Abstract
A cohort of genes associated with embryonic stem (ES) cell behaviour, including NANOG, are expressed in a number of human cancers. They form an ES-like signature we first described in glioblastoma multiforme (GBM), a highly invasive and incurable brain tumour. We have also shown that HEDGEHOG-GLI (HH-GLI) signalling is required for GBM growth, stem cell expansion and the expression of this (ES)-like stemness signature. Here, we address the function of NANOG in human GBMs and its relationship with HH-GLI activity. We find that NANOG modulates gliomasphere clonogenicity, CD133+ stem cell cell behavior and proliferation, and is regulated by HH-GLI signalling. However, GLI1 also requires NANOG activity forming a positive loop, which is negatively controlled by p53 and vice versa. NANOG is essential for GBM tumourigenicity in orthotopic xenografts and it is epistatic to HH-GLI activity. Our data establish NANOG as a novel HH-GLI mediator essential for GBMs. We propose that this function is conserved and that tumour growth and stem cell behaviour rely on the status of a functional GLI1-NANOG-p53 network.
Keywords: cancer, GLI1, glioma, HEDGEHOG, NANOG, stem cell
Introduction
Glioblastoma multiforme (GBM) is a devastating invasive brain tumour able to give rise to many kinds of differentiated tumour cells (Furnari et al, 2007). GBM growth and persistence depend on cancer stem cells (Singh et al, 2003, 2004) with enhanced DNA damage repair programmes (Bao et al, 2006) that also induce recurrence and resist current chemo- and radiotherapies. One important pathway implicated in the control of GBM growth and stemness is HEDGEHOG-GLI (HH-GLI) signalling (Dahmane et al, 2001; Bar et al, 2007; Clement et al, 2007; Ehtesham et al, 2007). This function of HH-GLI parallels its control of normal brain growth stem cell behaviour (e.g. Dahmane and Ruiz i Altaba, 1999; Lai et al, 2003; Palma and Ruiz i Altaba, 2004). Interestingly, HH-GLI was shown to regulate a number of stemness genes, including NANOG (Clement et al, 2007; Stecca and Ruiz i Altaba, 2009).
The homeodomain protein NANOG is required for the pluripotency of inner mass cells of the blastocyst and of derived embryonic stem (ES) cells (Mitsui et al, 2003; Chambers et al, 2003, 2007; Silva et al, 2009). NANOG along with OCT4 and SOX2 forms a core ES cell network (Boyer et al, 2005). NANOG promotes mouse and human ES cell expansion (Chambers et al, 2003; Darr et al, 2006) by regulating self-renewal (Ivanova et al, 2006). It is also involved in the reprogramming of differentiated cells towards the ES-like phenotype of induced pluripotent stem (iPS) cells by reprogramming gene sets that include OCT4 and SOX2 (Takahashi and Yamanaka, 2006; Yu et al, 2007). Finally, exogenous NANOG function mimics nuclear reprogramming, being essential to induce pluripotency (Silva et al, 2006, 2009). Whereas the germline requires Nanog function in mice (Silva et al, 2009; Yamaguchi et al, 2009), it is not known if it is active in somatic adult tissues, in which it could have similar functions in controlling stemness and multipotency.
In addition to NANOG, GBMs and lower-grade gliomas express a core ES-like stemness signature that includes the expression of OCT4 and SOX2, and the levels of this signature, GBM growth and the number of GBM stem cells decrease on inhibition of HH-GLI signalling (Clement et al, 2007). Conversely, enhanced GLI1 function in transgenic mice drives increased neural stem cell self-renewal, CNS hyperplasia and much increased levels of Nanog (Stecca and Ruiz i Altaba, 2009). For example, Nanog is induced to higher levels than other Gli1-regulated genes, such as Nestin and Ptc1, in the cerebellum of dox-treated Nestin → rtTA,LacZ; GFP←biTetO → GLI1 mice (Stecca and Ruiz i Altaba, 2009). Together, these data raised the possibility that NANOG could be an essential gene and stemness regulator in GBMs downstream of HH-GLI.
Recent work has implicated Nanog in liver cancer in mice (Machida et al, 2009), NANOG coding mRNAs (NANOG and the retrogene NANOGP8) have been detected in different human cancers types (e.g. Zhang et al, 2006; Chiou et al, 2008; Ye et al, 2008) and NANOGP8 is expressed and has been involved in prostate cancer xenograft growth (Jeter et al, 2009). However, it is not known if NANOG is critical for GBMs, In addition, it is not clear how NANOG may interact with HH signalling and with GLI1 in particular, which acts in a negative-regulatory loop with p53 (Stecca and Ruiz i Altaba, 2009).
Results
Expression of NANOG-encoding genes in human GBMs
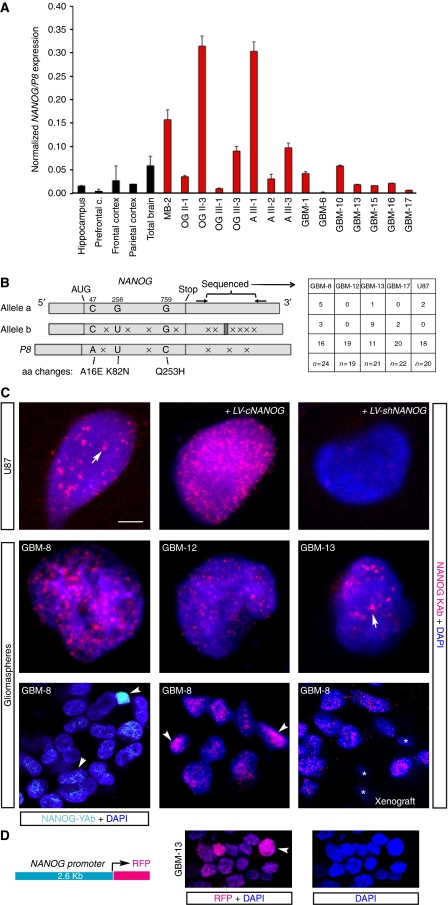
To test for the presence of the two NANOG-encoding transcripts in GBMs, we assayed for NANOG and NANOGP8 mRNAs (together referred to as NANOG/P8) by quantitative RT–PCR, normalizing the values with those of TBP and β ACTIN. NANOGP8 encodes NANOG protein with only 2 or 3 aa changes in comparison with each of the NANOG alleles, the existence of which is supported by the conserved polymorphisms (see Booth and Holland, 2004). In addition to NANOG and NANOGP8, there are 10 non-coding NANOG pseudogenes (Booth and Holland, 2004). Their sequences are not recognized by the PCR primers used here.
All primary GBMs (gliomas WHO grade IV), lower-grade astrocytomas and oligodendrogliomas (gliomas WHO grade III and II) tested expressed NANOG/P8 albeit to different levels (Figure 1A), consistent with our earlier data (Clement et al, 2007). Analysis of IDH1 and IDH2 in GBM-8 (GBM tumour sample #8), -12–14 and -17 showed a mutation, R132H (Yan et al, 2009), only in GBM-14. p53 mutant status was as described in Stecca and Ruiz i Altaba (2009) and GBM-14 had a C238Y change resulting from a GTA deletion in exon 7 known to cause loss of function (Epstein et al, 1998). A medulloblastoma included as control also expressed NANOG/P8 (Figure 1A), consistent with our data on the cerebellum (Stecca and Ruiz i Altaba, 2009). Normal brain samples also showed NANOG/P8 expression raising the possibility that NANOG could have a normal function in the adult human CNS (Figure 1A). Given the differences in tumour versus stroma content and amount of tumour within the various primary samples, it is difficult to correlate the actual levels of NANOG/P8 with other parameters. Nevertheless, the results show that all brain tumours tested express NANOG/P8.
Figure 1.
NANOG expression in human gliomas. (A) Quantification of NANOG/P8 expression by qRT–PCR in normal brain and in different normal brain regions (black bars), and in human brain tumours (red bars): A, astrocytoma; GBM, glioblastoma multiforme; OG, oligodendroglioma; MB, medulloblastoma. Roman numerals refer to WHO tumour grade. Arabic numerals refer to tumour sample as in Clement et al (2007) except GBM-15, -17. Expression values were normalized by using the geometrical mean of TBP and β ACTIN. (B) Diagram of the structure of NANOG alleles a (NW_001838051) and b (NC_000012.11), and of NANOGP8 (NT_010194.17). NANOG allele b contains 22 extra bp in the 3′UTR (dark grey box). These differences suggest the identification of these conserved polymorphic variants as alleles. The diagram also highlights the bp differences (crosses) and the 2 or 3 aa variants. Of these, K82N is not conserved in different species. The right part shows the frequency of alleles as determined from sequencing a portion of the 3′UTR that is diagnostic for the different alleles in different primary GBMs and in U87 cells. (C) Indirect immunofluorescence localization of NANOG protein with YAb (top left) or KAb antibody in patient-derived GBM cells grown as gliomaspheres and attached U87 cells as indicated with confocal microscopy. Nuclei are counterstained with DAPI (blue). Labelling is seen in multiple nuclear puncta (arrows). Cells express NANOG at different levels (arrowheads). Asterisks mark small cells with no labelling that likely represent mouse stromal cells in xenografts. Control staining was performed on LV-cNANOG- and LV-shNANOG-1-transduced cells (top right) and on cells that lacked primary antibodies (see Supplementary Figure S1). (D) Scheme of the NANOG reporter construct used to drive RFP expression (left) and the resulting RFP fluorescence (red) (middle; a high-expressing cell is denoted by an arrowhead) in a small GBM-13 gliomasphere derived from a transduced cell. The sphere was allowed to attach to the dish for 30 min before processing. The same image is shown only with DAPI staining (blue; right) to highlight nuclei. Scale bar=3 μm (C, top and middle rows), 10 μm (C, bottom row), 15 μm (D).
To discriminate whether the detected expression was due to NANOG or NANOGP8, we performed a sequence analysis of a diagnostic 3′UTR region, which varies among the NANOG alleles and NANOGP8. Sequencing of multiple clones derived from RT–PCR from each sample revealed the predominant expression of NANOGP8, although all but one GBM tested also expressed at least one NANOG allele (Figure 1B).
In contrast to ES cells in which NANOG protein is expressed at high levels, its immunodetection in GBM cells proved more challenging. Using a polyclonal anti-NANOG antibody kindly provided by the Yamanaka laboratory (YAb), we could detect high levels of NANOG protein in few nuclei of GBM neurospheres (stem cell-derived clones also called gliomaspheres) near the periphery, in which stem cells are located (Hall et al, 2006), but many more cells showed expression at lower levels (Figure 1C; Supplementary Figure S1A). As the stock of the YAb seems to be exhausted, we tested a mouse monoclonal anti-NANOG antibody from the same laboratory and also seven commercial antibodies (R&D rat mAb #1997; R&D goat #1997; AbCam rabbit #ab21603; AbCam rabbit #21624; Sigma mouse mAb #N3038; Cell Signaling rabbit #3580 and Kamiya rabbit#PC-102 hereafter referred as KAb). Whereas most of these antibodies recognized overexpressed NANOG protein by immunofluorescence and in western blots (Figure 1C, top; and not shown), only KAb identified the endogenous NANOG 42 kDa species (see below; Figure 2A).
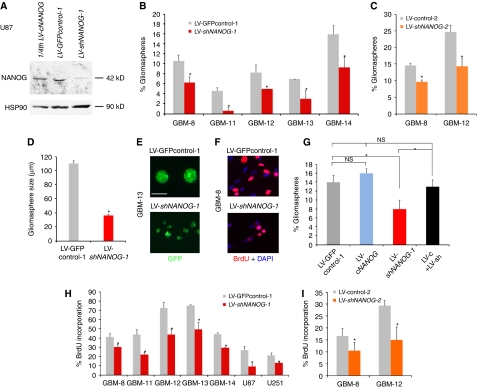
Figure 2.
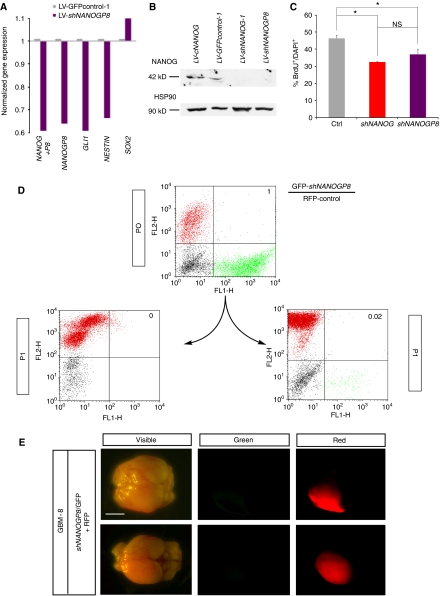
NANOG regulates stem cell self-renewal and GBM cell proliferation. (A) Western blot of U87 cells expressing LV-GFPcontrol-1, showing endogenous levels of NANOG protein (middle), LV-cNANOG (left; with only 1/4th, 20 μg, of the amount of total protein as in the other lanes loaded; expressing NANOG cDNA) and LV-shNANOG-1 (right, 80 μg), with greatly reduced NANOG protein levels. (B, C) Quantification of clonogenic single-cell gliomasphere formation by transduced GBM samples after expression of shNANOG-1 (B), shNANOG-2 (C) or controls (LV-GFPcontrol-1 (B) or LV-control-2 (C)); >900 clonogenic events were counted per condition, plated in 96-well plates. (D, E) Representative photomicrographs of GFP+ (green) gliomaspheres in suspension (E) and measurement of their sizes (D) from GBM-8 cells transduced with LV-GFPcontrol-1 or LV-shNANOG-1 as indicated. (F) Representative examples of anti-BrdU labelling (red) in GBM-8 cells transduced with LV-GFPcontrol-1 or LV-shNANOG-1 as indicated. GBM-8 gliomaspheres were dissociated and allowed to attach to facilitate immunohistochemistry and counting. (G) Histograms showing the quantification of the rescue of the inhibition of clonogenicity by shNANOG-1 through the overexpression of NANOG cDNA (LV-cNANOG) in GBM-12 gliomaspheres. (H, I) Quantification of BrdU incorporation in multiple primary GBM and GBM cell lines after NANOG kd with shNANOG-1 (H) or shNANOG-2 (I), shown in comparison with controls (GFP-control-1 (H) or control-2 (I)). Asterisks denote significative changes (P<0.05). ns, not significative. Error bars represent s.e.m. For clonogenic (sphere) assays, 900 clonogenic events were counted per condition. Scale bar=120 μm (E), 45 (F).
Using KAb, we could show near ubiquitous expression of NANOG protein in all U87 cells grown as attached cultures and in all primary GBM cells grown as gliomaspheres, as detected by confocal indirect immunofluorescent microscopy (Figure 1C; Supplementary Figure S1B and C). Exogenous and endogenous NANOG protein was mostly localized to nuclear microdomains or puncta and knock-down (kd) of NANOG (see below) greatly diminished staining (Figure 1C). Similar localization of NANOG was present also in GBM-8, -12 and -13 (Figure 1C), was absent from controls lacking primary antibody (Figure 1C; Supplementary Figure S1A and C) and was also detected in GMB-8 in vivo xenografts in which the smaller mouse nuclei were negative (asterisks in Figure 1C, bottom). Not all cells expressed the same levels of NANOG (Figure 1C), consistent with YAb labelling, although no special localization of high expressors was evident. NANOG puncta resemble transcriptional domains highlighted by nuclear dot antigens (Xie et al, 1993). They did not overlap with markers of other nuclear foci: promyelocytic leukaemia protein (PML) or 53BP1 (Supplementary Figure S1D).
Taken together, these results and the patterns of YAb and KAb indicate that NANOG is expressed in most if not all GBM tumour cells, but that a fraction of these cells express it at higher levels.
NANOG → RFP reporter reveals high and low expressors
As a complementary approach to characterize the identity of NANOG+ cells, and as NANOG protein can be made from both NANOG and NANOGP8, we used a promoter reporter construct from the NANOG locus driving the expression of red fluorescent protein (NANOG → RFP) (Figure 1D). High NANOG → RFP+ cells (Figure 1D) were detected by direct fluorescent microscopy at a frequency of ∼2 in small 32-cell attached spheres on average, consistent with clonogenic frequencies (Clement et al, 2007). Fluorescent-activated cell sorting (FACS) analyses revealed a smaller subpopulation of high RFP+ cells in large (∼100 cells) gliomaspheres (1.7% in GBM-8, 0.7% in GBM-12 and 0.5% in GBM-13). In contrast, weak RFP+ cells were ubiquitous (Figure 1D). Rare high RFP+ cells were also present in intracranial xenografts in immunocompromised mice (±1% for GBM-8; not shown), but no special localization (e.g. in clusters) was evident. NANOG → RFP thus partially mimics the expression of NANOG, revealing high and low expressors.
NANOG function modulates GBM clonogenicity and proliferation in vitro
To test for the function of NANOG, we have used two independent shRNAs expressed from replication incompetent lentivectors: shNANOG-1 targeting the 3′UTR in a GFP+ lentivector (Zaehres et al, 2005) and shNANOG-2-targeting exon 4, with 50–70% efficiencies for mRNA degradation and 80–90% for protein (Figures 2A and 5B). Each of these shRNAs inhibits both NANOG and NANOGP8. This strategy ensures the targeting of all NANOG-encoding genes. Indeed, western blot analyses showed that KAb identified the endogenous and exogenous 42 kDa NANOG protein in U87 cells and that NANOG kd led to a near complete loss of endogenous NANOG protein (Figure 2A).
The effects of NANOG kd were first investigated in gliomaspheres. Single-cell clonogenic assays over 2 weeks showed that NANOG kd reduced the number of GFP+ gliomaspheres by 20–80%, as compared with control parental lentivectors (Figure 2B and C). NANOG is thus required for normal clonogenic behaviour of GBM stem cells.
Gliomaspheres with NANOG kd were also ∼2.5-fold smaller than controls (Figure 2D and E). Consistently, BrdU incorporation analyses in plated gliomaspheres of five primary GBMs showed 20–40% reduction in cell proliferation, and 30–60% in the U87 and U251 GBM cell lines (Figure 2F, H and I), after NANOG kd. Therefore, as in the case of HH-GLI (Palma and Ruiz i Altaba, 2004; Palma et al, 2005) and GLI1 (Stecca and Ruiz i Altaba, 2009), NANOG is required both for normal cell proliferation, reflecting the widespread expression NANOG/P8, and for the clonogenicity of putative GBM stem cells.
Overexpression of NANOG cDNA, which has been shown to transform NIH3T3 cells (Piestun et al, 2006), produced a general deleterious effect, but at least it rescued the kd effect in clonogenicity of one GBM (Figure 2G). Such deleterious effect prevented us from performing other rescue and epistatic analyses.
NANOG is preferentially required in CD133+ GBM stem cells
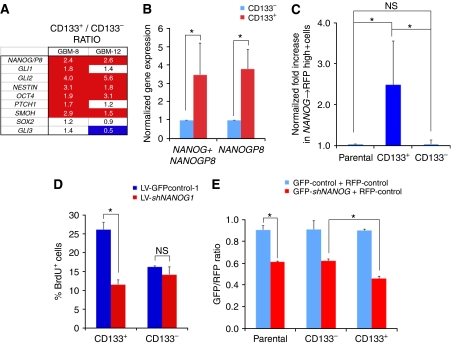
To test whether NANOG/P8 could be expressed in GBM stem cells, patient-derived GBM-8 cells were magnetic-activated cell sorted for CD133 and assayed by qRT–PCR. The expression of the AC133 epitope of CD133 (CD133+) marks a dynamic population of cells enriched for stem cells in a majority of tested GBMs (Singh et al, 2003, 2004; Chen et al, 2010). CD133+ cells were found to express ∼2.5-fold higher levels of NANOG/P8 than CD133− cells (Figure 3A; shown as the ratio of CD133+ over CD133− values after normalization). CD133+ cells also showed 1.4–5.6-fold higher levels of GLI1, GLI2, NESTIN and OCT4 (and pseudogenes; Liedtke et al, 2007) as compared with CD133− cells, consistent with earlier work (Clement et al, 2007).
Figure 3.
Expression and function of NANOG in CD133+ GBM cells. (A) Gene expression levels determined by RT–qPCR shown as the ratios of the expression values of CD133+ cells over those of CD133− cells, of GBM-8 and GBM-12 cells, after normalization with housekeeping genes. Enhanced levels ⩾1.5-fold are in red and repressed levels ⩽50% are in blue. CD133+ cells express higher levels of NANOG/P8 as compared with CD133− cells. (B) Quantification of the levels of mRNA expression of NANOG/P8 (NANOG + NANOGP8) and of NANOGP8 alone in freshly MACS-sorted CD133+ versus CD133− GBM-8 cells. Normalization was set so that CD133− levels equal 1. NANOGP8 shows a similar enrichment to that of the combined genes in CD133+ cells. (C) Normalized enrichment of NANOG → RFP+ cells in CD133+ versus CD133− GBM-8 cells. Normalization was performed by equating the level in the unsorted parental population to 1. (D) Acute (16 h) BrdU incorporation assay on freshly sorted CD133+ and CD133− cells of GBM-8 showing a selective effect of NANOG kd in CD133+ cells at this early time point. (E) Red/green competition assay in vitro testing for the changes in GFP+ experimental cells co-expressing shNANOG-1 or controlGFP-1 versus RFP+-control cells. Cells were transduced, FACS sorted to obtain 100% transduced cell populations, mixed and 5 days later sorted for CD133 by MACS. MACS-sorted cells were then FACS analysed to determine the relative GFP/RFP ratios per culture condition shown in the graph. Asterisks denote significative changes (P<0.05).
To differentiate between NANOG and NANOGP8, we performed an additional CD133 magnetic-activated cell sorting (MACS) analysis of GBM-8 and analysed the expression of combined NANOG/P8 versus NANOGP8 singly. In both cases, there was an ∼3.5-fold enrichment in CD133+ versus CD133− cell populations (Figure 3B), showing that NANOGP8 is enriched in CD133+ GBM stem cells.
Analyses of the behaviour of FACS-sorted NANOG → RFP+ cells revealed that these could indeed form gliomaspheres, although RFP+ and RFP− cells yielded similar clonogenic frequencies (not shown), suggesting that not all clonogenic cells are RFP+. We note that RFP− cells abundantly express NANOGP8 (not shown) and could thus be expected to include cells with stem cell properties, if indeed NANOG function is involved in aspects of GBM stemness. The situation is likely to be more complex than anticipated as cloning analyses with FACS-sorted cells revealed the production of RFP− clones from RFP+ cells, and the appearance of clones with RFP+ cells derived from the RFP− population, both at a frequency of ±30–40% over a period of 1 week. This situation, however, is reminiscent of the fluctuating nature of embryonic stemness (Hayashi et al, 2008).
To investigate a possible correlation of NANOG → RFP+ and CD133+ states, CD133 MACS-sorted GBM-8 cells were FACS analysed for RFP expression and the results normalized to the levels found in the parental unfractionated population (Figure 3C). CD133+ cells showed ∼2.5-fold higher NANOG → RFP expression as compared with CD133− cells (Figure 3C). Together with the data discussed above, these results suggest that the expression of NANOG/P8 is enhanced in but not restricted to CD133+ GBM stem cells, again paralleling the distribution of GLI1 (Clement et al, 2007).
To provide additional support for the idea that NANOG is required in GBM stem cells for their normal behaviour beyond the clonogenic assays, we tested whether NANOG kd could acutely impair the proliferation of sorted CD133+ versus CD133− cells. CD133-sorted GBM-8 and GBM-12 cells with NANOG kd were plated and immediately given a short BrdU pulse (16 h for these cells). Anti-BrdU immunohistochemistry revealed that CD133+ cells proliferated more than CD133− cells, consistent with earlier findings (Singh et al, 2004). It also revealed a differential effect in CD133+ versus CD133− cells. Only CD133+ cells showed reduced proliferation (50% for GBM-8 and 45% for GBM-12; Figure 3D and not shown), showing a preferential requirement of NANOG function in CD133+ GBM stem cells.
As an alternative method to test for preferential effects of NANOG in CD133+ GBM cells, we performed an in vitro red/green competition assay (Varnat et al, 2009) followed by MACS sorting. In this assay, GBM-8 cells transduced with GFP-control or GFP-shNANOG lentivectors were separately challenged in vitro with sibling cells transduced with a control lentivector-expressing RFP. After 5 days in culture, the mixed populations were MACS sorted for CD133 and the CD133+ and CD133− pools, then FACS analysed for the sizes of the GFP+ and RFP+ subpopulations. Prolonged (5 days) inhibition of NANOG function led to a decrease in the GFP+ population in both CD133+ and CD133− cells, as compared with GFP-control cells (Figure 3E). However, CD133+ cells were more affected than CD133− cells (Figure 3E). Together with the clonogenic data, the results with CD133 sorting support a function of NANOG in the control of GBM stem cell behaviour.
NANOG is essential for GBM tumourigenicity in vivo
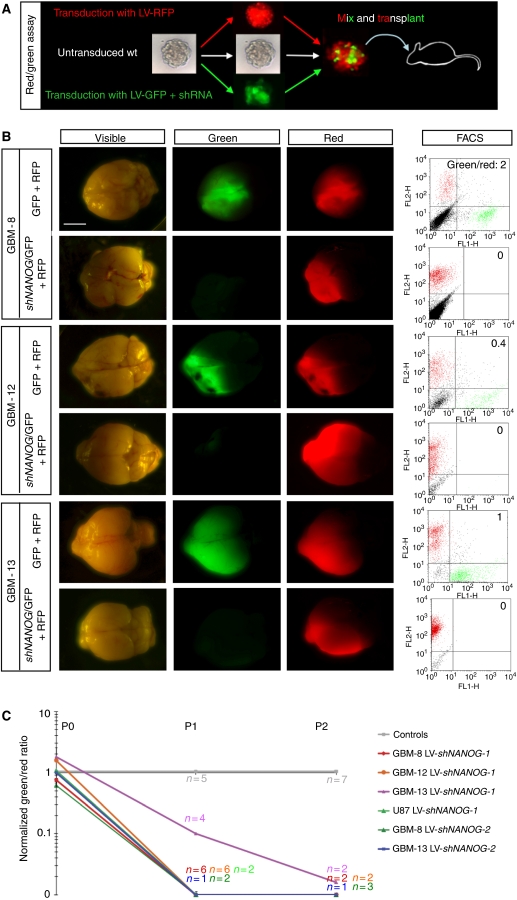
To test for the function of NANOG in vivo, we used the novel in vivo red/green assay (Varnat et al, 2009) in which tumour cells that are differently and indelibly marked compete within a tumour environment. Cancer cells comprising equivalent populations of transduced cells expressing different fluorescent proteins (see above) are mixed in xenografts in immunocompromised mice, thus allowing for competition in vivo (Figure 4A). The grown tumour is then isolated, cells dissociated, an aliquot is subjected to FACS quantification and the rest re-injected into a new host, repeating the cycle as long as required. Here, we have extended this assay to use it in orthotopic intracranial xenografts with primary GBM cells.
Figure 4.
NANOG function is essential for GBM growth in vivo. (A) Scheme of the red/green orthotopic assay with GBMs. For illustration purposes, gliomaspheres are shown as starting material, but adherent cells have also been used (see Supplementary Figure S2). In addition, also for illustration purposes, an aggregate of red and green cells is shown before intracerebral transplantation into immunocompromised mice, although FACS-sorted cells are routinely mixed and injected before aggregation. (B) Representative images of dissected whole brains with developed ‘red/green' brain tumours after orthotopic xenotransplantation of GBM gliomaspheres transduced with control red (RFP+) plus either control green (GFP+) or GFP+/shNANOG-expressing lentivectors as indicated. The same samples are shown in each row under visible and fluorescent light, the latter with filters to selectively detect green or red fluorescence. Far right panels show the FACS profiles and green/red ratios. (C) Quantification of FACS ratios in red/green competition assays in vivo after normalization with controls, which are equated to 1. The number of mice analysed at each passage (P) is also given (n) for each case. Scale bar=3.5 mm (B).
Injection of 105 GBM cells comprising mixed populations as described above showed that three patient-derived GBMs (GBM-8, GBM-12 and GBM-13) and U87 cells showed a rapid and massive loss of GFP+ cells expressing NANOG shRNAs in vivo, within the first passage, as compared with sibling RFP+ cells in the same tumours (Figure 4B and C). GFP-only controls showed limited variability and were used for normalization. Similar effects were obtained with a second shRNA confirming the specificity of the targeting (Figure 4C). The results were also recapitulated with adherent GBM cells (Supplementary Figure S2), bypassing thus any possible issues associated with the mode of culture before transplantation. These results identify NANOG as an essential factor for GBM tumour growth.
The ‘looser' phenotype of NANOG kd cells could be due, in principle, to cell competition (Morata and Ripoll, 1975) akin to that involving Myc and non-cell autonomous mechanisms in Drosophila (de la Cova et al, 2004; Moreno and Basler, 2004). To test for such non-autonomy and for the ability of NANOG to compromise the initiation of a whole tumour, rather than blocking a specific subpopulation, we performed intracranial xenografts as before but with FACS-sorted 100% shNANOG-GFP+ cells. GBM-8 cells with homogeneous NANOG kd were unable to initiate tumourigenesis, and recipient mice remained viable after >4 months, whereas control GFP+-only cells harbouring the parental lentivectors induced tumour formation and the mice had to be euthanized due to weight loss and early signs of neurological disease after 5 weeks (n=2/group). Analyses of these control mice confirmed the presence of tumours (not shown). NANOG is thus essential for GBM growth and its effects cannot be solely due to non-cell autonomous effects of wild-type siblings outcompeting NANOG kd cells.
NANOGP8 is essential for GBM growth in vivo
The complete requirement of NANOG protein function for GBM growth shown above raised the question of the functionality of the two NANOG-encoding genes: NANOG and NANOGP8. The few bp changes that differentiate the coding sequences of these two genes made it difficult to inhibit NANOG alone. However, the presence of a small, but unique 3′UTR sequence characteristic of NANOGP8 allowed us to design a lentivector expressing an NANOGP8-specific shRNA and to measure both the combined NANOG/P8 (NANOG+NANOGP8) expression levels and also the specific levels of NANOGP8.
shNANOGP8 greatly reduced the levels of NANOG+NANOGP8 (NANOG/P8) combined expression, as did shNANOG (see above), but also of NANOGP8 specifically, as well as those of GLI1 and NESTIN, but not of SOX2 in U87 cells (Figure 5A). Western blot analyses with KAb in U87 cells revealed the increased levels of NANOG protein after transduction with a lentivector expressing full-length NANOG cDNA, the normal levels of NANOG protein in cells transduced with LV-GFPcontrol-1 and the loss of NANOG protein in cells expressing either shNANOG-1 or shNANOGP8 (Figure 5B). This result confirms the targeting of NANOG by shNANOGP8, and together with the abundance of NANOGP8 as compared with NANOG (Figure 1B), it argues that the great majority of NANOG protein in GBMs derives from NANOGP8. Indeed, shNANOGP8 decreased U87 cell proliferation to the same extent as shNANOG (Figure 5C), further suggesting that NANOGP8 is the main source of functional NANOG activity in GBM cells.
Figure 5.
NANOGP8 is expressed and required in GBMs. (A) RT–qPCR quantification of the expression levels of NANOG, NANOGP8, GLI1 and NESTIN after NANOGP8 kd with a specific shRNA to NANOGP8 in a lentivector in U87 cells. Control U87 cells were transduced with a GFP-control1 lentivector. Values are shown after normalization. (B) Western blot analysis of NANOG protein with KAb in U87 GBM cells transfected with LV-cNANOG, overexpressing NANOG protein, control GFP-expressing LV-GFPcontrol-1 and two lentivectors-expressing shRNAs against NANOG plus NANOGP8 (LV-shNANOG-1) or against NANOGP8 (LV-shNANOGP8) separately. shNANOG-1 or shNANOGP8 abolishes detectable NANOG protein expression. HSP90 is shown as loading control; 80 μg of total protein were loaded per lane for SDS–PAGE. (C) Quantification of BrdU incorporation showing similar efficiencies of shNANOG-1 and shNANOGP8 in inhibiting cell proliferation in U87 cells. The graph shows the percentage of BrdU+ cells over the total number of DAPI-labelled cells. (D) FACS plots of in vivo red/green assays in orthotopic xenografts of GBM-8 with a GFP+ population also expressing shNANOGP8. Parental cells at passage 0 (P0) show about equal subpopulations of RFP+ and GFP+ cells. After the first in vivo passage (P1), the GFP+ cells with NANOGP8 kd have disappeared. (E) Dorsal views of dissected brain harbouring grafts of GFP+ cells co-expressing shNANOGP8 and RFP+ cells after tumour growth (at first passage). The tumours are composed exclusively of RFP+-control cells. Scale bar=3.5 mm (E). Asterisks denote significative changes (P<0.05).
To test for the contribution of the NANOGP8 locus to NANOG protein function in vivo, we performed red/green orthotopic xenografts with RFP+ control GBM-8 cells and sibling GFP+ cells co-expressing shNANOGP8. GFP+ cells were lost within the first passage (Figure 5D and E), mimicking the effects of shNANOG (Figure 4). NANOGP8 is thus the major source of functional NANOG protein in GBM cells.
Evidence of an NANOG-GLI1-p53 functional network
The similarity in the involvement of NANOG (this work) and GLI1 (Clement et al, 2007) both in GBM cell proliferation and stem cell clonogenicity, as well as their requirement for GBM tumour growth in vivo, prompted us to analyse their relationship. Cells with NANOG kd showed consistently diminished expression of GLI1, NESTIN and OCT4 as analysed 4 days post-transduction (Figure 6A). Analysis at 3 days post-transduction showed that PTCH1 was also inhibited, arguing for different dynamics of GLI target repression (Figure 6A). Importantly, NANOG kd greatly decreased GLI protein levels by western blot (of GLI1100 or GLI1130 isoforms; Stecca and Ruiz i Altaba, 2009) (Figure 6B). Consistently, the levels of activity of a GLI-binding site → luciferase reporter were reduced after kd of NANOG (Figure 6C). Together with our earlier data on the modulation of NANOG/P8 mRNA levels by HH signalling in different systems (Clement et al, 2007; Stecca and Ruiz i Altaba, 2009), these results suggest the presence of a positive loop between NANOG and GLI1.
Figure 6.
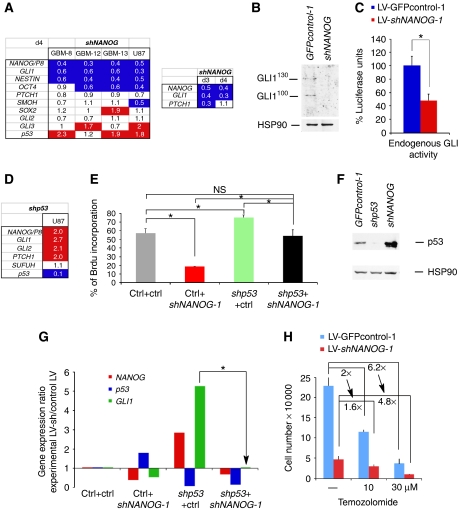
Functional NANOG, GLI1 and p53 interactions. (A) Gene expression changes at 4 days post-transduction in primary GBMs and U87 cells after NANOG kd with LV-shNANOG-1 shown as ratios over the levels obtained in cells transduced with control lentivectors and after normalization. Bottom panel shows the decrease of GLI1 and PTCH1 at 3 days post-transduction, whereas at 4 days PTCH1 levels recover, suggesting different dynamics for different GLI1 targets. (B) Inhibition of GLI1130 and GLI1100 protein isoforms detected with a specific affinity-purified polyclonal antibody (see Stecca and Ruiz i Altaba, 2009) in U87 cells after kd of NANOG. This confirms the relevance of 70% inhibition at the mRNA level (A). GLI1FL was not detectable. (C) GLI-binding site → luciferase reporter assays testing for the activity of endogenous GLI1 in U87 cells. Reporter activity is strongly reduced by NANOG kd. Note that luciferase levels were normalized so that normal endogenous level is set to 100%. Luciferase levels were firefly over control renilla ratios. (D) Enhancement of NANOG/P8 and the HH-GLI pathway after p53 kd with LV-shp53 in U87 cells. Values are ratios over control lentivectors after normalization. (E) Rescue of the proliferative (BrdU incorporation) defect induced by NANOG kd by simultaneous kd of p53, and rescue of the proliferative enhancement of p53 kd by simultaneous kd of NANOG in U87 cells. (F) Western blot analyses of p53 protein in U87 GBM cells after p53 kd or NANOG kd with appropriate lentivectors. HSP90 is shown as quantification control. Ctrl, control LV-GFPcontrol-1-transduced cells. (G) Antagonistic effects of NANOG kd and p53 kd on GLI1 mRNA levels and rescue by double kd in U87 cells. Gene expression levels (shown as fold changes) were quantified and normalized and are shown as ratios of the experimental condition over control (parental lentivector-transfected cells). The arrow points to the rescue of GLI1 levels by concomitant kd of p53 and NANOG. (H) Quantification of U87 cell numbers in control and NANOG kd populations after treatments with 0, 10 and 30 μM temozolomide for 5 days. NANOG kd dampens temozolomide effects, passing from a two-fold (2 × ) reduction in cell numbers in controls to 1.6-fold in shNANOG-expressing cells at 10 μM, and from 6.2- to 4.8-fold at 30 μM. Asterisks denote significative changes (P<0.05). NS, not significative. Error bars represent s.e.m.
Whereas we found three consensual GLI-binding sites in the NANOG and two in the NANOGP8-regulatory regions using MatInspector (Genomatix), the lack of reliable commercial antibodies against endogenous GLI1 and the very limited quantities of our own affinity-purified polyclonal serum (Figure 6B; Stecca and Ruiz i Altaba, 2009) prevented us from performing ChIP assays. We note that ChIP analyses of mouse cerebellar precursors expressing a tagged form of Gli1 and using reliable anti-tag antibodies did not identify Nanog as a direct target (M Scott personal communication; Lee et al, 2010). However, in the accompanying paper Po et al report evidence for direct regulation. Thus, independently of direct or indirect regulation, here we have focused on functional relationships.
An important repressor of Nanog in mouse ES cells is the tumour suppressor p53 (Lin et al, 2005), and p53 acts in a functional negative-regulatory loop with GLI1 in neural stem cells and tumours (Stecca and Ruiz i Altaba, 2009). To test whether p53 could also negatively regulate NANOG in GBMs, we used p53 wt U87 cells, as in our hands, all GBMs that grow well in vitro or in xenografts are p53 mutant (Stecca and Ruiz i Altaba, 2009). The kd of p53 resulted in a two-fold enhanced expression of NANOG/P8 as compared with sibling-control cells (Figure 6D). Consistent with the finding that p53 is also a negative regulator of GLI function (Abe et al, 2008; Stecca and Ruiz i Altaba, 2009), kd of p53 resulted in a two- to three-fold increase in GLI1, GLI2 and PTCH1 levels, whereas the levels of the GLI repressor SUFUH were unaltered (Figure 6D).
To probe into the functional relationship between NANOG and p53, BrdU incorporation analyses were performed in U87 cells after single or combined kd of NANOG and p53. The results showed that whereas NANOG kd decreased cell proliferation and kd of p53 increased it, they rescued each other's effects (Figure 6E). Thus, NANOG and p53 seem to have opposing functions on tumour cell behaviour. This result, together with the upregulation of NANOG/P8 after p53 kd (Figure 6D) and the upregulation of p53 after NANOG kd (Figure 6A and F), argues for mutual negative regulation. p53 (TP53) transcription was also enhanced in GBMs with mutant p53 (GBM-8 and -13), but not in GBM-12, which lacks it (Stecca and Ruiz i Altaba, 2009) (Figure 6A). This homeostatic loop may be operative in p53 wt GBM precursors, likely including neural stem cells, and once p53 is lost, tumours may acquire enhanced expansion rates. These results support the notion that NANOG and p53 establish a functional negative loop, much like that between GLI1 and p53 (Stecca and Ruiz i Altaba, 2009) and opposite to the functional positive loop we describe here between NANOG and GLI1.
To clarify the relationship of NANOG, GLI1 and p53, gene expression profiles were determined by RT–qPCR in U87 cells after kd of NANOG, kd of p53 or simultaneous kd of both. Expression levels were normalized with the levels in housekeeping genes and shown as ratios over those in control-transduced cells (Figure 6G): NANOG kd enhanced p53 and repressed GLI1, whereas p53 kd greatly boosted both NANOG and GLI1. Importantly, simultaneous kd of both NANOG and p53 restored GLI1 to control levels (Figure 6G arrow). Control lentivectors had no effect. This result suggests that the decrease of GLI1 after NANOG kd requires p53, and that its enhancement after kd of p53 requires NANOG. The decrease in reporter activity by exogenous GLI1 after NANOG kd (see above) is thus likely mediated, in part, by enhanced endogenous p53 levels, which antagonize GLI1 activity (Stecca and Ruiz i Altaba, 2009). Together, the data indicate that NANOG, p53 and GLI1 form functional cross-regulatory network.
As a further test for the interaction of NANOG and p53, we asked if NANOG kd, which enhances p53 levels (Figure 6F) in U87 cells, could reduce the effects of drugs that cause a reduction in cell number partly by activating p53 through the DNA damage response pathway. In particular, we have sought to test for a possible interaction between temozolomide (TMZ), the standard of patient care anti-GBM therapeutic agent that acts by alkylating DNA, and kd of NANOG. U87 GBM cells were treated with increasing doses of TMZ and cell number was scored after 5 days. We included the lowest doses able to reduce cell number by two-fold (Figure 6H). Whereas NANOG kd led to a marked reduction in BrdU incorporation, treatments with 10 and 30 μM TMZ resulted in dampened responses: from 2- to 1.6-fold reduction with 10 μM and from 6.2 to 4.8 reduction with 30 μM TMZ (Figure 6H). We interpret these results to suggest that the effect of TMZ, mediated in part by upregulation of p53, is diminished in cells with NANOG kd, which already elevates p53 levels. The data provides additional support for the functional NANOG-p53 loop described above and emphasize the potential of NANOG inhibition in combinatorial therapies.
Analyses of transcriptome data from the Cancer Genome Atlas (http://tcga-portal.nci.nih.gov/) did not reveal correlations between the levels of p53, NANOG or GLI1. This is likely due to the fact that (i) NANOGP8 is not included, making it unclear what is measured: NANOG alone or NANOGP8, or even possibly the additional 10 NANOG pseudogenes; (ii) p53 mutant alleles can express p53 at high levels, making mRNA levels per se without functional assays unreliable; (iii) GLI1 is expressed at low levels and genomic approaches have been uninformative (see Stecca et al, 2007; Varnat et al, 2009) and (iv) the GBM sequenced material represents the whole tumour, thus diluting epithelial signals with stromal contribution and masking the status and expression of tumour alleles.
NANOG function is essential for HH-GLI responses in GBMs in vivo
The existence of an NANOG-GLI1 positive functional loop (see above) does not clarify how NANOG acts in relation to the HH-GLI pathway: although it might be an important GLI1 target, NANOG kd also downregulates GLI1 (Figure 6A and B).
To clarify the regulation of NANOG by the HH-GLI pathway, we first reanalysed the expression of NANOG/P8 and HH-GLI pathway components after modulation of HH-GLI signalling. Blockade of HH-GLI through lentivector-mediated SMOOTHENED kd, with shSMOH (Clement et al, 2007), in GBM-8 and GBM-12 gliomaspheres, and U87 cells, resulted in the inhibition of NANOG/P8 expression by ∼30–99% (Figure 7A). In contrast, further enhancement of HH signalling through PATCHED1 kd, with shPTCH1, or enhanced GLI1 levels (Varnat et al, 2009) resulted in increased NANOG/P8 expression by 1.7–4.3-fold (Figure 7A). These manipulations similarly altered the levels of the ES stemness gene OCT4 (and pseudogenes) (Figure 7A), consistent with the coordinate regulation of the ES-like stemness signature genes by HH-GLI (see also Clement et al, 2007). Other HH-GLI pathway components were regulated in an expected manner and SOX2 was largely unaltered (Figure 7A). To verify that NANOG protein levels are dependent on the status of the HH-GLI pathway, U87 cells were transduced with LV-controlGFP-1, LV-GLI1, LV-shSMOH or LV-GLI3R lentivectors and extracts assayed for NANOG expression by western blotting. The endogenous levels of NANOG were boosted ∼two-fold by GLI1 and reduced >80% by GLI3R or SMOH kd (Figure 7B), confirming the regulation of NANOG by GBM-intrinsic HH-GLI signalling.
Figure 7.
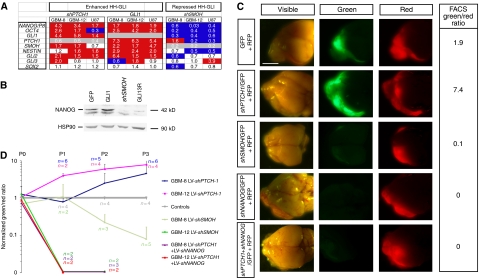
NANOG is regulated by HH-GLI signalling and NANOG function is epistatic to an active HH-GLI pathway. (A) Gene expression changes after enhancement (through PTCH1 kd or GLI1 expression) or repression (through SMOH kd) of HH-GLI signalling in GBM-8, -12 and U87 cells. Values are RT–qPCR ratios over controls after normalization at 4 days after transduction. (B) Western blot analysis of NANOG protein using KAb in U87 cells transduced with LV-GFPcontrol-1, LV-GLI1, LV-shSMOH or LV-GLI3R. HSP90 is shown as loading control. NANOG protein is greatly diminished in cells with compromised HH-GLI signalling. (C) Representative images of dissected whole brains with developed ‘red/green' brain tumours after orthotopic xenotransplantation of GBM-8 gliomaspheres transduced with control red (RFP+) plus green (GFP+) lentivectors expressing different shRNAs as indicated. Each row shows the images of dorsal brains with anterior to the left under visible, green or red fluorescence. Green/red FACS ratios are given in the right column. (D) Quantification and evolution of FACS ratios in red/green competition assays in vivo after normalization with controls, which are equated to 1. The number of mice analysed at each passage (P) is also given (n) for each case. Scale bar, 3.5 mm (C).
To test whether NANOG is a mediator of HH-GLI signalling in GBMs, acting downstream of GLI1, we performed in vivo epistatic analyses using orthotopic xenografts. Enhanced HH-GLI activity through shPTCH1 resulted in an increase in the population of GFP+/shPTCH1-expressing cells in intracerebral red/green assays (Figure 7C and D; Supplementary Figure S3). In contrast, shNANOG obliterated the expressing population (Figure 7C and D; Supplementary Figure S3). However, shNANOG was epistatic over shPTCH1 (Figure 7C and D; Supplementary Figure S3) in GBM-8 and GBM-12, indicating that NANOG is essential for HH-GLI responses.
Discussion
The expression of the homeobox gene NANOG forms part of an ES-like stemness signature we described in GBMs (Clement et al, 2007), later also found in other advanced cancer types (Ben-Porath et al, 2008). However, it is not known if this signature in general, and NANOG in particular, have functional relevance in these tumours or their stem cells. Given that homeobox genes control cell fates in vertebrates in many instances (e.g. Ruiz i Altaba and Melton, 1989; Duboule, 1994), and that NANOG is an archetypal embryonic stemness factor involved in maintaining pluripotency in the early embryo and in reprogramming differentiated cells to an ES-like state (e.g. Chambers et al, 2003, 2007; Mitsui et al, 2003; Silva et al, 2006, 2009; Takahashi and Yamanaka, 2006; Yu et al, 2007), we have initiated an analysis of the function of this ES-like cancer stemness signature by testing the function of NANOG in human GBMs. The most striking result we report here is that GBM growth in vivo shows a complete dependence on NANOG function.
The focus on NANOG also derives from our finding that the expression of the ES-like stemness signature in GBMs is dependent on sustained HH-GLI signalling (Clement et al, 2007), and from the strong and consistent upregulation of Nanog by GLI1 in neural precursors and stem cells in the mouse brain (Stecca and Ruiz i Altaba, 2009). For instance, Nanog was induced by GLI1 to higher levels (>40-fold) than known Gli-regulated genes such as Nestin and Ptch1 in CD133+ cerebellar stem cells (Stecca and Ruiz i Altaba, 2009). These results identified NANOG as a GLI-responsive factor and potential target. However, beyond resolving the functionality of consensus GLI-binding sequences that we have detected in the NANOG- and NANOGP8-regulatory regions, our data begged the question of the involvement of NANOG in brain tumours and its functional relationship with HH-GLI signalling. Here, we sought to address these functional issues. We find that NANOG is an essential HH-GLI1-dependent factor that is essential for HH-GLI responses in GBMs in vivo.
NANOG, mostly derived from NANOGP8, is expressed in GBM
In humans NANOG is made from two coding genes: NANOG and NANOGP8 (Booth and Holland, 2004), of which NANOGP8 has been detected in prostate and other cancers (e.g. Jeter et al, 2009). Although the exact expression of NANOG and NANOGP8 at single-cell resolution in patient samples remains to be explored, here we provide evidence that all GBM cells express NANOG-encoding genes. We find that NANOGP8 is the most abundantly expressed of the two NANOG-encoding genes in human GBMs, accounting for over 90% of all NANOG-encoding mRNAs in a number of tested cases. However, we find that there are different levels of NANOG protein present in individual GBM cells and that different antibodies may preferentially recognize varying levels of expression. This situation seems different from that reported for prostate cancer (Jeter et al, 2009), in which high NANOG+ cells were detected scattered throughout the tumour, but there was no mention of low expressors. Although we provide evidence that NANOG is present and active in, but not restricted to, GBM stem cells, the resolution of the nature of high NANOG+ cells, and the stability of NANOG expression, will require future lineage analyses.
NANOG is an essential GBM factor
We have probed for NANOG function in human GBMs by blocking its function through RNAi targeting both NANOG and NANOGP8, as well NANOGP8 alone. Given the high homology of these two genes, we were unable to only target NANOG. The most striking results reveal that NANOG function is essential for GBM tumourigenicity in a cell autonomous manner in vivo in immunocompromised mice. Such orthotopic grafting is a stringent test in which human GBM cells recapitulate the original human tumour (e.g. Singh et al, 2004). Patient-derived GBM cells with NANOG+NANOGP8 kd or NANOGP8 kd alone do not survive and do not form tumours, indicating that NANOG function, mostly derived from NANOGP8, is essential for GBM tumourigenicity in vivo. Further analyses should reveal whether the requirement of NANOG is a universal feature of human GBMs and other brain tumours, such as lower-grade gliomas and medulloblastomas, which also express NANOG/P8.
In vitro, we find that NANOG function is required to sustain normal high proliferative levels of GBM cells. In addition, we find that NANOG is also required for normal clonogenicity, suggesting an effect on GBM stem cells. This idea is supported by the finding that NANOG is enriched in CD133+ cells and that CD133+ cells are more sensitive than their CD133− counterparts, as assayed by both short- and longer-term proliferative behaviour. NANOG function, similar to that of GLI1 (Clement et al, 2007), thus seems to be present in all cells, including GBM stem cells, and interference with its function compromises both the growth of the tumour bulk and stem cell expansion. Given the more drastic effects of NANOG kd in vivo as compared with in vitro, the data support a more central function of NANOG in the former, raising the possibility that environmental factors regulate the requirement of NANOG. Although not directly tested, one possibility is that higher levels mark a subpopulation with stem cell properties. Some support for this idea derives from the enrichment of NANOG-RFP+ cells in the CD133+ fraction and the preferential effect of NANOG kd in CD133+ cells. Alternatively, however, the levels of NANOG in GBM cells may fluctuate, responding to so far uncharacterized cues. However it may be, we show that endogenous NANOG function is required for normal stem cell properties.
NANOG and GLI1 form a positive functional loop modulated by p53
To define the functional relationship of NANOG and HH-GLI, we have analysed the expression of NANOG, GLI1 and other genes under different experimental conditions in vitro and performed epistatic analyses in vivo. The results show that NANOG and GLI1 form a positive loop in which the normal levels of expression of one are dependent on the function of the other. In addition, as analyses in vivo show that NANOG is essential for HH-GLI responses, we conclude that NANOG function is an important GLI1 effector. NANOG and GLI1 thus form a functionally relevant positive module (Figure 8), the levels of which regulate stem cell behavior and tumour growth.
Figure 8.
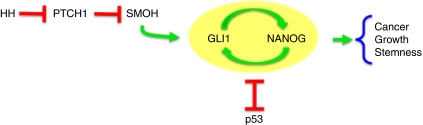
Model of the GLI1-NANOG module and cross-functional interactions with p53. Scheme of NANOG action downstream of HH-GLI signalling, establishing a positive feedback loop with GLI1 and being repressed by p53, which acts in a negative-regulatory loop with GLI1 in brain tumours (Stecca and Ruiz i Altaba, 2009). The outcome determines tumour growth and stem cell behaviour.
In an earlier study, we had determined that GLI1 and p53 establish a negative-regulatory loop, in which p53 negatively regulates GLI1 levels and status (Stecca and Ruiz i Altaba, 2009) and HH-GLI1, in turn, regulates p53 levels through Mdm2 (Abe et al, 2008; Stecca and Ruiz i Altaba, 2009). Given these regulatory relationships, we tested the influence of p53 on the NANOG-GLI1 positive loop. We find that p53 negatively regulates both NANOG and GLI1. This was established in U87 GBM cells, which are p53 wt unlike most primary GBMs (Furnari et al, 2007). Importantly, using RNAi, we have been able to show that the downregulation of GLI1 after NANOG kd is p53 dependent, that the upregulation of GLI1 after p53 kd is NANOG dependent and that the double kd of NANOG and p53 restores normal GLI1 levels. From this data, we propose the existence of a functional cross-regulatory network (Figure 8). The exact molecular nature of such interactions remains to be explored and may be direct or indirect. In addition, there must be other modes of interaction and/or additional players in such network. For instance, whereas the upregulation of GLI1 by NANOG kd is p53 dependent in U87 cells, this cannot possibly be so in GBM-12 cells, which lack p53 protein (Stecca and Ruiz i Altaba, 2009).
We suggest that a functional NANOG-GLI1-p53 network may be conserved in other cancers and that loss of p53, a common event in the genesis and progression of many human cancers including GBMs (Furnari et al, 2007), leads to the upregulation of the GLI1-NANOG module, which drive increased cancer growth and stem cell expansion. Similarly, the regulation of normal brain stem cell behaviour by HH-GLI1 and p53 (e.g. Lai et al, 2003; Palma and Ruiz i Altaba, 2004; Meletis et al, 2006) raises the possibility that the NANOG-GLI-p53 network is also active in normal stem cells.
In addition to loss of p53, the GLI1-NANOG module is predicted to be enhanced by the loss of other tumour suppressors such as PTEN, which also affects NANOG in a different context (Kuijk et al, 2009), and by oncogenes such as RAS-MEK and AKT that affect GLI1 (Stecca et al, 2007; Stecca and Ruiz i Altaba, 2010). Other inputs that may regulate NANOG independently of GLI1 would also be expected to impinge on the levels of the GLI1-NANOG module, with the balance of all inputs determining the levels of their targets, stem cell numbers and tumour growth (Ruiz i Altaba et al, 2002, 2007).
ES-like cancer signature: implications for tumour progression
The requirement of NANOG in GBMs provides support for the functional involvement of the HH-GLI-responsive ES-like stemness signature, present in different human cancers, including GBMs (Clement et al, 2007) and in cancers of the colon (Varnat, F and Ruiz i Altaba, personal communication), breast and bladder (Ben-Porath et al, 2008). Similarly, NANOG has also been implicated in the growth of prostate cancers (Jeter et al, 2009), and high NANOG levels have been suggested to correlate with poor prognosis of colon cancers and drive EMT (Meng et al, 2010), mimicking GLI1 (Varnat et al, 2009). In addition to NANOG, other ES-like signature factors, such as OCT4, may also be critical for GBMs and other cancers with ES-like signatures. Indeed, the levels of OCT4 are also regulated by HH-GLI signalling, OCT4 has been implicated in different human tumours (e.g. Jin et al, 1999; Gidekel et al, 2003; Yamaguchi et al, 2005) and can induce epithelial dysplasia in mice (Hochedlinger et al, 2005).
NANOG is essential for pluripotency (Silva et al, 2009) and important components of the ES-like signature, notably NANOG, OCT4 and SOX2, have been shown to reprogramme differentiated somatic cells to acquire ES status, becoming iPS cells (Takahashi and Yamanaka, 2006). We, therefore, raise the possibility that NANOG and the ES-like signature of cancers contribute to a GLI1-driven acquisition of advanced, more primitive and malignant states through a process akin to reprogramming.
Therapeutic implications
GBMs remain one of the most deadly cancers in adults, with an average period between diagnosis and death of ∼12 months (Furnari et al, 2007). The discovery of GBM stem cells (Singh et al, 2003, 2004) and their chemo- and radioresistance (Bao et al, 2006) have provided a plausible explanation for the difficulty in GBM treatment and the high rate of relapse. Indeed, as GBM cells are highly invasive, surgery is not routinely curative. New strategies to target GBM stem cells are thus required. HH-GLI signalling is essential for GBM stem cell self-renewal (Clement et al, 2007), suggesting that its targeting may be of therapeutic use. However, given the many suggested GLI1 downstream genes, it remained unclear if there would be essential mediators that could provide additional intervention strategies. Here, we show that NANOG fulfils these criteria: its expression depends on endogenous HH-GLI activity and its function is absolutely required for GBM growth in vivo. Moreover, we provide several lines of evidence in favour of a function not only in the tumour bulk, but also in the control of GBM stem cell behaviour. Therefore, in addition to its use as a biomarker, the requirement of NANOG in GBMs and its apparent highly restricted normal expression in adults suggest that blockade of NANOG, and thus of the GLI1-NANOG module, directly or indirectly will be beneficial to treat GBMs.
Materials and methods
Tumour dissociation, cell culture and FACS analyses
Fresh tumour samples or xenografts were manually dissociated after incubation in papain-containing 5.5 mM L-cystein, 1.1 mM EDTA at 37°C for 1 h, followed by treatment with DNAse (Roche) and Ovomucoid (BD bioscience), filtered with a 70 μm filter (Millipore), washed in PBS and cultured in gliomaspheres media (2/3 DMEM F12, 20% BIT 9500 (stem cell technology), FGF 10 ng/μl, EGF 10 ng/μl and 1% pen/strep, plus 1/3 filtered conditioned media). Primary glioma adherent cells were cultured on Laminin as described (Pollard et al, 2009). U87MG and U251 were cultured as described (Stecca and Ruiz i Altaba, 2009). For FACS analyses, cells were manually dissociated, resuspended in PBS-EDTA 5 mM and analysed for their red and green fluorescence using an FACSCalibur machine (BD bioscience). Primary tumours are described earlier (Clement et al, 2007). Additional tumours were GBM-14: right rolandic plus corpus callosum, female, 83 years old; GBM-15: right fronto-parietal, male, 45; GBM-16: left temporo-occipital, male, 62 and GBM-17: left frontal, female, 73. IDH1 and IDH2 sequencing of GBMs used primers described in Hartmann et al (2009). The p53 sequencing of exons 5–9 was as described (Stecca and Ruiz i Altaba, 2009). U87 were infected with lentivectors and 20000 cells/well were plated 48 h later in medium containing increasing concentrations of TMZ. Cells were harvested and counted 5 days later. All tumours were obtained with patient consent locally in Geneva (Clement et al, 2007) and under approved protocols at La Salpetrière Hospital, Paris, France. CD133 MACS and FACS were as described in Varnat et al, (2009).
RT–PCR and quantitative RT–PCR
Total RNA extracted with micro- or miniRNA easy kits (Qiagen) or Trizol (Invitrogen) was treated with DNAseI and cDNA synthetized by random priming. Quantitative real time (q) PCR used iQTm SYBR green mix (Bio Rad). Reactions were at 60°C using an Opticon PCR apparatus from MJ Research. The level of each target gene was normalized using the geometrical mean of TBP and β ACTIN. OCT4 (POU5F1) primers pick OCT4 plus 12/13 pseudogenes. Other PCRs were performed using Phusion polymerase (Finnzymes). RT–PCR primers were as described (Clement et al, 2007; Varnat et al, 2009) with the exception of the following written 5′ to 3′:
- NANOG/P8—fw
AAATTGGTGATGAAGATGTATTCG
- NANOG/P8—rev
GCAAAACAGAGCCAAAAACG
- NANOGP8—fw
GCTGCCTTCAAGCATCTGTT
- NANOGP8—rev
TTGTTTGCCTTTGGGACTGGT
- NANOG/P8 3′UTR—fw
GGATGGTCTCGATCTCCTGA
- NANOG/P8 3′UTR—rev
CCCAATCCCAAACAATACGA
- β ACTIN—fw
TGGAGAAAATCTGGCACCACACC
- β ACTIN—rev
GATGGGCACAGTGTGGGTGACCC
- TBP—fw
TGCACAGGAGCCAAGAGTGAA
- TBP—rev
CACATCACAGCTCCCCACCA
- PTCH1—fw
GGCAGCGGTAGTAGTGGTGTTC
- PTCH1—rev
TGTAGCGGGTATTGTCGTGTGTG
- SMOH—fw
GGGAGGCTACTTCCTCATCC
- SMOH—rev
GGCAGCTGAAGGTAATGAGC
- TP53—fw
GTGGAAGGAAATTTGCGTGT
- TP53—rev
CCAGTGTGATGATGGTGAGG
- SUFUH—fw
GGCTTTGAGTTGACCTTTCG
- SUFUH—rev
CATCTGTGGGTCCTCTGTCA
Primers for 3′UTR sequencing were 3′UTR—fw GAGACGGGGTTTCACTGTGT and 3′UTR—rev CACTCGGTGAAATCAGGGTAA. PCR products were then cloned in pCRII-TOPO vector (Invitrogen) and ±20 individual clones were grown and sequenced.
Lentivectors
The 293T cells were transfected with calcium chloride using the VSV-G envelope plasmid pMD2G plasmid, packaging R8.74 plasmid, and the following lentivectors: parental pLL3.7, pLL3.7-shNANOG-1 (GGGTTAAGCTGTAACATACTT; Zaehres et al, 2005), pLKO-shNANOG-2 (CCTGGAACAGTCCCTTCTATA; Biocat), pLL3.7-shNANOGP8 (AACAAAGCACATCTTGCCAGGA); pTween-NANOG; pRZ NANOG → Red (System Biosciences), pTW-GLI1, pTW-GLI3R, pLVCTH-shPTCH1 (Varnat et al, 2009), pLVCTH-shSMOH (Clement et al, 2007) and pLV-WPXL-shp53 (targeting human p53). A cDNA from the ATG to the stop codon of NANOG was synthesized from human foetal brain RNA and cloned in frame with a Flag tag in pFLAG-CMV2 (Sigma) vector. Flag-NANOG was then XbaI-XhoI cloned behind the CMV promoter in the pRRL-CMV-PGK-GFP-WPRE (pTWEEN) lentivector. Supernatants were harvested and concentrated by ultracentrifugation. Concentrated viruses were titrated and added to U87 cells, attached or dissociated gliomaspheres for 2 days to achieve >80% infection corresponding to MOI of ∼2. Transduced cells were then washed and collected 2–3 days later for analysis.
Clonogenic assays
Transduced cells were dissociated and plated at 1 cell/well in 96-well plates in gliomasphere media, in triplicate, for each experiment. The number of total and of GFP+ clones was determined using an inverted optical microscope with epifluorescence (Zeiss).
BrdU incorporation assays and immunodetection
BrdU pulses were performed for 16 h for gliomaspheres and for 1 h for U87 and U251 cells. Gliomaspheres were dissociated and plated on matrigel 1:1000 (BD bioscience) for 30 min to allow attachment and fixed with ice-cold PFA (4%) for 30 s, followed by extensive washing with ice-cold PBS and PBT (PBS-0.1% Triton). Anti-NANOG (see text), anti-PML rabbit SC-5621 and mouse SC-996, Santa Cruz; 54BP1, a kind gift of Thanos Halazonetis, U Geneva) or anti-FLAG epitope (SIGMA M2 clone) antibodies were applied after blocking with PBT plus 10% heat inactivated goat serum (HINGS) for O/N at 4°C. Secondary anti-rabbit Cy3 labelled was applied at 1/1000. For BrdU assays (anti-BrdU Ab used at 1/5000; University of Iowa Hybridoma Bank), cells were incubated first with 10% HCl for 15 min at room temperature and then blocked with borax 0.1 M for 10 min at room temperature before blocking. Secondary anti-mouse rodamine labelled (Santa Cruz) (1:500) was diluted in PBT-10% HINGS for 45 min at room temperature. After washing, cells were stained with DAPI (Sigma) 1:10 000 for 2 min, mounted in PBS/glycerol with a pinch of PPDA and analysed under fluorescent Axiphot or confocal LSM-meta microscopes (Zeiss). For proliferation assays, 10 independent fields of BrdU/DAPI-labelled cells were counted per condition.
Red/green in vitro assays and orthotopic xenografts
For in vitro red/green assays (Varnat et al, 2009), GBM-8 gliomaspheres or U87 cells were infected with LV-shNANOG-1 or LV-GFPcontrol-1 and mixed with sibling LV-RFP-transduced cells at ratio 1:1. After 5 days, magnetic CD133 sorting was performed (CD133 MicroBead kit, Myltenyi Biotec), and the GFP/Red ratio was determined by FACS analysis on the different fractions. For in vivo red/green assays, 105 dissociated cells were resuspended in 5 μl of HBSS and injected intracranially at coordinates {x, y, z=−2, −1, −2.5} relative to the bregma point using a stereotaxic apparatus. Fluorescence of xenografts was visualized in situ using dual red and green fluorescence excitation lasers in a special dark chamber with a colour CCD camera (Lightools Research), and digitally recorded. Mice were collected at the first signs of neurological disease.
Western blotting
Proteins were harvested in cold RIPA buffer 4 days after U87 cell transduction, incubated on ice for 20 min and centrifuged at 13 000 r.p.m. at 4°C for 20 min. Supernatants were collected and measured for protein concentration (BCA protein Assay, Pierce); 20 μg of total protein for HSP90 and p53 and 80 μg for GLI1 were run on an SDS–PAGE gel and transferred on a nitrocellulose membrane O/N at 4°C. Membranes were blocked in PBT-5% skimmed milk and blotted with an anti-p53 (1/2000) (mouse, Santa-Cruz, Clone DO-1) or HSP90 (1/4000) (mouse, Santa-Cruz, Clone F-8) antibodies for 1 h at room temperature, or GLI1 affinity-purified polyclonal antibodies (Stecca and Ruiz i Altaba, 2009) O/N at 4°C. Secondary antibodies (anti-mouse HRP (1/6000) (Promega) or anti-rabbit HRP (1/2000) (Promega)) were incubated for 1 h at room temperature. Signal on membranes was revealed with ECL (Thermo Scientific) for HSP90 and p53 or with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) for GLI1.
Luciferase reporter assay
Gli-binding site luciferase reporter and lentiviral/plasmid constructs (e.g. Stecca and Ruiz i Altaba, 2009; Varnat et al, 2009) were transfected in U87 cells with Fugene (Roche). Renilla controls were included in all cases and luciferase units were firefly/renilla ratios. Luminescence was analysed with the dual-glo luciferase reporters system (Promega) and read with a luminometer.
Supplementary Material
Acknowledgments
We thank Web Cavenee, Christophe Mas, Irene Siegl-Cachedenier, Alice Melotti and all Ruiz i Altaba laboratory members for comments on the paper and discussion. We are grateful to S Yamanaka for anti-NANOG antibodies and M Scott for sharing unpublished data. MZ, AD, ALT, IB and ARA designed and performed the experiments. ARA wrote the paper with MZ, AD and ALT. SNN and Jean-Yves Delattre supplied GBM-15, -16 and -17. This work was supported by postdoctoral grants from the Fondation pour la Recherche Médicale, France, to AD, and from Ikertzaileak Hobetzeko Laguntzen Programak, Eusko Jaurlaritza, to ALT; and by Swissbridge and Leenaards Awards and grants from the NCCR Frontiers in Genetics, Oncosuisse and the Swiss National Science Foundation to ARA.
Footnotes
ARA is an advisor to Phistem. The other authors declare that they have no conflict of interest.
References
- Abe Y, Oda-Sato E, Tobiume K, Kawauchi K, Taya Y, Okamoto K, Oren M, Tanaka N (2008) Hedgehog signaling overrides p53-mediated tumor suppression by activating Mdm2. Proc Natl Acad Sci USA 105: 4838–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar EE, Chaudhry A, Lin A, Fan X, Schreck K, Matsui W, Piccirillo S, Vescovi AL, DiMeco F, Olivi A, Eberhart CG (2007) Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells 25: 2524–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN (2006) Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444: 756–760 [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA (2008) An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40: 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth HA, Holland PW (2004) Eleven daughters of NANOG. Genomics 84: 229–238 [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122: 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A (2003) Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 113: 643–655 [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A (2007) Nanog safeguards pluripotency and mediates germline development. Nature 450: 1230–1234 [DOI] [PubMed] [Google Scholar]
- Chen R, Nishimura MC, Bumbaca SM, Kharbanda S, Forrest WF, Kasman IM, Greve JM, Soriano RH, Gilmour LL, Rivers CS, Modrusan Z, Nacu S, Guerrero S, Edgar KA, Wallin JJ, Lamszus K, Westphal M, Heim S, James CD, VandenBerg SR et al. (2010) A hirearchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell 17: 362–375 [DOI] [PubMed] [Google Scholar]
- Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ, Tsai TH, Chou SH, Chien CS, Ku HH, Lo JF (2008) Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res 14: 4085–4095 [DOI] [PubMed] [Google Scholar]
- Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A (2007) HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol 17: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmane N, Ruiz i Altaba A (1999) Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 126: 3089–3100 [DOI] [PubMed] [Google Scholar]
- Dahmane N, Sánchez P, Gitton Y, Palma V, Sun T, Beyna M, Weiner H, Ruiz i Altaba A (2001) The Sonic Hedgehog-Gli pathway regulates dorsal brain growth and tumorigenesis. Development 128: 5201–5212 [DOI] [PubMed] [Google Scholar]
- Darr H, Mayshar Y, Benvenisty N (2006) Overexpression of NANOG in human ES cells enables feeder-free growth while inducing primitive ectoderm features. Development 133: 1193–1201 [DOI] [PubMed] [Google Scholar]
- de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA (2004) Drosophila myc regulates organ size by inducing cell competition. Cell 117: 107–116 [DOI] [PubMed] [Google Scholar]
- Duboule D ed (1994) Homeobox Genes. Oxford, UK: Oxford University Press [Google Scholar]
- Ehtesham M, Sarangi A, Valadez JG, Chanthaphaychith S, Becher MW, Abel TW, Thompson RC, Cooper MK (2007) Ligand-dependent activation of the hedgehog pathway in glioma progenitor cells. Oncogene 26: 5752–5761 [DOI] [PubMed] [Google Scholar]
- Epstein CB, Attiyeh EF, Hobson DA, Silver AL, Broach JR, Levine AJ (1998) p53 mutations isolated in yeast based on loss of transcription factor activity: similarities and differences from p53 mutations detected in human tumors. Oncogene 16: 2115–2122 [DOI] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK (2007) Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 21: 2683–2710 [DOI] [PubMed] [Google Scholar]
- Gidekel S, Pizov G, Bergman Y, Pikarsky E (2003) Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell 4: 361–370 [DOI] [PubMed] [Google Scholar]
- Hall PE, Lathia JD, Miller NG, Caldwell MA, ffrench-Constant C (2006) Integrins are markers of human neural stem cells. Stem Cells 24: 2078–2084 [DOI] [PubMed] [Google Scholar]
- Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, Weller M, Herold-Mende C, Unterberg A, Jeuken JW, Wesseling P, Reifenberger G, von Deimling A (2009) Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol 118: 469–474 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Lopes SM, Tang F, Surani MA (2008) Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 3: 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R (2005) Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 121: 465–477 [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR (2006) Dissecting self-renewal in stem cells with RNA interference. Nature 442: 533–538 [DOI] [PubMed] [Google Scholar]
- Jeter CR, Badeaux M, Choy G, Chandra D, Patrawala L, Liu C, Calhoun-Davis T, Zaehres H, Daley GQ, Tang DG (2009) Functional evidence that the self-renewal gene NANOG regulates human tumor development. Stem Cells 27: 993–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T, Branch DR, Zhang X, Qi S, Youngson B, Goss PE (1999) Examination of POU homeobox gene expression in human breast cancer cells. Int J Cancer 81: 104–112 [DOI] [PubMed] [Google Scholar]
- Kuijk EW, van Mil A, Brinkhof B, Penning LC, Colenbrander B, Roelen B (2009) PTEN and TRP53 independently suppress Nanog expression in spermatogonial stem cells. Stem Cells Dev (in press) [DOI] [PubMed] [Google Scholar]
- Lai K, Kaspar BK, Gage FH, Schaffer DV (2003) Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat Neurosci 6: 21–27 [DOI] [PubMed] [Google Scholar]
- Lee EY, Ji H, Ouyang Z, Zhou B, Ma W, Vokes SA, McMahon AP, Wong WH, Scott MP (2010) Hedgehog pathway-regulated gene networks in cerebellum development and tumorigenesis. Proc Natl Acad Sci USA 107: 9736–9741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke S, Enczmann J, Waclawczyk S, Wernet P, Kogler G (2007) Oct4 and its pseudogenes confuse stem cell research. Cell Stem Cell 1: 364–366 [DOI] [PubMed] [Google Scholar]
- Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y (2005) p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol 7: 165–171 [DOI] [PubMed] [Google Scholar]
- Machida K, Tsukamoto H, Mkrtchyan H, Duan L, Dynnyk A, Liu HM, Asahina K, Govindarajan S, Ray R, Ou JH, Seki E, Deshaies R, Miyake K, Lai MM (2009) Toll-like receptor 4 mediates synergism between alcohol and HCV in hepatic oncogenesis involving stem cell marker Nanog. Proc Natl Acad Sci USA 106: 1548–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletis K, Wirta V, Hede SM, Nistér M, Lundeberg J, Frisén J (2006) p53 suppresses the self-renewal of adult neural stem cells. Development 133: 363–369 [DOI] [PubMed] [Google Scholar]
- Meng HM, Zheng P, Wang XY, Liu C, Sui HM, Wu SJ, Zhou J, Ding YQ, Li JM (2010) Overexpression of nanog predicts tumor progression and poor prognosis in colorectal cancer. Cancer Biol Ther 9: 295–302 [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113: 631–642 [DOI] [PubMed] [Google Scholar]
- Morata G, Ripoll P (1975) Minutes: mutants of drosophila autonomously affecting cell division rate. Dev Biol 42: 211–221 [DOI] [PubMed] [Google Scholar]
- Moreno E, Basler K (2004) dMyc transforms cells into super-competitors. Cell 117: 117–129 [DOI] [PubMed] [Google Scholar]
- Palma V, Ruiz i Altaba A (2004) Hedgehog-GLI signaling regulates the behavior of cells with stem cell properties in the developing neocortex. Development 131: 337–345 [DOI] [PubMed] [Google Scholar]
- Palma V, Lim DA, Dahmane N, Sánchez P, Brionne TC, Herzberg CD, Gitton Y, Carleton A, Alvarez-Buylla A, Ruiz i Altaba A (2005) Sonic hedgehog controls stem cell behavior in the postnatal and adult brain. Development 132: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piestun D, Kochupurakkal BS, Jacob-Hirsch J, Zeligson S, Koudritsky M, Domany E, Amariglio N, Rechavi G, Givol D (2006) Nanog transforms NIH3T3 cells and targets cell-type restricted genes. Biochem Biophys Res Commun 343: 279–285 [DOI] [PubMed] [Google Scholar]
- Pollard SM, Yoshikawa K, Clarke ID, Danovi D, Stricker S, Russell R, Bayani J, Head R, Lee M, Bernstein M, Squire JA, Smith A, Dirks P (2009) Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell 4: 568–580 [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Melton DA (1989) Interaction between peptide growth factors and homoeobox genes in the establishment of antero-posterior polarity in frog embryos. Nature 341: 33–38 [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Sánchez P, Dahmane N (2002) Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer 2: 361–372 [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A, Mas C, Stecca B (2007) The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol 17: 438–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Chambers I, Pollard S, Smith A (2006) Nanog promotes transfer of pluripotency after cell fusion. Nature 441: 997–1001 [DOI] [PubMed] [Google Scholar]
- Silva J, Nichols J, Theunissen TW, Guo G, van Oosten AL, Barrandon O, Wray J, Yamanaka S, Chambers I, Smith A (2009) Nanog is the gateway to the pluripotent ground state. Cell 138: 722–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB (2003) Identification of a cancer stem cell in human brain tumors. Cancer Res 63: 5821–5828 [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB (2004) Identification of human brain tumour initiating cells. Nature 432: 396–401 [DOI] [PubMed] [Google Scholar]
- Stecca B, Mas C, Clement V, Zbinden M, Correa R, Piguet V, Beermann F, Ruiz i Altaba A (2007) Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci USA 104: 5895–5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B, Ruiz i Altaba A (2009) A GLI1-p53 inhibitory loop controls neural stem cell and tumour cell numbers. EMBO J 28: 663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecca B, Ruiz i Altaba A (2010) Context-dependent regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J Mol Cell Biol 2: 84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676 [DOI] [PubMed] [Google Scholar]
- Varnat F, Duquet A, Malerba M, Zbinden M, Mas C, Gervaz P, Ruiz i Altaba A (2009) Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO Mol Med 1: 338–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Lambie EJ, Snyder M (1993) Nuclear dot antigens may specify transcriptional domains in the nucleus. Mol Cel Biol 13: 6170–6179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Kimura H, Tada M, Nakatsuji N, Tada T (2005) Nanog expression in mouse germ cell development. Gene Expr Patterns 5: 639–646 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kurimoto K, Yabuta Y, Sasaki H, Nakatsuji N, Saitou M, Tada T (2009) Conditional knockdown of Nanog induces apoptotic cell death in mouse migrating primordial germ cells. Development 136: 4011–4020 [DOI] [PubMed] [Google Scholar]
- Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360: 765–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Zhou C, Cheng Q, Shen J, Chen H (2008) Stem-cell-abundant proteins Nanog, Nucleostemin and Musashi1 are highly expressed in malignant cervical epithelial cells. BMC Cancer 8: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920 [DOI] [PubMed] [Google Scholar]
- Zaehres H, Lensch MW, Daheron L, Stewart SA, Itskovitz-Eldor J, Daley GQ (2005) High-efficiency RNA interference in human embryonic stem cells. Stem Cells 23: 299–305 [DOI] [PubMed] [Google Scholar]
- Zhang J, Wang X, Li M, Han J, Chen B, Wang B, Dai J (2006) NANOGP8 is a retrogene expressed in cancers. FEBS J 273: 1723–1730 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.