Abstract
Complexes between the quorum-sensing regulator TraR and its inducing ligand autoinducer (AAI) are soluble in Escherichia coli, whereas apo-TraR is almost completely insoluble. Here we show that the lack of soluble TraR is due in large part to rapid proteolysis, inasmuch as apo-TraR accumulated to high levels in an E. coli strain deficient in Clp and Lon proteases. In pulse labeling experiments, AAI protected TraR against proteolysis only when it was added before the radiolabel. This observation indicates that TraR proteins can productively bind AAI only during their own synthesis on polysomes, whereas fully synthesized apo-TraR proteins are not functional AAI receptors. Purified apo-TraR was rapidly degraded by trypsin to oligopeptides, whereas TraR–AAI complexes were more resistant to trypsin and were cleaved at discrete interdomain linkers, indicating that TraR requires AAI to attain its mature tertiary structure. TraR–AAI complexes eluted from a gel filtration column as dimers and bound DNA as dimers. In contrast, apo-TraR was monomeric, and incubation with AAI under a variety of conditions did not cause dimerization. We conclude that AAI is critical for the folding of nascent TraR protein into its mature tertiary structure and that full-length apo-TraR cannot productively bind AAI and is consequently targeted for rapid proteolysis.
Signal transduction systems are fundamental to all organisms, and many of these include receptor proteins that bind to chemical signal molecules. These receptors generally are synthesized in the absence of these signal molecules and reside in the liganded or nonliganded form either cytoplasmically or on cell surfaces (1–3). At least one receptor, phytochrome of higher plants, is destabilized to proteolysis by its cognate signal, visible light, so that it can transmit information only for the brief time interval between signal detection and receptor destruction (4). We now document the opposite phenomenon, in which a receptor protein, the TraR protein of Agrobacterium tumefaciens, attains its native conformation and resists cellular proteolysis only when synthesized in the presence of its cognate signal molecule, N-3-oxooctanoyl-l-homoserine lactone (here designated Agrobacterium autoinducer, or AAI).
During the past decade it has become clear that many groups of bacteria communicate by releasing and detecting diffusible chemical pheromones (5). A widely disseminated family of these so-called quorum-sensing regulatory systems resembles the LuxR and LuxI proteins of Vibrio fischeri, where LuxI-type proteins synthesize autoinducers (N-acylhomoserine lactones) that diffuse from the bacteria that produce them and form complexes with LuxR-type receptor proteins only at high population densities (6, 7). Among the bacteria containing such a system is A. tumefaciens, a plant pathogen that is best known for genetically transforming higher plants (8). Pathogenesis requires a large plasmid called the Ti plasmid, the interbacterial conjugal transfer of which is positively regulated by two proteins: TraI, which synthesizes AAI, and TraR, which is an AAI-dependent transcriptional activator of plasmid conjugal transfer genes (9).
Relatively few biochemical studies have been done on LuxR-type proteins (10–13). Among these studies, TraR was previously shown to bind one molecule of AAI per protein monomer and to bind with high affinity and specificity to specific DNA sites called tra boxes that are found directly upstream of several TraR-regulated promoters (12). Purified TraR activated transcription of these promoters on supercoiled DNA templates but was far less active on linear templates. In another study, binding of TraR to these binding sites required AAI in vivo (14). Here we provide evidence that AAI acts as a scaffold for the correct folding of the TraR protein and that TraR synthesized in the absence of AAI is irrevocably targeted for rapid proteolysis.
Materials and Methods
Measurements of TraR Turnover in Escherichia coli.
To radiolabel TraR protein, E. coli strain BL21/DE3(pJZ358) (12) was cultured at 28°C in AT medium (15) containing 1 mg/ml ampicillin, 10 μg/ml thiamin, and 20 μg/ml of each amino acid except methionine, in the absence or presence of 1 μM AAI. At an OD600 = 0.4, the culture was treated with isopropyl β-d-thiogalactoside (IPTG) to a final concentration of 0.5 mM. After 30 min, rifampicin was added to a final concentration of 200 μg/ml to inactivate host RNA polymerase. Forty minutes after the addition of IPTG, [35S]methionine was added to each culture to a final concentration of 5 μCi/ml (1 Ci = 37 GBq). After 1 min, nonlabeled methionine was added to a final concentration of 5 mM. To test the effect of AAI on protein stability, AAI was added to a final concentration of 1 μM at the times indicated. Samples (100 μl) were withdrawn at various intervals and rapidly chilled at −80°C. Frozen cells were thawed at 0°C and centrifuged, and the cell pellets were suspended in 100 μl of a buffer containing 50 mM Tris⋅HCl (pH 7.9), 200 mM NaCl, 1 mM DTT, 10 mM EDTA, 0.5 mM PMSF, and 300 μg/ml lysozyme and incubated on ice for 30 min. Cell debris was removed by ultracentrifugation (150,000 × g for 15 min), and the supernatant was size-fractionated by SDS/PAGE and analyzed with the use of a Storm B840 PhosphorImager (Molecular Dynamics).
TraR turnover rates and accumulation were compared in the following protease-deficient strains of E. coli: SG22163 (malP∷lacIQ), SG22174 (SG22163 clpP∷cat), SG22186 (SG22163 Δlon rcsA51∷kan) (16); CAG39118 (W3110 ΔhflB3∷Km) (17), KY2347 (MG1655 Δ(clpPX-lon)1196∷cat), KY2966 (MG1655 ΔhslVU1172∷tet), and KY2981 (MG1655 Δ(clpPX-lon)1196∷cat ΔhslVU1172∷tet sulA2981) (18). To quantitate TraR accumulation, plasmid pJZ335, which contains a Plac-traR fusion (19), was introduced into these strains by transformation, and the resulting strains were cultured in LB broth containing appropriate antibiotics and IPTG in the absence or presence of AAI to late logarithmic phase at 28°C. The cells were concentrated and lysed with the use of lysozyme and EDTA, the lysates were cleared by ultracentrifugation, and the supernatant fractions were subjected to Western immunoblotting with a polyclonal antiserum (20).
To measure the half-life of TraR in the protease-deficient strains described above, two plasmids were introduced into each strain. Plasmid pJZ405, which harbors PT7-traR, was constructed by subcloning a 2.1-kb ScaI–EcoRI fragment of pJZ358 (12) into SmaI–EcoRI-digested pBluescript SK+ (Stratagene). Plasmid pJZ410, which has a gene encoding T7 RNA polymerase, was constructed by ligating BamHI-digested pGP1-2 (21) to BamHI-digested pBBR1MCS5 (22), which confers resistance to gentamycin. Cells were cultured at 28°C to an OD600 = 0.4, shifted to 45°C for 20 min to induce expression of T7 RNA polymerase, and treated with rifampicin to a final concentration of 200 μg/ml. After 10 min at 45°C, the cultures were shifted to 28°C for 30 min, and [35S]methionine was then added to a final concentration of 5 μCi/ml. Radiolabeling was terminated after 3 min by the addition of nonlabeled methionine to a final concentration of 5 mM.
Trypsin Proteolysis.
Radiolabeled apo-TraR and TraR–AAI complexes were incubated with trypsin at the indicated concentrations for 30 min at room temperature and size-fractionated by SDS/PAGE. Radioactive fragments were detected with the use of a Storm PhosphorImager.
Radiolabeling and Gel Filtration Chromatography.
TraR protein was radiolabeled for chromatography by methods described above, except that radiolabeling of 25-ml cultures was carried out for 30 min in the absence of nonlabeled methionine. The cells were then centrifuged and suspended in 1 ml of a buffer containing 50 mM Tris⋅HCl (pH 7.9), 200 mM NaCl, 1 mM DTT, 10 mM EDTA, 0.5 mM PMSF, and 300 μg/ml lysozyme and incubated on ice for 30 min. Cellular debris was removed by ultracentrifugation (150,000 × g for 15 min), and the supernatant was purified by column chromatography with 5 ml of SP-Sepharose (Amersham Pharmacia) equilibrated with TEDG buffer (50 mM Tris⋅HCl, pH 7.9/0.5 mM EDTA/1 mM DTT/5% glycerol) plus 200 mM NaCl. Bound proteins were eluted with the use of a 5-ml linear gradient of NaCl from 0.2 to 1 M. Gel filtration chromatography was performed with a FPLC chromatograph and a Superdex-75 column (Amersham Pharmacia) with TEDG buffer supplemented with 200 mM NaCl at a flow rate of 0.25 ml/min. The eluted fractions were analyzed with the use of an LS 5000CE scintillation counter (Beckman Coulter).
Purification of Maltose-Binding Protein (MBP)-TraR Proteins and Gel Retardation Assays.
To construct a MBP-TraR protein fusion, the traR gene was PCR amplified with the use of plasmid pCF222 (15) as a template and oligonucleotides 5′-GGCGGATCCCAGCACTGGCTGGACAAGCTG-3′ and 5′-GCGAAGCTTCGAACTCTCAGATGAGTT-3′ as primers. The resulting DNA fragment was digested with BamHI and HindIII and cloned into pMal-c2 (New England Biolabs) digested with the same enzymes, resulting in plasmid pJZ301. Strain DH5α(pJZ301) was cultured at 28°C in LB broth containing 1 mg/ml ampicillin, in the presence or absence of 1 μM AAI to an OD600 = 0.4, treated with IPTG to a final concentration of 0.5 mM, and incubated for 4 h. Cells were concentrated by centrifugation and lysed with a French press, and the lysates were cleared by ultracentrifugation. The resulting supernatant was chromatographed with the use of 10 ml of amylose affinity resin (New England Biolabs). MBP-TraR protein was step eluted with a buffer containing 10 mM maltose.
For gel mobility shift assays, a radiolabeled fragment containing the traA-traC intergenic region was synthesized by PCR amplification with the use of pJZ304 (12) as a template and oligonucleotides 5′-GCATCTAGAGCCCGGTCTCACCGGGCCGAG-3′ and 5′-GCCGTCGACGATTTCTTCCCGGATTTTCGATGA-3′ as primers. The former primer was radiolabeled with the use of [γ-32P]dATP and T4 polynucleotide kinase before PCR amplification. When TraR and MBP-TraR were combined, these proteins were incubated at 28°C overnight to allow the formation of heterodimers. Binding reactions contained 10−12 M DNA and indicated amounts of TraR and MBP-TraR in a buffer containing 10 mM Tris⋅HCl (pH 7.9), 1 mM EDTA, 1 mM DTT, 60 mM potassium glutamate, 30 μg/ml calf thymus DNA, 20 μg/ml BSA, and 10% glycerol. After 20 min of incubation on ice, samples were size-fractionated with the use of 5% polyacrylamide gels in 0.5× TAE buffer (20 mM Tris–acetate, pH 8.5/1 mM EDTA). Radioactive bands were quantitated with a Storm PhosphorImager.
Results
Clp and Lon Proteases Cause Rapid Proteolysis of apo-TraR.
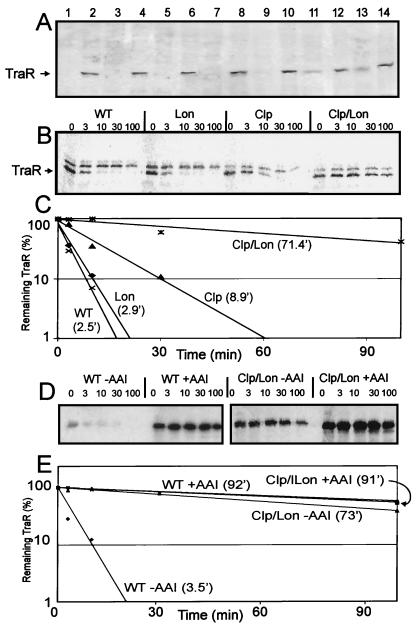
In a previous study, we showed that when TraR is overexpressed in E. coli in the absence of AAI, most of this protein forms insoluble inclusion bodies, whereas when AAI is provided, approximately half of the TraR protein was soluble (12). It was not clear whether all TraR molecules made in the absence of AAI formed inclusion bodies, or whether some fraction of the apo-TraR was soluble but did not accumulate because it was highly unstable to cytoplasmic proteases. If the latter hypothesis is correct, then soluble apo-TraR should accumulate in protease-deficient bacteria. We therefore tested protease-deficient E. coli strains for TraR accumulation with the use of Western immunoblots. Single mutations in hflB, hslVU, clpP, or lon did not detectably increase apo-TraR accumulation (Fig. 1A). In contrast, a clpP, lon double mutant accumulated soluble apo-TraR at levels slightly lower than those found in the presence of AAI (Fig. 1A, lane 11). A clpP, lon, hslVU triple mutant showed similar results (Fig. 1A, lane 13). We conclude that in wild-type cells, cytoplasmic proteases play a significant role in preventing TraR accumulation in the soluble fraction.
Figure 1.
Apo-TraR accumulation and stability in protease-deficient strains. (A) TraR accumulation assayed with the use of Western immunoblots. Lanes 1 and 2, wild type; lanes 3 and 4, hflB; lanes 5 and 6, hslVU ; lanes 7 and 8, clpP; lanes 9 and 10, lon; lanes 11 and 12, clp, lon; lanes 13 and 14, clp, lon, hslVU. Samples in odd- and even-numbered lanes are from cells cultured in the absence and presence of AAI, respectively. (B) Pulse-labeled TraR in wild-type and protease-deficient E. coli strains. The upper bands are probably due to translation of a particularly stable mRNA from chromosome or a gene containing a T7-like promoter. (C) TraR turnover rates obtained from the data in B. Calculated half-lives are indicated in parentheses. (D) Stabilization of TraR by AAI in wild-type strain and in a clp, lon mutant. (E) TraR turnover rates calculated from data in D.
We also conducted pulse–chase experiments to measure the turnover rates of apo-TraR in these same strains. A single mutation in lon had little if any affect on the stability of apo-TraR (Fig. 1 B and C), whereas a mutation in clpP caused a slight increase in apo-TraR stability. However, this half-life, 8.9 min, is still considerably shorter than that of TraR–AAI complexes in a wild-type strain (92 min, Fig. 1E). In contrast, a clpP, lon double mutant strongly decreased TraR proteolysis, such that the half life was increased to over 70 min (Fig. 1 B and C). Addition of AAI to this strain increased the stability of TraR only slightly (Fig. 1 D and E). Differing absolute amounts of TraR are due to unequal cell concentrations (data not shown). We conclude that Clp is the primary protease responsible for apo-TraR turnover, whereas Lon plays a secondary but significant role. These properties are similar to those found in A. tumefaciens, where apo-TraR is rapidly degraded by cytoplasmic proteases (12).
AAI Protects Only Nascent TraR Protein.
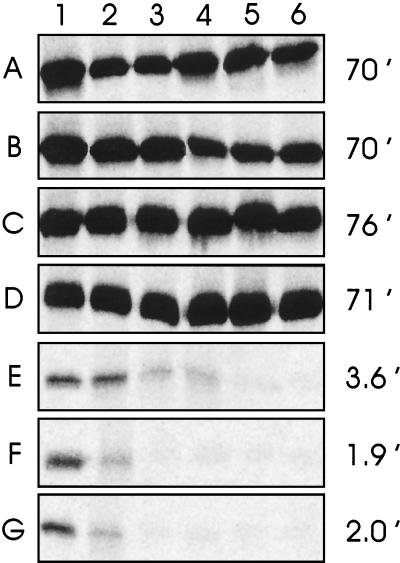
Soluble TraR protein synthesized during a 1-min pulse with radiolabeled methionine in the presence of AAI had a half-life of 70 min (Fig. 2A), whereas TraR made in the absence of AAI had a half-life of 2 min (Fig. 2G). These proteolysis rates are similar to those found in A. tumefaciens (12), despite the fact that TraR is overexpressed in these experiments.
Figure 2.
TraR stability in E. coli in the presence and absence of AAI. Cells expressing TraR from a phage T7 promoter were treated with rifampicin to block host transcription, [35S]methionine, and excess nonlabeled methionine 1 min later to inhibit radiolabeling. AAI was added to a final concentration of 1 μM 20 min (A), 2 min (B), 1 min (C), or 0 min (D) before the addition of the radiolabel, or 1 min (E) or 2 min (F) after the addition of label. In G, AAI was omitted. At various time intervals (0, 2, 4, 8, 16, and 64 min after the addition of nonlabeled methionine in lanes 1–6, respectively), aliquots were frozen at −80°C to terminate proteolysis, lysed, and cleared by ultracentrifugation. Radioactivities of soluble TraR were quantitated with the use of a Storm PhosphorImager. Calculated TraR half-lives are indicated at the right of each panel.
To determine whether AAI can bind to and stabilize fully synthesized apo-TraR protein, we added AAI at various intervals before or after the methionine pulse label. Addition of AAI 2 min, 1 min, or 0 min before the beginning of the pulse label fully stabilized the protein against proteolysis (Fig. 2 B–D). These findings indicate that AAI can diffuse into the cells and productively bind TraR at least as rapidly as exogenous methionine becomes incorporated into protein. In contrast, when AAI was added at the termination of the pulse label or 1 min later, it had virtually no effect on TraR stability (Fig. 2 E and F). We conclude that AAI can protect only nascent TraR from proteolysis. Soluble TraR proteins that fail to bind AAI during their own synthesis are refractory to AAI and are targeted for rapid proteolysis.
Apo-TraR Is Highly Sensitive to Proteolysis in Vitro.
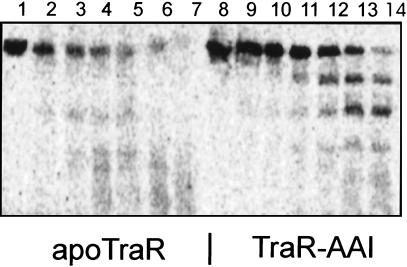
When TraR is synthesized in the absence of AAI, most of it is found in insoluble inclusion bodies (12). However, when radiolabeled, soluble apo-TraR was detected and was purified by ion exchange chromatography by the same protocols as used for TraR–AAI complexes. We treated apo-TraR and TraR–AAI complexes with varying concentrations of trypsin. TraR–AAI complexes were cleaved into three discrete fragments (Fig. 3, lanes 8–14). The largest of these had an apparent mass equivalent to that of the amino-terminal AAI binding domain, whereas the second largest had an apparent mass equivalent to that of the carboxyl-terminal DNA binding domain. These fragments were moderately resistant to further proteolysis. In contrast, apo-TraR was rapidly degraded to oligopeptides by the same concentrations of protease (Fig. 3, lanes 1–7). We conclude that apo-TraR lacks sufficient tertiary structure to resist trypsin-mediated proteolysis.
Figure 3.
Trypsin-mediated proteolysis of apo-TraR and TraR–AAI complexes. Radiolabeled apo-TraR (1 μM) (lanes 1–7) or TraR–AAI (1 μM) (lanes 8–14) complexes were combined with trypsin at the following concentrations: 0 μM (lanes 1 and 8), 0.125 μM (lanes 2 and 9), 0.25 μM (lanes 3 and 10), 0.5 μM (lanes 4 and 11), 1 μM (lanes 5 and 12), 2 μM (lanes 6 and 13), and 4 μM (lanes 7 and 14); incubated for 30 min at room temperature; and size fractionated by SDS/PAGE.
TraR–AAI Complexes Are Dimeric in Solution, Whereas apo-TraR Is Monomeric.
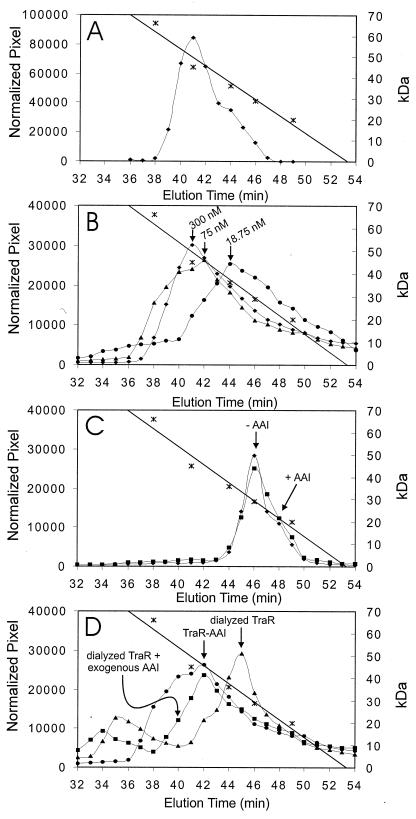
We used gel filtration chromatography to estimate the native molecular masses of TraR–AAI complexes and of apo-TraR. TraR–AAI complexes eluted with a single peak having an apparent mass of approximately 52 kDa (Fig. 4A), in close agreement with an independent study (23). Because the monomer molecular mass of TraR–AAI complexes is 26.5 kDa, we conclude that these complexes are predominantly dimeric under the conditions of this assay. When TraR was preincubated overnight at concentrations of 75 nM or less, its elution profile showed a lower apparent molecular mass (Fig. 4B), indicating, first, that at this concentration, the protein exists as a mixture of monomers and dimers in a rapid, dynamic equilibrium, and second, that the dissociation constant for TraR dimer formation lies somewhere in this range. Because 75 nM is equivalent to just a few molecules per bacterial cell, we conclude that TraR–AAI complexes are predominantly dimeric in vivo.
Figure 4.
Gel filtration chromatography of TraR. (A) TraR was radiolabeled in the presence of AAI and purified by ion exchange chromatography. One hundred microliters of an 0.3 μM TraR solution was size fractionated by gel filtration chromatography. Elution of molecular mass standards is indicated (∗). (B) Same as A, but TraR–AAI complexes were preincubated at the indicated concentrations for 4 h at room temperature before size fractionation. (C) Soluble, radiolabeled apo-TraR, purified by ultracentrifugation and ion exchange chromatography. A 100-μl sample of a 0.3 μM solution of apo-TraR was loaded onto the column in the absence of AAI (⧫) or after preincubation with 1 μM AAI for 4 h (■). In the latter case, the elution buffer also contained 1 μM AAI. (D) Gel filtration of TraR–AAI complexes before and after dialysis to remove AAI. ●, TraR–AAI complexes; ▴, TraR after dialysis in the presence of 3% Tween to remove AAI; ■, TraR dialyzed against 3% Tween and then incubated with 1 μM AAI. All three experiments were conducted with 100 μl of a 75 nM TraR solution.
Apo-TraR eluted from the gel filtration column as a monomer (Fig. 4C). Preincubation of apo-TraR with 1 μM AAI for 4 h did not stimulate TraR dimerization (Fig. 4C). TraR–AAI complexes tolerate dialysis against mild detergents such as 3% Tween if the protein is subsequently dialyzed against 0.01% Tween (12). Dialysis of apo-TraR against 3% Tween containing 1 μM AAI followed by dialysis against 0.01% Tween containing 1 μM AAI also did not promote dimerization (data not shown). This treatment also failed to promote specific DNA binding by this preparation of protein (data not shown). These data suggested that apo-TraR proteins remain as monomers and cannot form functional dimers in the presence of AAI in vitro.
Removal of AAI from TraR–AAI Complexes Does Not Irreversibly Denature the Protein.
Because AAI appears to be essential for the proper folding of TraR and the addition of AAI to apo-TraR does not help it to fold, it seemed plausible that the removal of AAI by dialysis would irreversibly denature the protein. However, when 90% of AAI was removed by dialysis in the presence of 3% Tween, DNA binding was restored by the addition of exogenous AAI (12). This observation indicates that TraR is not irreversibly denatured by the removal of AAI. Here we provide further evidence for this conclusion through the use of gel filtration chromatography to size-fractionate nondialyzed and dialyzed TraR in the presence and absence of AAI. A 75-nM solution of TraR–AAI complexes migrated at a mass of 45 kDa (see above and Fig. 4B). Removal of AAI by dialysis caused these complexes to migrate with an apparent mass of 32 kDa, indicating an increased tendency to dissociate (Fig. 4D), agreeing with an independent study (23). The addition of AAI to dialyzed TraR before and during chromatography shifted the apparent mass back to 45 kDa. These data, combined with the DNA binding data described previously (12), indicate that removal of AAI by dialysis does not irreversibly denature the protein. Once folded, TraR can retain sufficient tertiary structure to bind exogenous AAI when its own AAI has been removed by dialysis.
Apo-MBP-TraR Is Soluble but Inactive.
In an effort to express soluble apo-TraR in protease-proficient E. coli strains, we constructed a malE-traR fusion. When the resulting MBP-TraR fusion protein was expressed in A. tumefaciens, it activated tra gene expression in an AAI-dependent fashion (data not shown). An E. coli strain expressing this protein was cultured with or without AAI and disrupted, and the lysate was cleared by ultracentrifugation. This fusion protein accumulated at very high levels, and virtually all apo-MBP-TraR and MBP-TraR–AAI complexes were found in the supernatant fraction (data not shown). These proteins were purified by amylose affinity chromatography and tested for the ability to bind to the traA-traC intergenic region. MBP-TraR–AAI complexes shifted these fragments with a dissociation constant of 10 nM (Fig. 5A, Lanes 1–8), which is similar to that of native TraR (12). In contrast, apo-MBP-TraR did not shift the gel mobility of the same fragment at any protein concentration tested (Fig. 5B, Lane 2, and data not shown). Therefore, solubility of this protein is not sufficient for DNA binding. Preincubation of this protein with AAI did not promote DNA binding (Fig. 5B, Lane 3). Dialysis against a solution containing 3% Tween and AAI, followed by dialysis against 0.01% Tween and AAI, also did not promote DNA binding (Fig. 5B, Lane 4). Furthermore, MBP-TraR–AAI complexes and apo-MBP-TraR were denatured with 6 M guanidine⋅HCl containing 1 μM AAI. The denaturant was then removed by stepwise dialysis against native buffers containing 1 μM AAI and 0.01% Tween. The preparation originally containing MBP-TraR–AAI complexes bound DNA with the same affinity as a nondenatured control, whereas the preparation originally containing apo-MBP-TraR did not detectably bind DNA (data not shown). The same experiment was repeated with the use of 8 M urea in place of guanidine⋅HCl, and the same results were obtained. We conclude that, at least under these in vitro conditions, purified apo-MBP-TraR is unable to productively bind AAI.
Figure 5.
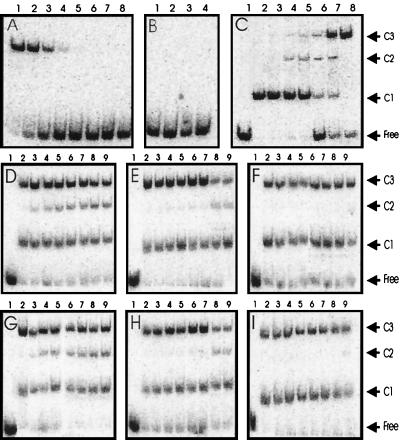
Gel mobility shift assays with MBP-TraR. (A) MBP-TraR–AAI complexes in the following concentrations: 100, 30, 10, 3, 1, 0.3, 0.1, and 0.03 nM. (B) Lane 1, no protein added; lane 2, 1 μM apo-MBP–TraR without AAI; lane 3, 1 μM apo-TraR after incubation with 10 μM AAI for 4 h; lane 4, 1 μM apo-TraR after dialysis in the presence of 3% Tween and 10 μM AAI, followed by dialysis against 0.01% Tween and 10 μM AAI. (C) Gel mobility shifts by mixtures of TraR-AAI and MBP-TraR–AAI. TraR was incubated with MBP-TraR (both complexed with AAI) overnight, combined with DNA fragments, and size-fractionated. Lane 1, no protein; lanes 2–8, TraR and MBP-TraR combined at ratios of 1:0, 1:0.1, 1:0.33, 1:1, 1:3, 1:10, and 0:1, respectively. The total protein concentration was 20 nM. (D) TraR and MBP-TraR (500 nM each) were combined in binding buffer at 28°C for 0, 2, 4, 6, 8, 10, 12, and 14 h (lanes 2–9, respectively) and then stored at −20°C. Samples were thawed and combined with DNA at 0°C for 20 min and size-fractionated. (E and F) Similar to D, except that proteins were combined at 16°C and 4°C, respectively. (G–I) Similar to D–F, respectively, except that proteins were combined at concentrations of 100 nM. Free, unbound DNA; C1, DNA complexed with TraR homodimers; C2, DNA complexed with TraR–MBP-TraR heterodimers; C3, DNA complexed with MBP-TraR homodimers.
The finding that MBP-TraR–AAI complexes were active in vivo and in vitro allowed us to determine the number of TraR protomers per TraR–DNA complex (24). MBP-TraR–AAI complexes were incubated overnight with TraR–AAI complexes in varying ratios. Radiolabeled DNA fragments containing the traA–traC intergenic region were then added, and the resulting complexes were size-fractionated by native PAGE. Native TraR formed relatively rapidly migrating complexes on these gels (Fig. 5C, complex C1), whereas MBP-TraR formed much more slowly migrating complexes (Fig. 5C, complex C3), because of the larger mass of the fusion protein. When native TraR and MBP-TraR were combined, one complex of intermediate mobility was detected (Fig. 5C, complex C2). The fact that only one intermediate complex was seen indicates that this complex contains one protomer of native TraR and one protomer of MBP-TraR. We repeated this experiment, combining these proteins in the presence of 3% Tween followed by dialysis against 0.01% Tween, and obtained identical results (data not shown). We conclude that TraR binds this DNA fragment as a dimer.
Detection of heterodimers was highly dependent on the duration and temperature at which these two proteins were combined before the addition of DNA. When the two proteins were combined at 28°C (at a concentration of 500 nM each), they began to form heteromeric complexes within 2 h, as judged by the detection of complex C2 (Fig. 5D). This observation indicates that TraR–AAI dimers reversibly dissociate to monomers under physiological conditions. In contrast, when the two proteins were combined at 16°C, complex C2 was barely detectable until 12 h of incubation (Fig. 5E), and when the proteins were combined at 4°C, complex C2 was almost undetectable (Fig. 5F). These experiments were repeated with the use of a 100-nM concentration of each protein, and the rate of heterodimer formation was virtually identical to the rates observed with 500 nM (Fig. 5 G, H, and I). Because the rate is largely independent of protein concentration, we conclude that the rate-limiting step in heterodimer formation is the dissociation of each homodimer.
Discussion
We have previously shown that TraR synthesized in A. tumefaciens in the absence of AAI is a highly unstable protein in vivo, whereas TraR–AAI complexes are far more stable (12). There are many examples of unstable regulatory proteins and several examples of regulators whose stability is altered by interaction with other proteins. For example, the stationary phase sigma factor RpoS is destabilized by binding to the response regulator protein RssB (25). Similarly, RpoH is turned over rapidly under normal growth conditions but is more stable during heat stresses, primarily because cytoplasmic proteases are diverted to other substrates (26). The LexA and CI repressor proteins are autoproteolyzed by interaction with RecA under SOS-inducing conditions (27, 28). However, we are not aware of other examples of regulators that must bind inducing ligands to acquire their native conformation and protease resistance.
Our finding that AAI stabilizes TraR against proteolysis in E. coli parallels earlier studies on A. tumefaciens (12). The addition of AAI after termination of the pulse label did not rescue soluble apo-TraR from proteolysis (Fig. 2F). For technical reasons, this experiment was far more feasible in E. coli than in A. tumefaciens. However, given the other similarities in TraR turnover in these two bacteria, it is highly probable that this finding applies to both organisms. The addition of AAI concomitantly with nonlabeled methionine appeared to cause a very slight increase in its stability (Fig. 2E). However, interpretation of this experiment is complicated by two factors. First, AAI may well be imported into the cells more rapidly than methionine. If so, then the added AAI would stabilize nascent TraR molecules that were still incorporating labeled methionine. Second, synthesis of some TraR molecules must have been initiated before the addition of AAI and nonlabeled methionine and terminated after their addition. These TraR proteins would therefore have incorporated radiolabel during the labeling interval and were still nascent and able to bind AAI when the labeling interval was terminated.
Cytoplasmic proteases are generally thought to bind hydrophobic amino acids that are exposed in unfolded proteins but are buried in native proteins (29). The Clp protease sometimes detects hydrophobic amino acids at the carboxyl termini of target proteins (29), and TraR has the dipeptide Leu-Ile at its carboxyl terminus. However, the TrlR protein, which closely resembles TraR but is truncated at its carboxyl terminus, is also stabilized by AAI (Y. Chai, J.Z., and S.C.W., unpublished observations). TrlR does not have hydrophobic residues at its carboxyl terminus, indicating that the carboxyl terminus cannot be the unique determinant detected by this protease. Furthermore, AAI binds the amino-terminal domain of TraR (Y. Chai, J.Z., and S.C.W., unpublished observations) and might not be expected to alter the properties of the carboxyl terminus.
Because AAI is a rather hydrophobic molecule, it is plausible that the AAI binding site would also be hydrophobic. It is tempting to speculate that the hydrophobic AAI-binding pocket of apo-TraR might be detected by Clp and/or Lon and that this site is protected from these proteases by AAI. It is equally possible that this binding site is less exposed in a TraR dimer than in a monomer. On the other hand, our finding that apo-TraR is rapidly degraded in vitro by trypsin without preferential cleavage at the predicted interdomain hinge suggests that apo-TraR may be completely unfolded in vivo, exposing many hydrophobic regions to proteolysis.
A model for TraR function was recently presented by another group in which apo-TraR was monomeric and membrane-associated, whereas TraR–AAI complexes were soluble and dimeric (23). That study used an A. tumefaciens strain in which TraR was strongly overexpressed with the trc promoter. In our studies in which TraR was mildly overexpressed, apo-TraR was undetectable by Western immunoblots because of its rapid proteolytic turnover (12). Our data would seem to preclude models in which apo-TraR is membrane localized. We have also found that when apo-TraR is strongly overexpressed in A. tumefaciens with the T7 promoter, it forms insoluble inclusion bodies (unpublished data). It would be extremely interesting to attempt to detect and localize apo-TraR in strains that express wild-type levels of this protein.
We previously showed that AAI strongly increases the cellular abundance of TraR by two mechanisms. First, TraR–AAI complexes activate transcription of the traR gene, causing a positive autoregulatory loop (15). Second, TraR–AAI complexes have a 20-fold longer half life than apo-TraR (12), indicating that AAI reduces the rate at which TraR is degraded by cytoplasmic proteases. This description of a regulatory protein that requires its coinducer for protease stability may be without precedent, and it is not clear what useful role this proteolysis would play in cell–cell communication. However, positive autoregulation could help to set this regulatory system in two stable states, either completely uninduced or fully induced. The stabilization of TraR by AAI can be interpreted as a form of posttranscriptional positive autoregulation, because it would help ensure that TraR pool sizes will be much larger under inducing conditions than under noninducing conditions. Many other LuxR/LuxI-type systems also display positive autoregulation, and it will be interesting to determine whether other LuxR homologs are stabilized by their cognate autoinducers.
The data described in this study lead us to speculate about how this system, once activated, can be returned to a down-regulated state. TraR binds AAI extremely tightly in vitro, such that the protein remains saturated during purification. TraR releases AAI only in the presence of detergents, and even then only over a period of several days. Therefore, dissociation of AAI is unlikely to play a significant role in down-regulating the tra regulon. In contrast, proteolysis of TraR–AAI complexes may play an important role in terminating induction. These complexes, while far more stable than apo-TraR, are nevertheless turned over at readily detectable rates. We propose that, after a shift from high cell density to low density, existing TraR–AAI complexes remain functional but are eventually proteolyzed, while newly synthesized TraR proteins cannot bind AAI and are rapidly degraded.
Acknowledgments
We thank Susan Gottesman, Christophe Herman, and Takashi Yura for providing protease-deficient strains of E. coli, and Susan Gottesman, Joe Calvo, John Helmann, Jim Shapleigh, and the members of our laboratory for helpful discussions and for critical evaluation of this manuscript. This work was supported by a National Research Service Award (GM 42893) from the National Institutes of Health.
Abbreviations
- AAI
Agrobacterium autoinducer
- MBP
maltose-binding protein
- IPTG
isopropyl β-d-thiogalactoside
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Schleif R. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1300–1309. [Google Scholar]
- 2.Choy H, Adhya S. In: in Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1287–1299. [Google Scholar]
- 3.Hoch J A, Silhavy T J. Two-Component Signal Transduction. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 4.Vierstra R D. Plant Mol Biol. 1996;32:275–302. doi: 10.1007/BF00039386. [DOI] [PubMed] [Google Scholar]
- 5.Dunny G M, Winans S C, editors. Cell–Cell Signaling in Bacteria. Washington, DC: Am. Soc. Microbiol.; 1999. [Google Scholar]
- 6.Fuqua C, Greenberg E P. Curr Opin Microbiol. 1998;1:183–189. doi: 10.1016/s1369-5274(98)80009-x. [DOI] [PubMed] [Google Scholar]
- 7.Hardman A M, Stewart G S, Williams P. Antonie Van Leeuwenhoek. 1998;74:199–210. doi: 10.1023/a:1001178702503. [DOI] [PubMed] [Google Scholar]
- 8.Zhu J, Oger P M, Schrammeijer B, Hooykaas P J J, Farrand S K, Winans S C. J Bacteriol. 2000;182:3885–3895. doi: 10.1128/jb.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winans S C, Zhu J, Moré M I. In: Cell–Cell Signaling in Bacteria. Dunny G M, Winans S C, editors. Washington, DC: Am. Soc. Microbiol.; 1999. pp. 117–128. [Google Scholar]
- 10.Nasser W, Bouilant M L, Salmond G, Reverchon S. Mol Microbiol. 1998;29:1391–1405. doi: 10.1046/j.1365-2958.1998.01022.x. [DOI] [PubMed] [Google Scholar]
- 11.Stevens A M, Dolan K M, Greenberg E P. Proc Natl Acad Sci USA. 1994;91:12619–12623. doi: 10.1073/pnas.91.26.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J, Winans S C. Proc Natl Acad Sci USA. 1999;96:4832–4837. doi: 10.1073/pnas.96.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welch M, Todd D E, Whitehead N A, McGowan S J, Bycroft B W, Salmond G P. EMBO J. 2000;19:631–641. doi: 10.1093/emboj/19.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo Z Q, Farrand S K. Proc Natl Acad Sci USA. 1999;96:9009–9014. doi: 10.1073/pnas.96.16.9009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu J, Winans S C. Mol Microbiol. 1998;27:289–297. doi: 10.1046/j.1365-2958.1998.00672.x. [DOI] [PubMed] [Google Scholar]
- 16.Fuqua W C, Winans S C. J Bacteriol. 1994;176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman S, Roche E, Zhou Y, Sauer R T. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herman C, Thévenet D, Bouloc P, Walker G C, D'Ari R. Genes Dev. 1998;12:1348–1355. doi: 10.1101/gad.12.9.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanemori M, Yanagi H, Yura T. J Bacteriol. 1999;181:3674–3680. doi: 10.1128/jb.181.12.3674-3680.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, Beaber J W, Moré M I, Fuqua C, Eberhard A, Winans S C. J Bacteriol. 1998;180:5398–5405. doi: 10.1128/jb.180.20.5398-5405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabor S, Richardson C C. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kavach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop R M, II, Peterson K M. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 23.Qin Y, Luo Z-Q, Smyth A J, Gao P, Beck von Bodman S, Farrand S K. EMBO J. 2000;19:5212–5221. doi: 10.1093/emboj/19.19.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hope I A, Struhl K. EMBO J. 1987;6:2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker G, Klauck E, Hengge-Aronis R. Proc Natl Acad Sci USA. 1999;96:6439–6444. doi: 10.1073/pnas.96.11.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottesman S. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 27.Little J W, Esmiston S H, Pacelli L Z, Mount D W. Proc Natl Acad Sci USA. 1980;77:3225–3280. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts J W, Roberts C W. Proc Natl Acad Sci USA. 1975;72:4714–4751. [Google Scholar]
- 29.Wickner S, Maurizi M R, Gottesman S. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]