Abstract
CCR4, an evolutionarily conserved member of the CCR4–NOT complex, is the main cytoplasmic deadenylase. It contains a C-terminal nuclease domain with homology to the endonuclease-exonuclease-phosphatase (EEP) family of enzymes. We have determined the high-resolution three-dimensional structure of the nuclease domain of CNOT6L, a human homologue of CCR4, by X-ray crystallography using the single-wavelength anomalous dispersion method. This first structure of a deadenylase belonging to the EEP family adopts a complete α/β sandwich fold typical of hydrolases with highly conserved active site residues similar to APE1. The active site of CNOT6L should recognize the RNA substrate through its negatively charged surface. In vitro deadenylase assays confirm the critical active site residues and show that the nuclease domain of CNOT6L exhibits full Mg2+-dependent deadenylase activity with strict poly(A) RNA substrate specificity. To understand the structural basis for poly(A) RNA substrate binding, crystal structures of the CNOT6L nuclease domain have also been determined in complex with AMP and poly(A) DNA. The resulting structures suggest a molecular deadenylase mechanism involving a pentacovalent phosphate transition.
Keywords: CCR4, crystal structure, deadenylase, mRNA degradation, poly(A) substrate
Introduction
The CCR4–NOT complex from Saccharomyces cerevisiae is a multi-functional machinery controlling mRNA metabolism. Its components are implicated in several aspects of mRNA and protein expression, including transcription initiation, elongation, mRNA degradation, ubiquitination and protein modification (Collart, 2003; Lau et al, 2009). The complex is common to most eukaryotes (Draper et al, 1995; Albert et al, 2000; Gavin et al, 2002) and exists in two predominant forms in vivo, with molecular weights of 0.9–1.2 and 1.9–2.0 MDa in yeast. The identified smaller core complex (Liu et al, 1998; Bai et al, 1999; Chen et al, 2001; Cui et al, 2008) contains the following proteins: five NOT proteins (Not1p to Not5p), Caf1p, Caf40p, Caf130p, Ccr4p and BTT1. A number of other proteins known to interact with Ccr4p, Caf1p or NOT proteins, such as Dhh1p Caf4p, Caf16p, Dbf2p and Mob1p, may also be part of the larger complex (Liu et al, 1997; Hata et al, 1998; Komarnitsky et al, 1998). In mammals, including human beings, their homologues form a similar multi-subunit complex, which has a significant function in the regulation of several cellular machines (Mahadevan and Struhl, 1990; Seufert and Jentsch, 1990; Collart and Struhl, 1993, 1994; Moqtaderi et al, 1996; Lee et al, 1998; Oberholzer and Collart, 1998; Badarinarayana et al, 2000; Tucker et al, 2001, 2002; Albert et al, 2002; Deluen et al, 2002; Lenssen et al, 2002; Maillet and Collart, 2002; Viswanathan et al, 2003).
Among the various functions of the CCR4–NOT complex, deadenylation has the greatest importance as it is crucial for gene expression and many biological processes (Garneau et al, 2007; Bartlam and Yamamoto, 2010). Regulation of mRNA turnover rates is essential in determining the abundance and translational efficiencies of mRNAs. Removal of the 3′ poly(A) tail triggers degradation of mRNAs, and thus deadenylation is a critical step in mRNA degradation and regulates the abundance of mRNA (Wilusz et al, 2001; Parker and Song, 2004). As a highly conserved member of the CCR4–NOT complex, yCcr4p is the major cytoplasmic deadenylase in yeast and acts as the main catalytic component (Chen et al, 2002). Yeast Ccr4p features three major functional domains and has been categorized as a member of the endonuclease-exonuclease-phosphatase (EEP) family of proteins through biochemical studies (Draper et al, 1994). The N-terminal glutamine/asparagine-rich region is believed to have a transcriptional activation domain that interacts with the transcriptional machinery. A central domain featuring several tandem copies of a leucine-rich-repeat (LRR) domain (Malvar et al, 1992) has been reported to interact with yCaf1p (Draper et al, 1995), with other putative components of the core CCR4–NOT complex (Liu et al, 2001) and with potential-binding ligands of the whole complex. This LRR domain is thus considered to be the link that connects yCcr4p to the remainder of the complex (Draper et al, 1994; Liu et al, 1998) and other ligands, and distinguishes all Ccr4p orthologues from other EEP family members and CCR4-like proteins (Dupressoir et al, 2001; Chen et al, 2002). The C-terminal region contains a deadenylase domain characteristic of the exonuclease-endonuclease-phosphatase superfamily with conserved catalytic Asp and His residues. Within the EEP family, the C-terminal region of yCcr4p shares significant homology to an Mg2+-dependent exonuclease III (Exo III) group of proteins, also known as apurinic (AP) endonucleases, which are critical for maintenance of genome integrity in the DNA base excision repair pathway. The AP endonucleases include the Exo III from Escherichia coli, human APE1 (HAP1) and APN2 from S. cerevisiae (Chen et al, 2002), and are homologous to inositol polyphosphate-5′-phosphatases and sphingomyelinases (Dlakic, 2000; Whisstock et al, 2000).
As the main catalytic deadenylase in the yeast CCR4–NOT complex, yCcr4p was found to have both RNase and single-stranded DNA 3′–5′ exonuclease activity in vitro (Chen et al, 2002), and to have a strong preference for poly(A) substrates (Viswanathan et al, 2003). Mutations in yCcr4p putative catalytic residues abolish CCR4–NOT deadenylase function in vivo (Chen et al, 2002; Tucker et al, 2002). According to earlier results, the CCR4p is active as a monomer and retains activity in the absence of CAF1 and the other components of the CCR4–NOT complex. Recent observations that the LRR domain of yCcr4p is vital for CCR4 deadenylase activity (Clark et al, 2004) suggest that it provides an interaction surface for binding partners and/or substrate RNA and may also be required for its own deadenylase activity. The transition of the distributive-to-processive manner of deadenylation for yCcr4p and its ability to contact a certain length or type of RNA sequence are identified as well to be sites by which other factors may regulate yCcr4p deadenylase activity in vivo (Viswanathan et al, 2003). Interestingly, Ccr4 homologues do not appear to exist in trypanosomes, suggesting that deadenylation is mediated exclusively by Caf1 (Schwede et al, 2008).
In humans, two genes are homologous to yCcr4p: CNOT6 (also known as CCR4a) and the recently identified CNOT6L (also known as CCR4b). CNOT6L, such as CNOT6 and yCcr4p, exhibits deadenylase activity both in vitro and in vivo, and associates with human CNOT1, CNOT2, CNOT3, CNOT7 (CAF1a), CNOT8 (CAF1b) and CNOT9 (CAF40, Rcd1) to form a multi-subunit complex acting as a regulatory platform (Morita et al, 2007). Although significant data suggests that the deadenylase activity of yCcr4p contributes to cellular homeostasis and a number of other important biological processes, the function and catalytic mechanism of human CNOT6L still remain unclear. In contrast to yeast Caf1, which replaces the consensus DEDD nuclease motif with an SEDQ motif and thus lacks two essential catalytic residues (Thore et al, 2003), its human orthologues CNOT7 and CNOT8 have been shown to contain a conserved catalytic site and to be highly active in deadenylation in vitro. Furthermore, in vivo analysis has shown that CAF1 has a critical function in deadenylation in mammalian cells (Mauxion et al, 2008; Schwede et al, 2008), although mutual functions of CAF1 and CCR4 remain unclear.
To gain insight into the substrate RNA recognition, deadenylation mechanism and related functional processes, we have determined the high-resolution crystal structure of the C-terminal catalytic domain of CNOT6L. We have also performed site-directed mutagenesis and assayed both the activity and substrate specificity. Our results show that the catalytic domain of CNOT6L folds into an active, magnesium-dependent monomer and remains functional without the presence of the N-terminal LRR domain. Amino acids equivalent to those critical for the deadenylase activity of CCR4p are vital for the activity of CNOT6L, which shows its substrate preference for RNA with poly(A) sequence. The mechanism of deadenylation, a poly(A)-binding model and how the catalytic manner affects the whole reaction are discussed.
Results and discussion
Overall structure
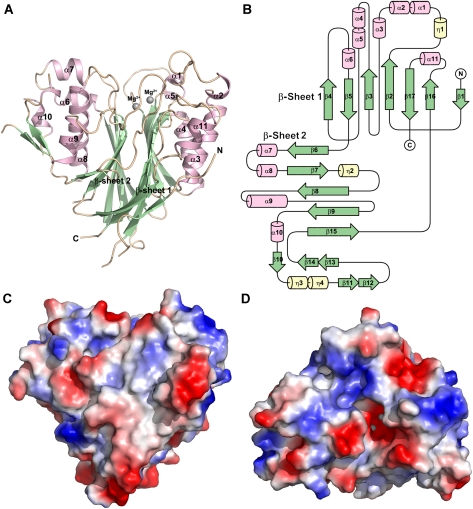
In this study, the full-length human CNOT6L protein, consisting of LRR and nuclease domains, could not be expressed and purified well in E. coli. Only by truncating the human CNOT6L protein to its nuclease domain could we produce crystals, and the structure of a SeMet derivative was determined at 1.94 ˚ resolution. The crystal structure of the catalytic domain of CNOT6L, with a molecular mass of around 45 kDa, is to the best of our knowledge the first representative deadenylase from the EEP superfamily. The overall structure of the CNOT6L catalytic domain features a two-layered α–β sandwich with approximate two-fold internal symmetry (Figure 1A). Two mostly antiparallel sheets sit on the interior, flanked by two α-helical layers on the outside. The heart-shaped molecule has approximate dimensions of 58 × 55 × 54 ˚ and shares the typical topology within the EEP family (Supplementary Figure 1). The present model covers amino-acid residues from 169 to 539 of CNOT6L and includes a total of 11 α-helices and 17 β-strands. The interior β-strands are arranged in the two sheets in the order β4-β5-β3-β2-β17-β16 and β6-β7-β8-β9-β15, respectively. The outer α-helices and small β-strands interrupt the interior sheets and form the wings of the heart-shaped structure (Figure 1A and B). Two magnesium ions were identified in the structure and are buried in the cleft formed by several loops on the top of the molecule (Figure 1C and D). Furthermore, the cleft extends throughout the surface and is generally positively charged, with selected negative charge around the metal ion-binding site, suggesting a preformed cradle for RNA substrate binding.
Figure 1.
The overall structure of the human CNOT6L nuclease domain. (A) Cartoon representation of the CNOT6L nuclease domain. α-helices, β-strands and loops are coloured in light pink, pale green and wheat, respectively. The bound Mg2+ ions are shown as grey sphere. (B) Schematic of the secondary structure components of CNOT6LT. The diagram is generated by using the program of TopDraw with α-helices, β-strands and other secondary structure components coloured by purple, green and yellow, respectively. (C, D) Surface vacuum electrostatics of the Mg2+-bound CNOT6L nuclease domain. Positively charged and negatively charged regions are presented by blue and red, respectively. Mg2+ ion is buried in the substrate-binding cleft on the top of the molecule (C).
Similarity to other EEP members
CCR4 proteins from different species have a strictly conserved catalytic domain and share high overall sequence identity (Supplementary Figure 2). As predicted by protein-fold recognition, but not confirmed before this study, the structure of human CNOT6L also shares strong similarity with other members of the EEP family of nucleases. The DALI server identified a number of related structures, including Sphingomyelin phosphodiesterase (PDB code 2DDS, Z-score 19.7, RMSD 3.2 ˚ for 235 residues), the L1-EN reverse transcriptase domain (PDB code 1VYB, Z-score 19.0, RMSD 2.7 ˚ for 209 residues), DNA apurinic or apyrimidinic site lyase (PDB code 2ISI, Z-score 18.9, RMSD 3.2 ˚ for 226 residues), human APE1 (PDB code 1DEW/1DE8, Z-score 18.6, RMSD 3.2 ˚ for 228 residues) and E. coli. Exo III (PDB code 1KAO, Z-score 18.3, RMSD 3.2 ˚ for 215 residues). These proteins only share 15–18% sequence identity with the catalytic domain of CNOT6L.
Superposition of the CNOT6L structure with those of L1-EN and human APE1 shows that the interior core β-sheets, each composed of at least five strands, are very similar, whereas the outer ‘wings’ exhibit significant diversity in size and shape (Supplementary Figure 1). Despite the rather low sequence identity and the differences in molecular weight, the core of CNOT6L superimposes well with the cores of L1-EN and APE1 to yield an RMSD <2.0 ˚. Another striking similarity appears around the coordinated magnesium ion with several strictly conserved residues, including the putative catalytic Asp (Asp489 in CNOT6L) and His (His529 in CNOT6L), indicating that the EEP family members share a rigid active site and common mechanism in their various functions.
Active site and magnesium ion binding
The APE1-like nucleases typically contain five highly conserved motifs representing the active site and which have important functions during the substrate binding and reaction, including (i) YNV, (ii) SLQE, (iii) ADLN, (iv) IDY and (v) SDH (Chen et al, 2002) (Supplementary Figure 2). Bold characters represent highly conserved residues in the active site of APE1-like nucleases. On the basis of structural comparison with APE1 and site-directed mutagenesis (to be discussed below), five conserved residues in human CNOT6L are, respectively, located within the five highly conserved motifs: Asn195, Glu240, Asp410, Asp489 and His529. These five residues were found to be essential for magnesium ion binding (Figure 2). All five residues are spatially close to each other and clustered within the negatively charged pocket around the metal ions, providing a suitable environment to carry out the specific hydrolysation.
Figure 2.
Deadenylase activity of human CNOT6LT in vitro. (A) The purified truncated CNOT6L and CNOT6 proteins with no tag were incubated with 5′-fluorescein isothiocyanate-labelled RNA substrate (RNA 3′-7N+20 As) and then, the labelled RNA was then analysed on a denaturing sequencing gel. A Coomassie stained gel of purified proteins were set up for the control. (B) Magnesium dependence of the CNOT6L deadenylase activity. The reaction was performed as described for the panel A, except the concentration of the Mg2+ ion was increased. The other reaction conditions were regulated to the optimal for the appearance of degraded ladders of RNA 3′–20 As. (C) The active site of CNOT6L is composed of five conserved residues. Ribbon representation of the molecule is coloured according to the scheme in Figure 1A. Active site residues are shown in stick representation and labelled, whereas the Mg2+ ions and waters are shown as grey and red spheres, respectively. (D) Enzymatic activities of three CNOT6L active site mutants (E240A, D489A, H529A) were tested as described in panel A. The wild-type CNOT6L and CNOT6 proteins were used as positive control.
As shown in Figure 2C, two bound magnesium ions were identified in the active site. The first, more stable ion is coordinated by the indispensable residue Glu240. Mutation of Glu240 and the absence of Mg2+ both abolished the enzymatic activity (Figure 2, discussed below), which indicates that Mg2+ is a necessary cofactor for the activity of CNOT6L. In contrast to the typical hexagonal coordination system observed in Mg2+-bound enzymes, the Mg2+ ion in the CNOT6L structure is only coordinated tightly by the Oɛ2 atom of Glu240 and by three water molecules with distances of 2.31, 2.40, 2.47 and 2.45 ˚, respectively. All coordinating ligands occupy one side of the substrate-binding cleft, leaving the other two positions available for waters or substrate during the reaction. Asp410, Asn412 and His529 coordinate a second, weakly bound Mg2+ ion approximately 4 ˚ from the first Mg2+.
CNOT6L catalytic domain exhibits Mg2+-dependent deadenylase activity
As earlier reported, the full-length CNOT6L is a catalytic component of the cytoplasmic deadenylase complex CCR4–NOT and exhibits deadenylase activity (Chen et al, 2002), as do all homologues of CCR4 proteins (Chen et al, 2002). To examine whether truncated CNOT6L, which was crystallized in this study, possesses deadenylase activity, a series of in vitro deadenylase assays with single-stranded poly(A) RNA as substrate was carried out. First, the nuclease domains of human CNOT6L and CNOT6 expressed in E. coli (Figure 2A) were tested and the reaction products were analysed on the denaturing sequencing gel. As the substrate was 5′-terminal labelled, the generation of an RNA ladder indicated trimming of the substrate from the 3′-terminal suggested that truncated forms of CNOT6L and CNOT6 have deadenylase activity in vitro despite the absence of the LRR domain (Figure 2A). This is consistent with an earlier study on nocturnin, a CCR4-like deadenylase from Xenopus laevis (Baggs and Green, 2003).
Generally, divalent metal ion binding will affect the enzymatic activity, and particularly so for the metal-dependent hydrolases. In this study, to examine the metal dependence of CNOT6L, exogenous Mg2+ was added into the reaction system in the concentration range 0.001–10 mM to detect changes in activity. As predicted, CNOT6L exhibited no deadenylase activity in the absence of Mg2+ (Figure 2B). After adding Mg2+, a gradual increase in activity was observed as the concentration of magnesium was increased (Figure 2B). Therefore, the deadenylase activity of CNOT6L was strictly dependent on the presence of Mg2+, with an approximately linear dependence on the concentration of magnesium.
Five single point mutations were introduced into truncated CNOT6L based on alignment of the active site residues: N195A, E240A, D410A, D489A and H529A (Figure 2C). Three mutants (E240A, D489A, H529A) were successfully purified and used for subsequent activity assays. With wild-type CNOT6L and CNOT6 nuclease domains as positive control, none of the three CNOT6L mutants tested exhibited deadenylase activity (Figure 2D), confirming that each of the conserved residues is crucial for deadenylase activity and formation of the rigid active site.
Substrate specificity and preference
yCcr4p was earlier reported to have both RNase and single-stranded DNA 3′–5′ exonuclease activity in vitro (Chen et al, 2002) with a strong preference for poly(A) substrates (Viswanathan et al, 2003). Full-length CNOT6L also showed 3′–5′ deadenylase activity (Morita et al, 2007). In addition to the RNA substrate with a 3′ 20mer poly(A) tail used for deadenylase activity assays (Morita et al, 2007), 12mer poly(A) RNA, 12mer poly(C) RNA, 12mer poly(G) RNA, 12mer poly(U) RNA and 14mer poly(A) DNA were also synthesized for activity assays to examine the substrate specificity and substrate preference. The concentration gradient for CNOT6L was settled at 0–0.08 μg/μl to gradually show the degraded RNA ladder. In the results of poly(A) RNA against poly(C), poly(G) and poly(U) RNA, the degraded RNA ladder appeared only in the poly(A) RNA profile, whereas poly(C), poly(G) and poly(U) RNA remained intact during the reaction (Figure 3A), indicating that CNOT6L prefers poly(A) RNA to poly(C), poly(G) and poly(U) RNA as a substrate. Another set of results comparing similar lengths of poly(A) RNA and poly(A) DNA as substrate (Figure 3B) shows the gradual appearance of the degraded ladder in the RNA profile, but exhibits only a trace of activity in the DNA profile, confirming earlier reports that CNOT6L prefers poly(A) RNA to poly(A) DNA.
Figure 3.
Substrate specificity and preference of human CNOT6L. (A) CNOT6L prefers poly(A) RNA to poly (C), poly(G) and poly(U) RNA as a substrate. The reactions were performed as described for the panels in Figure 2. The gradient concentrations of CNOT6L revealed the gradual appearance of the RNA ladder characteristic of deadenylase activity. (B) CNOT6L prefers poly(A) RNA to poly(A) DNA as a substrate. 12mer poly(A) RNA and 14mer poly(A) DNA substrates were used in the left and the right profiles, respectively. The concentration of CNOT6L was graded as well.
As all assays were performed on the catalytic domain of CNOT6L in the absence of the LRR domain, these data suggest that the nuclease domain of the CNOT6L protein recognizes poly(A) RNA and interacts with poly(A) RNA. The function of the LRR domain in substrate recognition and conferring deadenylase activity appears to be dispensable in the case of human CNOT6L. The 3′–5′ deadenylase activity of the CNOT6L catalytic domain alone for poly(A) RNA substrates appears to be increased with 7 nucleotides at the 5′ end, or reduced with 25 nucleotides at the 5′ end (Supplementary Figure 3). This suggests that the LRR domain might be a determining factor in the length of the poly(A) RNA substrates, either directly through substrate binding or indirectly through recruitment of other proteins into a poly(A) RNA-binding complex. Further work is required to determine the function of the LRR domain of CNOT6L. In the case of the larger substrate with 25 nucleotides at the 5′ end, a number of unexpected products were detected, even at low concentrations in which the 20A tail is not efficiently cleaved. As CNOT6L is a member of the EEP family of enzymes, it may be that CNOT6L catalytic domain has artificial endonuclease activity, although further experiments are needed to verify this and to identify the products.
Complex structures with ribo-adenine-5′-monophosphate and poly(A) DNA
To further understand the possible modes of binding and interaction of CNOT6L with poly(A) RNA substrates, we determined the structures of the CNOT6L nuclease domain in complex with ribo-adenine-5′-monophosphate (rAMP) to 2.4 Å, and with a 5A stretch of poly(A) DNA to 2.2 Å. Although the results above indicate that CNOT6L has a strong preference for poly(A) RNA over poly(A) DNA, we were unable to obtain complex crystal structures with poly(A) RNA substrates because of stability issues. As the protein–RNA contact should be similar to the protein–ssDNA contact, and because CNOT6L has trace activity for a poly(A) DNA substrate (Figure 3B), we, therefore, synthesized a short 5A stretch of poly(A) DNA to mimic the preferred substrate of CNOT6L. Data collection and refinement statistics for the two structures are summarized in Table I.
Table 1. Data collection and refinement statistics.
| Dataset | SeMet-CNOT6L (peak) | CNOT6L-AMP | CNOT6L-DNA (3A) |
|---|---|---|---|
| Data collection statistics | |||
| Wavelength | 0.9790 | 0.9796 | 0.9796 |
| Space group | P3221 | P3221 | P3221 |
| Cell dimensions a/b/c (Å) | 77.2/77.2/165.9 | 77.0/77.0/165.7 | 77.4/77.4/167.2 |
| Resolution (Å) | 50–1.94 (2.03–1.94) | 50–2.4 (2.44–2.40) | 50–2.2 (2.24–2.20) |
| Average I/σ (I)a | 47.7 (2.9) | 54.4 (4.6) | 52.7 (4.7) |
| Completeness (%)a | 85.1 (75.2) | 99.6 (96.0) | 99.7 (99.2) |
| Redundancya | 12.3 (9.2) | 10.2 (7.2) | 10.5 (9.2) |
| Rmerge (%)a,b | 7.6 (46.3) | 7.1 (38.1) | 6.6 (41.8) |
| Structure refinement statistics | |||
| Resolution (Å) | 50–1.94 | 50–2.4 | 50–2.2 |
| Average B-factor (Åb) | 44.6 | 48.8 | 45.0 |
| Rwork/Rfree (%)c | 22.0/25.4 | 20.0/25.3 | 21.5/26.3 |
| RMSD bond lengths (Å) | 0.018 | 0.011 | 0.016 |
| RMSD bond angles (deg) | 1.793 | 1.428 | 1.629 |
| Ramachandran plot | |||
| Most favoured (%) | 91.0 | 92.3 | 90.8 |
| Allowed (%) | 8.3 | 7.7 | 8.2 |
| Generously allowed (%) | 0.7 | 0 | 1.0 |
| Disallowed (%) | 0 | 0 | 0 |
| aValues in parentheses correspond to the highest-resolution shell. | |||
| bRmerge=∑i ∣I-〈I〉∣/∑〈I〉, where Ii is an individual intensity measurement and 〈I〉 is the average intensity for all the reflections. | |||
| cRwork/Rfree=∑∣∣Fo∣-∣Fc∣∣/∑∣Fo∣, where Fo and Fc are the observed and calculated structure factors, respectively. | |||
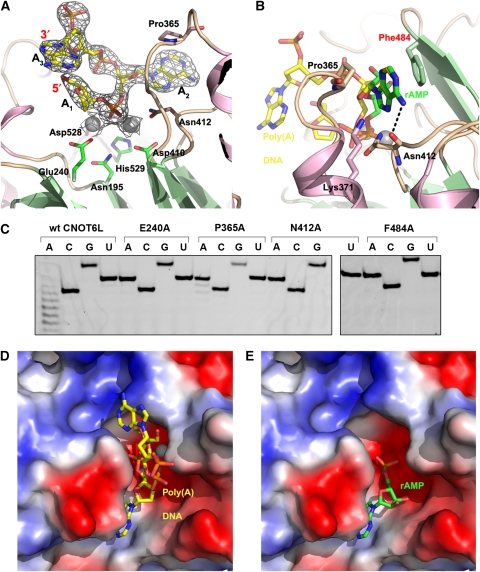
From the crystal structure of CNOT6L with poly(A) DNA, we could clearly identify two complete nucleotides (A2 and A3), and the deoxyribose moiety of a third nucleotide (A1) (Figure 4A; Supplementary Figure 4A). The 3A trinucleotide substrate fits comfortably into the binding cleft, with the scissile phosphate of nucleotide A2 fixed by three bonds, including two coordinated bonds with the two Mg2+ ions and a third bond between Asn412 and the dissociative O atom of the phosphate moiety. The adenine base stacks between the phenyl ring of Phe484 and the ring of Pro365 (Figure 4B). The 6′-NH2 group of the adenine base forms an additional 3.1 Å bond with the hydroxyl group of Asn412 and a 3.0 Å with a water molecule. A near-identical conformation of the adenine group of AMP suggests that Phe484, Pro365 and Asn412 are important for binding the poly(A) substrate. Mutating Asn412 or Phe484 abolishes deadenylase activity, whereas mutating Pro365 reduces the activity (Figure 4C; Supplementary Figure 5). It should be noted that mutating residues near the active site, but not interacting directly with the poly(A) DNA substrate, does not significantly impact deadenylase activity (Supplementary Figure 5). From our structural analysis and deadenylase assays, therefore, the base-stacking interaction with Phe484 and the specific interaction between Asn412 and the –NH2 group in the 6′ position of the adenine base suggest why adenine bases fit more comfortably into the active site than other nucleotide bases. The purine base G is most similar to A, but features a carbonyl oxygen in the 6′ position that would clash with the carbonyl oxygen of Asn412. The pyrimidine bases C and U have either an –NH2 or carbonyl group at the 4′ position. In our complex structure, the carbonyl oxygen of Asn412 interacts with the nitrogen atom at the 7′ position of the imidazole ring (within 3 ˚). If the pyrimidine ring of C or U bases occupied this position, the larger ring and the additional –NH2 or carbonyl group would point towards the carbonyl oxygen of Asn412 (within 1.5 ˚), thus preventing pyrimidine bases from accessing the recognition pocket. Nucleotides A1 and A3 do not appear to form any specific interactions with CNOT6L. Only the deoxyribose moiety of A1 is visible in the electron density maps; it interacts loosely with Tyr201 and Leu206. A3 is oriented such that the ribose moiety stacks against the ribose of A1, whereas the adenine base interacts largely with Trp363.
Figure 4.
Poly(A) substrate binding by human CNOT6L. (A) Structure of the poly(A) DNA substrate. The DNA molecule is shown in yellow stick representation and covered by a 2Fo-Fc difference map contoured at 1.5σ. Active site residues of CNOT6L are shown in stick representation and labelled. (B) Superposition of the AMP (green) and poly(A) DNA (yellow) ligands in the substrate-binding site of CNOT6L. The adenine bases of rAMP and poly(A) DNA bind in identical positions. Residues interacting with the adenine base are shown in stick representation and labelled. (C) Effect of mutations in the A2-binding pocket on deadenylase activity. The wild-type CNOT6L was used as positive control and the E240A mutant as negative control. Activities for the P365A, N412A and F484A mutants were assayed as described in Figures 2 and 3, using a substrate concentration of 0.02 μg/μl. Three gels were run simultaneously under identical conditions to accommodate all of the samples (see also Supplementary Figure 5). (D, E) Electrostatic surface potential representations showing poly(A) DNA (yellow) and rAMP (green), respectively, in the substrate-binding site of CNOT6L.
As detailed above, rAMP was identified from clear electron density in the complex structure (Supplementary Figure 4B) and adopts a largely similar orientation to nucleotide A2 in the poly(A) DNA complex structure (Figure 4B, D and E). The adenine base is in an identical position to the adenine of A2, stacked between Pro365 and Phe484, with the same interaction between Asn412 and the 6′ –NH2 group of the adenine base (Figure 4B). The phosphate moiety of rAMP, however, is in a different position to the equivalent A2 phosphate in the poly(A) DNA complex. In the rAMP structure, the phosphate group occupies the position of the Mg2+ ions in the wild-type and poly(A) DNA complex structure. Indeed, no Mg2+ ions are observed in the rAMP complex structure. The 2′OH group of rAMP forms a hydrogen bond to a water molecule stabilized by the -OH group of Thr481 (all within 3 ˚), whereas the terminal 3′OH group can also hydrogen bond to this water molecule (3.3 ˚), but interacts more strongly (2.6 ˚) with the Nδ2 atom of Asn479 (Supplementary Figure 6). Therefore, the rAMP structure provides some clues for the recognition of the 2′OH and 3′OH groups and might explain how the 3′ most residue of RNA fits into the catalytic site. A second rAMP nucleotide is observed in the structure about 25 ˚ from the first rAMP in the substrate-binding pocket (Supplementary Figure 7). This second rAMP nucleotide occupies a site formed by helix α6 and the β4-α6 loop, and interacts with the residues Lys270, Ile271, Ser273, Glu274 and Glu276. The two rAMP nucleotides are linked by a stretch of positively charged residues, suggesting that the second rAMP may delineate an additional RNA-binding site. However, we cannot rule out the possibility that the second rAMP nucleotide is a crystallization artefact and further work is needed to confirm the validity of this second site.
Proposed catalytic mechanism for CNOT6L deadenylation
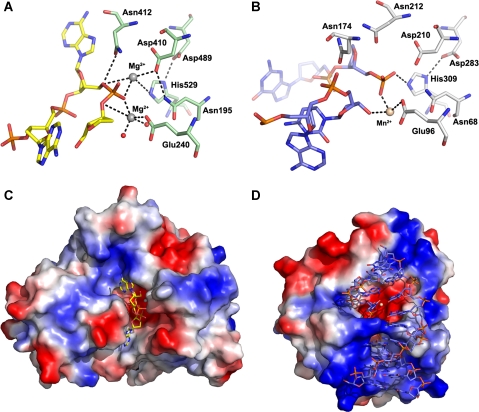
CNOT6L is related to APE1, particularly in the active site, which includes the same conserved residues, with similar orientations and binding pockets, except that the manganese ion in APE1 is replaced by a smaller magnesium ion in CNOT6L. In contrast to CNOT6L, the DNA substrate and product complexes of APE1 have been structurally determined, and a detailed reaction mechanism for this enzyme has been described (Mol et al, 2000). As the nature and position of all side chains in APE1 that are proposed to be involved in the orientation and phosphoester bond cleavage are strictly conserved in CNOT6L (Figure 5), the catalytic mechanism of hydrolase is likely to be conserved between APE1 and CNOT6L, although CNOT6L acts as a phosphatase instead of a phosphoesterase. In strict analogy to APE1, residue Asp410 in CNOT6L can act as an aspartate-activated hydroxyl nucleophile to attack the target phosphate polarized about the scissile bond.
Figure 5.
Close-up view of poly(A) DNA in the CNOT6L catalytic site. (A) The interaction of poly(A) DNA with the active site residues of CNOT6L. The poly(A) DNA is shown in yellow stick representation. Active site residues are shown in pale green stick representation. Mg2+ ions and waters are shown as grey and red spheres, respectively. (B) The APE1 (PDB code: 1DE8) active site interacts with the AP site of binding DNA, including an aspartate-activated hydroxyl nucleophilic attack and formation of a pentacovalent phosphate transition state. Part of the DNA molecule is shown in blue stick representation. Active site residues are shown in light grey, and the Mn2+ ion is shown in wheat. (C, D) Surface representations of the DNA-bound structures of CNOT6L and APE1, respectively.
No other contact is found to stabilize another O atom of the phosphate moiety adjacent to the adenosine, equivalent to the bond between APE1-Asn174 and its substrate. From the CNOT6L structures, there are no residues in the corresponding position to Asn174 of APE1 with equivalent function or properties. Nevertheless, the above chemical bonds are capable of orienting the reactive phosphate and indirectly polarizing it, making it more prone to hydroxyl nucleophilic attack. In CNOT6L, the potential nucleophile Asp410 is directed towards the target phosphate by forming hydrogen bonds with the main chain amino group of Asn412 and the side chain acylamino group of Asn195 through its side chain carbonyl and carboxyl groups, respectively. The distance is within 4 ˚, making hydroxyl nucleophilic attack possible. Residue Glu240 takes part in the coordination of Mg2+, and Asp489 stabilizes His529 through a hydrogen bond with its pentagonal ring. All of the above contacts provide an optimal fit for the 3A substrate and place it ready for reaction. It is evident from the CNOT6L-3A substrate complex structure that the phosphodiester bond aligned for cleavage is the one joining nucleotides A1 and A2. In this configuration, cleavage of poly(A) DNA would release a dinucleotide rather than the expected mononucleotide, as observed in the RNA ladder from enzymatic assays with poly(A) RNA. It should be noted that poly(A) DNA is not the natural substrate for CNOT6L and nucleotide A3 is likely to be irrelevant for natural substrates. Indeed, our assays show that CNOT6L has very low deadenylase activity towards poly(A) DNA (Figure 3B).
Asp410 is buried at the bottom of the active site, positioned by hydrogen bonds with the backbone amide of Asn412 and Asn195 and inclined to protonation. As a result of the bonds described above, the scissile P–O bond will be oriented and polarized with the buried Asp410 side chain favourably aligned to activate an attacking hydroxyl nucleophile. The aspartate-activated hydroxyl nucleophile will attack the target phosphate and form a new P–O bond, resulting in a pentacovalent phosphate transition state. Collapse of the transition state will result in cleavage of the scissile P–O bond. The transition state and leaving group of poly(A) will then be stabilized by the metal, with the inversion of the phosphate configuration after the bond cleavage.
Although the rAMP and poly(A) DNA complex structures reported here mimic the optimal poly(A) binding in the reaction pocket and imply a reasonable deadenylase mechanism of phosphate cleavage, more precise positioning of the poly(A) RNA substrate in the active site will require further structural studies using soaking or co-crystallization techniques to obtain complexes of the enzyme with its substrate or product. A structure-based deadenylation scheme will then be derived from the actual model.
Conclusions
The crystal structure of the truncated nuclease domain of human CNOT6L is the first representative structure of a deadenylase member from the EEP family, and reveals how a conserved APE1-like catalytic core has been expanded to yield a diverse family of EEP enzymes. The crystal structure of the catalytic domain of human CNOT6L, combined with mutational and biochemical data, indicate that CNOT6L displays full Mg2+-dependent deadenylase activity in vitro, and has strict substrate recognition and preference for poly(A) RNA. A similar catalytic mechanism and substrate-binding mode shared between CNOT6L and APE1 is supported by their conserved active site residues and structure-based superposition. A complex structures of the CNOT6L nuclease domain with a poly(A) DNA trinucleotide enables us to propose a deadenylase mechanism for CNOT6L, in which the target phosphate is polarized about the scissile bond and attacked by an aspartate-activated hydroxyl nucleophile. A pentacovalent phosphate transition state will collapse, with the metal ions stabilizing both the 3′-OH group of the uncleaved poly(A) chain and the leaving phosphate group of the rAMP product. The structures reported here provide an important starting point for the study of deadenylases and will benefit further studies aimed at dissecting the molecular mechanisms of other RNA-related enzymes.
Materials and methods
Protein expression, purification and site-directed mutagenesis
The truncated catalytic domain of human CNOT6L (residues 158–555) was cloned into the pGEX-6p-1 vector (GE Healthcare) and expressed as a glutathione-S-transferase fusion protein in E. coli. The CNOT6L catalytic domain was purified using glutathione-Sepharose 4B, ResourceQ and Superdex 200 gel filtration columns (GE Healthcare). The purity of samples was verified using SDS–PAGE stained with Coomassie Brilliant Blue. The CNOT6L catalytic domain was observed as a single band at 45 kDa. The protein was then concentrated to 15 mg/ml for crystallization. A selenomethionyl (SeMet) derivative of the CNOT6L catalytic domain was expressed in a minimal media containing 20 mg/ml SeMet (Neidhardt et al, 1974), and purified in the same way as the native protein, except that the dithiothreitol (DTT) concentration used was 2 mM. The catalytic domain of the wild-type human CNOT6 protein used for deadenylase assays was cloned, purified and concentrated in the same way.
All site-directed mutants for deadenylase assays were prepared according to their recombinant expression in mammalian cells (Morita et al, 2007). The CNOT6L catalytic domain mutants (L197A, L216A, L414A, E240A, P365A, N412A, F484A, D489A, H529A, P365A+F484A and P365A+N412A+F484A, numbered according to the full-length sequence) were amplified using the corresponding mammalian vector as a template and inserted into the pGEX-6p-1 vector (GE Healthcare). The mutants were then expressed and purified after the protocol used for the wild-type protein.
Crystallization, data collection and structure determination
Initial crystals of native CNOT6L catalytic domain were initially obtained using the hanging drop vapour diffusion method and further improved by adding 10 mM DTT into the condition consisting of 1.1 M ammonium tartrate dibasic, pH 7.0. A total of 1 μl of protein solution (15 mg/ml) was mixed with 1 μl of reservoir solution and equilibrated over 300 μl reservoir solution at 16°C. For the SeMet-substituted CNOT6L crystals, the condition was optimized into 0.1 M Hepes pH 7.5, 1.1 M ammonium tartrate and 0.2 M NDSB-201 as additive. The concentration of protein solution was also increased to 20 mg/ml. The crystals grew to maximum dimensions of 0.1 × 0.1 × 0.2 mm within 2 weeks. Diffraction quality crystals of both native and SeMet-substituted CNOT6L were transferred to cryoprotectant (reservoir solution mixed with 4 M sodium formate) and immediately flash cooled in liquid nitrogen.
Data for the Se-Met-derivative crystal were collected by synchrotron radiation on beamline BL-5A of the Photon Factory (Tsukuba, Japan) to 1.94 ˚. The derivative protein crystal belongs to the space group P3221 and contains only one molecule per asymmetric unit. All diffraction data were integrated, scaled and merged by the HKL2000 suite (Otwinowski and Minor, 1997). The structure of SeMet-substituted CNOT6L was solved by the single-wavelength anomalous dispersion method. The selenium sites were located with SHELX (Schneider and Sheldrick, 2002) and refined with SOLVE/RESOLVE (Terwilliger and Berendzen, 1999; Terwilliger, 2000). About 80% of the final model was automatically built with ARP/wARP (Perrakis et al, 2001). The remainder of the model was built manually with COOT (Emsley and Cowtan, 2004). Crystallographic refinement was carried out with REFMAC5 (Murshudov et al, 1997) in the CCP4 package. All data collection and refinement statistics are summarized in Table I.
To obtain ligand-bound complex structures, native CNOT6L crystals were soaked for 5 h at 20°C in a solution containing rAMP or poly(A) DNA (5′-AAAAA-3′) at a final concentration of 2 mM in 0.1 M Hepes, pH 7.5, 1.1 M ammonium tartrate and with 0.2 M NDSB-201 as additive. Immediately before data collection, crystals were soaked in cryoprotectant solution consisting of 4 M sodium formate. X-ray diffraction data were collected at 100 K on beam line BL17U1 of the Shanghai Synchrotron Radiation Facility (SSRF). All diffraction data were integrated, scaled and merged by the HKL2000 suite (Otwinowski and Minor, 1997).
The program PHASER (McCoy et al, 2007) was used for molecular replacement with the native structure of CNOT6L as a search model. The metal ions, AMP and poly(A) (DNA) were manually built and adjusted under the guidance of Fo-Fc difference maps using the program Coot (Emsley and Cowtan, 2004). Structural refinement was carried out with REFMAC5 (Murshudov et al, 1997).
Similarity alignments
The overall structure-based alignment of CCR4 deadenylases was performed for members of the family from different origins based on sequence comparison of their catalytic domains. The amino-acid sequences of human CCR4b (NP_653172), human CCR4a (NP_056270), CCR4b from Mus musculus (NP_659159), CCR4a from M. musculus (NP_997649), CCR4 from Drosophila melanogaster (AAK85705) in full-length and partial CCR4 from S. cerevisiae (AAB24455) were aligned using CLUSTALW (Thompson et al, 1994) and the sequence alignment was drawn with ESPript (Gouet et al, 1999).
The detailed alignment of the conserved motifs was generated using CLUSTALW (Thompson et al, 1994) and the sequence alignment was drawn with ESPript (Gouet et al, 1999). The aligned sequences are human CCR4b/CNOT6L, human CCR4a/CNOT6, INP54 (a representative yeast inositol 5′-phosphatase), ISC1 (yeast sphingomyelinase), APN2 (yeast orthologue of AP endonuclease), HAP1 (human APE1) and Exo III from E. coli.
Deadenylation assays
The synthesized RNA substrate (5′-UCUAAAUAAAAAAAAAAAAAAAAAAAA-3′) with final concentration of 0.1 μM was labelled with fluorescein isothiocyanate at the 5′ end. The purified CNOT6LT/CNOT6T (human CCR4b/CCR4a truncated form) proteins with no tag were used at a concentration of 0.1 μg/μl in the assays. The proteins in the reaction buffer (50 mM Hepes-NaOH, 150 mM NaCl, 10% glycerol, 1 mM DTT, 2 mM MgCl2, pH 7.4) were incubated at 37°C for 30 to 60 min together with the substrate. The reaction mixtures were fractionated on a 7 M urea, 25% sequence polyacrylamide denaturing gel. The products were analysed and quantified with an FLA-5000 (Fujifilm) fluorescence imager. The site-directed CNOT6LT mutants (L197A, L216A, L414A, E240A, P365A, N412A, F484A, D489A, H529A, P365A+F484A and P365A+N412A+F484A) were used for activity assays. The concentration of MgCl2 was regulated to determine the metal ion dependence. Poly(C), poly(G), poly(U) RNA and poly(A) DNA substrates were also used for further investigation of substrate specificity.
Structural data
Atomic coordinates and structure-factor amplitudes have been deposited in the Protein Data Bank for the CNOT6L nuclease domain (accession number: 3NGQ), the CNOT6L–AMP complex (accession number: 3NGN) and the CNOT6L–DNA complex (accession number: 3NGO).
Supplementary Material
Acknowledgments
We thank Yunhai Luo, Mingxiao He, Ya Wang, Mitsuhiro Yoneda and Takeshi Inoue for their technical assistance; Zhiyong Lou for help with data collection and processing and Yiwei Liu, Xuehui Chen and Liu Li for useful discussion. We also thank all of the beamline scientists at the Photon Factory in Japan and the SSRF in Shanghai for technical support. This work was supported by the Ministry of Science and Technology International Cooperation Project (grant number 2006DFB32420), the Ministry of Science and Technology 973 Project (grant number 2007CB914301), the National Natural Science Foundation of China (grant numbers 30221003 and 30770438) and the Tianjin Municipal Science and Technology Commission (grant number 08SYSYTC00200). This work was also supported in part by the Global COE Program (Integrative Life Science Based on the Study of Biosignaling Mechanisms), MEXT, Japan.
Footnotes
The authors declare that they have no conflict of interest.
References
- Albert TK, Hanzawa H, Legtenberg YI, de Ruwe MJ, van den Heuvel FA, Collart MA, Boelens R, Timmers HT (2002) Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J 21: 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert TK, Lemaire M, van Berkum NL, Gentz R, Collart MA, Timmers HT (2000) Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucleic Acids Res 28: 809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badarinarayana V, Chiang YC, Denis CL (2000) Functional interaction of CCR4-NOT proteins with TATAA-binding protein (TBP) and its associated factors in yeast. Genetics 155: 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs JE, Green CB (2003) Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol 13: 189–198 [DOI] [PubMed] [Google Scholar]
- Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, Denis CL (1999) The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol Cell Biol 19: 6642–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlam M, Yamamoto T (2010) The structural basis for deadenylation by the CCR4-NOT complex. Protein & Cell 5: 443–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chiang YC, Denis CL (2002) CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J 21: 1414–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Rappsilber J, Chiang YC, Russell P, Mann M, Denis CL (2001) Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J Mol Biol 314: 683–694 [DOI] [PubMed] [Google Scholar]
- Clark LB, Viswanathan P, Quigley G, Chiang YC, McMahon JS, Yao G, Chen J, Nelsbach A, Denis CL (2004) Systematic mutagenesis of the leucine-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J Biol Chem 279: 13616–13623 [DOI] [PubMed] [Google Scholar]
- Collart MA (2003) Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313: 1–16 [DOI] [PubMed] [Google Scholar]
- Collart MA, Struhl K (1993) CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J 12: 177–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Struhl K (1994) NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev 8: 525–537 [DOI] [PubMed] [Google Scholar]
- Cui Y, Ramnarain DB, Chiang YC, Ding LH, McMahon JS, Denis CL (2008) Genome wide expression analysis of the CCR4-NOT complex indicates that it consists of three modules with the NOT module controlling SAGA-responsive genes. Mol Genet Genomics 279: 323–337 [DOI] [PubMed] [Google Scholar]
- Deluen C, James N, Maillet L, Molinete M, Theiler G, Lemaire M, Paquet N, Collart MA (2002) The Ccr4-not complex and yTAF1 (yTaf(II)130p/yTaf(II)145p) show physical and functional interactions. Mol Cell Biol 22: 6735–6749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakic M (2000) Functionally unrelated signalling proteins contain a fold similar to Mg2+-dependent endonucleases. Trends Biochem Sci 25: 272–273 [DOI] [PubMed] [Google Scholar]
- Draper MP, Liu HY, Nelsbach AH, Mosley SP, Denis CL (1994) CCR4 is a glucose-regulated transcription factor whose leucine-rich repeat binds several proteins important for placing CCR4 in its proper promoter context. Mol Cell Biol 14: 4522–4531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper MP, Salvadore C, Denis CL (1995) Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol Cell Biol 15: 3487–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Morel AP, Barbot W, Loireau MP, Corbo L, Heidmann T (2001) Identification of four families of yCCR4- and Mg2+-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ (2007) The highways and byways of mRNA decay. Nat Rev 8: 113–126 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308 [DOI] [PubMed] [Google Scholar]
- Hata H, Mitsui H, Liu H, Bai Y, Denis CL, Shimizu Y, Sakai A (1998) Dhh1p, a putative RNA helicase, associates with the general transcription factors Pop2p and Ccr4p from Saccharomyces cerevisiae. Genetics 148: 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky SI, Chiang YC, Luca FC, Chen J, Toyn JH, Winey M, Johnston LH, Denis CL (1998) DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol Cell Biol 18: 2100–2107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Kolkman A, van Schaik FM, Mulder KW, Pijnappel WW, Heck AJ, Timmers HT (2009) Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem J 422: 443–453 [DOI] [PubMed] [Google Scholar]
- Lee TI, Wyrick JJ, Koh SS, Jennings EG, Gadbois EL, Young RA (1998) Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol Cell Biol 18: 4455–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenssen E, Oberholzer U, Labarre J, De Virgilio C, Collart MA (2002) Saccharomyces cerevisiae Ccr4-not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Mol Microbiol 43: 1023–1037 [DOI] [PubMed] [Google Scholar]
- Liu HY, Badarinarayana V, Audino DC, Rappsilber J, Mann M, Denis CL (1998) The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J 17: 1096–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HY, Chiang YC, Pan J, Chen J, Salvadore C, Audino DC, Badarinarayana V, Palaniswamy V, Anderson B, Denis CL (2001) Characterization of CAF4 and CAF16 reveals a functional connection between the CCR4-NOT complex and a subset of SRB proteins of the RNA polymerase II holoenzyme. J Biol Chem 276: 7541–7548 [DOI] [PubMed] [Google Scholar]
- Liu HY, Toyn JH, Chiang YC, Draper MP, Johnston LH, Denis CL (1997) DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J 16: 5289–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan S, Struhl K (1990) Tc, an unusual promoter element required for constitutive transcription of the yeast HIS3 gene. Mol Cell Biol 10: 4447–4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillet L, Collart MA (2002) Interaction between Not1p, a component of the Ccr4-not complex, a global regulator of transcription, and Dhh1p, a putative RNA helicase. J Biol Chem 277: 2835–2842 [DOI] [PubMed] [Google Scholar]
- Malvar T, Biron RW, Kaback DB, Denis CL (1992) The CCR4 protein from Saccharomyces cerevisiae contains a leucine-rich repeat region which is required for its control of ADH2 gene expression. Genetics 132: 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauxion F, Faux C, Seraphin B (2008) The BTG2 protein is a general activator of mRNA deadenylation. EMBO J 27: 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Cryst 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol CD, Izumi T, Mitra S, Tainer JA (2000) DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature 403: 451–456 [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z, Bai Y, Poon D, Weil PA, Struhl K (1996) TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 383: 188–191 [DOI] [PubMed] [Google Scholar]
- Morita M, Suzuki T, Nakamura T, Yokoyama K, Miyasaka T, Yamamoto T (2007) Depletion of mammalian CCR4b deadenylase triggers elevation of the p27Kip1 mRNA level and impairs cell growth. Mol Cell Biol 27: 4980–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF (1974) Culture medium for enterobacteria. J Bacteriol 119: 736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer U, Collart MA (1998) Characterization of NOT5 that encodes a new component of the Not protein complex. Gene 207: 61–69 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of x-ray diffraction data collected in oscillation mode. In Methods in Enzymology, Carter CW Jr, Sweet RM (eds), Vol. 276, pp 307–326. New York: Academic Press [DOI] [PubMed]
- Parker R, Song H (2004) The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS (2001) ARP/wARP and molecular replacement. Acta Crystallogr 57: 1445–1450 [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM (2002) Substructure solution with SHELXD. Acta Crystallogr 58: 1772–1779 [DOI] [PubMed] [Google Scholar]
- Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C (2008) A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res 36: 3374–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W, Jentsch S (1990) Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J 9: 543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC (2000) Maximum-likelihood density modification. Acta Crystallogr 56: 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore S, Mauxion F, Seraphin B, Suck D (2003) X-ray structure and activity of the yeast Pop2 protein: a nuclease subunit of the mRNA deadenylase complex. EMBO Rep 4: 1150–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R (2002) Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J 21: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R (2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386 [DOI] [PubMed] [Google Scholar]
- Viswanathan P, Chen J, Chiang YC, Denis CL (2003) Identification of multiple RNA features that influence CCR4 deadenylation activity. J Biol Chem 278: 14949–14955 [DOI] [PubMed] [Google Scholar]
- Whisstock JC, Romero S, Gurung R, Nandurkar H, Ooms LM, Bottomley SP, Mitchell CA (2000) The inositol polyphosphate 5-phosphatases and the apurinic/apyrimidinic base excision repair endonucleases share a common mechanism for catalysis. J Biol Chem 275: 37055–37061 [DOI] [PubMed] [Google Scholar]
- Wilusz CJ, Wormington M, Peltz SW (2001) The cap-to-tail guide to mRNA turnover. Nat Rev 2: 237–246 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.