Abstract
Background
A mean of 9–10 years of human immunodeficiency virus type 1 (HIV-1) infection elapse before clinical AIDS develops in untreated persons, but this rate of disease progression varies substantially among individuals. To investigate host genetic determinants of the rate of progression to clinical AIDS, we performed a multistage genomewide association study.
Methods
The discovery stage comprised 156 individuals from the Multicenter AIDS Cohort Study, enriched with rapid and long-term nonprogressors to increase statistical power. This was followed by replication tests of putatively associated genotypes in an independent population of 590 HIV-1–infected seroconverters.
Results
Significant associations with delayed AIDS progression were observed in a haplotype located at 1q41, 36 kb upstream of PROX1 on chromosome 1 (relative hazard ratio, 0.69; Fisher’s combined P = 6.23 × 10−7). This association was replicated further in an analysis stratified by transmission mode, with the effect consistent in sexual or mucosal and parenteral transmission (relative hazard ratios, 0.72 and 0.63, respectively; combined P = 1.63 × 10−6).
Conclusions
This study identified and replicated a locus upstream of PROX1 that is associated with delayed progression to clinical AIDS. PROX1 is a negative regulator of interferon-γ expression in T cells and also mitigates the advancement of vascular neoplasms, such as Kaposi sarcoma, a common AIDS-defining malignancy. This study adds to the cumulative polygenic host component that effectively regulates the progression to clinical AIDS among HIV-1–infected individuals, raising prospects for potential new avenues for therapy and improvements in AIDS prognosis.
Polymorphisms in numerous human genes have been reported to confer differential susceptibility to human immunodeficiency virus (HIV) infection and rates of progression to AIDS [1, 2]. Genes encoding the major HIV-1 coreceptor chemokine (C-C motif) receptor 5 (CCR5), its ligands, and HLA class I genes are well documented and consistently replicated AIDS restriction genes. Homozygosity for the CCR5 Δ32 allele provides near absolute protection against HIV-1 infection, whereas Δ32 heterozygosity delays progression to clinical AIDS [3]. Certain HLA-B alleles are variously associated with increased or decreased rates of progression [3]. For example, B★5701 is strongly and consistently associated with slower disease progression [4, 5] and elite viral control [6].
Recent genomewide association studies (GWAS) involving HIV-infected individuals that use Illumina genotyping platforms have confirmed the strong associations between variation in HLA genes and the surrogate markers of plasma viral load and CD4+ T cell count [7–9]. However, the statistical noise introduced by a large number of tests generally require P values ≤5 × 10−8 for genomewide significance; in GWAS, true-positive signals with P values above this threshold cannot be distinguished from false-positive signals by purely statistical methods. This was observed in the failure of early HIV-related GWAS to identify previously identified AIDS restriction genes, such as CCR5 Δ32 or RANTES (CCL5). However, the effects of CCR5 Δ32 on disease progression were confirmed in a meta-analysis [10]. Approaches using replication or meta-analysis of GWAS results can help overcome these problems [11].
Although HIV-1 load is a robust prognostic marker for clinical disease progression, studies show that viral load explains <50% of the variation in time from primary infection to the development of clinical AIDS [12–14]. Therefore, it is important to assess the direct host genetic contribution to the actual clinical end point of HIV infection: AIDS or AIDS-related death. To address the potential differences between the virologic and clinical end points, we used a 2-stage strategy to identify host common genetic polymorphisms associated with variation among HIV-infected individuals in their rate of progression to clinical AIDS (Centers for Disease Control and Prevention [CDC] 1987 definition). First, we conducted a GWAS involving a population of HIV-1–infected men from the Multicenter AIDS Cohort Study (MACS) who were chosen to be enriched with participants representing the extreme ends of phenotypic distribution rates of HIV-1 disease progression to clinical AIDS: rapid progressors and long-term nonprogressors. A focus on extreme genotypes improves our power to detect differences between these readily discernible groups [15, 16], as seen elsewhere with AIDS phenotypes [17, 18]. Second, we selected the top-ranking single-nucleotide polymorphisms (SNPs) from the initial GWAS for replication tests in an independent cohort of 590 HIV-1 seroconverters. Third, we stratified the replication cohort by transmission mode (sexual or parenteral) and tested for consistent effects across these distinct populations.
SUBJECTS AND METHODS
Stage 1 GWAS population
In stage 1 of the study (discover stage), we conducted a GWAS involving HIV-1–infected unrelated men who have sex with men from the MACS, a longitudinal prospective cohort conducted since 1984 in 4 US cities: Baltimore, Chicago, Los Angeles, and Pittsburgh [19]. A total of 6973 men have been enrolled. From April 1984 through March 1985, 4954 men were enrolled; 668 more men were enrolled from April 1987 through September 1991. A third enrollment of 1351 men occurred from October 2001 through August 2003. The overwhelming majority, if not all, of the participants in our study were infected with HIV-1 subtype B, and they were eligible for inclusion if they were naive to highly active antiretroviral therapy (HAART) or treated only with zidovudine monotherapy.
Progression to clinical AIDS phenotype
We attempted to select equal numbers of individuals in 3 distinct categories of AIDS-free interval: 51 rapid progressors, 57 moderate progressors, and 48 long-term nonprogressors (Table 1). Study participants were chosen to be enriched with those who had HIV-1 disease progression rates from the extreme ends of this phenotypic distribution (rapid progressors and long-term non-progressors), because inclusion of extreme participants has been shown to increase power in genetic association analyses [15–18]. Rapid progressors were seroconverters for whom the interval from the estimated date of seroconversion to the date of the first clinical AIDS diagnosis or death due to an AIDS-related disease [20] was <5 years; it is estimated that ~10% of the MACS participants are rapid progressors [21]. Moderate progressors were seroconverters for whom this interval was close to the median AIDS-free interval of 9.2 years [21]. Long-term nonprogressors included seronconverters and those already infected at entry in the cohort. These individuals had no 2 consecutive CD4+ T cell counts <500 cells/mm3, no AIDS diagnosis, and no HAART use for ≥14.8 years after seroconversion (or after enrollment, if they were seropositive at entry). Dates of seroconversion were estimated as the midpoint between the last seronegative visit and the first seropositive visit; only seroconverters with <1 year between these visits were included in our study. For long-term nonprogressors, the end point was defined as the date of the last follow-up visit or the date of first HAART treatment, if applicable. Approximately 10% of the MACS participants are long-term nonprogressors [21]. These phenotype criteria yielded 194 individuals for genotyping.
Table 1.
Distributions of Age and HIV-1 Disease Phenotypes among the 156 Multicenter AIDS Cohort Study Participants of European Descent Included in This Study
| Characteristic | All | RPs (n = 51) | MPs (n = 57) | LTNPs (n = 48) |
|---|---|---|---|---|
| Age, years | ||||
| Mean ± SD | 32.8 ± 6.1 | 34.6 ± 5.8 | 31.1 ± 6.5 | 33.0 ± 5.6 |
| Range | 19.1–47.6 | 19.1–47.6 | 19.5–45.9 | 22.5–46.8 |
| Time from seroconversion to AIDS, years | ||||
| Mean ± SD | 10.4 ± 6.9 | 3.3 ± 1.1 | 8.4 ± 1.3 | 18.2 ± 1.7 |
| Range | 0.7–21.2 | 0.7–4.9 | 6.5–11.3 | 13.4–21.2 |
| Viral set point, log10 RNA copies/mL plasma | ||||
| Mean ± SD | 3.9 ± 0.8 | 4.9 ± 0.7 | 4.3 ± 0.5 | 3.2 ± 0.7 |
| Range | 1.6–6.4 | 1.6–6.4 | 2.6–5.1 | 2.1–4.9 |
NOTE. The time of progression to AIDS for rapid progressors (RPs) and moderate progressors (MPs) was calculated as the interval from the estimated date of seroconversion to the first clinical AIDS diagnosis or death from an AIDS-related disease. Disease end points for long-term nonprogressors (LTNPs) were defined as the interval from the date of seroconversion or enrollment to the date of the last follow-up visit or start of highly active antiretroviral therapy(if applicable). SD, standard deviation.
Genotyping
For the stage 1 discovery analysis, we successfully genotyped 189 individuals with use of the Affymetrix GeneChip Human Mapping 500K Array Set (http://www.affymetrix.com; 5 individuals either did not pass the minimal call rate threshold [>95%] or were found to have potential sample errors in subsequent quality control). DNA was obtained from peripheral blood mononuclear cells (PBMCs) for 118 individuals and from lymphoblastoid cell lines for 71 individuals with limited PBMC availability. Genotypic fidelity between PBMCs and lymphoblastoid cell lines genotypes was validated using paired genotypic samples from 16 individuals [22]. SNP genotypes were called using the Affymetrix BRLMM algorithm [23]; the mean call rate for all SNPs was 98.5%. For SNP quality assurance, we repeated the genotyping of 471,000 SNPs for 151 individuals with use of the Affymetrix Genome-Wide Human SNP Array (version 6.0; J.L.T., in preparation) in a different laboratory (Laboratory of Genomic Diversity, National Cancer Institute) and achieved >99.5% genotyping concordance between the genotype calls for the 2 laboratories (unfiltered for SNP call rate; 71,121,000 genotypes were compared).
Correction for potential population stratification
The stage 1 discovery study population included individuals of 4 self-reported ethnicities: white, non-Hispanic; black, non-Hispanic; white, Hispanic; and Asian or Pacific Islander. To confirm ancestries, we combined our 189 sample genotypes with 30 HapMap reference samples [24] were also genotyped on Affymetrix 500K arrays, including 10 individuals from each of the following populations: Yoruba in Ibadan, Nigeria; Japanese in Tokyo, Japan, and Chinese in Beijing, China; and Utah residents with ancestry from northern and western Europe. Using this combined data set, and after removing regions of known high linkage disequilibrium, we estimated identity-by-state pairwise distances with the genomewide association software PLINK (version 1.07) [25]; we used these estimates for multidimensional scaling analysis. Among the 189 MACS participants analyzed, self-reported ethnicities were consistent with observed HapMap ancestry in all but 2 instances. To avoid spurious associations resulting from population stratification, we restricted subsequent genomewide association analyses to 156 individuals whose MACS samples were grouped with the HapMap European American population (51 rapid progressors, 57 moderate progressors, and 48 long-term nonprogressors) (Table 2). In this population, we also corrected for potential population stratification with use of a modified Eigenstrat method [26]; 14 significant principal component axes were identified and included as covariates in the regression models described below. Q-Q plots testing the normality of the P value distribution after SNP filtering and Eigenstrat correction showed no significant deviations from what would be expected with a null hypothesis (λ = 1.0056; λ = 1.0 is expected with a null hypothesis), indicating little effect of population stratification.
Table 2.
Multistage Analysis of the Association with Delayed Progression to Clinical AIDS Diagnosis and AIDS-Related Death for 4 Linked Single-Nucleotide Polymorphisms (SNPs) 36 kb Upstream of PROX1 on Chromosome 1
| SNP (minor allele) |
|||
|---|---|---|---|
| Variable | rs17762192 (C) | rs1367951 (G) | rs17762150 (T) |
| Stage 1 (discovery): AIDS in 1987, P | 7.13 × 10−5 | 6.69 × 10−5 | 3.64 × 10−5 |
| Stage 2 (replication): AIDS in 1987 | |||
| Overall | |||
| P | 4.80 × 10−4 | 5.90 × 10−4 | 8.30 × 10−4 |
| Corrected Pa | 7.20 × 10−3 | 8.85 × 10−3 | 1.25 × 10−2 |
| RH | 0.69 | 0.69 | 0.7 |
| Sexual transmission | |||
| P | 1.7 × 10−2 | ||
| RH | 0.72 | … | … |
| Parenteral transmission | |||
| P | 7.0 × 10−3 | ||
| RH | 0.63 | … | … |
| AIDS death | |||
| P | 2.00 × 10−3 | 2.00 × 10−3 | 3.00 × 10−3 |
| RH | 0.68 | 0.69 | 0.69 |
| CD4+ T cell count <200 cells/mm3 | |||
| P | 4.50 × 10−2 | 5.40 × 10−2 | 8.20 × 10−2 |
| RH | 0.82 | 0.83 | 0.84 |
NOTE. For the 4 linked SNPs, r2 > 0.9. In stage 1, n = 156; in stage 2, n = 590; for sexual transmission, n = 405; and for parenteral transmission, n = 182. RH, relative hazard.
Corrected for 15 tests with use of the Bonferroni correction.
Stage 1 statistical analysis
We tested for associations between individual SNP genotypes and phenotypes for time to clinical AIDS by using 2 approaches: implemented with PLINK software [25]. First, we used logistic regression and an additive genetic model and included as covariates age at seroconversion and the 14 significant principal component axes identified by the Eigenstrat method. In this categorical approach, phenotypes were coded as follows: rapid progression (3), moderate progression (2), long-term nonprogression (1). Next, we used quantitative progression phenotypes (log10 transformed) in a linear regression model (with the same genetic model and 15 covariates). Results were overlapping for both analyses; only the results for logistic regression (categorical progression phenotypes) are shown. To avoid excessive artifacts of small samples and for quality control screening of genotypes before testing, we excluded all SNPs with minor allele frequencies of <5% (n = 113,205), all SNPs that deviated from Hardy-Weinberg equilibrium at a significance level of P < .001 (n = 4459), and all SNPs that yielded unambiguous genotype calls in <95% of samples (n = 54,739). These quality control measures yielded 345,926 SNPs. We located SNPs in a gene or gene region with use of the Ensembl database [27], implemented with WGA-Viewer software (version 1.25) [28]; SNPs were mapped to 5′ upstream, 5′ untranslated, coding, intronic, 3′ untranslated, or 3′ downstream gene regions, as well as intergenic regions. The Affymetrix signal intensity plots for all top-ranking SNPs were examined to confirm genotype calls. We show P values corrected using the false discovery rate procedure [29].
Stage 2 replication study population
From the stage 1 analysis, we selected the 25 top-ranking SNPs with P values of <1 ×10−5 and q values <.70, representing 15 loci with SNPs with r2 values >0.90, to test for replication in an independent replication cohort of 590 seroconverters. These stage 2 participants were enrolled in 5 natural history cohorts of patients with HIV infection or AIDS: AIDS Link to the Intravenous Experience (n = 13) [30], MACS (n = 291, excluding the 156 MACS individuals used in our stage 1 GWAS) [19], the San Francisco City Clinic Cohort Study (n = 76) [31], and the Multicenter Hemophilia Cohort Study (n = 169) [32]. Details of these cohorts have been described elsewhere [33]. The date of seroconversion after study enrollment was estimated as the midpoint between the last seronegative and the first seropositive HIV-1 antibody test result; only individuals of European ancestry with <2 years between the 2 tests were included. The censoring date was either the date of the last follow-up visit or 31 December 1995, the date HAART became the standard of care, to avoid potential confounding by virus-suppressive therapy. Genotyping was performed using the Affymetrix Genome-Wide Human SNP Array, version 6.0.
Replication study statistical analysis
In the stage 2 analysis, we analyzed the 25 top-ranking stage 1 SNPs for genotypic association with progression to clinical AIDS (CDC 1987 definition), time to AIDS-related death, and time to CD4+ T cell count <200 cells/mm3. Cox model P values were subjected to a Bonferroni correction for 15 independent tests, representing all SNPs tested for replication and pruned for linkage disequilibrium at r2 > 0.9 (25 SNPs in 15 loci). Next, we stratified the replication cohort by viral transmission mode and tested the significant associations in each resulting population. The 2 discrete populations represented sexual transmission (men who have sex with men; n = 405) and parenteral transmission (hemophiliacs and injection drug users; n = 182).
RESULTS
Association between locus upstream of PROX1 on chromosome 1 and progression rate
Our discovery-stage genomewide association analysis of 345,926 validated SNPs with rate of progression to clinical AIDS resulted in 25 SNPs with statistically significant associations at P < 1 ×10−5 and corrected P values of <.70 (q values, corrected for false discovery rate) (Table 3). These SNPs were tested for replication in an independent cohort of 590 seroconverters. In this replication population, results with an additive (codominant) genetic model revealed a single haplotype segment including 3 linked SNPs (r2 > 0.9) 36 kb upstream of PROX1 on chromosome 1, with minor alleles that were associated with delayed progression to AIDS diagnosis as well as AIDS-related death (Figure 1) after Bonferroni correction (rs17762192; discovery-stage, P = 7.13 ×10−5; replication-stage, P = 4.8 ×10−4 and P = 7.2 ×10−3 [corrected]; relative hazard ratio, 0.69; Fisher’s combined P = 6.23 ×10−7) (Table 2). Effects were consistent in the replication analysis stratified by HIV-1 transmission mode, in which equivalent relative hazard ratios for AIDS diagnosis were found in the sexual and parenteral transmission populations (relative hazard ratios, 0.72 for sexual and 0.63 for parenteral transmission; Fisher’s combined P = 1.63 ×10−6) (Table 2).
Table 3.
Fifteen Top-Ranking Associations of Loci with Rate of Progression to Clinical AIDS (25 Single-Nucleotide Polymorphisms [SNPs]) from the Discovery-Stage Genomewide Association Study (Stage 1)
| SNP | Rank | P | qa | Replication Pb | Chromosome | Position, base pair | Type | Ancestral allele | Closest gene |
|---|---|---|---|---|---|---|---|---|---|
| rs6984368 | 1 | 8.76 × 10−6 | .6834 | .158 | 8 | 82055464 | Intronic | G | PAG1 |
| rs7006101 | 22 | 4.81 × 10−5 | .7105 | 6.58 × 10−2 | 8 | 82059755 | Synonymous | C | PAG1 |
| rs16899646 | 2 | 1.33 × 10−5 | .6834 | .114 | 6 | 31524899 | Intergenic | C | HCP5 |
| rs1691946 | 3 | 1.70 × 10−5 | .6834 | .995 | 10 | 29200463 | Intronic | G | C10orf126 |
| rs1775929 | 7 | 2.69 × 10−5 | .6834 | .881 | 10 | 29199354 | Intronic | T | C10orf126 |
| rs1691943 | 8 | 2.69 × 10−5 | .6834 | .881 | 10 | 29200842 | Intronic | G | C10orf126 |
| rs1775933 | 11 | 3.10 × 10−5 | .6834 | .844 | 10 | 29200882 | Intronic | G | C10orf126 |
| rs11648686 | 4 | 1.78 × 10−5 | .6834 | .175 | 16 | 50950897 | Intergenic | G | TOX3 |
| rs4800820 | 5 | 2.20 × 10−5 | .6834 | .710 | 18 | 23630471 | Intergenic | T | CDH2 |
| rs9957708 | 9 | 2.71 × 10−5 | .6834 | .523 | 18 | 23633388 | Intergenic | A | CDH2 |
| rs7102407 | 6 | 2.32 × 10−5 | .6834 | .828 | 11 | 9512693 | Intergenic | C | ZNF143 |
| rs10082685 | 38 | 7.89 × 10−5 | .7185 | .460 | 11 | 9500492 | Intronic | G | ZNF143 |
| rs4910466 | 39 | 8.28 × 10−5 | .7263 | 4.79 × 10−2 | 11 | 9465928 | Intronic | G | ZNF143 |
| rs12332694 | 10 | 2.71 × 10−5 | .6834 | .942 | 5 | 79377745 | Intronic | C | THBS4 |
| rs775095 | 12 | 3.20 × 10−5 | .6834 | .346 | 20 | 15917077 | Intronic | T | MACROD2 |
| rs4910052 | 13 | 3.31 × 10−5 | .6834 | 1.50 × 10−2 | 11 | 9411950 | Synonymous | T | IPO7 |
| rs36048375 | 14 | 3.31 × 10−5 | .6834 | 1.49 × 10−2 | 11 | 9419893 | Intronic | G | IPO7 |
| rs7983751 | 15 | 3.37 × 10−5 | .6834 | .673 | 13 | 29992021 | Intronic | T | HMGB1 |
| rs11622505 | 16 | 3.46 × 10−5 | .6834 | .980 | 14 | 92387319 | Intergenic | A | GOLGA5 |
| rs17762150 | 17 | 3.64 × 10−5 | .6834 | 8.30 × 10−4 | 1 | 212191441 | Intergenic | C | PROX1 |
| rs1367951 | 30 | 6.69 × 10−5 | .7105 | 5.90 × 10−4 | 1 | 212192136 | Intergenic | A | PROX1 |
| rs17762192 | 34 | 7.13 × 10−5 | .7105 | 4.80 × 10−4 | 1 | 212191750 | Intergenic | G | PROX1 |
| rs2267420 | 18 | 3.73 × 10−5 | .6834 | .220 | 22 | 38678466 | Intronic | A | GRAP2 |
| rs498280 | 19 | 3.84 × 10−5 | .6834 | 7.88 × 10−2 | 7 | 105188251 | 3′_utr | C | ATXN7L1 |
| rs17050206 | 20 | 3.95 × 10−5 | .6834 | .700 | 4 | 139372068 | Intronic | C | SLC7A11 |
NOTE. Boldface type SNPs in adjacent rows represent linked SNPs (r2 > 0.09).
P values corrected for false discovery rate (q values).
P values for AIDS 1987 diagnoses in the replication cohort, uncorrected for multiple tests.
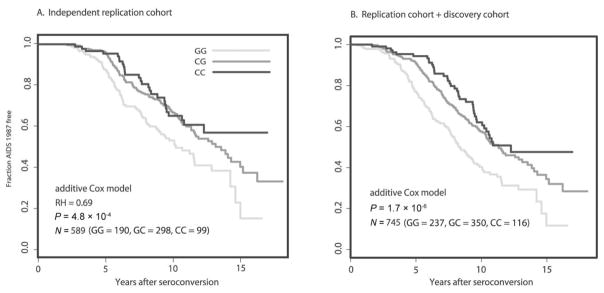
Figure 1.
Survival analysis of a locus associated with delayed progression to AIDS. Kaplan-Meier survival curves for genotypes of single-nucleotide polymorphism rs17762192, representing a haplotype located 36 kb upstream of PROX1 on chromosome 1, showing strong associations with differing rates of progression to clinical AIDS (see Table 3 for P values and genomic location). This analysis incorporates seroconverters from the stage 2 replication analysis that were typed for rs17762192 (n = 587). GG, GC, and CC genotype counts are shown for the independent replication cohort (A) and the independent replication cohort combined with the cohort used in the stage 1 discovery genomewide association study (B). RH, relative hazard.
The 3 associated SNPs were examined for linkage disequilibrium in relation to PROX1. There was a strong backbone of linkage around the associated SNPs, and for 2 SNPs, the linkage disequilibrium extended, albeit weakly, into the PROX1 coding region (Figure 2, which appear only in the electronic version of the Journal). We are presently dissecting the haplotype structure of the region, which includes genotyping additional SNPs and resequencing the region, to further refine the association with progression to clinical AIDS.
Figure 2.

Location of the 3 single-nucleotide polymorphisms (SNPs) in the upstream region of the PROX1 gene (1q32) and linkage disequilibrium matrix; the linkage disequilibrium matrix was shown by Haploview for the HapMap Utah European population. The intensity of the red color in each box is proportional to the strength of the linkage disequilibrium estimates (D′) for the SNP pair. Linkage disequilibrium blocks depicted by black triangles were based on 95% confidence interval criteria [34].
Replication of previously identified genes associated with AIDS progression
Our stage 1 genomewide association analysis provides an opportunity to replicate previously identified candidate AIDS restriction genes in these cohorts. In the analysis of progression to clinical AIDS, we found clear associations for SNPs within 50 kb of HLA-B (the most significant was rs16899646; P = 1.33 ×10−5). Likewise, we found associations for SNPs within or in linkage disequilibrium with SNPs within the following genes, reported elsewhere to influence AIDS progression: CCR2/CCR5 (rs916093; P = 1.8 ×10−3) and CXCR1 (Table 4).
Table 4.
Associations of HIV-1 Disease Progression from the Stage 1 Genomewide Association Study for Single-Nucleotide Polymorphisms (SNPs) in or within 50 kb of Genes that Produce Proteins Reportedly Involved in HIV-1 Infection
| Gene | Chromosome | SNP | P | No. of SNPs | Corrected Pa |
|---|---|---|---|---|---|
| APOBEC3G | 22 | rs139314 | 1.63 × 10−1 | 10 | 1.63 |
| CCL2 | 17 | rs1024612 | 7.10 × 10−3 | 27 | 1.9 × 10−1 |
| CCL3L1 | 17 | rs9910447 | 3.00 × 10−1 | 3 | 9.0 × 10−1 |
| CCL5 | 17 | rs9303692 | 1.10 × 10−1 | 18 | 1.82 |
| CCR2/CCR5 | 3 | rs916093 | 1.80 × 10−3 | 31 | 5.6 × 10−2 |
| CUL5 | 11 | rs2089174 | 1.79 × 10−2 | 26 | 4.7 × 10−1 |
| CXCR1 | 2 | rs13027120 | 4.20 × 10−3 | 10 | 4.2 × 10−2 |
| CX3CR1 | 3 | rs2271489 | 6.62 × 10−2 | 26 | 1.72 |
| CXCL12 | 10 | rs928565 | 7.50 × 10−3 | 50 | 3.8 × 10−1 |
| HLA-B | 6 | rs16899646 | 1.33 × 10−5 | 81 | 1.1 × 10−3 |
| IFNG | 12 | rs4913277 | 5.13 × 10−1 | 35 | 17.96 |
| IL10 | 1 | rs7228082 | 4.85 × 10−2 | 24 | 1.16 |
| IL4 | 5 | rs3798135 | 3.83 × 10−2 | 23 | 8.8 × 10−1 |
| MBL | 10 | rs10824779 | 1.34 × 10−1 | 38 | 5.09 |
| PPIA | 7 | rs7785231 | 2.18 × 10−1 | 11 | 2.40 |
| TRIM5 | 11 | rs10769180 | 6.80 × 10−3 | 106 | 7.2 × 10−1 |
| TSG101 | 11 | rs11024717 | 2.83 × 10−1 | 12 | 3.40 |
NOTE. Categorical progression rate phenotypes were used for this analysis; the SNPs shown are those with the smallest uncorrected P values.
Bonferroni-corrected P values.
The associations found by Fellay et al [7] between RNF39 and ZNRD1 and disease progression were not replicated in our stage 1 analysis (assessing the SNPs in strong linkage disequilibrium with those identified by Fellay and colleagues). The discrepancies between these 2 progression GWAS may reflect small sample sizes (n = 486 in the study by Fellay et al [7] and n = 156 in the present study) or the differences in disease progression phenotypes. Fellay et al used time from seroconversion to the start of antiretroviral treatment or time to the predicted or observed first CD4+ T cell count <350 cells/mm3, whereas both stage 1 (discovery) and stage 2 (replication) of our study tracked time from seroconversion to a clinical AIDS diagnosis or AIDS-related death.
Replication of previously identified genes associated with viral load
Because recent HIV-related GWAS have focused on viral load phenotypes [7–9], we sought to replicate these associations with use of viral load measurements for the 156 individuals from stage 1. Set point viral load was calculated according to the methods of Fellay et al [7] as the mean of log10-transformed viral load (measured in viral RNA copies per milliliter) from 6 months to 3 years after the first visit with a positive HIV-1 antibody test result, including only visits before any antiretroviral therapy. To avoid measurements obtained during the initial peak viremia or during the accelerating phase of chronic disease characterized by increasing viral load, we removed all measurements > 0.5 log10 higher or lower than the mean viral load during visits from 6 months to 3 years after seroconversion, according to the methods of Fellay et al [7].
We tested for associations between SNP genotypes and viral load phenotypes with use of the aforementioned SNP filtering and linear regression methods. Fellay et al [7] examined viral RNA set point in 486 European individuals and found 2 SNPs strongly associated with set point HIV-1 load. One polymorphism was found in a nonsynonymous coding nucleotide of HLA complex P5 (HCP5); this SNP is in high linkage disequilibrium with HLA-B★5701 on chromosome 6. The other SNP was found in the 5′ region of HLA-C. Although there was minimal overlap in SNPs tested between our genotyping platform (Affymetrix 500K) and that used by Fellay and colleagues (Illumina 550K), certain relationships were replicated (Table 5) and reaffirmed the dominant role of the HLA in controlling viral load. For example, SNP rs2248462, intergenic to HCP5 on chromosome 6 and associated with viral set point by Fellay and colleagues (P = 3.61 ×10−6) [7], was also significantly associated with viral set point in our study (P = 4.15 ×10−4; corrected, P = 1.16 ×10−2); this SNP is in strong linkage disequilibrium (r2 = 1) with 2 other associated SNPs from our study: rs2516422 (P = 5.12 ×10−4) and rs2395034 (P = 5.13 ×10−4). The minor alleles of these 3 SNPs were associated with decreased viral set point. SNP rs9348876, 7.8 kb upstream of AIF1 on chromosome 6, was significantly associated with viral set point both by Fellay et al [7] (P = 4.09 ×10−5) and in our data set (P = 1.3 ×10−3; corrected, P = 3.75 ×10−2).
Table 5.
Association Analysis of Set Point HIV-1 Load in the Stage 1 Genomewide Association Study (GWAS) Population
| HIV load set point in GWAS by Fellay et al [7] |
Current study |
|||||
|---|---|---|---|---|---|---|
| SNP | Rank | P | Chromosome | Closest gene | P | Corrected Pa |
| rs2248462 | 10 | 3.61 × 10−6 | 6 | HCP5 | 4.51 × 10−4 | 1.16 × 10−2 |
| rs9263715 | 17 | 7.08 × 10−6 | 6 | PSORS1C1 | 4.00 × 10−2 | 1.12 |
| rs9348876 | 51 | 4.09 × 10−5 | 6 | AIF1 | 1.34 × 10−3 | 3.75 × 10−2 |
| rs10736862 | 53 | 4.24 × 10−5 | 9 | ASS | 5.55 × 10−1 | 15.54 |
| rs919214 | 74 | 7.02 × 10−5 | 12 | GNPTAB | 5.10 × 10−1 | 14.28 |
| rs7894582 | 79 | 7.41 × 10−5 | 10 | NHLRC2 | 7.31 × 10−1 | 20.47 |
| rs10738377 | 83 | 7.78 × 10−5 | 9 | FREM1 | 7.29 × 10−1 | 20.41 |
| rs12037583 | 88 | 8.26 × 10−5 | 1 | TNR | 1.81 × 10−2 | 5.07 × 10−1 |
| rs3749971 | 90 | 8.32 × 10−5 | 6 | OR12D3 | 3.08 × 10−1 | 8.62 |
| rs2248617 | 97 | 9.23 × 10−5 | 6 | HCP5 | 1.62 × 10−1 | 4.54 |
| rs6674438 | 106 | 1.00 × 10−4 | 1 | GNG4 | 6.78 × 10−1 | 18.98 |
| rs633265 | 109 | 1.00 × 10−4 | 18 | MC4R | 6.87 × 10−1 | 19.24 |
| rs1119911 | 111 | 1.00 × 10−4 | 13 | RXFP2 | 7.96 × 10−2 | 2.23 |
| rs2326434 | 120 | 1.00 × 10−4 | 8 | DEPDC6 | 3.09 × 10−1 | 8.65 |
| rs2355164 | 121 | 1.00 × 10−4 | 2 | UPP2 | 4.64 × 10−1 | 12.99 |
| rs1980360 | 128 | 1.00 × 10−4 | 4 | SCD5 | 8.36 × 10−1 | 23.41 |
| rs4626416 | 134 | 1.00 × 10−4 | 6 | NA | 5.54 × 10−1 | 15.51 |
| rs2860975 | 141 | 1.00 × 10−4 | 10 | CYP2C8 | 7.74 × 10−1 | 21.67 |
| rs10104973 | 151 | 2.00 × 10−4 | 8 | COL14A1 | 9.81 × 10−1 | 27.47 |
| rs1420198 | 160 | 2.00 × 10−4 | 16 | TOX3 | 8.24 × 10−1 | 23.07 |
| rs10087151 | 168 | 2.00 × 10−4 | 8 | COL14A1 | 8.55 × 10−1 | 23.94 |
| rs1544149 | 171 | 2.00 × 10−4 | 1 | SMYD3 | 4.81 × 10−1 | 13.47 |
| rs6910183 | 173 | 2.00 × 10−4 | 6 | THBS2 | 1.99 × 10−1 | 5.57 |
| rs11772832 | 177 | 2.00 × 10−4 | 7 | CNOT4 | 2.52 × 10−1 | 7.06 |
| rs9266409 | 187 | 2.00 × 10−4 | 6 | Q5SS58_HUMAN | 8.35 × 10−3 | 2.34 × 10−1 |
| rs600130 | 193 | 2.00 × 10−4 | 9 | FBP2 | 2.29 × 10−1 | 6.41 |
| rs3130544 | 194 | 2.00 × 10−4 | 6 | C6orf15 | 7.01 × 10−2 | 1.96 |
| rs10509689 | 198 | 2.00 × 10−4 | 10 | ALDH18A1 | 4.56 × 10−1 | 1.96 |
NOTE. Boldface type single nucleotide polymorphisms (SNPs) are SNPs found in the top 200 by Fellay et al [7] that were also tested in this study (direct overlap between Illumina Hap550 and Affymetrix 500K arrays). NA, not available.
Bonferroni-corrected P values.
DISCUSSION
In our stage 1 GWAS of viral set point, we observed significant associations with HIV-1 load for several SNPs in the HLA region, as reported elsewhere [7]. This portion of our study confirms the dominant role of HLA genes in controlling viral load.
In our 2-stage association study of host genetic polymorphisms and rate of progression to clinical AIDS, we found SNPs near the transcription factor PROX1 that were confirmed in replication tests, with use of a larger and independent set of seroconverters. Human PROX1 is involved in biologic functions closely tied to HIV infection, most notably as a negative regulator of interferon (IFN) γ expression in T cells [35]. IFN-γ plays an important role in HIV disease progression through its activity as a regulatory cytokine and inflammatory effector. Furthermore, PROX1 encodes a transcription factor in which differential expression has been shown to mediate the progression of Kaposi sarcoma [36], a common sequela of HIV-1 infection. The exact role of this locus in HIV disease progression is unclear, although the regulation of IFN-γ in T cells by PROX1 presents a possible mechanism of action. In our stage 1 population, the association between this locus and disease progression is not explained by an association with Kaposi sarcoma as the AIDS-defining illness. Additional studies will be necessary to determine the association between these SNPs and differential levels of expression of PROX1 and/or IFN-γ.
These data suggest that, beyond the major role of HLA in viral control, a cumulative polygenic host component may be involved in the regulation of rate of progression to clinical AIDS. Our results should prove to be valuable in informing larger host genetics studies of HIV-1 and progression to AIDS and in guiding future replication and follow-up studies in other cohorts and populations.
Supplementary Material
Acknowledgments
We thank Rob Hall, Trevor King, and Roger Bumgarner at the Center for Array Technologies, University of Washington; Janet Schollenberger at the Center for the Analysis of MACS Data, Johns Hopkins University; and all the individuals who participated in the cohorts used for this study.
Financial support: National Institutes of Health (NIH; R37 AI47734 to J.I.M. and T32 AI07140 to J.T.H.) and University of Washington Center for AIDS Research Genomics Core (P30 AI27757 to J.T.H. and J.I.M.). The Multicenter AIDS Cohort Study is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute (NCI) and the National Heart, Lung and Blood Institute (UO1 AI35042, 5MO1 RR00722 [GCRC], UO1 AI35043, UO1 AI37984, UO1 AI35039, UO1 AI35040, UO1 AI37613, and UO1 AI35041); partial support was provided by the Intramural Research Program, NCI, NIH. The AIDS Link to the Intravenous Experience study was supported by the National Institute on Drug Abuse (R01-DA04334 and R01–12586). The San Francisco City Clinic Cohort Study was supported by the Centers for Disease Control and Prevention (U64/CCU900523–08).
Footnotes
Presented in part: 16th Conference on Retroviruses and Opportunistic Infections, Montreal, Quebec, February 2009 (poster 544).
Potential conflicts of interest: none reported.
References
- 1.O’Brien SJ, Nelson GW. Human genes that limit AIDS. Nat Genet. 2004;36:565–574. doi: 10.1038/ng1369. [DOI] [PubMed] [Google Scholar]
- 2.O’Brien T, Welzel TM, Kaslow RA. HIV-1 and AIDS. In: Kaslow RA, McNicholl J, Hill AVS, editors. Genetic susceptibility to infectious diseases. New York: Oxford; 2008. [Google Scholar]
- 3.Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 4.Kaslow RA, Carrington M, Apple R, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 5.Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 6.Migueles SA, Sabbaghian MS, Shupert WL, et al. HLA B★5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Limou S, Le Clerc S, Coulonges C, et al. Genomewide association study of an AIDS-nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02) J Infect Dis. 2009;199:419–426. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 9.de Bakker PI, Pereyra F, Jia X, et al. A genome-wide association study in HIV-1 controllers. Presented at: Conference on Retroviruses and Opportunistic Infections; 2009; Montreal. [Google Scholar]
- 10.Ioannidis JP, Rosenberg PS, Goedert JJ, et al. Effects of CCR5-Delta32, CCR2–64I, and SDF-1 3′A alleles on HIV-1 disease progression: an international meta-analysis of individual-patient data. Ann Intern Med. 2001;135:782–795. doi: 10.7326/0003-4819-135-9-200111060-00008. [DOI] [PubMed] [Google Scholar]
- 11.Chanock SJ, Manolio T, Boehnke M, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 12.Mellors JW, Rinaldo CR, Jr, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma [published correction appears in Science 1997; 275(5296): 14] Science. 1996;272(5265):1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 13.Mellors JW, Margolick JB, Phair JP, et al. Prognostic value of HIV-1 RNA, CD4 cell count, and CD4 cell count slope for progression to AIDS and death in untreated HIV-1 infection. JAMA. 2007;297:2349–2350. doi: 10.1001/jama.297.21.2349. [DOI] [PubMed] [Google Scholar]
- 14.Mellors JW, Kingsley LA, Rinaldo CR, Jr, et al. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann Intern Med. 1995;122:573–579. doi: 10.7326/0003-4819-122-8-199504150-00003. [DOI] [PubMed] [Google Scholar]
- 15.Van Gestel S, Houwing-Duistermaat JJ, Adolfsson R, van Duijn CM, Van Broeckhoven C. Power of selective genotyping in genetic association analyses of quantitative traits. Behav Genet. 2000;30:141–146. doi: 10.1023/a:1001907321955. [DOI] [PubMed] [Google Scholar]
- 16.Huang BE, Lin DY. Efficient association mapping of quantitative trait loci with selective genotyping. Am J Hum Genet. 2007;80:567–576. doi: 10.1086/512727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rappaport J, Cho YY, Hendel H, Schwartz EJ, Schachter F, Zagury JF. 32 bp CCR-5 gene deletion and resistance to fast progression in HIV-1 infected heterozygotes. Lancet. 1997;349:922–923. doi: 10.1016/S0140-6736(05)62697-9. [DOI] [PubMed] [Google Scholar]
- 18.Hendel H, Caillat-Zucman S, Lebuanec H, et al. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J Immunol. 1999;162:6942–6946. [PubMed] [Google Scholar]
- 19.Kaslow RA, Ostrow DG, Detels R, Phair JP, Polk BF, Rinaldo CR., Jr The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. Am J Epidemiol. 1987;126:310–318. doi: 10.1093/aje/126.2.310. [DOI] [PubMed] [Google Scholar]
- 20.Centers for Disease Control and Prevention. 1993 revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. MMWR Recomm Rep. 1992;41:1–19. [PubMed] [Google Scholar]
- 21.Muñoz A, Sabin CA, Phillips AN. The incubation period of AIDS. AIDS. 1997;11:S69–S76. [PubMed] [Google Scholar]
- 22.Herbeck JT, Gottlieb GS, Wong K, et al. Fidelity of SNP array genotyping using Epstein Barr virus-transformed B-lymphocyte cell lines: implications for genome-wide association studies. PLoS ONE. 2009;4(9):e6915. doi: 10.1371/journal.pone.0006915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Affymetrix. BRLMM: an improved genotype calling method for the GeneChip human mapping 500K array set. [Accessed 1 July 2009];2007 Published 14 April 2006. Available at: http://www.affymetrix.com/support/technical/whitepapers/brlmm_whitepaper.pdf.
- 24. [Accessed 1 July 2009];International HapMap Project. Available at: http://www.hapmap.org.
- 25.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 27.Hubbard TJ, Aken BL, Beal K, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge D, Zhang K, Need AC, et al. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 2008;18:640–643. doi: 10.1101/gr.071571.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 30.Vlahov D, Graham N, Hoover D, et al. Prognostic indicators for AIDS and infectious disease death in HIV-infected injection drug users: plasma viral load and CD4+ cell count. JAMA. 1998;279:35–40. doi: 10.1001/jama.279.1.35. [DOI] [PubMed] [Google Scholar]
- 31.Buchbinder SP, Katz MH, Hessol NA, O’Malley PM, Holmberg SD. Long-term HIV-1 infection without immunologic progression. AIDS. 1994;8:1123–1128. doi: 10.1097/00002030-199408000-00014. [DOI] [PubMed] [Google Scholar]
- 32.Goedert JJ, Kessler CM, Aledort LM, et al. A prospective study of human immunodeficiency virus type 1 infection and the development of AIDS in subjects with hemophilia. N Engl J Med. 1989;321:1141–1148. doi: 10.1056/NEJM198910263211701. [DOI] [PubMed] [Google Scholar]
- 33.O’Brien SJ, Nelson GW, Winkler CA, Smith MW. Polygenic and multifactorial disease gene association in man: lessons from AIDS. Annu Rev Genet. 2000;34:563–591. doi: 10.1146/annurev.genet.34.1.563. [DOI] [PubMed] [Google Scholar]
- 34.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Zhu J, Shan S, et al. Repression of interferon-gamma expression in T cells by Prospero-related homeobox protein. Cell Res. 2008;18:911–920. doi: 10.1038/cr.2008.275. [DOI] [PubMed] [Google Scholar]
- 36.Dadras SS, Skrzypek A, Nguyen L, et al. Prox-1 promotes invasion of kaposiform hemangioendotheliomas. J Invest Dermatol. 2008;128(12):2798–2806. doi: 10.1038/jid.2008.176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.