Abstract
High levels of VEGF and leptin are strongly linked to worse prognosis of breast cancer. Leptin signalling up-regulates VEGF in human and mouse mammary tumor cells (MT), but the specific molecular mechanisms are largely unknown. Pharmacologic and genetic approaches were used to dissect the mechanism of leptin regulation of VEGF protein and mRNA in MT (4T1, EMT6 and MMT). A series of VEGF-promoter Luc-reporters (full-length and transcription factor-binding deletions) were transfected into MT to analyze leptin regulation of VEGF transcription. Deletion analysis of VEGF promoter and RNA knockdown shows that HIF-1α and NFκB are essentials for leptin regulation of VEGF. Leptin activation of HIF-1α was mainly linked to canonic (MAPK, PI-3K) and non-canonic (PKC, JNK and p38 MAP) signalling pathways. Leptin non-canonic signalling pathways (JNK, p38 MAP and to less extent PKC) were linked to NFκB activation. SP1 was involved in leptin regulation of VEGF in 4T1 cells. AP1 was not involved and AP2 repressed leptin-induced increase of VEGF. Overall, these data suggest that leptin signalling regulates VEGF mainly through HIF-1α and NFκB. These results delineate a comprehensive mechanism for leptin regulation of VEGF in MT. Disruption of leptin signalling could be used as a novel way to treat breast cancer.
1. INTRODUCTION
VEGF is a master regulator of the growth of blood vessels required for tissue differentiation and function. VEGF has several isoforms resulted from alternative splicing of VEGF mRNA. VEGF-A, the most abundant isoform, binds to transmembrane tyrosine kinase receptors, VEGFR1 (Flt-1) and VEGFR2 (Flk-1/KDR) to exert its cellular functions. VEGFR2 signalling is linked to the main known cellular responses to VEGF. Other VEGF isoforms bind to VEGFR3 (Flt-4), which mediates lymphangiogenesis [1]. Remarkably, the aberrant expression of VEGF is a hallmark of malignant tumor development required for the colonization of endothelial cells that allow tumor nutrition. Therefore, VEGF is a viable target for pharmacological intervention in cancer [2].
Leptin, a pleiotropic cytokine, is mainly secreted by adipose but also overexpressed by cancer cells. Strikingly, leptin/OB-R [3–5] and VEGF [6–8] overexpressions in breast cancer are strongly linked to rapid growth of tumors and worse prognosis. Leptin has absolute specificity to bind and signal through its receptor, OB-R. However, several OB-R isoforms supposedly derived from alternative splicing are expressed in different tissues [9] OB-Rb (full length isoform) is mainly expressed by the hypothalamus where leptin signalling plays a role in neuroendocrine function and controls the appetite and energy balance in normal-weight individuals. OB-R short isoforms expressed by peripheral tissues have diminished signalling capabilities but not well defined biological roles [9]. Leptin/OB-R binding activates several canonic (JAK2/STAT; MAPK/ERK 1/2 and PI-3K/AKT1) and non-canonic signalling pathways (protein kinase C, PKC; stress-activated protein kinase c-Jun N-terminal kinase, JNK and p38 MAP kinase) to exert an increasing number of biological effects in diverse cells [11]. In addition, leptin activates the 5'-AMP-protein kinase (AMPK) that stimulates fatty-acid oxidation in skeletal muscle. Leptin also activates cyclic nucleotide phosphodiesterase 3B (PDE3B) [11]. Importantly, leptin signalling upregulates the expression of several molecules involved in proliferation, survival, inflammation and angiogenesis, i.e., c-Myc [12], cdk2 [13], cyclin D1 [12–14], BCL2 and surviving [15], β3 integrin [16], leukemia inhibitory factor (LIF), LIF receptor (LIF-R), interleukin–1 (IL-1), IL-1 receptor (IL-1R tI), IL-1 receptor antagonist (IL-1Ra) [17–19].
Leptin initially identified as a pro-angiogenic factor [20] is also a positive regulator of VEGF in endometrial [17, 18] and breast cancer cells [14, 21]. Furthermore, the blockade of leptin signaling markedly reduced the growth of tumors and the expression of VEGF/VEGFR2 in mouse models of syngeneic and human breast cancer xenografts [14, 21]. Many growth factors/cytokines and oncogenes have been shown to induce the expression of VEGF through the activation and binding of several transcription factors to the VEGF promoter [22]. Transcriptional upregulation of VEGF by hypoxia inducible factor (HIF-1) under hypoxic conditions has been extensively characterized in many cells and tissues [22]. The mechanisms involved in VEGF regulation include positive regulation of gene transcription by a series of activated transcription factors (HIF-1, AP1, NFκB, SP1/SP3) and steroid hormone receptors, and negative effects of AP2 [23] and p53 [24]. VEGF expression is also under post-transcriptional control of mRNA stability [25]. However, the specific mechanisms involved in leptin regulation of VEGF gene are largely unknown.
Here we describe how leptin activates canonic and non-canonic signalling pathways and transcription factors for the upregulation of VEGF in mouse mammary cancer cells (MT). Basal expressions of VEGF/VEGFR2, leptin/OB-R and estrogen receptor, ERα were initially investigated in MT. Pharmacologic and genetic approaches for key leptin-target kinases and transcription factors were used to dissect the signalling pathways involved in leptin-induced levels of VEGF protein and mRNA. MAPK, PI-3K, PKC, JNK and p38 MAP kinase signalling pathways were generally involved in leptin regulation of VEGF. Examination of transcription factor activation, transcription factor knockdown, effects of inhibitors and deletion analysis of VEGF-promoter Luc-reporters suggest that HIF-1α and NFκB were essential for leptin activation of VEGF transcription in MT. These results delineate for first time the mechanisms for leptin regulation of VEGF and reinforce the idea that the disruption of leptin signalling could impact breast cancer growth by inhibiting proliferation and angiogenesis.
2. MATERIALS AND METHODS
2.1. Materials and Reagents
Anti-STAT3 (F-2), anti-pSTAT3 (B-7), anti-VEGFR2 (A-3) and anti-estrogen receptor-alpha (ERα) antibodies, non-specific species-matched IgGs, HeLa, RAW 264.7 and Caco-2 cell lysates for Western blot positive controls were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Anti-p42/44 MAPK was from Cell Signalling Technology (Danvers, MA). Anti-phospho-p42/44 MAPK (Thr202/Tyr204) was obtained from New England BioLabs Inc (Beverly, MA). Anti-AKT1/PKBα (PH Domain), anti-phospho-AKT1/PKBα (Ser473) antibodies and wortmannin were from Upstate Inc. (Lake Placid, NY). Anti-mouse leptin receptor (OB-R) antibody was from BioVision Inc. (Mountain View, CA). Anti-β-actin antibody was from Abcam Inc (Cambridge, MA). Anti-mouse and anti-rabbit horseradish peroxidase (HRP) conjugates, and iScript cDNA Synthesis, IQ SYBR Green Supermix and protein determination kits were from Bio-Rad Lab. (Hercules, CA). Streptavidin-HRP conjugate was from GE Healthcare UK Limited (Chalfont, UK). Biotinylated anti-mouse IgG/anti-Rabbit IgG (H+L) and normal horse serum were from Vector Laboratories Inc. (Burlingame, CA). ECL Western Blotting Substrate and Restor™ Western blot stripping buffer were from Thermo Scientific (Rockford, IL). Immobilized recombinant Protein G was obtained from Pierce Biotechnology Inc. (Rockford, IL). Fetal bovine serum was obtained from Gemini Bioproducts (West Sacramento, CA). Penicillin-streptomycin cocktails were purchased from Mediatech, Inc. (Manassas, VA). RPMI 1640 medium was from American Type Culture Collection (ATCC, Manassas, VA). Mouse recombinant leptin, mouse VEGF and leptin enzyme-linked immunosorbent assay (ELISA) Quantikine kits and DuoSet IC human/mouse active hypoxia-inducing factor-1 alpha (HIF-1α) activity assay kit were obtained from R & D Systems Inc. (Minneapolis, MN). Dual-luciferase assay system and pLR-TK plasmid were obtained from Promega (Madison, WI). Tanshinone IIA was from BIOMAL International (Plymouth Meeting, PA). NS-398 was from EMD Chemicals Inc. (Darmstadt, Germany). IKKγ NEMO binding domain (NBD) inhibitory and control peptides were from IMGENEX (San Diego, CA). RNeasy Mini kits, DNase kits and Superfect transfect reagents were obtained from Qiagen (Valencia, CA). Nuclear extract kit, TransAM™ AP1 c-Jun, NFκB and SP1 Transcription factor assays kits were obtained from Active Motif (Carlsbad, CA). Tyrphostin AG490, PD98059, Mithramycin A (from Streptomyces plicatus), YC-1, Gö6976, SB203580, SP600125, RIPA buffer, Endofree plasmid maxiprep kit, protease inhibitor and phosphatase inhibitor cocktails 1 and 2 and other chemicals were obtained from Sigma (St. Louis, MO).
2.2. Cell cultures and treatments
Mouse mammary tumor cell lines (MT): 4T1 (CRL-2539), EMT6 (CRL-2755) and MMT060562 (MMT; CCL-51) were purchased from ATCC. 4T1 and MMT were cultured in RPMI-1640 medium containing 10% fetal bovine serum (FBS), 100 units/ml penicillin and 100 µg/ml streptomycin. EMT6 cells were cultured in RPMI-1640 medium containing 15% FBS and 2 mM L-glutamine. All cell cultures were grown as sub-confluent monolayers in a humidified atmosphere containing 5% CO2 at 37°C. Generally, for treatment experiments cells were grown until they were 70% confluent in 60 mm dishes, 6-well plates, 12 well plates or 24 well plates. Cells were subsequently serum-starved for 16–24 h and treated with different compounds in RPMI-1640 medium FBS free (basal medium, BM).
2.3. Endogenous levels of leptin and targeted proteins
To determine the endogenous levels of expression of selected proteins the cells were cultured in 12 well plates (1.5 × 105 cells/well) with BM for 24–48h. Culture supernatants were harvested for VEGF and leptin determinations by ELISA and cell lysates were obtained to determine VEGFR2, ERα and OB-R levels using Western blot (WB).
2.4. Leptin-dose effects
Semi-confluent cells (70 %) were starved for 24h in BM and incubated with mouse leptin (0, 0.6, 1.2 and 6.2 nM) to determine the dose-effects on levels of VEGF protein (ELISA) and mRNA (Real-time RT-PCR).
2.5. Involvement of specific kinases and transcription factors (TF) in leptin-mediated effects on VEGF
Cells starved as described above were incubated for 24h with mouse leptin (0 or 1.2 nM) in the presence or absence of inhibitors of JAK2/STAT3 (AG490, 30µM), ERK1/2/MAPK (PD98059, 30µM), PI3K/AKT1 (Wortmannin, 20µM), PKC (Gö6976, 20µM), p38/MAPK (SB203580, 20µM) and JNK (SP600125, 20µM) signalling pathways. The supernatants were harvested and cell lysates and total RNA were prepared for VEGF protein (ELISA) and mRNA quantification (Real-Time RT-PCR). In other series of experiments, the relationships between leptin-activated kinases and specific DNA binding activity of TF was determined. To this end, the cells were cultured in BM containing the above described kinase inhibitors for 1h in 6 well plates (5×105 cells/well) or 60 mm dishes (106 cells/dish). Then, leptin 1.2 nM was added to the wells containing the kinase inhibitors and cells were incubated for 30 min. After treatments, nuclear extracts were prepared with nuclear extraction kit. Levels of activated TF were determined at 5 µg protein/well using TransAM SP1, AP1 c-Jun, and NFκB p65 ELISAs. Nuclear levels of activated HIF-1α were determined at 10 µg protein/well using the DuoSet IC human/mouse HIF-1α activity assay kit. Assay specificities and sensitivities were verified using positive controls and in competition assays by testing non-labeled oligonucleotides provided by the kits. Positive control nuclear extract for HIF-1α was prepared from 4T1 cells treated with CoCl2 (100µM) for 6h. Protein concentrations were determined by the Bio-Rad kit.
2.6. Impact of TF inhibition on leptin-mediated effects on VEGF
To further determine the role of specific TF on leptin-mediated increase in VEGF expression the cells were incubated in BM containing leptin (0 or 1.2nM) in the presence or absence of inhibitors for HIF-1α (NS398 and YC-1, 10µM), AP1 (Tanshinone IIA, 50µM), NFκB (IKK inhibitor, 100µM) and SP1 (Mithramycin A, 0.2µM) for 24h. Culture supernatants were harvested to determine VEGF protein (ELISA) and cell lysates were used to determine VEGF mRNA (Real-Time RT-PCR).
2.6.1. Short Hairpin RNA Knockdown
Short hairpin RNA (shRNA) constructs against Mus musculus HIF-1α, NFκB, as well as control plasmid (catalog number TR517255, TR516641, TR30012) were purchased from Origene, Technologies, Inc. (Rockville, MD). Plasmid DNA were transformed into competent E. coli, amplified and purified from the culture using PureLinK HQ mini plasmid purification kit (Catalog number K2100-01, Invitrogen, Carlsbad, CA). MT were maintained in complete medium and plated onto 6-well culture dishes (60,000 cells/well for 4T1; 150,000/well for EMT6 and 200,000/well for MMT). After 24 h the culture medium was changed and cells were transfected with shRNA plasmids (1µg) or with control plasmid (1µg) using the Superfect transfect reagents following the manufacture’s guidelines. After 24 h cells were starved for additional 24 h and treated with leptin (6.25nM). Cell culture supernatants were used to quantify VEGF levels by ELISA. Total RNA was extracted for VEGF mRNA quantification by real-time RT-PCR. Nuclear extracts were prepared as described above and used for immunoblot analyses of HIF-1α and NFκB (p65 and p50).
2.7. Western Blot (WB)
Thirty µg of cell lysate protein were incubated with anti-VEGFR-2 antibody, immunoprecipitated using Protein G agarose beads and resolved by WB. To detect ERα and OB-R isoforms 30 µg cell lysate proteins were analyzed by WB. To detect the impact of kinase inhibitors of signalling intermediaries 40 µg of cell lysate protein were analyzed by WB for STAT3, p-STAT3, ERK1/2, p-ERK1/2, AKT1 and p-AKT1 kinases. β-actin and non-phosphorylated proteins were used as loading controls for OB-R, VEGFR2 and ERα and phosphorylated kinases, respectively. Positive controls included HeLa, RAW 264.7 and Caco-2 cells. Nonspecific mouse, rabbit, and goat IgGs were used as negative controls for Western blot analysis. HIF-1α (MAB 1536, R & D Systems) and, NFκB-p65, NFκB-p50 and PCNA (proliferation cell nuclear antigen) for loading control (Sc-109, Sc-114 and Sc-7907 antibodies, Santa Cruz Biotech., respectively); were used for immunoblot analyses of nuclear extracts. The ECL-chemiluminescent assay was used to detect the specific protein bands. To quantitatively assess the effects of cytokines and inhibitors of leptin/OB-R signalling on antigen expression, the X-ray films were analyzed using the NIH Image program (http://rsb.info.nih.gov/ij/).
2.8. Quantification of VEGF and leptin in cell culture supernatants
Mouse leptin and VEGF levels in culture supernatants as determined by ELISA were within the dynamic range of the ELISA standard curves and expressed as pg/ml/mg of total protein. Standards, controls, and samples were assayed in duplicate. According to the manufacturer, the performance characteristics of the mouse VEGF-ELISA were as follows: sensitivity, 3pg/ml, and 100% specificity for mouse VEGF-A (164 and 120 amino acid residue forms) and no significant cross-reactivity with any of several tested cytokines or growth factors, including other VEGF isoforms and their receptors. The mouse leptin-ELISA’s performance characteristics were as follows: sensitivity 22 pg/ml, 100 % specificity for mouse leptin, no-cross-reactivity with any of several tested cytokines.
2.9. Real-time RT-PCR detection of VEGF mRNAs in MT
Total RNA from MT were extracted using RNeasy Mini Kit according to the manufacturer’s protocol. cDNA was synthesized by using iScript cDNA Synthesis kit. Total RNA (0.7–1.0 µg) was used as template for cDNA synthesis. For quantitative comparisons, cDNA samples were analyzed by real-time PCR using the IQ SYBR Green Supermix on the Bio-Rad’s I cycler. Relative expression values (R) were calculated using the equation R = 2–(ΔCt target – ΔCt reference), where Ct target is the fractional threshold cycle of the target gene and Ct reference is the fractional threshold cycle of the reference gene. Primers for VEGF were forward: 5’-TACCTCCACCATGCCAAGTGGT-3’ and reverse: 5’-AGGACGGCTTGAAGATGTAC-3’, which amplify 180bp DNA fragment. The 18s rRNA was used as internal control or reference and was detected using the following primers: forward 5’-GGAAGGGCACCACCAGGAGT-3’ and reverse 5’-TGCAGCCCCGGACATCTAAG-3’, which amplify 320bp DNA fragment. The VEGF PCR conditions were: 1 cycle, 95°C for 3 min; 45 cycles, 95°C for 30 sec; 52°C for 30 sec and 72°C for 30 sec. Different annealing temperatures of 18s rRNA was 61°C. Real-time PCR determinations were duplicated for each cell preparation.
2.10. Immunocytochemistry (ICC)
4T1, MMT and EMT6 cells (5×105 cells/chamber) were cultured in ICC-treated glass slides (BD Falcon™, Belford, MA). Semi-confluent cells (~ 70%) were starved in BM for 48h, fixed with 2% paraformaldehyde solution and incubated with anti-OB-R, ERα and VEGFR2 antibodies (1:100). Negative controls were also included in which the primary antibody was omitted. Specific staining was developed using the ABC staining system (Santa Cruz Biotechnology Inc.). Cells were counterstained with hematoxylin and treated with permanent mounting medium. Staining intensity for each antigen was assigned by two independent observers using a semiquantitative HSCORE. The HSCORE was calculated using the following equation: HSCORE = Σ Pi (i +1) as previously described [26].
2.11. Reporter gene plasmids and constructs
Plasmids of luciferase-reporters for mouse VEGF promoter (full-length) and 5’ deletions of cis elements for hypoxia response element (HRE), AP1, AP2, NFκB and SP1 were prepared as previously described [27]. Briefly, pGL3-basic (promoterless) luciferase vector (Promega, Madison, WI) was used to prepare the reporter constructs. A 1091-bp fragment of the mouse VEGF promoter region (−975 to +116; full-length pLUC-VEGF-975) (GenBank TM/EMBL accession number U41393) and a series of its 5'-end deletions pLUC-VEGF-923, pLUC-VEGF-857, pLUC-VEGF641, pLUC-VEGF-107 and pLUC-VEGF-40 were generated by PCR using oligonucleotide primers designed with sites for specific restriction enzymes. All PCR products were sequenced and confirmed to be identical to published sequences within the mouse VEGF promoter.
2.11.1. Transient transfection experiments and reporter assays
Plasmid pRL-TK renilla luciferase reporter construct was used for internal control. All plasmids were prepared using Endofree plasmid maxiprep kit according to the protocol provided. MT were transiently transfected with plasmids using Superfect following the manufacturer’s guidelines. Briefly, MT were plated at density of 2×104/well in 96 well plates and cultured until they were ~ 70% confluent. Cells were starved in BM for 24h and transfected with 0.3 µg of plasmid DNA per well. To determine the transfection efficiencies of the test plasmids, normalization was performed by cotransfection 0.03 µg of pRL-TK renilla luciferase expression plasmid per well. After transfection cells were washed, incubated in BM for 18–24h, and treated with 0.6nM of leptin for 24h. Cell extracts were prepared and subsequently analyzed for luciferase activity by Dual-luciferase reporter assay system according to the manufacturer’s protocol. Luciferase activity was expressed as relative light units (RLU).
2.12. Statistical analyses
One-way analysis of variance and student t tests were used from Prism 5 software. P<0.05 was designated as a statistically significant difference. All experiments were performed in triplicate and repeated at least three times, and all samples were analyzed in duplicate. The data were expressed as means± SEM.
3. RESULTS
3.1. Endogenous levels of cytokines and receptors in MT
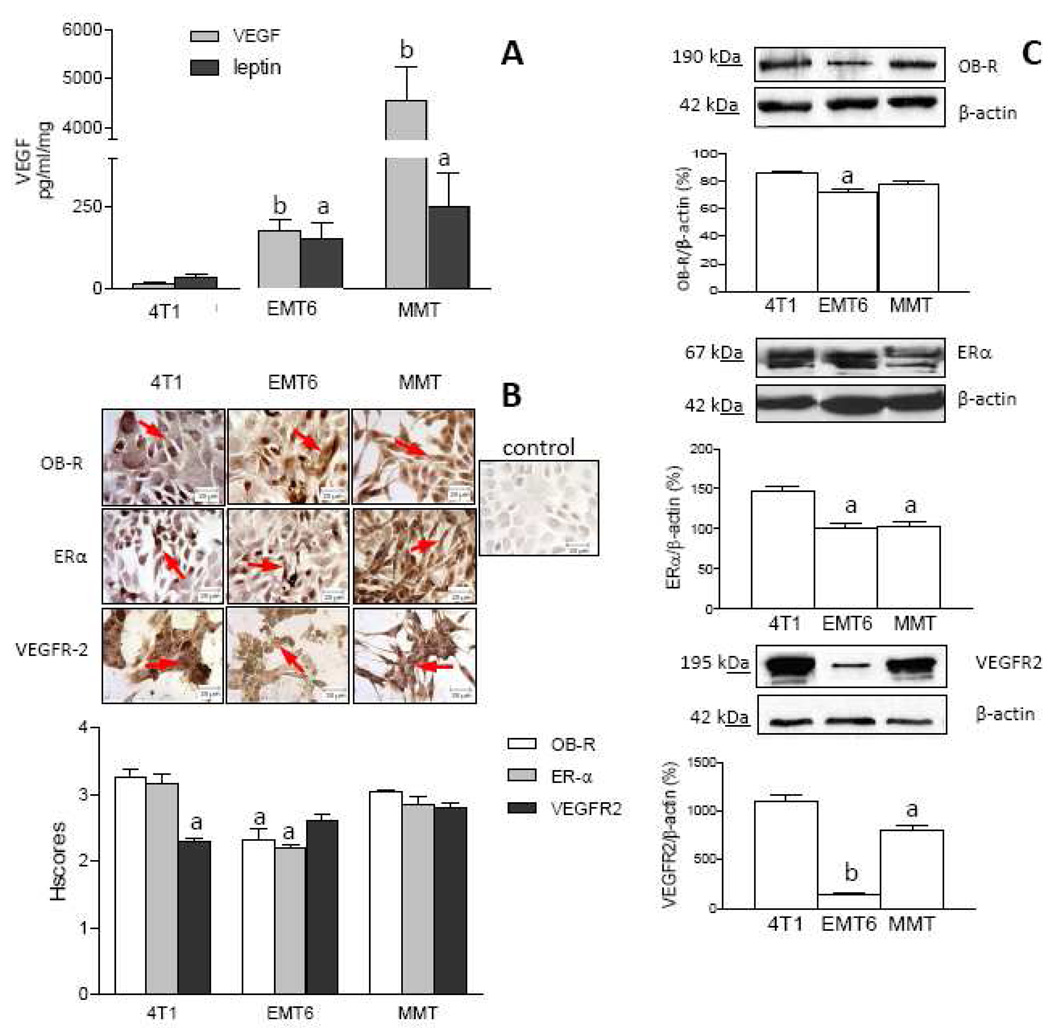
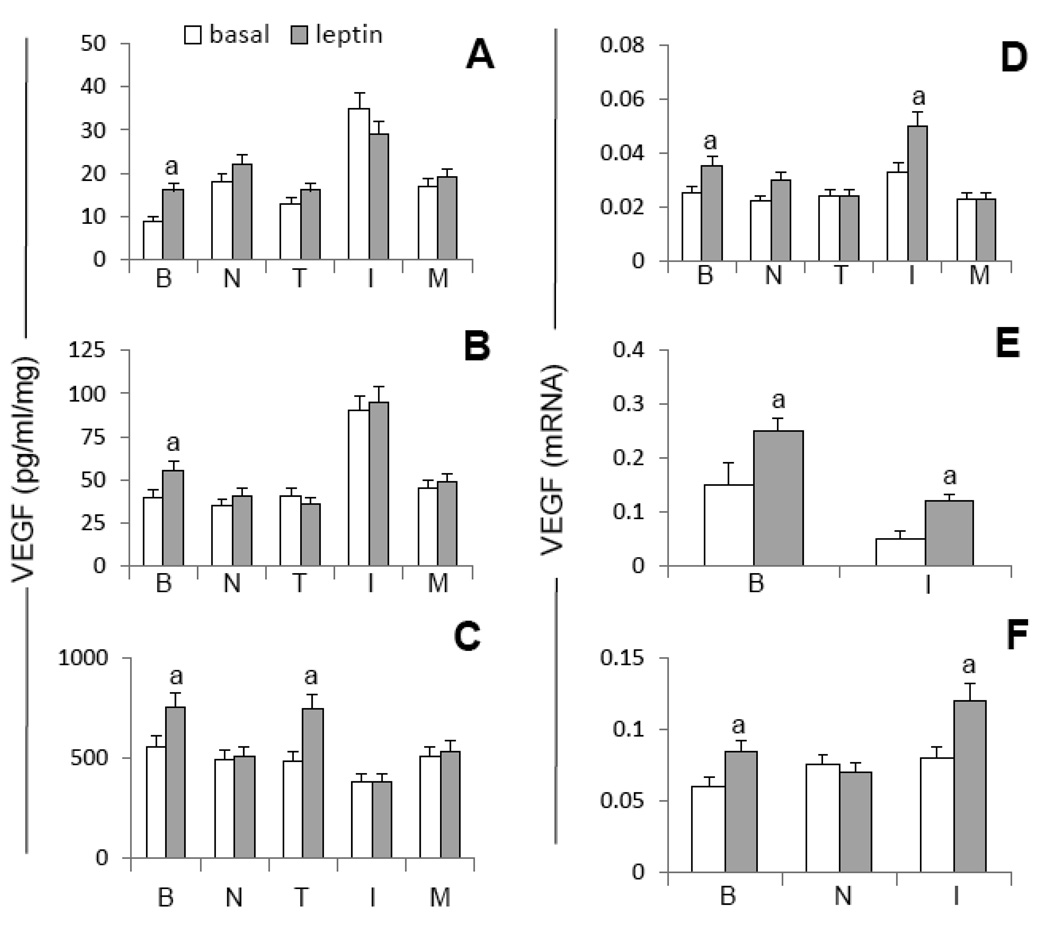
To initially characterize MT used in this research the basal expressions of leptin and VEGF ligands and their corresponding receptors (OB-R and VEGFR2, respectively) and estrogen receptor alpha (ERα) were investigated. 4T1, EMT6 and MMT secrete leptin and VEGF (Fig 1A) in basal conditions. Interestingly, similar patterns for the basal levels of leptin and VEGF correlated among these cell lines. The highest basal levels of leptin and VEGF were secreted by MMT while the lowest levels of these factors were detected in 4T1 cell cultures. The basal secretion patterns for leptin and VEGF were as follows: MMT>>EMT6>4T1 (Fig 1A). Results from ICC show that in basal conditions all cells expressed OB-R and VEGFR2 (Fig 1B). HSCORE analysis suggests that, 4T1 and MMT had higher basal levels for OB-R and VEGFR2 expression (Fig 1B). These results were further corroborated by WB (Fig 1C). Basal expression of OB-R and VEGFR2 as detected by WB correlated to ICC findings in all cells. However, MMT and 4T1 cells showed higher expression of VEGFR2 than OB-R (Fig 1C). Thus, according to WB the basal expression of OB-R and VEGFR2 were as follows: 4T1>MMT>>EMT6. All cells assayed express ERα as detected by ICC (Fig 1B) and WB (Fig 1C). HSCORE analysis suggests that in basal conditions EMT6 cells express lower levels of ERα compared to 4T1 and MMT (Fig 1B). Thus, according to ICC the ERα basal expression follows a similar pattern than VEGFR2 and OB-R (4T1, MMT>EMT6). WB analysis corroborated the basal expression of ERα in all cells and shows that it is higher expressed by 4T1 cells (4T1>EMT6, MMT; Fig 1C).
Fig. 1. Endogenous expression of leptin and target molecules in MT.
A VEGF and leptin levels in MT as determined by ELISA (pg/ml/mg-protein). B and C, OB-R, ERα and VEGFR2 expression (arrows) in MT as determined by immunocytochemistry (ICC) and western blot (WB), respectively. Pictures show representative results from ICC (magnification ×63). Relative expression of receptors was determined by HSCORE (see Section 2.10). Quantitative WB data were calculated from densitometric analysis of bands with the NIH image program. The values were normalized to β-actin as a control. Cells were cultured for 48h in basal medium (BM). (a) p<0.05 and (b) p<0.01 when comparing levels between cells. Data (mean ± standard error) representative results derived from a minimum of 3 independent experiments.
3.2. Leptin induces increased levels of VEGF protein and mRNA in MT
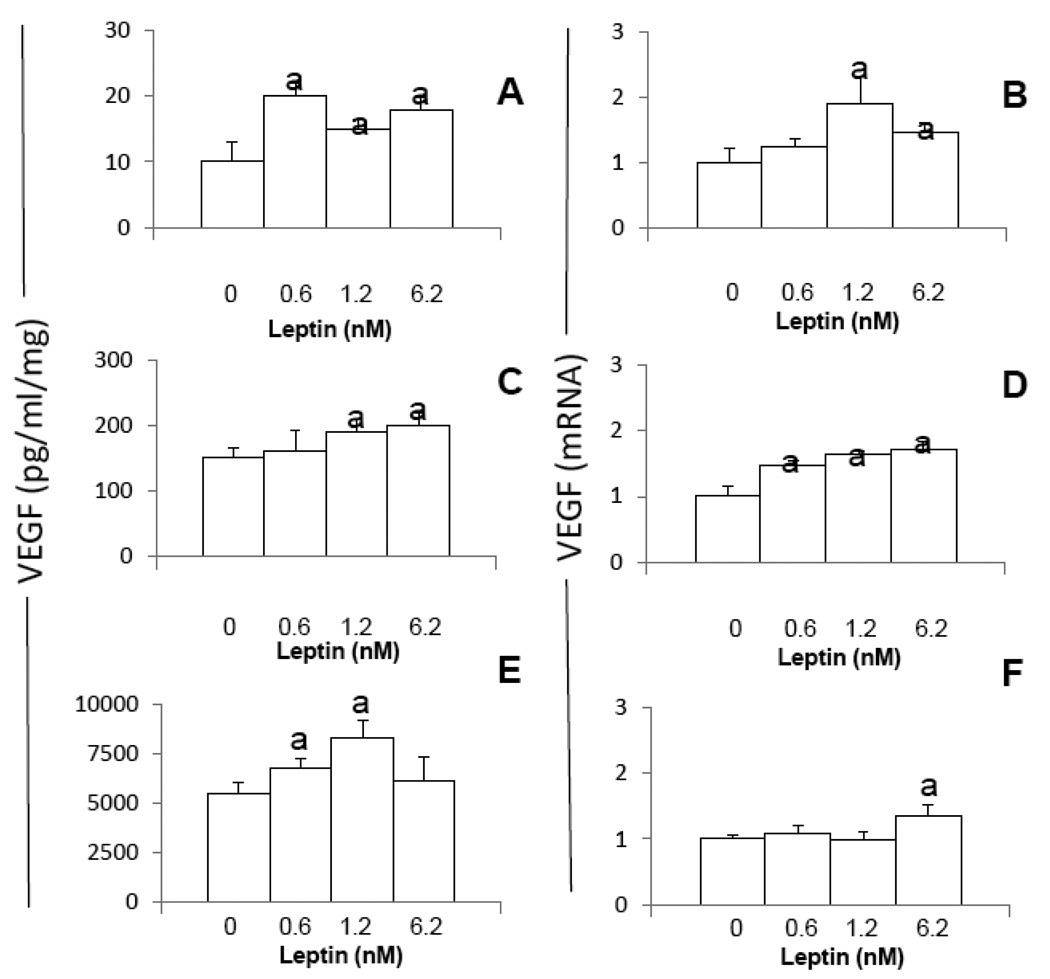
Addition of leptin to MT cultures increases VEGF protein and/or mRNA in all cell lines but not dose-effects were found (Fig 2A–F). VEGF protein was increased in all MT by leptin (Fig 2A, C and D). In comparison, only leptin higher dose (6.25 nM) was able to increase VEGF protein in 4T1 cells (Fig 2A). Leptin at all doses increased VEGF mRNA in EMT6 (Fig 2D) but the higher doses of leptin were required to increase VEGF mRNA in 4T1 (Fig 2B) and MMT (Fig 2F).
Fig. 2. Dose-effects of leptin on the levels of VEGF protein and mRNA in MT.
A, C and E VEGF protein and B, D and F, VEGF mRNA levels induced by leptin (0, 0.6, 1.2 and 6.2nM) in 4T1, EMT6 and MMT, respectively. Cells were cultured for 48h. VEGF levels were determined by ELISA. VEGF mRNA levels were quantified by real-time RT-PCR. (a) p<0.05 when comparing levels of VEGF protein (pg/ml/mg-protein) or mRNA to control (basal). Data (mean ± standard error) representative results derived from a minimum of 3 independent experiments.
3.3. Multiple signal pathways are involved in leptin-mediated regulation of VEGF in MT
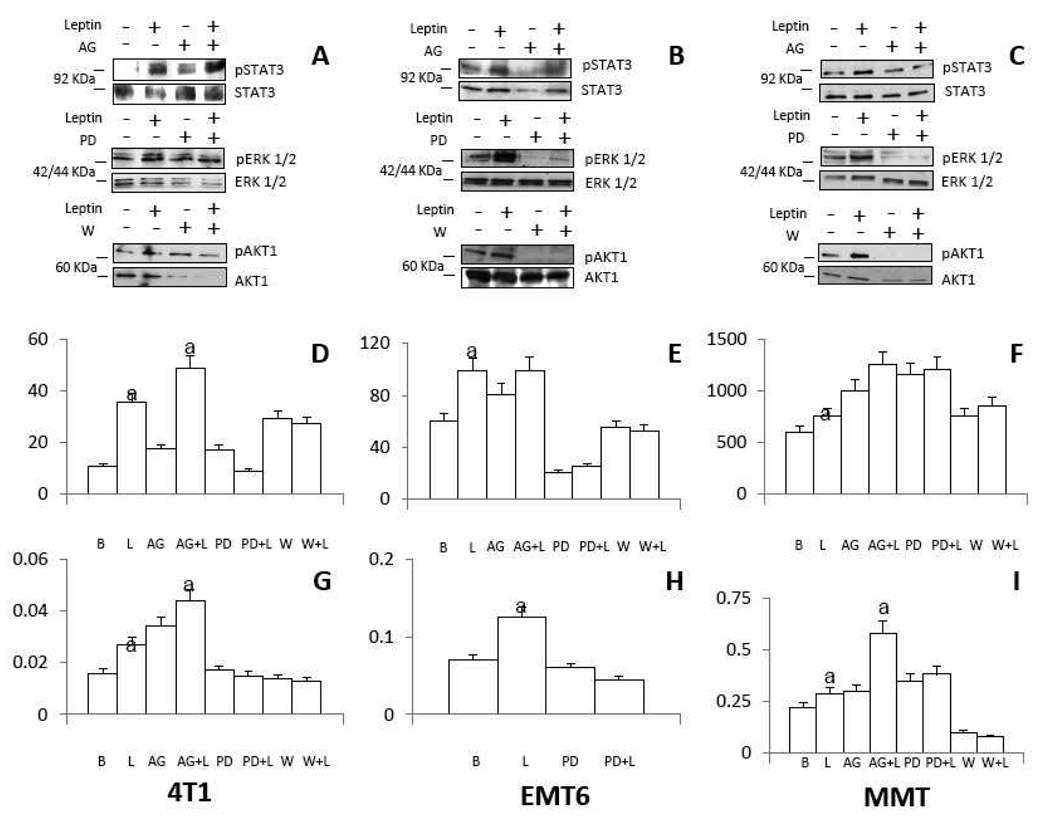
To determine which signalling pathways activated by leptin are involved in the regulation of VEGF in MT several pharmacological inhibitors of specific kinases were used. WB analysis was used to determine the impact of leptin and inhibitors on the phosphorylation of kinases and VEGF levels in 4T1, EMT6 and MMT (Fig. 3). Addition of leptin promoted the activation of leptin canonic signalling kinases (JAK2/STAT3, MAPK/ERK 1/2 and PI-3K/AKT1) in all MT. The co-incubation of cells with leptin and pharmacological inhibitors of JAK2/STAT3 (AG490), MAPK (PD98059) and PI-3K/AKT1 (wortmannin) abrogated the leptin-mediated increase of phosphorylated kinases (Fig 2A, B and C). However, leptin-mediated STAT3 phosphorylation was not inhibited by AG490 in 4T1 (Fig 3A). The analysis of leptin and kinase inhibitor effects suggest that leptin-mediated increase of VEGF protein (Fig 3D) and mRNA (Fig 3G) was mainly related to the activation of PI-3K and MAPK kinases in 4T. Similar results were found for leptin regulation of VEGF protein in MMT (Fig 3F and I) and EMT6 (Fig 3E and 3H) but leptin upregulation of VEGF mRNA only involved MAPK kinase in EMT6 (Fig 3H). Interestingly, inhibition of JAK2/STAT3 signalling increased the basal levels of VEGF protein and mRNA in 4T1 (Fig 3D and G) and MMT (Fig 3F and I). To further examine how leptin could regulate VEGF expression in MT a series of pharmacological inhibitors for leptin non-canonic signalling pathways were used (i.e., Gö6976 for PKC; SP600125 for JNK and SB203585 for p38 kinase; Fig 4). Remarkably, PKC and p38 kinases were linked to leptin-induction of VEGF in all MT (Fig 4A, B and C). Intriguingly, p38 MAPK inhibition in basal conditions increased the basal levels of VEGF in 4T1 cells (Fig 4A).
Fig. 3. Leptin induced canonic signalling pathways involved in the regulation of VEGF in MT.
A, B and C, representative western blots from leptin induction of phosphorylated proteins in 4T1, EMT6 and MMT, respectively. D, E and F, effects of leptin and canonic signalling inhibitors on VEGF protein levels in 4T1, EMT6, and MMT, respectively. G, H and I, effects of leptin and canonic signalling inhibitors on VEGF mRNA levels in 4T1, EMT6, and MMT, respectively. MT were incubated for 24h with mouse leptin (0 or 1.2 nM) in the presence of inhibitors of JAK2/STAT3 (AG490, 30µM), ERK1/2/MAPK (PD98059, 30µM) and PI3K/AKT1 (Wortmannin, 20µM) signalling pathways. VEGF levels were determined by ELISA (pg/ml/mgprotein). VEGF mRNA levels were quantified by real-time RT-PCR. (a) p<0.05 when comparing levels of protein or mRNA of leptin-treated cells to control (basal) with or without inhibitors. Data (mean ± standard error) representative results derived from a minimum of 3 independent experiments. B: Basal, L: leptin, AG: AG490, PD: PD98059, W: Wortmannin.
Fig. 4. Leptin induced non-canonic signalling pathways involved in the regulation of VEGF in MT.
A, B and C, effects of leptin and kinase inhibitors on VEGF levels in 4T1, EMT6 and MMT, respectively. MT were incubated for 24h with mouse leptin (0 or 1.2 nM) in the presence of inhibitors of PKC (Gö6976, 20µM), p38/MAPK (SB203580, 20µM) and JNK (SP600125, 20µM) signalling pathways. VEGF levels were determined by ELISA (pg/ml/mg-protein). (a) p<0.05 when comparing levels of VEGF protein of leptin-treated cells to control (basal) with or without inhibitors. Data (mean ± standard error) representative results derived from a minimum of 3 independent experiments. B: Basal, L: leptin, Gö: Gö6976, SB: SB203580, SP: SP600125.
In addition, JNK activity was related to leptin-mediated increase of VEGF protein in 4T1 cells (Fig 4A) and EMT6 (Fig 4B). In general, these results suggest that leptin-canonic (MAPK and PI-3K) and non-canonic signalling (PKC and p38 kinase and to less extent JNK) are involved in the regulation of VEGF in MT.
3.4. Leptin signalling pathways involved in transcription factor (TF) activation
The VEGF promoter has specific cis-elements for the binding of TF that regulate VEGF expression. To explore how leptin signalling pathways could be linked to transcriptional regulation of VEGF gene the activation of several TF (HIF-1α, AP1, NFκB and SP1) was determined in MT incubated with leptin. To define which leptin-induced signalling pathways were linked to the specific activation of TF several kinase inhibitors were used (Fig 5).
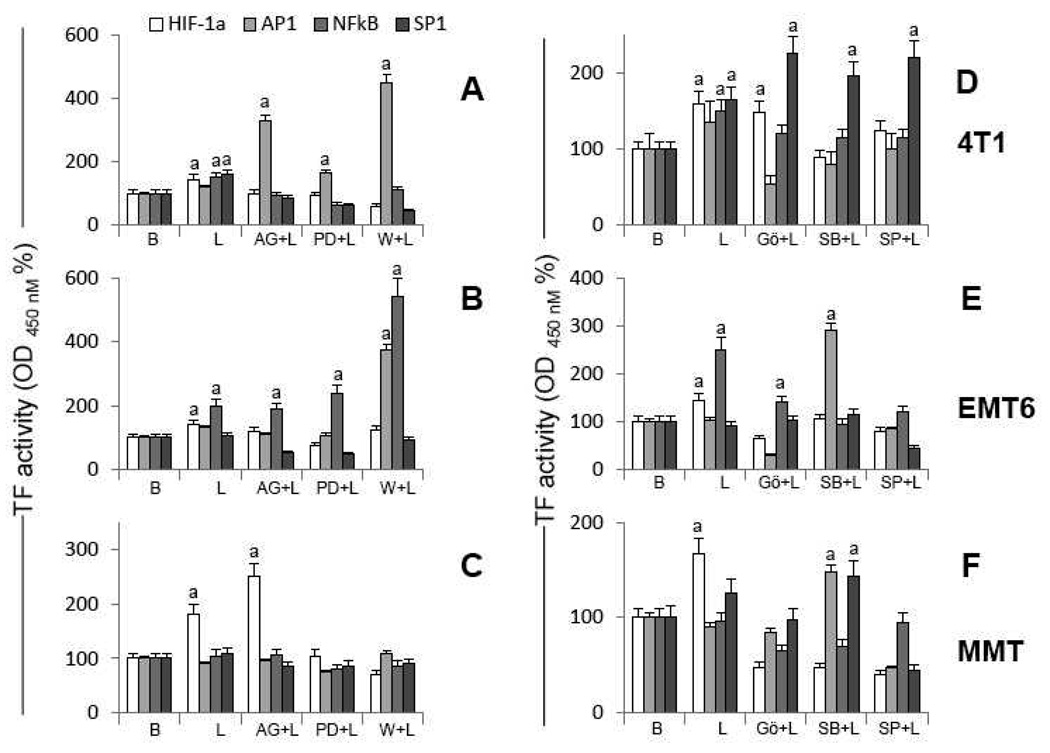
Fig. 5. Leptin-induced signal pathways involved in transcription factor activation in MT.
A, B and C, transcription factor activities in 4T1, EMT6 and MMT, respectively, treated with leptin and inhibitors of canonic signalling pathways. D, E and F, transcription factor activities in 4T1, EMT6 and MMT, respectively, treated with leptin and inhibitors of non-canonic signaling pathways. MT were cultured for 30 min with leptin (0 or1.2 nM) and inhibitors of canonic [JAK2/STAT3 (AG490, 30µM), ERK1/2/MAPK (PD98059, 30µM) and PI3K/AKT1 (Wortmannin, 20µM)] and non-canonic [PKC (Gö6976, 20µM), p38/MAPK (SB203580, 20µM) and JNK (SP600125, 20µM)] signalling pathways. AP1, NFκB and SP1 activities were determined by TransAM ELISA and HIF-1α activity by DuoSet-IC ELISA in nuclear extracts. TF activity levels were expressed as a percent of basal (vehicle treated) in response to the various treatments. (a) p<0.05 when comparing levels of TF activity of leptin-treated cells to control (basal) with or without inhibitors. Data (mean ± standard error) representative results derived from a minimum of 3 independent experiments. B: Basal, L: leptin, AG: AG490, PD: PD98059, W: Wortmannin, Gö: Gö6976, SB: SB203580, SP: SP600125.
Leptin signalling activated HIF-1α in all cells tested (Fig 5A–F). In addition, NFκB was activated by leptin in 4T1 (Fig 5A and D) and EMT6 (Fig 5B and E). Analysis of kinase inhibition shows that HIF-1α was mainly linked to the activation of specific leptin canonic (MAKP and PI-3K; Fig 5A, B and C) and non-canonic (PKC, JNK and p38; Fig 5D, E and F) signalling pathways in MT. However, JAK2/STAT3 pathway was also linked to leptin activation of HIF-1α in 4T1 (Fig 5A) and EMT6 cells (Fig 5B). Interestingly, the inhibition of JAK2/STAT3 increased the levels of activated HIF-1α in MMT (Fig 5C). In comparison, NFκB was mainly linked to leptin activation of non-canonic signalling pathways (Fig 5D and E). Though, leptin activation of JAK2/STAT3, MAPK/ERK1/2 and PI-3K were also linked to NFκB activation in 4T1 (Fig 5A) and MMT (Fig 3C). Intriguingly, leptin did not activated AP1 in MT but the inhibition of JAK2/STAT3 or PI-3K increased the levels of activated AP1 in 4T1 cells (Fig 5A). Similarly, inhibition of PI-3K (Fig 5B) or p38 (Fig 5E) increased the levels of AP1 in EMT6 cells. Leptin also activated SP1 in 4T1 cells that was related to all leptin canonic signalling pathways (Fig 5A).
Overall, these results suggest that leptin signalling mainly activates HIF-1α and NFκB to regulate VEGF gene expression in MT. Leptin-mediated activation of HIF-1α and NFκB were mainly related to MAPK, PI-3K, PKC, JNK and p38 signalling pathways.
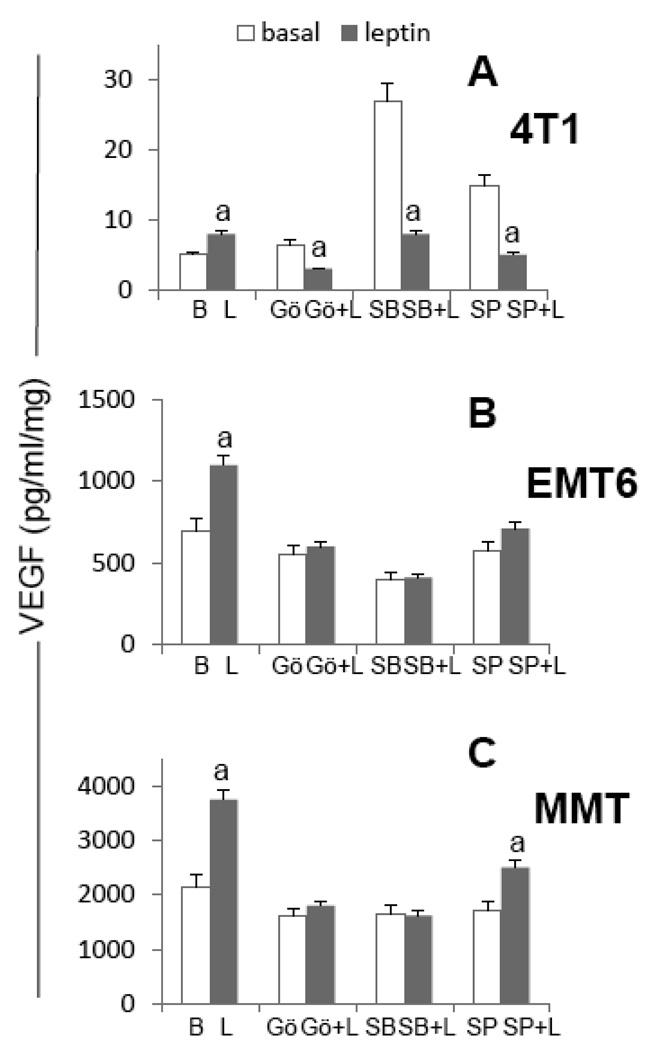
3.5. Impact of pharmacologic inhibition of TF on leptin-mediated induction of VEGF in MT
To further elucidate how leptin regulate VEGF in MT several pharmacological inhibitors of TF that can potentially bind to the VEGF promoter were used. The effects of the inhibitors NS398 (for HIF-1α), Tanshinone IIA (for AP1), inhibitor IKK antagonist (for NFκB) and Mithramycin A (for SP1) on leptin-induction of VEGF protein and mRNA were determined (Fig 6). Inhibition of HIF-1α negatively impact leptin-induced levels of VEGF protein in all cells (Fig 6A, B and C) and negatively affect leptin-mediated induction of VEGF mRNA in 4T1 (Fig 6D) and MMT (Fig 6F). On the other hand, inhibition of NFκB abrogated leptin-mediated induction of VEGF protein in all MT (Fig 6A, B and C). However, the IKK antagonist only inhibited leptin-induced VEGF mRNA in EMT6 cells (Fig 6E). Results from Tanshinone and Mithramycin treatment suggest that AP1 and SP1 are related to leptin regulation of VEGF protein in all MT (Fig 6A–C) and mRNA in 4T1 cells (Fig 6D). AP1 and SP1 were not previously identified as a target for leptin in all MT (see Fig 5). However, previous observations also suggest leptin induces SP1 activation to regulate VEGF in 4T1 cells (see Fig 5A). These results further confirm previous findings on the essential role of HIF-1α and NFκB activation for the leptin upregulation of VEGF in MT.
Fig. 6. Effects of transcription factor inhibitors on leptin-induced VEGF protein and mRNA in MT.
A, B and C, VEGF protein levels and D, E and F, VEGF mRNA levels in 4T1, EMT6 and MMT, respectively. MT were treated with leptin (0 or 1.2nM) in the presence of inhibitors for HIF-1α (NS398, 10µM), AP1 (Tanshinone IIA, 50µM), NFκB (IKK inhibitor, 100µM) and SP1 (Mithramycin A, 0.2µ µM) for 24h. VEGF protein levels were determined by ELISA (pg/ml/mg-protein) and VEGF mRNA levels were quantified by real-time RT-PCR. (a) p<0.05 when comparing levels of protein or mRNA of leptin-treated cells to control (basal) with or without inhibitors. Data (mean ± standard error) representative results derived from a minimum of 3 independent experiments. B: basal, L: leptin, N: NS398, T: Tanshinone IIA, I: IKK inhibitor, M: Mithramycin A.
3.6. Cis-acting elements involved in leptin regulation of VEGF promoter in MT
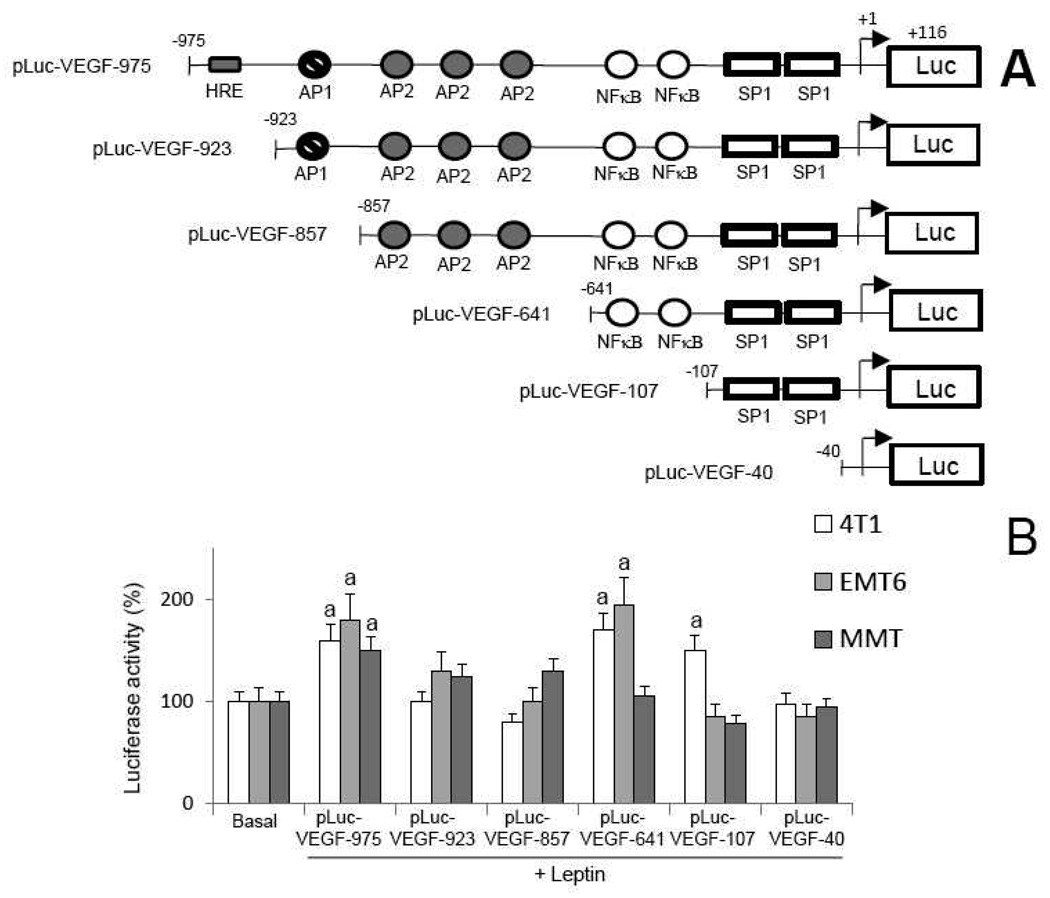
To closely determine which cis-acting elements within the mouse VEGF promoter are involved in leptin regulation of VEGF transcription, full-length and truncated VEGF promoter constructs linked to luciferase reporter gene were transiently introduced into MT (Fig 7A). Luciferase assay was conducted to determine the promoter activity after leptin challenge. Deletion analysis of luciferase reporter activities shows that leptin significantly increases the transcriptional activity of full-length VEGF promoter in all MT (Fig 7B). The analysis of cells transfected with a construct lacking the hypoxia response element (HRE) showed a significant decrease of the leptin-mediated activation of VEGF promoter in all MT. This suggests that the HRE region (specific for HIF-1α binding) of VEGF promoter is essential for leptin-induction of VEGF in MT. In contrast, the deletion of AP1 binding region did not affect leptin-mediated regulation of VEGF promoter. However, the deletion of AP2 binding region recovered the ability of leptin to induce the VEGF promoter in 4T1 and EMT6 cells but had no effects on the leptin regulation of VEGF promoter activity in MMT (Fig 7B). This suggests that AP2 activity could be involved in feed-back regulation of VEGF promoter activity by leptin in MT. Remarkably, the deletion of NFκB binding sites reduced leptin-mediated activation of VEGF promoter in EMT6 and MMT (Fig 7B). Finally, the deletion of SP1 binding sites blocked leptin-induction of VEGF promoter activity in 4T1 cells but had no effects in EMT6 and MMT (Fig 6B). This suggests that SP1 sites are critical for leptin-induction of VEGF promoter activity in 4T1 cells but do not play an important role in leptin regulation of VEGF in EMT6 and MMT. In agreement with these observations leptin activation of SP1 in 4T1 cells was linked to VEGF upregulation (see Fig 5A and Fig 6A and D). These results further suggest that HIF-1α and NFκB are mainly activated by leptin for upregulation of VEGF in MT.
Fig. 7. Leptin regulation of VEGF promoter in MT.
A, schematic representation of the full-length mouse VEGF promoter-luciferase reporter construct and serial 5’-end deletions. A 1091-bp fragment (−975 to +116) of the murine VEGF promoter, and a series of sequential 5’-end deletion fragments were inserted into a promoterless luciferase reporter vector (pGL3 basic). B, deletion analysis of VEGF promoter. MT were transiently transfected with VEGF reporter constructs and treated with leptin (0, 0.6 or 1.2nM), and luciferase activity was determined (see Section 2.11.) and expressed as a percent of basal (vehicle treated) in response to the leptin treatment. (a) p<0.05 when comparing levels of luciferase activity to control (basal). Data (mean ± standard error) representative results derived from a minimum of 3 independent experiments.
3.7. Short Hairpin RNA Knockdown of HIF-1α and NFκB
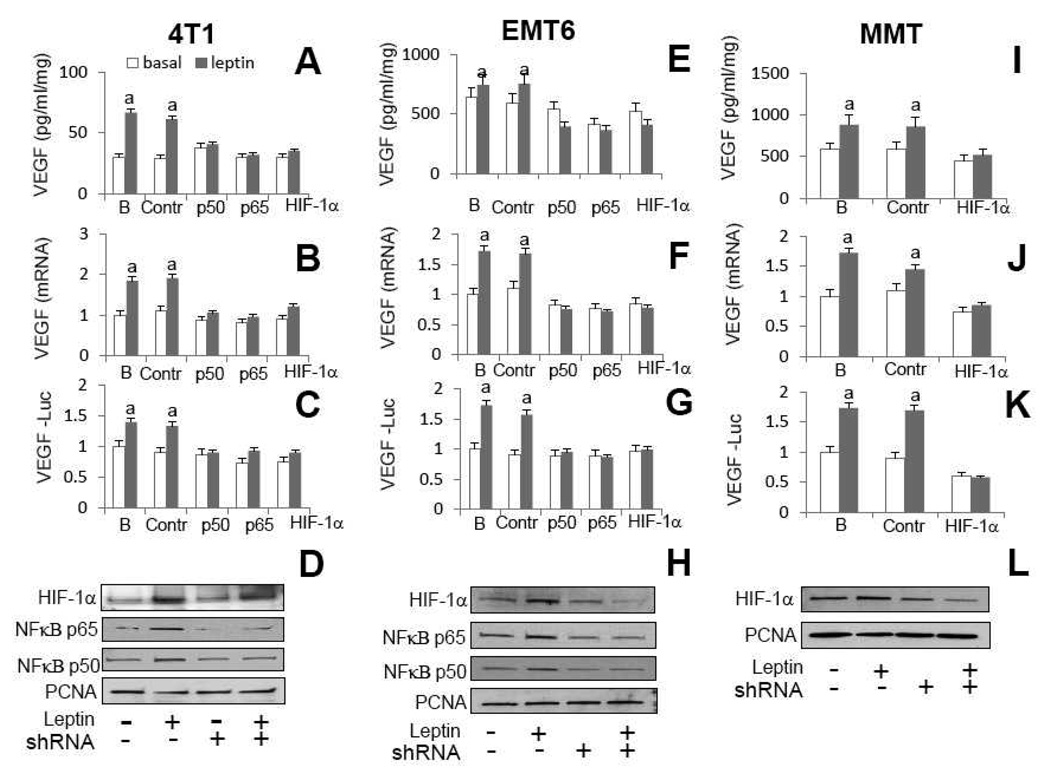
To further ascertain the roles of leptin-induced TF on VEGF upregulation in MT shRNA were used. To confirm whether leptin upregulation of HIF-1α and NFkB is linked to increased levels of VEGF (see Fig 5) 4T1 and EMT6 cells were transfected with shRNA for HIF-1α and NFκB and MMT were transfected with NFκB shRNA. Knockdown of HIF-1α or NFκB (p50 or p65) genes completely abrogated leptin-induced increase in VEGF protein (Fig 8A, E and I) and mRNA in all MT (Fig 8B, F and J). Moreover, shRNA treatment for HIF-1α or NFκB (p50 or p65) completely inhibited leptin-mediated upregulation of VEGF promoter in MT transfected with full-length pLUC-VEGF-975 (Fig 8C, G and K). WB analysis confirmed that leptin increased and shRNA treatment reduced the levels of nuclear of HIF-1α and NFκB in MT (Fig 8D, H and L). These data strongly suggest that leptin uses HIF-1α and NFκB to upregulate VEGF in MT.
Fig. 8. Effects of shRNA knockdown of HIF-1α and NFκB on leptin regulation of VEGF in MT.
A, E and I, VEGF protein levels and B, F and J, VEGF mRNA levels in 4T1, EMT6 and MMT, respectively. C, G and K, luciferase activity of VEGF-LUC transfected cells in 4T1, EMT6 and MMT, respectively. D, H and L, WB analysis for HIF-1a and NFkB (p50 and p65) in nuclear extracts from 4T1, EMT6 and MMT, respectively. PCNA antigen was used as loading control. MT were treated with leptin (0 or 6.25 nM) in the presence of shRNA for HIF-1α and NFκB (p50 and p65) for 24h. VEGF protein levels were determined by ELISA (pg/ml/mg-protein) and VEGF mRNA levels were quantified by real-time RT-PCR. MT were transiently transfected with full-length VEGF-LUC reporter constructs and treated with leptin (0, 6.25 nM), and luciferase activity was determined (see Section 2.11.) and expressed as a percent of basal (vehicle treated) in response to the leptin treatment. (a) p<0.05 when comparing levels of protein, mRNA and luciferase activity of leptin-treated cells to control (basal) with or without shRNA. Data (mean ± standard error) representative results derived from a minimum of 3 independent experiments. B: untransfected cells; Contr: cells transfected with control plasmid; p50, p65 and HIF-1α (cells transfected with specific shRNA)
4. DISCUSSION
Overweight and obesity are pandemic, especially in western countries, and strongly linked to breast cancer incidence. These relationships are probably associated to the activity of adipose tissue that secretes an array of cytokines and growth factors impacting tumor growth. Among these factors, leptin has been suggested a major player that has several pleiotropic effects promoting tumor growth. Moreover, leptin and its receptor, OB-R, are overexpressed in breast cancer. An increased number of reports support a critical role for leptin signalling in tumor angiogenesis [20, 28]. This could be linked to leptin’s ability to increase the levels of VEGF [14, 19, 21]. However, the precise mechanisms linking leptin signalling and VEGF levels in breast cancer are not well understood. Here we describe for the first time a comprehensive molecular mechanism for leptin regulation of VEGF in breast cancer cells.
VEGF regulation is complex and occurs at both transcriptional and post-transcriptional levels in a cell-specific manner. Several cis-regulatory elements for transcription factor binding sites have been identified within the 5’-flanking promoter region of VEGF promoter [22, 29]. Hypoxia is a very well-known factor implicated in the transcriptional upregulation of VEGF in many tissues. The VEGF promoter contains distal enhancer sites that bind HIF-1α (hypoxia response elements, HRE) [29] and AP1 [30]. The proximal GC-rich region in the VEGF promoter contains binding sites for AP2 [23, 31], Egr-1 (early growth response-1), WT1 [32], NFκB [29] and SP1/SP3 [31]. The activation of many of these TF by hypoxia and cytokines/growth factors is linked to the constitutive and induced expression of VEGF in different cancer cell lines [30–34]. In addition, ERα-SP1 and -SP3 complexes bind to an imperfect ER site within the VEGF promoter to regulate VEGF levels in breast (34) and endometrial cancer cells [35].
In the present studies, we examined the regulation of VEGF in response to leptin within mammary cancer cells. Initially 4T1, EMT6 and MMT were characterized for the expression of leptin and targeted molecules. 4T1 and EMT6 cells are derived from BALB/c background. The 4T1 mouse cell model is commonly used in research because it closely resembles breast cancer [36]. However, limited information is available on EMT6 (hyperplastic mammary alveolar nodule origin) and MMT (epithelial origin) cell models. Therefore, all MT were initially investigated for their basal expression of ligands and receptors. All MT investigated secreted leptin and VEGF and, co-expressed OB-R, VEGFR2 and ERα in basal conditions. MMT showed higher levels of leptin and VEGF when compared to EMT6 and 4T1 cells. Interestingly, the basal levels of leptin and VEGF were harmonized in all MT. These data confirm our previously results on the synchronized levels of leptin and VEGF in human breast cancer cell cultures [21] and reinforce the idea that increased levels of leptin derived either from body, mammary adipose tissues and/or tumor cells can upregulate VEGF in breast cancer.
To dissect the mechanisms of leptin-induced levels of VEGF protein and mRNA, MT were challenged with leptin and several pharmacological and genetic inhibitors for key leptin-targeted kinases and TF and, transfected with reporters containing serial deletions of VEGF-promoter. Leptin uses a network of canonic and non-canonic signalling pathways to upregulate VEGF in MT. Leptin induces PI3K, MAPK (ERK 1/2, p38 and JNK) and PKC signalling pathways to activate HIF-1α and NFκB (see Fig 9). These leptin-mediated events were strikingly linked to upregulation of VEGF protein and mRNA levels and VEGF-Luc activity. In addition, JAK2/STAT3 and SP1 were involved in leptin-induced effects on VEGF levels in 4T1 cells. Interestingly, incubation of 4T1 cells with AG490 (a JAK2/STAT3 inhibitor) did not completely abolish STAT3 phosphorylation. This data suggest that leptin-mediated phosphorylation of STAT3 in 4T1 could also occur through a JAK2-independent mechanism. It raises the possibility that leptin/OB-R crosstalk to other kinases, i.e., Src that has been shown to phosphorylate STAT3 upon leptin challenge in a JAK2-independent manner [37].
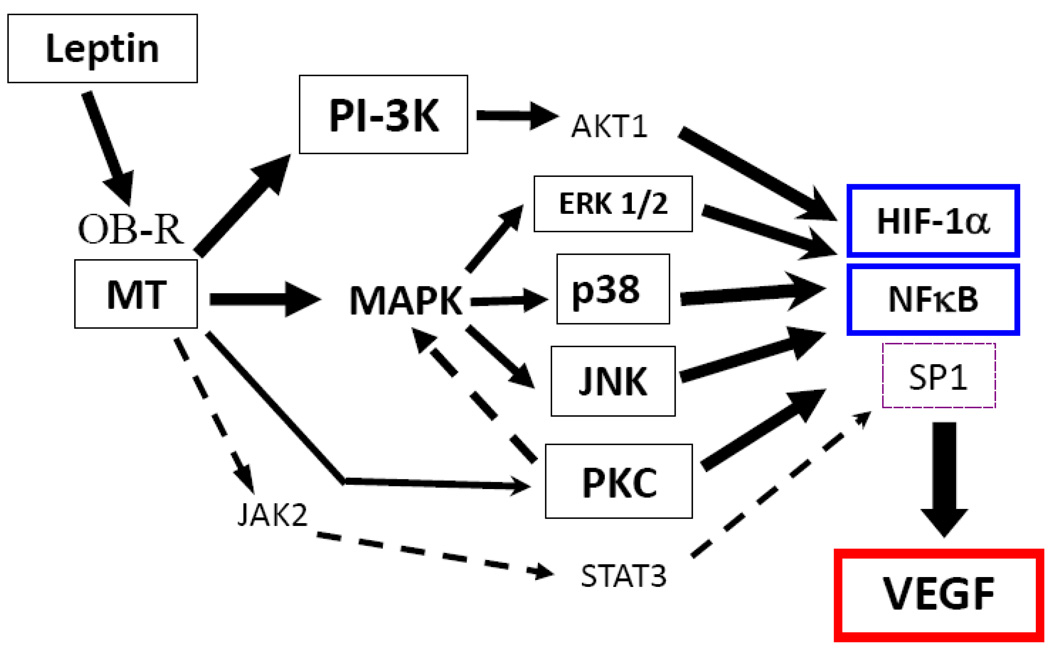
Fig. 9. Schematic representation of the molecular mechanism for leptin regulation of VEGF in MT.
Leptin-induced upregulation of VEGF expression in MT mainly involves the activation of HIF-1α and NFκB through leptin canonic (PI-3K/AKT1 and MAPK/ERK1) and non-canonic (JNK, p38 and to less extent PKC) signalling pathways. However, leptin-induced SP1 through leptin canonic signalling pathways (JAK2/STAT3, MAPK and PI-3K) signalling pathway is also involved in VEGF gene regulation in 4T1 cells. Leptin activation of PKC may be upstream to MAPK/ERK and p38 kinases that in turn activate HIF-1α and NFκB for the regulation of VEGF expression in MT.
We have previously reported that inhibition of leptin signalling significantly impacted on the levels of VEGF in syngeneic mouse mammary tumors [14] and in human breast cancer xenografts hosted by immunodeficient mice [21]. In this investigation, we confirmed our previous finding that leptin induces an increase in the levels of VEGF in 4T1 cells [14]. Furthermore, we found that leptin also induces the increase in VEGF protein levels in EMT6 and MMT and VEGF mRNA in all MT. These results were found in parallel to the leptin-induction of VEGF promoter activity in all MT assayed. Overall, these data strongly suggest that leptin induces the transcription and translation of VEGF in breast cancer.
HIF-1 is a heterodimer transcription factor considered a master regulator of hypoxic-gene expression and promoter of angiogenesis. HIF-1 is composed of two subunits: HIF-1α (induced by hypoxia) and HIF-1β (constitutively expressed). In normoxia, HIF-1α is targeted to proteasome degradation via ubiquitinylation [38]. However, under normoxic conditions HIF-1 is upregulated by growth factors, cytokines, oncogenes and hormones [29]. HIF-1 binds to HRE sites in the VEGF promoter [29] for VEGF regulation. Our present results from deletion analysis of VEGF promoter suggest that HRE binding sites are critical for leptin regulation of VEGF gene in MT. Leptin induces the increase of HIF-1α DNA binding activity. Moreover, leptin–induction of VEGF protein and mRNA was blocked by an inhibitor (NS398) of HIF-1α. NS398 can reduce HIF-1α mRNA and protein in a COX-2 dependent way but it can also decrease HIF-1 levels by increasing ubiquitination and the clearance of ubiquitylated protein [39]. To further confirm these results HIF-1α gene was knocked down with shRNA. Treatment of cells with HIF-1α shRNA completely abrogated leptin-mediated induction of VEGF protein, mRNA, promoter-LUC activity and HIF-1α accumulation in the nucleus of all MT. Present results suggest that leptin-increased VEGF transcription involves the enhanced ability of HIF-1α to bind HRE within VEGF promoter in MT.
Present findings show that leptin induced VEGF expression through HIF-1α was linked to the activation of canonic (JAK2/STAT3, PI3K/AKT1 and MAPK/ERK 1/2) and non- canonic (p38, JNK and to less extent PKC) signalling pathways. This was in agreement to previous reports showing the involvement of ERK [40], PI-3K [41], p38 MAPK and JNK [40] in the regulation of VEGF by diverse factors in different cells. In hamster fibroblasts, ERK 1/2 kinases trigger the activation/binding of SP1/AP2 complexes to VEGF promoter [40]. However, ERK but not JNK was essential for HIF-1 activation in human hepatocellular liver carcinoma cell line, HepG2 [42]. However, present results suggest that leptin-induced JAK2/STAT3 signals were not essential for VEGF upregulation (see Fig 3). This was in contrast to findings reported for MCF-7 cells where STAT3 blockade abrogated both HIF-1α and VEGF expression [43]. HIF-1α is constitutively degraded by ubiquitination under normoxic conditions [37]. Thus, it is probable that the increased HIF-1α activity by leptin in MT is related to decreased HIF-1α ubiquitination and proteosome degradation. Therefore, leptin activation of HIF-1α could occur in MT through phosphorylation or stabilization of HIF-1α by PKC/MAPK and PI-3K/AKT1.
Leptin-induced NFκB activation was linked to VEGF expression in MT. Interestingly, leptin induced increase in NFκB activity in 4T1 and EMT6 but not in MMT. However, deletion analysis of VEGF promoter suggests that NFκB binding regions were involved in leptin upregulation of the VEGF promoter in EMT6 and MMT but were not essential in 4T1 cells. This was further assessed by abrogation of NFκB activation. The IKK inhibitor and NFκB shRNA negatively impacted the leptin induction of VEGF protein and mRNA in MMT and EMT6, respectively. Leptin canonic signals (JAK/STAT3, ERK 1/2 and PI-3K) were only linked to NFκB activation in 4T1 cells. In contrast, leptin non-canonic signals (JNK and p38) were only involved in leptin activation of NFκB in EMT6 cells. NFκB comprises a complex family of hetero- or homodimer proteins (i.e., RelA or p65, RlB, cRel, p50, p1005 and p105) that are kept in inactive forms in the cytoplasm mainly linked to inhibitor proteins (IκB). Diverse stimuli can activate NFκB that is transported to the nucleus to activate the expression of genes involved in inflammation, immune regulation, survival and proliferation [44]. The precise mechanisms involved in leptin activation of NFκB in MT are unknown but may be related to IKK activation by PKC as it was previously reported in immune cells [44]. Our previous results on VEGF transcription in macrophages show that HRE is essential for the VEGF transcription. In these cells, HIF-1α mRNA levels were increased in LPS-treated macrophages in an NFκB–dependent manner but deletion of putative NFκB–binding sites from the VEGF promoter did not affect LPS induced VEGF promoter activity, suggesting that NFκB is not directly involved in VEGF transcription [45]. In contrast, present data from MT suggest that leptin induce VEGF expression in a HIF-1α and NFκB–dependent way.
Stress-activated protein kinases (p38 MAPK and JNK) increased expression of VEGF by stabilizing the VEGF mRNA in human embryonic kidney and hamster fibroblasts [43]. On the other hand, PKC activity was previously found linked to the activation of HIF-1α mainly through Ras/Raf-1/ERK signalling pathways in bovine aortic endothelial [46] and fibrosarcoma cells [47]. PKC can activate several members of MAPK family, including p38 MAPK [48]. Moreover, PKC-mediated activation of NFκB was linked to the effects of inflammatory cytokines and growth factors, i.e., TNF-α, IL-1 and EGF in human kidney cells [49]. Then, leptin activation of PKC may be upstream to MAPK/ERK and p38 kinases that in turn activate HIF-1α and NFκB for the regulation of VEGF expression in MT. In agreement with this notion, both PKC and MAPK/ERK/p38 inhibitors completely blocked the ability of leptin to activate HIF-1α and upregulate VEGF in MT.
Pro-angiogenic factors show differential mechanisms for VEGF regulation. Present results show that AP1 is not involved and AP2 is a repressor of leptin-mediated regulation of VEGF gene expression in MT. In contrast, TNFα can activate VEGF gene expression through SP1 [50], AP2 [31] and HIF-1α [51]. bFGF activated VEGF expression through SP1 [50] and TGFβ through AP1 and HIF-1α [30]. Interestingly, AP2 is a positive regulator of many genes, including VEGF in human epidermoid cancer cells [23, 31] but can repress VEGF expression in prostate cancer cells [23] and is a suspected repressor of VEGF in breast cancer [52, 53]. These reports are in agreement with our present findings that suggest AP2 represses leptin-mediated VEGF gene expression in MT.
Diverse tyrosine kinases and growth factors are involved in VEGF regulation in different cells [41]. Leptin actions leading to VEGF upregulation could be reinforced through crosstalk to cytokines and growth factors. Indeed, leptin is an up-stream regulator of a number of angiogenic molecules [17–19]. Various pro-angiogenic/pro-inflammatory factors involved in endometrial cancer are regulated by leptin [19]. Moreover, leptin is an activator of IL-1 receptor type I gene expression in endometrial [19] and breast cancer cells (unpublished results). Remarkably, leptin can crosstalk to some factors that activate HIF-1 in breast cancer and other cells, i.e., IGF and epidermal growth factor receptor-2 (HER2/neu; erbB2) [54, 55]. Leptin can also transactivate ERα [56] that in turn can upregulate VEGF expression through an imperfect estrogen-responsive element (ERE) and AP1 binding sites in the VEGF promoter [57]. On the other hand, many growth factors and inflammatory cytokines can activate NFκB in cancer cells [58]. Therefore, leptin signalling could directly and indirectly (through estrogen and growth factor/cytokine crosstalk) induce the activation of HIF-1α and NFκB to upregulate the VEGF gene. Leptin signalling could give an additional advantage to tumors by upregulating VEGF before hypoxia is manifested. Remarkably, HIF-1α can also induce leptin expression in choriocarcinoma [59] and breast cancer cells [60]. This suggests the existence of a feedback loop for leptin activation of HIF-1α and regulation of VEGF and leptin genes. On the other hand, NFκB can also induce the expression of HIF-1α [61]. Hence, the NFκB-mediated regulation of HIF-1α, the regulation of leptin by HIF-1α and, leptin upregulation of VEGF through activation of HIF-1α and NFκB suggest that complex mechanisms for regulation of VEGF and leptin expression occur in breast cancer. This provides an interesting paradigm for leptin/VEGF relationships in signal transduction in cancer. Moreover, we have previously reported that leptin increased the levels of Cyclin D1 in 4T1 cells [14] and MCF-7 and MD-MBA231 cells [21]. In line with these results, leptin-mediated activation of NFκB could activate the cyclin D1 promoter [62]. These data support the idea for a dual role of leptin in breast cancer through the activation of NFκB that can impact both angiogenesis (increased VEGF) and tumor growth (increased cyclin D1).
4.1. Conclusions
4.1.1. Our data suggest that mechanisms for leptin upregulation of VEGF are cell specific. In MT leptin regulation of VEGF involves PI-3K/AKT1 and MAPK/ERK 1/2 signalling pathways. In contrast, leptin-induction of VEGF levels in endometrial cancer cells was related to the activation of MAPK/ERK1/2 and mTOR but not to PI-3K/AKT1 signalling pathway [19].
4.1.2. Taken together, our findings add new evidence on biological diversity of leptin signalling in the regulation of VEGF to promote tumor angiogenesis in normoxic conditions. This also highlights the importance of studying leptin signalling crosstalk with other factors in the context of TF activation and gene expression.
4.1.3. Present data together with the previously in vivo findings showing an impressive impact of leptin signalling inhibition on tumor growth, angiogenesis and reactive stroma [14, 21] emphasize the idea that leptin is an important regulator of the tumor microenvironment and angiogenesis. Leptin signalling and crosstalk could lead to the promotion of angiogenesis, growth and survival of breast cancer cells that could be further sustained by increased adiposity and the associated higher levels of leptin.
ACKNOWLEDGMENTS
This work was supported in part by Grants from NIH/NCI 1SC1CA138658-01; NIH/ARRA/3SC1CA138658-02S1; NIH/UAB Breast SPORE Career Development Award; the Georgia Cancer Coalition Distinguished Cancer Scholar Award; CIG-07-114 Consortium for Industrial Collaboration in Contraceptive Research (CICCR), a program of Contraceptive Research and Development Program (CONRAD), Eastern Virginia Medical School (to R.R.G-P.); facilities and support services at Morehouse School of Medicine (NIH RR03034 and 1C06 RR18386).
The abbreviations used are
- MT
mammary tumor cells
- VEGF
Vascular endothelial growth factor
- VEGFR2
Vascular endothelial growth factor–receptor 2
- ER
Estrogen receptor
- JAK2
Janus kinase 2
- STAT3
Signal transducer and activator of transcription 3
- ERK 1/2
Extracellular signal-regulated kinases 1 and 2
- PI-3K
Phosphatidylinositol-3 kinase
- AKT1
Protein kinase B
- AMPK
5'-AMP-protein kinase
- IGF-1
Insulin like growth factor-1
- MT
mouse mammary tumor cells
- PKC
Protein kinase C
- MAPK
Mitogen-activated protein kinase
- p38
p38 MAP kinase
- JNK
c-Jun N-terminal kinase
- PDE3B
cyclic nucleotide phosphodiesterase 3B
- HIF-1
hypoxia inducible factor
- NFκ B
Nuclear factor-κ B
- AP1
activating protein 1
- AP2
activating protein-2
- erbB2 or HER2/neu
Erythroblastic leukemia viral oncogene homolog 2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ferrara N, Gerber HP. Acta Haematol. 2001;106:148–156. doi: 10.1159/000046610. [DOI] [PubMed] [Google Scholar]
- 2.Ferrara N, Hillan KJ, Novotny W. Biochem. Biophys. Res. Comm. 2005;333:328–335. doi: 10.1016/j.bbrc.2005.05.132. [DOI] [PubMed] [Google Scholar]
- 3.Hu X, Juneja SC, Maihle NJ, Cleary MP. J. Natl. Cancer Inst. 2002;94:1704–1711. doi: 10.1093/jnci/94.22.1704. [DOI] [PubMed] [Google Scholar]
- 4.Miyoshi Y, Funahashi T, Tanaka S, Taguchi T, Tamaki Y, Shimomura I, Noguchi S. Int. J. Cancer. 2005;118:1414–1419. doi: 10.1002/ijc.21543. [DOI] [PubMed] [Google Scholar]
- 5.Ishikawa M, Kitayama J, Nagawa H. Clin. Cancer Res. 2004;10:4325–4331. doi: 10.1158/1078-0432.CCR-03-0749. [DOI] [PubMed] [Google Scholar]
- 6.Yoshiji H, Gomez DE, Shibuya M, Thorgeirsson UP. Cancer Res. 1996;56:2013–2016. [PubMed] [Google Scholar]
- 7.Adams J, Carder PJ, Downey S, Forbes MA, MacLennan K, Allgar V, Kaufman S, Hallam S, Bicknell R, Walker JJ, Cairnduff F, Selby PJ, Perren TJ, Lansdown M, Banks RE. Cancer Res. 2000;60:2898–2905. [PubMed] [Google Scholar]
- 8.Foekens JA, Peters HA, Grebenchtchikov N, Look MP, Meijer-van Gelder ME, Geurts-Moespot A, van der Kwast TH, Sweep CGJ, Klijn JGM. Cancer Res. 2001;61:5407–5414. [PubMed] [Google Scholar]
- 9.Tartaglia LA. J. Biol. Chem. 1997;272:6093–6096. doi: 10.1074/jbc.272.10.6093. [DOI] [PubMed] [Google Scholar]
- 10.Sweeney G. Cell Signal. 2002;14:655–663. doi: 10.1016/s0898-6568(02)00006-2. [DOI] [PubMed] [Google Scholar]
- 11.Minokoshi Y, Kim YB, Peroni OD, Fryer LG, Müller C, Carling D, Kahn BB. Nature. 2002;415:339–343. doi: 10.1038/415339a. [DOI] [PubMed] [Google Scholar]
- 12.Chen C, Chang YC, Liu CL, Chang KL, Gno IC. Breast Cancer Res. Treat. 2006;98:121–132. doi: 10.1007/s10549-005-9139-y. [DOI] [PubMed] [Google Scholar]
- 13.Okumura M, Yamamoto M, Sakuma H, Kojima T, Maruyama T, Jamali M, Cooper D, Yasuda K. Biochim. Biophys. Acta. 2002;1592:107–116. doi: 10.1016/s0167-4889(02)00276-8. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez RR, Cherfils S, Escobar M, Yoo JH, Carino C, Styer AK, Sullivan BT, Sakamoto H, Olawaiye A, Serikawa T, Lynch MP, Rueda BR. J. Biol. Chem. 2006;281:26320–26328. doi: 10.1074/jbc.M601991200. [DOI] [PubMed] [Google Scholar]
- 15.Perera CN, Chin HG, Durul N, Camarillo IG. J. Endocrinol. 2007;199:221–233. doi: 10.1677/JOE-08-0215. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez RR, Leavis P. Endocrine. 2001;16:21–28. doi: 10.1385/ENDO:16:1:21. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez RR, Rueda BR, Ramos MP, Littell RD, Glasser S, Leavis PC. Endocrinology. 2004;145:3850–3857. doi: 10.1210/en.2004-0383. [DOI] [PubMed] [Google Scholar]
- 18.Ramos MP, Rueda BR, Leavis PC, Gonzalez RR. Endocrinology. 2005;146:694–701. doi: 10.1210/en.2004-1186. [DOI] [PubMed] [Google Scholar]
- 19.Carino C, Olawaiye AB, Cherfils S, Serikawa T, Lynch MP, Rueda BR, Gonzalez RR. Int. J. Cancer. 2008;123:2782–2790. doi: 10.1002/ijc.23887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sierra-Honigmann MR, Nath AK, Murakami C, García-Cardeña G, Papapetropoulos A, Sessa WC, Madge LA, Schechner JS, Schwabb MB, Polverini PJ, Flores-Riveros JR. Science. 1998;281:1683–1686. doi: 10.1126/science.281.5383.1683. [DOI] [PubMed] [Google Scholar]
- 21.Gonzalez RR, Watters A, Xu Y, Singh UP, Mann DR, Rueda BR, Penichet ML. Breast Cancer Res. 2009;11:R36. doi: 10.1186/bcr2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carmeliet P, Dor Y, Herbert J-M, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, Koch CJ, Ratcliffe P, Moons L, Jain RK, Collen D, Keshet E. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 23.Ruiz M, Pettaway C, Song R, Stoeltzing O, Ellis L, Bar-Eli M. Cancer Res. 2004;64:631–638. doi: 10.1158/0008-5472.can-03-2751. [DOI] [PubMed] [Google Scholar]
- 24.Pal S, Datta K, Mukhopadhyay D. Cancer Res. 2001;61:6952–6957. [PubMed] [Google Scholar]
- 25.Levy AP. T.C.M. 1998;8:246–250. doi: 10.1016/s1050-1738(98)00020-6. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez RR, Leary K, Petrozza JC, Leavis PC. Mol. Hum. Reprod. 2003;9:151–158. doi: 10.1093/molehr/gag022. [DOI] [PubMed] [Google Scholar]
- 27.Ramanathan M, Giladi A, Leibovich SJ. Exp. Biol. Med. (Maywood) 2003;228:697–705. doi: 10.1177/153537020322800608. [DOI] [PubMed] [Google Scholar]
- 28.Ribatti D, Conconi MT, Nussdorfer GG. Pharmacol. Rev. 2007;59:185–205. doi: 10.1124/pr.59.2.3. [DOI] [PubMed] [Google Scholar]
- 29.Shima DT, Kuroki M, Deutsch U, Ng Y-S, Adamis AP, D’Amore AP. J. Biol. Chem. 1996;271:3877–3883. doi: 10.1074/jbc.271.7.3877. [DOI] [PubMed] [Google Scholar]
- 30.Shih S-C, Claffey KP. Growth Factors. 2001;19:19–34. doi: 10.3109/08977190109001073. [DOI] [PubMed] [Google Scholar]
- 31.Gille J, Swerlick RA, Caughman SW. EMBO J. 1997;16:750–759. doi: 10.1093/emboj/16.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson J, Gorman J, Reese J, Fraizer G. Front. Biosci. 2007;12:2279–2290. doi: 10.2741/2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schafer G, Cramer T, Suske G, Kemmner W, Wiedenmann B, Hocker M. J. Biol Chem. 2003;278:8190–8198. doi: 10.1074/jbc.M211999200. [DOI] [PubMed] [Google Scholar]
- 34.Stoner M, Wormke M, Saville B, Samudio I, Qin C, Abdelrahim M, Safe S. Oncogene. 2004;23:1052–1063. doi: 10.1038/sj.onc.1207201. [DOI] [PubMed] [Google Scholar]
- 35.Mueller MD, Vigne J-L, Minchenko A, Lebovic DI, Leitman DC, Taylor RN. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10972–10977. doi: 10.1073/pnas.200377097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dexter DL, Kowalski HM, Blazar BA, Fligiel Z, Vogel R, Heppner G. Cancer Res. 1978;38:3174–3181. [PubMed] [Google Scholar]
- 37.Jiang L, Li Z, Rui L. J. Biol. Chem. 2008;283:28066–28073. doi: 10.1074/jbc.M805545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang LE, Gu J, Schau M, Bunn HF. Proc. Natl. Acad. Sci. U.S.A. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong H, Willard M, Simons J. Int. J. Cancer. 2004;112:585–595. doi: 10.1002/ijc.20438. [DOI] [PubMed] [Google Scholar]
- 40.Berra E, Pages G, Pouysségur J. Cancer Metastasis Rev. 2000;19:139–145. doi: 10.1023/a:1026506011458. [DOI] [PubMed] [Google Scholar]
- 41.Aleffi S, Petrai I, Bertolali C, Parola M, Colombatto S, Novo E, Vizmtti F, Anania FA, Milani S, Rombouts K, Lath G, Pinznli M, Marra F. Hepatology. 2005;42:39–38. doi: 10.1002/hep.20965. [DOI] [PubMed] [Google Scholar]
- 42.Mottet D, Michel G, Renard P, Ninane N, Raes M, Michelis C. J. Cell. Physiol. 2003;194:30–44. doi: 10.1002/jcp.10176. [DOI] [PubMed] [Google Scholar]
- 43.Xu Q, Briggs J, Park S, Niu G, Kortylewski M, Zhang S, Gritsko T, Turkson J, Kay H, Semenza GL, Cheng JQ, Jove R, Yu H. Oncogene. 2005;24:5552–5560. doi: 10.1038/sj.onc.1208719. [DOI] [PubMed] [Google Scholar]
- 44.Schulze-Luehrmann J, Ghosh S. Immunity. 2006;25:701–715. doi: 10.1016/j.immuni.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 45.Ramanathan M, Pinhal-Enfield G, Hao I, Leibovich SJ. Mol. Biol. Cell. 2007;18:14–23. doi: 10.1091/mbc.E06-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lo LW, Cheng JJ, Chiu JJ, Wung BS, Liu YC, Wang DL. J. Cell Physiol. 2001;188:304–312. doi: 10.1002/jcp.1124. [DOI] [PubMed] [Google Scholar]
- 47.Baek SH, Lee UY, Park EM, Han MY, Lee YS, Park YM. J. Cell Physiol. 2001;188:223–235. doi: 10.1002/jcp.1117. [DOI] [PubMed] [Google Scholar]
- 48.Lee MR, Dominguez C. Curr. Med. Chem. 2005;12:2979–2994. doi: 10.2174/092986705774462914. [DOI] [PubMed] [Google Scholar]
- 49.Daniel W, Civoli F, Rogers MA, Smitherman PK, Raju PA, Roederer M. Cancer Res. 1995;55:4844–4849. [PubMed] [Google Scholar]
- 50.Ryuto M, Ono M, Izumi H, Yoshida S, Weich HA, Kohno K, Kuwano M. J. Biol. Chem. 1996;271:28220–28228. doi: 10.1074/jbc.271.45.28220. [DOI] [PubMed] [Google Scholar]
- 51.Hellwig-Bürgel T, Rutkowski K, Metzen E, Fandrey J, Jelkmann W. Blood. 1999;94:1561–1567. [PubMed] [Google Scholar]
- 52.Gee JM, Robertson JF, Ellis IO, Nicholson RI, Hurst HC. J. Pathol. 1999;189:514–520. doi: 10.1002/(SICI)1096-9896(199912)189:4<514::AID-PATH463>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 53.Pellikainen JM, Ropponen KM, Kataja VV, Kellokoski JK, Eskelinen MJ, Kosma VM. Clin. Cancer Res. 2002;8:3487–3495. [PubMed] [Google Scholar]
- 54.Saxena NK, Taliaferro-Smith L, Knight BB, Merlin D, Anania FA, O'Regan RM, Sharma D. Cancer Res. 2008;68:9712–9722. doi: 10.1158/0008-5472.CAN-08-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eisenberg A, Biener E, Charlier M, Krishnan RV, Djiane J, Herman B, Gertler A. FEBS Lett. 2004;565:139–142. doi: 10.1016/j.febslet.2004.03.089. [DOI] [PubMed] [Google Scholar]
- 56.Catalano S, Mauro L, Marsico S, Giordano C, Rizza P, Rago V, Montanaro D, Maggiolini M, Panno ML, Ando S. J. Biol. Chem. 2004;279:19908–19915. doi: 10.1074/jbc.M313191200. [DOI] [PubMed] [Google Scholar]
- 57.Buteau-Lozano H, Ancelin M, Lardeux B, Milanini J, Perrot-Applanat M. Cancer Res. 2002;62:4977–4984. [PubMed] [Google Scholar]
- 58.Pirarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E, Ben-Neriah Y. Nature. 2004;431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 59.Grosfeld A, Andre J, Hauguel-De Mouzon S, Berra E, Pouyssegur J, Guerre-Millo M. J. Biol. Chem. 2002;277:42953–42957. doi: 10.1074/jbc.M206775200. [DOI] [PubMed] [Google Scholar]
- 60.Cascio S, Bartella V, Auriemma A, Johannes GJ, Russo A, Giordano A, Surmacz E. Oncogene. 2008;27:540–547. doi: 10.1038/sj.onc.1210660. [DOI] [PubMed] [Google Scholar]
- 61.Van Uden P, Kenneth NS, Rocha S. Biochem J. 2008;412:477–484. doi: 10.1042/BJ20080476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saitoh M, Ohmichi M, Takahashi K, Kawagoe J, Ohta T, Doshida M, Takahashi T, Igarashi H, Mori-Abe A, Du B, Tsutsumi S, Kurachi H. Endocrinology. 2006;146:4917–4925. doi: 10.1210/en.2004-1535. [DOI] [PubMed] [Google Scholar]