excess of RF1; catalytic rates measured for 2- and 10-fold excess of RF1 were similar for each of the tested reactions, suggesting that the reactions proceeded under release-factor saturating conditions.
Supplementary Material
Acknowledgments
We thank John Paul Donohue for assistance with data processing and computational support, Dmitri Ermolenko, Iraj Ali and Laura Lancaster for reagents and helpful discussions regarding biochemical experiments, Alexei Korennykh for helpful discussions regarding kinetic measurements and the staffs at SSRL beamlines 7-1, 9-1, 9-2, 11-1 and 12-2 and APS beamline 23ID-D for their expert support during screening and data collection. Structural coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID 3MR8, 3 MRZ, 3 MS0, 3MS1). These studies were supported by grants (to HFN) from the NIH, the NSF and the Agouron Foundation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Agrawal RK, Penczek P, Grassucci RA, Burkhardt N, Nierhaus KH, Frank J (1999) Effect of buffer conditions on the position of tRNA on the 70 S ribosome as visualized by cryoelectron microscopy. J Biol Chem 274: 8723–8729 [DOI] [PubMed] [Google Scholar]
- Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF (2006) Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol Cell 23: 865–874 [DOI] [PubMed] [Google Scholar]
- Brenner S, Barnett L, Katz ER, Crick FH (1967) UGA: a third nonsense triplet in the genetic code. Nature 213: 449–450 [DOI] [PubMed] [Google Scholar]
- Brenner S, Stretton AO, Kaplan S (1965) Genetic code: the ‘nonsense' triplets for chain termination and their suppression. Nature 206: 994–998 [DOI] [PubMed] [Google Scholar]
- Brunelle JL, Shaw JJ, Youngman EM, Green R (2008) Peptide release on the ribosome depends critically on the 2′ OH of the peptidyl-tRNA substrate. RNA 14: 1526–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Bürgi HB, Dunitz JD, Lehn JM, Wipff G (1974) Stereochemistry of reaction paths at carbonyl centres. Tetrahedron 30: 1563–1572 [Google Scholar]
- Capecchi MR (1967) Polypeptide chain termination in vitro: isolation of a release factor. Proc Natl Acad Sci USA 58: 1144–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR, Klein HA (1969) Characterization of three proteins involved in polypeptide chain termination. Cold Spring Harb Symp Quant Biol 34: 469–477 [DOI] [PubMed] [Google Scholar]
- Caskey CT, Beaudet AL, Scolnick EM, Rosman M (1971) Hydrolysis of fMet-tRNA by peptidyl transferase. Proc Natl Acad Sci USA 68: 3163–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T (2009) Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci USA 106: 2571–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific [Google Scholar]
- Dincbas-Renqvist V, Engstrom A, Mora L, Heurgue-Hamard V, Buckingham R, Ehrenberg M (2000) A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J 19: 6900–6907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr E, Jelesarov I (2000) Thermodynamic analysis of cavity creating mutations in an engineered leucine zipper and energetics of glycerol-induced coiled coil stabilization. Biochemistry 39: 4472–4482 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fei J, Kosuri P, MacDougall DD, Gonzalez RL (2008) Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell 30: 348–359 [DOI] [PubMed] [Google Scholar]
- Freistroffer DV, Kwiatkowski M, Buckingham RH, Ehrenberg M (2000) The accuracy of codon recognition by polypeptide release factors. Proc Natl Acad Sci USA 97: 2046–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova LY, Tsivkovskii RY, Sivolobova GF, Oparina NY, Serpinsky OI, Blinov VM, Tatkov SI, Kisselev LL (1999) Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5: 1014–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Lorenzo MG, Spahn CM, Agrawal RK, Grassucci RA, Penczek P, Chakraburtty K, Ballesta JP, Lavandera JL, Garcia-Bustos JF, Frank J (2000) Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 A resolution. EMBO J 19: 2710–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graille M, Heurgue-Hamard V, Champ S, Mora L, Scrima N, Ulryck N, van Tilbeurgh H, Buckingham RH (2005) Molecular basis for bacterial class I release factor methylation by PrmC. Mol Cell 20: 917–927 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Pt 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst 26: 795–800 [Google Scholar]
- Kellis JT Jr, Nyberg K, Sali D, Fersht AR (1988) Contribution of hydrophobic interactions to protein stability. Nature 333: 784–786 [DOI] [PubMed] [Google Scholar]
- Klaholz BP, Pape T, Zavialov AV, Myasnikov AG, Orlova EV, Vestergaard B, Ehrenberg M, van Heel M (2003) Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature 421: 90–94 [DOI] [PubMed] [Google Scholar]
- Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF (2008) Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci USA 105: 19684–19689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Bertram R, Chapman MS (2002) Simulated-annealing real-space refinement as a tool in model building. Acta Crystallogr D Biol Crystallogr 58: 761–767 [DOI] [PubMed] [Google Scholar]
- Lancaster L, Noller HF (2005) Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol Cell 20: 623–632 [DOI] [PubMed] [Google Scholar]
- Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF (2008) Structural basis for translation termination on the 70S ribosome. Nature 454: 852–857 [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF (1986) Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell 47: 985–994 [DOI] [PubMed] [Google Scholar]
- Moazed D, Noller HF (1989) Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell 57: 585–597 [DOI] [PubMed] [Google Scholar]
- Mora L, Heurgue-Hamard V, Champ S, Ehrenberg M, Kisselev LL, Buckingham RH (2003) The essential role of the invariant GGQ motif in the function and stability in vivo of bacterial release factors RF1 and RF2. Mol Microbiol 47: 267–275 [DOI] [PubMed] [Google Scholar]
- Mora L, Heurgue-Hamard V, de Zamaroczy M, Kervestin S, Buckingham RH (2007) Methylation of bacterial release factors RF1 and RF2 is required for normal translation termination in vivo. J Biol Chem 282: 35638–35645 [DOI] [PubMed] [Google Scholar]
- Pavlov MY, Freistroffer DV, Dincbas V, MacDougall J, Buckingham RH, Ehrenberg M (1998) A direct estimation of the context effect on the efficiency of termination. J Mol Biol 284: 579–590 [DOI] [PubMed] [Google Scholar]
- Petry S, Brodersen DE, Murphy FVt, Dunham CM, Selmer M, Tarry MJ, Kelley AC, Ramakrishnan V (2005) Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 123: 1255–1266 [DOI] [PubMed] [Google Scholar]
- Rawat U, Gao H, Zavialov A, Gursky R, Ehrenberg M, Frank J (2006) Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM. J Mol Biol 357: 1144–1153 [DOI] [PubMed] [Google Scholar]
- Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J (2003) A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature 421: 87–90 [DOI] [PubMed] [Google Scholar]
- Rodnina MV, Beringer M, Wintermeyer W (2006) Mechanism of peptide bond formation on the ribosome. Q Rev Biophys 39: 203–225 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Strobel SA, Steitz TA (2005) An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438: 520–524 [DOI] [PubMed] [Google Scholar]
- Scolnick E, Tompkins R, Caskey T, Nirenberg M (1968) Release factors differing in specificity for terminator codons. Proc Natl Acad Sci USA 61: 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scolnick EM, Caskey CT (1969) Peptide chain termination. V. The role of release factors in mRNA terminator codon recognition. Proc Natl Acad Sci USA 64: 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seit Nebi A, Frolova L, Ivanova N, Poltaraus A, Kiselev L (2000) [Mutation of a glutamine residue in the universal tripeptide GGQ in human eRF1 termination factor does not cause complete loss of its activity]. Mol Biol (Mosk) 34: 899–900 [PubMed] [Google Scholar]
- Seit-Nebi A, Frolova L, Justesen J, Kisselev L (2001) Class-1 translation termination factors: invariant GGQ minidomain is essential for release activity and ribosome binding but not for stop codon recognition. Nucleic Acids Res 29: 3982–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Serrano L, Neira JL, Sancho J, Fersht AR (1992) Effect of alanine versus glycine in alpha-helices on protein stability. Nature 356: 453–455 [DOI] [PubMed] [Google Scholar]
- Shaw JJ, Green R (2007) Two distinct components of release factor function uncovered by nucleophile partitioning analysis. Mol Cell 28: 458–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Brandsen J, Jancarik J, Yokota H, Kim R, Kim SH (2004) Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome. J Mol Biol 341: 227–239 [DOI] [PubMed] [Google Scholar]
- Tama F, Valle M, Frank J, Brooks CL III (2003) Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc Natl Acad Sci USA 100: 9319–9323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkins RK, Scolnick EM, Caskey CT (1970) Peptide chain termination. VII. The ribosomal and release factor requirements for peptide release. Proc Natl Acad Sci USA 65: 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobro S, Aqvist J (2007) A model for how ribosomal release factors induce peptidyl-tRNA cleavage in termination of protein synthesis. Mol Cell 27: 758–766 [DOI] [PubMed] [Google Scholar]
- Vestergaard B, Sanyal S, Roessle M, Mora L, Buckingham RH, Kastrup JS, Gajhede M, Svergun DI, Ehrenberg M (2005) The SAXS solution structure of RF1 differs from its crystal structure and is similar to its ribosome bound cryo-EM structure. Mol Cell 20: 929–938 [DOI] [PubMed] [Google Scholar]
- Vestergaard B, Van LB, Andersen GR, Nyborg J, Buckingham RH, Kjeldgaard M (2001) Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol Cell 8: 1375–1382 [DOI] [PubMed] [Google Scholar]
- Vogel Z, Zamir A, Elson D (1969) The possible involvement of peptidyl transferase in the termination step of protein biosynthesis. Biochemistry 8: 5161–5168 [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V (2009) Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol 16: 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V (2008) Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman EM, McDonald ME, Green R (2008) Peptide release on the ribosome: mechanism and implications for translational control. Annu Rev Microbiol 62: 353–373 [DOI] [PubMed] [Google Scholar]
- Zhang W, Dunkle JA, Cate JH (2009) Structures of the ribosome in intermediate states of ratcheting. Science 325: 1014–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoldak G, Redecke L, Svergun DI, Konarev PV, Voertler CS, Dobbek H, Sedlak E, Sprinzl M (2007) Release factors 2 from Escherichia coli and Thermus thermophilus: structural, spectroscopic and microcalorimetric studies. Nucleic Acids Res 35: 1343–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.