Abstract
Background
Identification of pathogen DNA from archaeological human remains is a powerful tool in demonstrating that the infectious disease existed in the past. However, it is very difficult to detect trace amounts of DNA remnants attached to the human skeleton, especially from those buried in a humid atmosphere with a relatively high environmental temperature such as in Asia.
Methodology/Principal Findings
Here we demonstrate Mycobacterium leprae DNA from archaeological skeletal remains in Japan by polymerase chain reaction, DNA sequencing and single nucleotide polymorphism (SNP) analysis. In addition, we have established a highly sensitive method of detecting DNA using a combination of whole genome amplification and polymerase chain reaction, or WGA-PCR, which provides superior sensitivity and specificity in detecting DNA from trace amounts of skeletal materials.
Conclusion/Significance
We have detected M. leprae DNA in archaeological skeletal remains for the first time in the Far East. Its SNP genotype corresponded to type 1; the first detected case worldwide of ancient M. leprae DNA. We also developed a highly sensitive method to detect ancient DNA by utilizing whole genome amplification.
Introduction
Leprosy is a chronic infectious disease caused by Mycobacterium leprae (M. leprae) and has affected humans for millennia. Whole genome sequence analysis of M. leprae has revealed that the genome is 3.3 Mbp in size, with only 1,605 genes that encode proteins and 1,115 pseudogenes [1]. Trough comparative genome analysis of M. leprae strains from Brazil, India, Thailand and United States, remarkably little genomic diversity has been uncovered suggesting that leprosy has a single clonal origin [2], [3]. Phylogeographic analysis of single nucleotide polymorphisms (SNPs) has revealed that M. leprae originated in Africa and spread to European and Asian countries and then worldwide along with human migrations and trade routes. Such geographic and temporal migration has also been demonstrated by SNPs analysis of ancient M. leprae DNA present in skeletal remains as old as 1,500 years ago [2], [3].
Leprosy results in multiple deformities including progressive bone defects secondary to the peripheral neuropathy caused by M. leprae infection [4], [5]. Additionally, in advanced cases M. leprae infection causes specific osteological deformations in the areas of the nasal aperture, anterior nasal spine and alveolar process on the premaxilla, cortical areas of the tibia and fibula, distal ends of the metatarsals and diaphyses of the phalanges that may include both direct and reactive changes [6], [7], [8], [9], [10]. Paleopathological diagnosis of leprosy has been made solely based on these macroscopic changes in skeletal remnants. However, it is occasionally difficult to make a definitive paleopathological diagnosis of leprosy solely based on osseous lesions, because syphilis, tuberculosis and other infectious and granulomatous diseases as well as carcinomas of nasal or oral origin will cause rhinomaxillary lesions similar to leprosy.
Direct detection of DNA remnants of pathogenic microorganisms from ancient human skeletons using polymerase chain reaction (PCR) is becoming a powerful molecular tool for clearly demonstrating the presence of pathogens within the skeletal remains [11]. This method provides an inestimable tool for investigating the temporal and geographical spread of infectious diseases in human history, in addition to the anthropological interest in diagnosing infectious diseases in ancient remains. By the use of so-called palaeomicrobiology, it may be possible to elucidate the epidemiology of past infectious disease by reconstituting the distribution of infected individuals, which is especially important in the field of emergence and re-emergence of infectious diseases [11]. Moreover, it may also be possible to track the genetic evolution of the pathogenic microorganisms.
A study employing paleomicrobiology was first reported in 1993, demonstrating the presence of Mycobacterium tuberculosis (M. tuberculosis) DNA using PCR from an ancient human skeleton [12]. Detection of M. leprae DNA was reported the next year [13] from ancient bone dating from 600 AD, followed closely by several other reports [14], [15], [16], [17], [18], [19], [20], [21]. However, all of these studies used European and Middle Eastern archaeological materials from countries such as England, Germany, Denmark, Poland, Hungary, Czech, Croatia, Turkey, Israel and Egypt, but not from the Far East Asian countries. This may be related to the superior preservation of ancient buried skeletons in these countries compared to that in more a humid atmosphere with a relatively high environmental temperature, such as is commonly found in Asia [22].
Recently, we have obtained permission to examine skeletal remains excavated in Japan with paleopathological evidence of leprosy In the present study, we demonstrate M. leprae DNA and SNPs from skeletal remains. We also describe a highly sensitive whole genome amplification (WGA)-PCR method that may be suitable for detecting trace amounts of ancient microbial DNA.
Materials and Methods
Archaeological background and osteological description
The materials used in this study were archaeological skeletal remains excavated from the Hatanai site (N40°22′, E141°29′) in Aomori prefecture, in the northeastern part of Honshu Island in Japan (Fig. 1A). The Hatanai site consists of several strata including artifacts and remnants from the prehistoric Initial-Middle Jomon (ca. 8000-4500 BP) to pre-modern Edo period (1603–1867). All skeletal materials are stored in the Tohoku University Museum, Sendai, Japan.
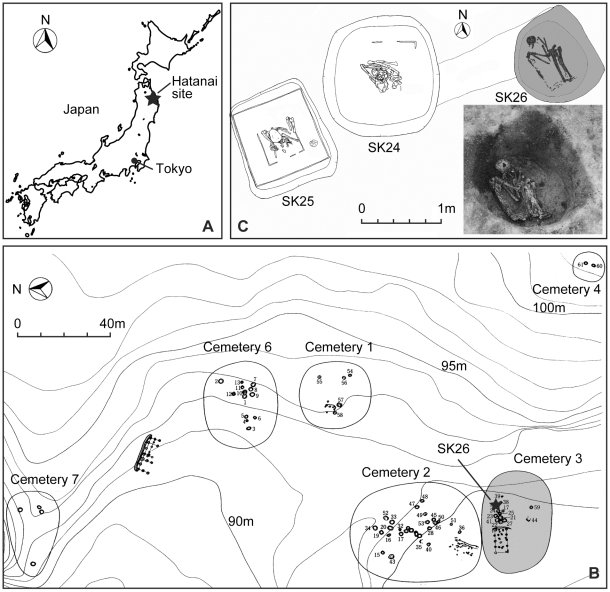
Figure 1. Geographical representation of excavation site.
Location of Hatanai site in northeastern Japan (A). A map of seven cemeteries and the site where SK26 was excavated (B). A grave pit of SK26 at excavated (C) Cemetery 5 (not shown in this map) locates 50 m south of cemetery 2. All these figures were modified with permission from reference [23].
The present material (Grave No. SK26) was buried at a gravel pit of an Edo-period cemetery in a farm village, in which was found about 50 human skeletal remains (Fig. 1B). This corpse was lying on its left side and hunching its back, with the elbow, hip and knee joints closely flexed inside the burial pit (Fig. 1C). It is not clear whether or not SK26 was placed in a wooden coffin. The SK26 skeleton had only one Kouki-tsuhou (Kangxi-tongbao), a metallic currency used during the rule of the Emperor Kangxi (reign in 1661–1722) of the Qing dynasty (1616–1912) in China, as grave goods. Accordingly, the age of this burial is estimated as the middle 18th to the early 19th century, as supposed by evidences from other graves [23].
SK26's skull was almost perfectly intact except for part of the cranial base and parietal bones (Fig. 2). Although its main long bones were intact, many of their epiphyses and all the hand/foot bones were missing within the burial matrix of the postmortem environment. The sex of this skeleton was assessed as male on the basis of cranial morphologies, such as anterior decline of the frontal bone, projection of the superciliary ridge, inferior prominence of the mastoids, and developments of the superior nuchal line and the external occipital protuberance. But its bilateral pelvis was incompletely preserved and therefore unavailable for sex and age determination.
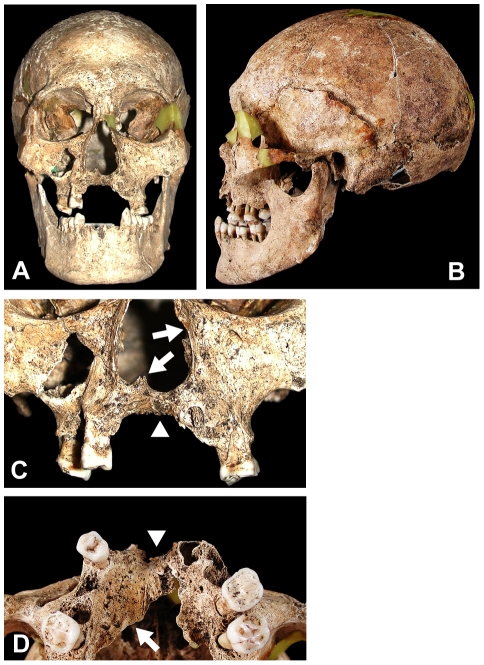
Figure 2. Macroscopic view of the skeletal lesion in the skull of sample SK26.
Frontal view (A) and left side view (B) of the skull. Closer view of the nasal aperture and maxillary palate (C) and upward view of the palate (D). Arrows in panel C indicate erosive deformity of the nasal aperture and disappearance of the anterior nasal spine. Arrowheads in panels C and D indicate severe atrophy of alveolar bone in the maxilla/palatal process with loss of anterior teeth. An arrow in panel D denotes the locus of sample No. 1 shown in Table 1 and Figure 2.
The age at death was estimated from dental attrition and fusions of the cranial sutures. Surviving permanent teeth were worn to dentine, including the 3rd molar. In the calvarium, the unions of all sutures in the internal table were finished; coronal and lambdoid sutures of the external table had incomplete fusing. These dental and skeletal evidences suggest that of the age of death, SK26 was middle-aged, in the range of 30–50 years old.
Palaeopathological observations and diagnosis
The facial cranium of SK26 showed several typical symptoms of leprosy, suggestive of lepromatous type. In the rhinomaxillary area, rounding deformation and abnormal enlargement of the nasal aperture, disappearance of the anterior nasal spine, and severe atrophy of the alveolar process were clearly observed (Fig. 2). The palatal process showed degenerative changes, and alveolar bones at the anterior teeth were closed by antemortem loss of all upper incisors and the right canine. The incisor foramen in the hard palate had also disappeared due to osseous resorption; for maxillary atrophy, the vertical thickness of the palatal bone was about nine millimeters on the right side of the alveolar process, and at the same portion on the left, only six millimeters at the maximum value.
These features of craniofacial malformation, especially the rhinomaxillary syndrome, are specific to lepromatous leprosy but essentially distinct from other facial-deforming diseases such as craniofacial tuberculosis and syphilis, a treponematoses [7], [8]. Although the nasal, maxillary and palatal lesions in craniofacial tuberculosis closely resemble those in lepromatous leprosy, cranial involvements of tuberculosis are extremely rare in adult patients and mainly appear in young children, often with lesions of the mandible. In tertiary syphilis, rhinomaxillary deformations accompany several osteo-periostitic lesions and focal destructive remodeling (caries sicca) caused by the bone gummas in the cranial vault [7], [8]. From differences in these pathological features, the facial deformities of SK26 were diagnosed as leprous symptoms.
In the bilateral tibiae and fibulae of this skeleton, slight periostitic changes were seen on the cortical surfaces of the diaphyses. The SK26 skeleton had lost its hand and foot bones from the long-term burial conditions, making it impossible to confirm any pencil-shaped recessions in the distal ends of the metacarpals, metatarsals or phalanges, which are typical leprous lesions.
Sampling of skeletal remains
Sampling was performed at the Tohoku University Museum, where no other leprous materials were stored. Skeletal samples were taken from the affected area as summarized in Table 1 using a small-sized rotating electric saw (Mini router, Kiso Power Tool, Osaka, Japan). Two samples were also taken from another skeleton (SK20; middle-aged male) found in the same cemetery, which had no leprous changes as a negative control for DNA purification and PCR analysis. One premolar root was extracted from another skeleton (SK16; middle-aged female) as a positive control for human DNA recovery. Sterile materials were used for the sampling to avoid possible contamination. Ethical approval to work with the material was obtained from the review board at National Institute of Infectious Diseases, Japan. A permission to obtain the sample materials was granted by Tohoku University.
Table 1. Skeletal samples.
| No. | Material reference | Sampling site | Paleopathological evidence | Sample weight (mg) |
| 1 | SK26 | Maxillary palate, right | Erosion/atrophy | 9.7 |
| 2 | SK26 | Inner surface of nasal cavity, right | Erosion/atrophy | 6.0 |
| 3 | SK26 | Maxillary palate, left | Erosion/atrophy | 4.6 |
| 4 | SK26 | Fibular diaphysis, right | Periostitis | 22.1 |
| 5 | SK20 | Maxillary palate, right | N.R. | 4.5 |
| 6 | SK20 | Inner surface of nasal cavity, right | N.R. | 6.8 |
| 7 | SK16 | Lower 1st premolar root, right | N.R. | 3.4 |
N.R.: No remarkable change.
DNA preparations
DNA was purified using a QIAamp DNA Micro Kit (Qiagen, Valencia, CA) according to the manufacturer's protocol with some modification. Briefly, 500 µl of Buffer ATL was added to the skeletal samples placed in a 2 ml test tube. The tube was then filled with 1.0 mm diameter Zicronia beads (Bio Space Products, Inc, South Lancaster, MA) and homogenized using Micro Smash MS-100 (TOMY, Tokyo, Japan) at 3,000 rpm for 5 min at 4°C [24], [25]. Samples were then frozen at −80°C, thawed at 37°C and homogenized for five times. The supernatant was subsequently treated with 40 µl of Proteinase K (20 mg/ml) and incubated overnight at 56°C. The samples were frozen and thawed, vortexed in 400 µl of Buffer AL then 400 µl of ethanol was added and incubated for 5 min. Then the mixture was applied to a QIAamp MinElute column and centrifuged at 6,000× g for 1 min at room temperature. Flow-through was discarded and the column was subsequently washed and DNA was eluted by adding 20 µl of Buffer AE and centrifugation at 20,000× g for 1 min.
Polymerase chain reaction (PCR) and DNA sequencing
Touchdown PCR was performed using a PCR Thermal Cycler DICE (TaKaRa, Kyoto, Japan) as described previously [26]. PCR primers were designed to be specific to the M. leprae genome and were used in previous studies [24], [27], [28]; all the primers are listed in Table 2. The PCR products were analyzed by 2% agarose gel electrophoresis. Nested PCR was used to amplify hsp-70, 16S rRNA and SNPs. Some of the results were confirmed in an independent laboratory.
Table 2. PCR primer sequences.
| Gene name | Synonyma) | Forward (5′-3′) | Reverse (5′-3′) | Length (bp) |
| Nested primer | ||||
| hsp-70, 1st | ML2496 | gggctgtccaaggaagag | cgtcaaccacatcgtcagtag | 391 |
| (dnaK) 2nd | tcgtcaaggaacaacggg | cgtcaaccacatcgtcagtaga | 261 | |
| 16S rRNA, 1st | MLP000016 | agagtttgatcctggctcag | tgcacacaggccacaaggga | 1039 |
| (rrs) 2nd | cggaaaggtctctaaaaaatctt | catcctgcaccgcaaaaagctt | 171 | |
| SNP1, 1st | 14,676 | atttccaggtcttgtgcg | aggtcttcccaggacacc | 351 |
| 2nd | aatggaatgctggtgagagc | caatgcatgctagccttaatga | 194 | |
| SNP2, 1st | 1,642,875 | ttagtcaacatcgttagcagccc | actcataagcacggtgtctttgc | 403 |
| 2nd | tgctagtttaaccgagtactgcta | gtagtagtcttccaagttgtggtg | 189 | |
| SNP3, 1st | 2,935,685 | cgagcataatcgtaggcg | aaatgtggtcacctgggc | 510 |
| 2nd | atctggtccgggtaggaatc | accggtgagcgcactaag | 180 | |
| Gene | ||||
| cspA | ML0198 | gaactgtgaagtggttcaacg | agcgaactccagtggcttg | 186 |
| hisE | ML1309 | gaccttcgaggatctgttcg | atagacgtcatcgagcgaca | 247 |
| purM | ML2205 | aaactcacgtccgtaccttc | agcacggtcagtatcttcag | 233 |
| cpsA | ML2247 | ggcaccttcaccaacctaga | ccaacttaggatccgcttga | 511 |
| Pseudogene | ||||
| pseudo scoB | ML0434 | tggaacacctcgtcgtatgtgg | tataagtggcaccgccgaactc | 201 |
| pseudo speE | ML0476 | tcgcaacttcactgatcgtc | gtctggcaccaataccgagt | 465 |
| pseudo REP | ML0794 | aaagacggagactacgatg | gtttagaaggttggtcgttg | 191 |
| pseudo ahpD | ML2043 | tcaacatggcgatctgcattc | tgcgtgaccttacaacgct | 200 |
| Non-coding region | ||||
| 348,457 | tggactcgatgttgaagtg | tgcttagctatgcagtgag | 202 | |
| 1,593,211 | catcgagtccaagctcaac | tgccgatgattacatcatcc | 191 | |
| 2,134,972 | cggaatcctgttgacgtgtt | cggcgctaacaactatcctc | 242 | |
| 2,152,288 | ccgatatgttcggtagtcgt | gcatcgatatcgccttcag | 241 | |
| 2,307,322 | ggttcaccggaagagttgg | cgcgacgactaagccagtag | 242 | |
| 2,546,884 | tcaatatggcttcctatgttgc | gctgcattaatccatgattcg | 202 | |
| 2,551,060 | acattcgagaccagctaccg | ttccgcttggaggataattg | 194 | |
| 2,664,658 | tgagcttgccgattacgatt | gccattgaactggccatc | 227 | |
| 2,858,681 | atgttggttgagcttggac | ttgcttagctatgcagtgag | 217 |
Numbers in SNP nested primer indicate the coordinate location of the SNP. Numbers in non-coding regions indicate coordinate position of the primer within M. leprae genome (http://genolist.pasteur.fr/Leproma/).
The DNA sequence was analyzed using an ABI PRISM 310 Genetic Analyzer and GeneScan Collection software (Applied Biosystems).
Whole genome amplification (WGA)
A GenomePlex Whole Genome Amplification kit (Sigma, St Louis, MO) was used to amplify genomic DNA samples according to the manufacturer's protocol [29]. Briefly, 1 µl of 10× Fragmentation Buffer was mixed with 10 µl of DNA solution and incubated for 4 min at 95°C. After cool down, 2 µl of 1× Library Preparation Buffer and 1 µl of Library Stabilization Solution were added and incubated for 2 min at 95°C. Then 1 µl of 1× Library Preparation Enzyme was added and incubated at 16°C for 20 min, at 24°C for 20 min, at 37°C for 20 min and at 75°C for 5 min to ensure library preparation. To amplify genomic DNA, 7.5 µl of 10× Amplification Master Mix, 47.5 µl of Nuclease-free Water and 5 µl of Jumpstart Taq DNA polymerase were added and heated at 95°C for 3 min, then 14 cycles of two-temperature amplification was employed at 94°C for 15 sec and 65°C for 5 min using a PCR Thermal Cycler DICE (TaKaRa). The quality of the WGA products was evaluated by 1% agarose gel electrophoresis.
Others
All experiments were performed in a biosafety level 2 (BSL2) laboratory where samples were handled in a safety cabinet using disposable sterile materials to avoid any contamination.
Results
Demonstration of M. leprae DNA by PCR and DNA sequencing
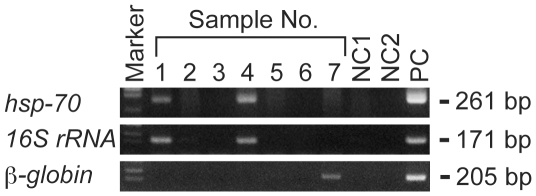
The purified DNA from seven different sites of skeletons from three individuals was subjected to PCR amplification of the M. leprae-specific hsp-70 gene and 16S rRNA genes. A nested PCR protocol to amplify the M. leprae-specific 16S rRNA region was developed in our laboratory. Among four samples taken from leprous lesions, a specific positive signal was derived from sample No. 1 (right maxillary palate of SK26) and No. 4 (right fibula of SK26), but not from the others (Fig. 3). Samples taken from control bones were all negative. Human β-globin DNA was detected by PCR only from sample No. 7 (lower premolar root of SK16).
Figure 3. PCR detection of M. leprae DNA from skeletal samples.
PCR analysis was performed using M. leprae-specific hsp-70 and 16S-rRNA primers for the DNA samples listed in Table 1. PCR products were evaluated by 2% agarose gel electrophoresis. Human β-globin gene was also PCR amplified as a control for DNA preparation from a premolar root. NC1: a negative control for DNA purification in which DNase/RNase-free water was used as a sample for DNA extraction; NC2: a negative control for PCR in which DNase/RNase-free water was used instead of a DNA sample for PCR reaction; and PC: positive control DNA from Thai 53 strain of M. leprae.
The specificity of these PCR amplifications was confirmed by DNA sequencing of the PCR products purified from agarose gel. Basic Local Alignment Search Tool (BLAST) search of the DNA sequence was a 100% match with the reported M. leprae sequence for both hsp-70 and 16S rRNA as expected (data not shown). The results clearly confirmed that M. leprae was involved in the skeletal lesions of SK26 diagnosed as lepromatous leprosy.
SNP analysis of the M. leprae DNA
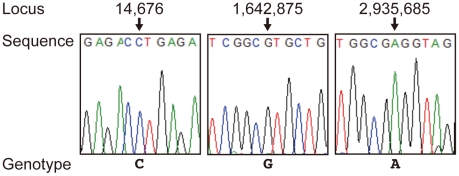
To determine the possible origin of the M. leprae found in this skeleton, we analyzed single-nucleotide polymorphisms (SNPs) at 3 reported loci in the M. leprae genome [2], [3]. PCR amplification followed by direct sequencing identified the sequence at the 3 loci. Positions 14,676, 1,642,875 and 2,935,685, were “C”, “G” and “A”, respectively (Fig. 4). This genotype of M. leprae DNA is reported as SNP type 1, which is the dominant type in India and Southeast Asia [2], [3].
Figure 4. Sequence analysis of the three reported SNPs of M. leprae DNA.
Each locus was PCR amplified and sequenced as described in the Materials and Methods.
Evaluation of WGA-PCR method
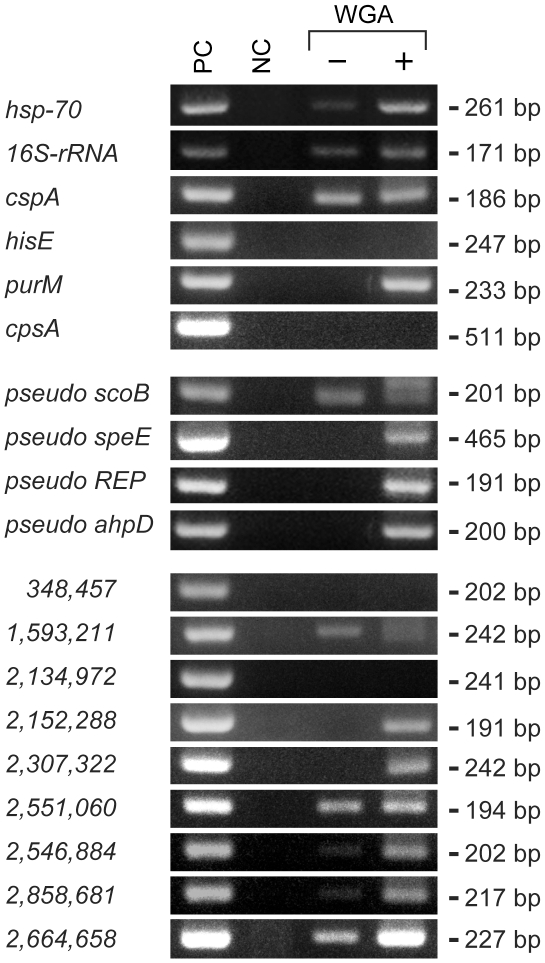
We have attempted to amplify other regions of the M. leprae genome using PCR primers we have designed previously [27] and found that the sensitivity to detect M. leprae DNA from ancient samples is significantly lower than from the clinical samples we analyze in our laboratory. To solve this problem, we have applied WGA to amplify whole genomic DNA purified from skeletal samples. Purified DNA from sample No. 1 was subjected to WGA and the DNA before and after WGA was compared by PCR using 19 different PCR primers targeting open reading frames, pseudogenes and non-coding regions of the M. leprae genome [24], [27]. WGA-amplified DNA was positive for 15 sets of PCR primers, while M. leprae genomic DNA was detected by only 7 primer sets in the original DNA sample (Fig. 5), showing that WGA-PCR has superior sensitivity over conventional PCR.
Figure 5. Comparison of conventional PCR and WGA-PCR.
PCR was performed using a DNA sample derived from the present study (sample No. 1 shown in Table 1 and Fig. 3). The DNA before and after WGA (“−” and “+”, respectively) was amplified using specific nested primers and direct primers targeted for the genes, pseudogenes and non-coding regions of M. leprae genome as listed in Table 2. Names of the genes and pseudogenes are indicated and the coordinate positions of non-coding regions in the genome (http://genolist.pasteur.fr/Leproma/) are indicated numerically. PC: positive control DNA from Thai 53 strain of M. leprae; and NC: negative control using DNase/RNase-free water instead of a DNA sample for PCR reaction.
Discussion
We have demonstrated M. leprae DNA in excavated human skeletal remnants from Japan by both PCR analysis and DNA sequencing. We also sequenced three SNPs in the M. leprae DNA and confirmed that the bacilli are of the dominant type in Southeast Asia. Although M. leprae DNA has already been found in archaeological bone remains in Europe and the Middle East [13], [14], [15], [16], [17], [18], [19], [20], [21], there have been no reports from Far East Asia. This may be related to the geographical and environmental diversity between Europe and Asia, where archaeologists and anthropologists have not paid much attention to palaeopathological topics of leprosy and other infectious diseases [22]; Asia's predominantly hot and humid climate may cause difficulty in recovering skeletons buried for several hundred years.
The present case was excavated in the northern part of Japan and showed typical osteological changes of leprosy. We took skeletal samples from four different areas, the inner surface of the nasal cavity, maxillary palate and diaphysis of the fibula, that showed deformities by erosion and atrophy, or mild periostitis. M. leprae DNA was successfully demonstrated in the maxillary palate and fibular diaphysis.
Although subperiostal exostoses and/or hypertrophy accompanied by swelling or porotic hyperosteosis of the fibula are reported as the typical signs of leprosy [6], [7], the presence of M. leprae DNA in the fibula was somewhat surprising to us because apparent fibular lesion is not common in leprosy patients at present. It is well known that the common fibular nerve is one of the sites preferably affected by M. leprae, which causes foot drop in the patient [30]. It may therefore be possible to speculate that M. leprae affected the periosteum through this nerve. If that is the case, the present identification of M. leprae DNA in the fibula supports the possibility that M. leprae can spread significantly throughout the body when patients are left untreated.
The SNP genotype of M. leprae DNA in Hatanai SK26 was type 1, which is a major group in Southeast Asia and India, but rather minor in modern East Asia including Japan and China [2], [3]. All reported SNP types of ancient M. leprae DNA from Europe and Middle East are type 3, which is also a major genotype in the same regions at present [3]. However, this study confirmed the first case of SNP type 1 as ancient DNA samples. Although more evidence is required, this suggests the possibility that in the Far East inclusive of Japan, the dispersal process of the phylogenetic type of leprosy is historically different from the occidental world. In the Southeast Asia and India where the modern dominant SNP group is type 1, archaeological skeletal remains with lesions of lepromatous leprosy have been excavated and dated from ca. 2000 BC (India) to ca. 300 BC-500 AD (Thailand) [31], [32]; therefore the type 1 genotype in modern Japan might be traced back to Southeast Asia and probably originated in India. A historical record of ancient Japan mentioned that the oldest case suggestive of leprosy dated back to the 7th century, and the oldest positive palaeopathological evidence at present is dated to the 13th century. The genotypes of these ancient and medieval materials are still unknown, and further examination of ancient materials from Eastern and Southeast Asia are needed in order to determine the dispersal history of leprosy in Far East Asia.
In our laboratory, PCR detection of M. leprae DNA is routinely performed from small amounts of clinical samples of skin smears or paraffin sections as part of a government reference center. It is difficult, however, to apply the same PCR protocol to detect M. leprae from ancient DNA recovered from skeletal remains. To overcome this problem, we have utilized WGA, a technique that enables amplification of minute amounts of DNA [29], to increase the sensitivity for PCR detection. We have successfully utilized this method to amplify and detect environmental bacteria from small amounts of water taken from the sea or lakes in Japan (unpublished data).
By PCR reaction using WGA-amplified DNA (WGA-PCR method), large amounts of DNA could be generated from minute amounts of original DNA samples and the detection rate of M. leprae DNA and signal intensity were significantly improved. Thus, this method enables highly sensitive detection of small amounts of clinical, environmental and ancient DNA. On the other hand, it also carries the risk of contamination and false positives, particularly if performed by an individual lacking appropriate knowledge and training in molecular biology. Even using this method, however, some primers still did not work on ancient DNA. This might be due to DNA fragmentation in the ancient sample and/or because of the DNA fragmentation process employed during the library preparation step in the WGA protocol. In general, genomic DNA from dead organisms gradually degrades into short fragments over time mainly due to the activity of nucleases produced by microorganisms in the soil. PCR detection, even using the present WGA-PCR method, will be difficult to perform when such a fragmentation proceeds, especially for very old materials and those buried in conditions where the decomposition process is accelerated, e.g. a hot and humid environment. Nevertheless, WGA-PCR seems to be advantageous for improving the sensitivity of DNA recovery from these samples. We are now routinely utilizing this method to detect ancient DNA and believe that it will be applicable for detecting other kinds of DNA from archaeological samples, as well as other kinds of samples with trace amounts of DNA.
Acknowledgments
The authors wish to thank Drs. Toshio Yanagida and Jun Nemoto (Tohoku University Museum) for permission to obtain the sample materials. We also appreciate the contribution of Drs. Junya Sakurai (Shobi University), Aiko Saso (The University of Tokyo), and Keigo Hoshino and Kazuaki Hirata (St. Marianna University) for to discussion.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by Grant-in-Aid for Research on Emerging and Re-emerging Infectious Diseases from the Ministry of Health, Labour, and Welfare of Japan. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- 2.Monot M, Honore N, Garnier T, Araoz R, Coppee JY, et al. On the origin of leprosy. Science. 2005;308:1040–1042. doi: 10.1126/science/1109759. [DOI] [PubMed] [Google Scholar]
- 3.Monot M, Honore N, Garnier T, Zidane N, Sherafi D, et al. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet. 2009;41:1282–1289. doi: 10.1038/ng.477. [DOI] [PubMed] [Google Scholar]
- 4.Carpintero P, Logrono C, Carreto A, Carrascal A, Lluch C. Progression of bone lesions in cured leprosy patients. Acta Leprol. 1998;11:21–24. [PubMed] [Google Scholar]
- 5.Coutelier L. The bone lesions in leprosy. A study based on microradiography and fluorescence microscopy. Int J Lepr Other Mycobact Dis. 1971;39:231–243. [PubMed] [Google Scholar]
- 6.Boldsen JL. Leprosy in the early medieval Lauchheim community. Am J Phys Anthropol. 2008;135:301–310. doi: 10.1002/ajpa.20744. [DOI] [PubMed] [Google Scholar]
- 7.Boldsen JL, Mollerup L. Outside St. Jorgen: leprosy in the medieval Danish city of Odense. Am J Phys Anthropol. 2006;130:344–351. doi: 10.1002/ajpa.20363. [DOI] [PubMed] [Google Scholar]
- 8.Aufderheide AC, Rodríguez-Martín C. Cambridge: Cambridge University Press; 1998. The Cambridge Encyclopedia of Human Paleopathology. [Google Scholar]
- 9.Ortner DJ. London: Academic Press; 2003. Identification of Pathological Conditions in Human Skeletal Remains. [Google Scholar]
- 10.Roberts C, Manchester K. New York: Cornell University Press; 2007. The Archaeology of Disease. [Google Scholar]
- 11.Drancourt M, Raoult D. Palaeomicrobiology: current issues and perspectives. Nat Rev Microbiol. 2005;3:23–35. doi: 10.1038/nrmicro1063. [DOI] [PubMed] [Google Scholar]
- 12.Spigelman M, Lemma E. The use of the polymerase chain reaction to detect Mycobacterium tuberculosis in ancient skeletons. Int J Osteoarchaeol. 1993;3:143. [Google Scholar]
- 13.Rafi A, Spigelman M, Stanford J, Lemma E, Donoghue H, et al. Mycobacterium leprae DNA from ancient bone detected by PCR. Lancet. 1994;343:1360–1361. [PubMed] [Google Scholar]
- 14.Watson CL, Lockwood DN. Single nucleotide polymorphism analysis of European archaeological M. leprae DNA. PLoS One. 2009;4:e7547. doi: 10.1371/journal.pone.0007547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donoghue HD, Marcsik A, Matheson C, Vernon K, Nuorala E, et al. Co-infection of Mycobacterium tuberculosis and Mycobacterium leprae in human archaeological samples: a possible explanation for the historical decline of leprosy. Proc Biol Sci. 2005;272:389–394. doi: 10.1098/rspb.2004.2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montiel R, Garcia C, Canadas MP, Isidro A, Guijo JM, et al. DNA sequences of Mycobacterium leprae recovered from ancient bones. FEMS Microbiol Lett. 2003;226:413–414. doi: 10.1016/S0378-1097(03)00617-7. [DOI] [PubMed] [Google Scholar]
- 17.Haas CJ, Zink A, Palfi G, Szeimies U, Nerlich AG. Detection of leprosy in ancient human skeletal remains by molecular identification of Mycobacterium leprae. Am J Clin Pathol. 2000;114:428–436. doi: 10.1093/ajcp/114.3.428. [DOI] [PubMed] [Google Scholar]
- 18.Taylor GM, Widdison S, Brown IN, Young D. A mediaeval case of lepromatous leprosy from 13-14 century Orkney, Scotland. J Archaeol Sci. 2000;27:1133–1138. [Google Scholar]
- 19.Donoghue HD, Gladykowska-Rzeczycka J, Marscik A, Holton J, Spigelman M. Mycobacterium leprae in archaeological samples. In: Roberts CA, Lewis ME, Manchester K, editors. The Past and Present of Leprosy: Archaeological, Historical, Palaeopathological and Clinical Approaches. Oxford: British Archaeological Reports; 2002. pp. 271–285. [Google Scholar]
- 20.Taylor GM, Watson CL, Lockwood DNJ, Mays SA. Variable nucleotide tandem repeat (VNTR) typing of two cases of lepromatous leprosy from the archaeological record. J Archaeol Sci. 2006;33:1569–1579. [Google Scholar]
- 21.Likovský J, Urbanová M, Hájek M, Černý V, Čech P. Two cases of leprosy from Žatec (Bohemia), dated to the turn of the 12th century and confirmed by DNA analysis for Mycobacterium leprae. J Archaeol Sci. 2006;33:1276–1283. [Google Scholar]
- 22.Blau S, Yagodin V. Osteoarchaeological evidence for leprosy from western Central Asia. Am J Phys Anthropol. 2003;126:150–158. doi: 10.1002/ajpa.20121. [DOI] [PubMed] [Google Scholar]
- 23.Takigawa W, Kawakubo Y, Maeda T, Sakaue K, Saeki F, et al. Educational Board of Aomori Prefecture ed. Hatanai site VIII Archaeological Investigation Report of Aomori Prefecture 326. Aomori: Aomori Prefecture; 2002. Early modern skeletal remains excavated from Hatanai site in Nango Village, Aomori Prefecture. pp. 254–282. (in Japanese) [Google Scholar]
- 24.Akama T, Suzuki K, Tanigawa K, Kawashima A, Wu H, et al. Whole-genome tiling array analysis of Mycobacterium leprae RNA reveals high expression of pseudogenes and noncoding regions. J Bacteriol. 2009;191:3321–3327. doi: 10.1128/JB.00120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki K, Nakata N, Bang PD, Ishii N, Makino M. High-level expression of pseudogenes in Mycobacterium leprae. FEMS Microbiol Lett. 2006;259:208–214. doi: 10.1111/j.1574-6968.2006.00276.x. [DOI] [PubMed] [Google Scholar]
- 26.Tanigawa K, Suzuki K, Nakamura K, Akama T, Kawashima A, et al. Expression of adipose differentiation-related protein (ADRP) and perilipin in macrophages infected with Mycobacterium leprae. FEMS Microbiol Lett. 2008;289:72–79. doi: 10.1111/j.1574-6968.2008.01369.x. [DOI] [PubMed] [Google Scholar]
- 27.Nakamura K, Akama T, Bang PD, Sekimura S, Tanigawa K, et al. Detection of RNA expression from pseudogenes and non-coding genomic regions of Mycobacterium leprae. Microb Pathog. 2009;47:183–187. doi: 10.1016/j.micpath.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki K, Udono T, Fujisawa M, Tanigawa K, Idani G, et al. Infection during infancy and long incubation period of leprosy suggested in the case of a chimpanzee used for medical research. J Clin Microbiol. 2010 doi: 10.1128/JCM.00017-10. July 14 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barker DL, Hansen MS, Faruqi AF, Giannola D, Irsula OR, et al. Two methods of whole-genome amplification enable accurate genotyping across a 2320-SNP linkage panel. Genome Res. 2004;14:901–907. doi: 10.1101/gr.1949704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soysal A, Atay T, Ozu T, Arpaci B. Electrophysiological evaluation of peripheral and autonomic involvement in leprosy. Can J Neurol Sci. 2004;31:357–362. doi: 10.1017/s0317167100003449. [DOI] [PubMed] [Google Scholar]
- 31.Robbins G, Tripathy VM, Misra VN, Mohanty RK, Shinde VS, et al. Ancient skeletal evidence for leprosy in India (2000 B.C.). PLoS One. 2009;4:e5669. doi: 10.1371/journal.pone.0005669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tayles N, Buckley HR. Leprosy and tuberculosis in Iron Age Southeast Asia? Am J Phys Anthropol. 2004;125:239–256. doi: 10.1002/ajpa.10378. [DOI] [PubMed] [Google Scholar]