Abstract
Background
Epistasis is recognized as a fundamental part of the genetic architecture of individuals. Several computational approaches have been developed to model gene-gene interactions in case-control studies, however, none of them is suitable for time-dependent analysis. Herein we introduce the Survival Dimensionality Reduction (SDR) algorithm, a non-parametric method specifically designed to detect epistasis in lifetime datasets.
Results
The algorithm requires neither specification about the underlying survival distribution nor about the underlying interaction model and proved satisfactorily powerful to detect a set of causative genes in synthetic epistatic lifetime datasets with a limited number of samples and high degree of right-censorship (up to 70%). The SDR method was then applied to a series of 386 Dutch patients with active rheumatoid arthritis that were treated with anti-TNF biological agents. Among a set of 39 candidate genes, none of which showed a detectable marginal effect on anti-TNF responses, the SDR algorithm did find that the rs1801274 SNP in the FcγRIIa gene and the rs10954213 SNP in the IRF5 gene non-linearly interact to predict clinical remission after anti-TNF biologicals.
Conclusions
Simulation studies and application in a real-world setting support the capability of the SDR algorithm to model epistatic interactions in candidate-genes studies in presence of right-censored data.
Availability: http://sourceforge.net/projects/sdrproject/
Background
The complex nature of human disease has long been recognized and, with the exception of a limited number of examples which follow the rules of mendelian inheritance patterns, common disease results from the poorly understood interaction of genetic and environmental factors [1,2]. At the same time, gene-gene interactions that do not result in linearity between genotype and phenotype (epistasis), may involve several genes at time, dramatically increasing the complexity of the phenomenon. Epistasis can either be defined from a biological point of view as deviations from the simple inheritance patterns observed by Mendel [3] or, from a mathematical point of view, as deviations from additivity in a linear statistical model [4].
The study of statistical epistasis by traditional parametric models is challenging and hindered by several limitations. These include, the problem of the sparseness of data into the multidimensional space [5], the loss of power when adjusting for multiple testing to decrease type I error [6,7], the loss of power in presence of multicollinearity [8] or genetic heterogeneity [1]. To address these issues, several non-parameteric multi-locus methods, essentially based on machine-learning techniques, have been developed and/or applied to genetic association studies with positive results [9]. The application of data mining algorithms to detect non-linear high-order interactions in the context of survival analysis is more complex and thus far limited to a few examples [10-12]. However, the effective ability of these algorithms to model gene-gene interactions and their power to detect epistasis in survival analysis has yet to be determined.
At least two points in modelling non-linear interactions in survival analysis should be taken into account. The first, is the proper way to handle censored data, that is those cases for whom the outcome has not yet happened at the end of the observation time (survival time) or who did not have the event until the end of study (including lost cases and missing data), which are commonly referred to as right-censored cases [13]. The second, is the optimal performance measure to be used in assessing a learned model in survival analysis. In this paper we present an extension of the multifactor dimensionality reduction (MDR) algorithm [14,15], to detect and characterize epistatic interactions in the context of survival analysis which was specifically designed to address the abovementioned issues. Censored data were directly handled by estimating individual multilocus cells survival functions by the Kaplan-Meier method [16]. Multilocus genotypes were then pooled into high-risk and low-risk groups whose predictive accuracy was evaluated by the Brier score for censored samples proposed by Graf et al [17].
The power of the method we propose was at first evaluated in lifetime simulated datasets with epistatic effects which belonged to the most common survival distributions and with different degrees of right-censorship. The method was then applied to identification of single-nucleotide polymorphisms (SNPs) associated with responses to anti-tumor necrosis factor (TNF) agents in patients with rheumatoid arthritis (RA) and active disease.
The notion of pharmacogenetics is not anew in RA and several candidate-gene studies have demonstrated a genetically-based individual variability to treatment with methotrexate or anti-TNF therapy [18-20]. However, there is no consensus at present as to whether pharmacogenomics will allow prediction of anti-TNF therapy efficacy in RA. So far pharmacogenomics studies in RA have produced conflicting results and population stratification and linkage disequilibrium have been cited as potential causes for the inability to replicate results of genetic association studies [21]. Yet, as demonstrated by Greene et al [22] when main effects fail to replicate, gene-gene interaction analysis should also be considered as a potential source of variance.
Methods
Description of the survival dimensionality reduction (SDR) algorithm
The core of the SDR algorithm is the classification procedure used to label as "high-risk" or "low-risk" the multilocus cells that result from gene-gene interaction. This procedure will be used both for feature selection and for model validation as described in the forthcoming sections.
SDR assignments and evaluation
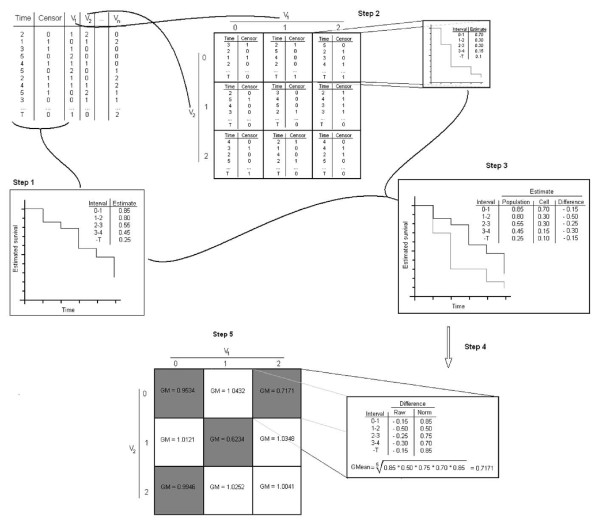
The SDR procedure for classification is illustrated in Figure 1 and it involves 5 steps.
Figure 1.
Survival Dimensionality Reduction (SDR) process. Step 1, survival estimates are calculated in the whole population by the Kaplan-Meier method. Step 2, survival estimates are computed in each multidimensional cell. Step 3, time-point differences in survival estimates between the whole population and each multidimensional cell are calculated. Step 4, time-point differences are normalized to 1 to take into account negative values and then averaged via the geometric mean (GM). Step 5, individual data from multidimensional cells with GM ≤ 1 ("high-risk") are pooled and compared via log rank-test statistic, to individual data from multidimensional cells with GM > 1 ("low-risk").
Step 1: We firstly calculate by the Kaplan-Meier method [16] the survival estimates Ŝ(t) for the whole population in a dataset or dataset partition:
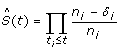 |
where, ni is the number of cases "at risk" of an event prior to ti, and δi, is the number of events at time ti.
Step 2: We then select n discrete variables from the dataset and represent all the possible multidimensional cells resulting from their interaction. For each multidimensional cell, survival estimates at each time interval Ŝc(ti) are calculated as described above.
Step 3: The difference Dc(ti) between the multilocus cell survival estimates and the whole population survival estimates, is calculated for each time interval ti:
Step 4: All the Dc(ti) for each multilocus cells are then averaged. As the product-limit estimator is de facto a geometric progression, Dc(ti) are averaged via the geometric mean (GM) rather than via the algebraic mean. As it is impossible to calculate the geometric with zero or negative data point values, these should be transformed to a meaningful equivalent positive number; being -1 <Dc(ti) < 1, transformation is made adding 1 to any Dc(ti) value. Considering a finite number n of time intervals and denoting tn as the survival time at the nth time-interval we thus have:
Step 5: Cells with GMc(tn) ≤ 1 are classified as "high-risk" and cells with GMc(tn) > 1 are classified as "low-risk". Examples from high-risk cells are pooled into one group and those from low-risk cells into another.
Once dimensionality has been reduced to one dimension, SDR predictions may be evaluated via different fitness measure. Herein, we employed the Brier score for censored data. The Brier score is a metric widely used for predicting the inaccuracy of a model and in its modified version proposed by Graf et al [17], it can incorporate censored samples. The Brier score BS(t) for censored samples for a given t > 0, is defined as:
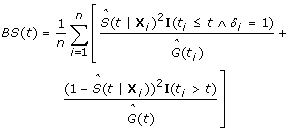 |
where Ĝ(t) denotes the Kaplan-Meier estimate of the censoring distribution G(t) which is based on the observations (ti, 1 - δi) and I stands for the indicator function. BS(t) depends on time t, hence it makes sense to use the integrated Brier score (IBS) as an overall measure for the prediction of the model at all times:
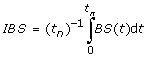 |
The lower the IBS the less inaccurate or, conversely, the more precise the prediction is.
Feature selection and model validation (k-fold Cross-Validation)
From a lifetime dataset, relevant features are extracted and validated via the k-fold cross-validation method, as described by Ritchie and co-workers [14].
For feature selection, the dataset is equally partitioned in k mutually-exclusive testing sets; one k set is retained for model validation whilst the remaining k-1 parts of the dataset are used as a training set. This process is then repeated k-times with each of the k testing samples never included in the feature selection process. In every training set, all the possible combinations of n-variables and the multilocus cells that result from their interactions are represented into the multidimensional space. SDR models are then iteratively built for each combination of n-variables and training IBS scores are calculated. For each n-combination of variables, the k training IBS are then averaged and the n-combination yielding the lowest mean IBS is selected and considered for model validation.
In the model validation phase, SDR assignments for the best n-combination of variables are determined in every training set. On the basis of training assignments, the instances from the corresponding k testing sets are labelled as "high-risk" or "low-risk". From these labels, a cross-validated IBS is then calculated; for this purpose, instead of mathematically averaging the k testing IBS, we merged the testing instances in a meta-analysis-based on individual patient data-fashion [23]. Let T1, T2, --- Tk be the k testing sets, the individual patients data together by their assigned labels are merged sort to produce a larger TM testing set. The IBS (henceforth labelled as meta-IBS) is computed in the TM set and the n-combination yielding the lowest meta-IBS is then chosen as the final model. This way both feature selection and model validation are used to determine the best epistatic model in lifetime datasets. Working on the merged dataset (TM), we: 1) still ensure the independence of the testing sets, as testing assignments are calculated during the feature selection phase; 2) reduce the bias in the calculation of the BS(t), as the Ĝ(t) and G(t) weights used in the BS(t) formula would otherwise be unreliably estimated in testing sets of limited size; 3) avoid to solely rely on a measure of central tendency (e.g. the mean) to estimate the predictive accuracy of our model, utterly ignoring any measure of variance.
Data simulation and power calculation
Simulated epistatic datasets were modelled upon five different survival distributions, described by the logistic-exponential equation [24]; exponential (EXP), bathtub-shaped failure rate (BT), upside-down bathtub-shaped failure rate (UBT), decreasing failure rate (DFR) and increasing failure rate (IFR):
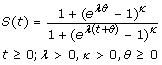 |
where, S(t) is the logistic-exponential survival distribution, t is the survival time, λ is a positive scale parameter and κ is a positive shape parameter and θ is a ≥ 0 parameter that shifts the distribution to the left. λ, κ and θ were adjusted so that the cumulative prevalence of the event at the end of the observation time tn or K(tn), was equal to an arbitrary value of 0.750 (see Additional file 1, Table S1). For data simulation tn was set to 5 time units.
According to Culverhouse et al [25], we then generated different epistatic models for two biallelic SNPs A and B, both in Hardy-Weinberg equilibrium (HWE) and with minor allele frequency (MAF) = 0.2, so that K(tn) = KA = KB, where KA and KB are the marginal penetrances for SNP A and SNP B. As these models were adjusted to fit the cumulative prevalence at tn, their broad-sense heritability (H2) was considered to be a cumulative estimate of H2 at tn or H2(tn). For data simulation H2(tn) was set to 0.10, 0.15, 0.20 or 0.25.
Time-point multilocus genotype penetrances- henceforth labelled as (Genotype)tI -, were adjusted so that the cumulative prevalence at each time-point K(ti) would fit the survival distribution. We assumed that the relation between SNP A and SNP B is purely epistatic at each time-interval ti, that is K(ti) = KA(ti) = KB(ti) with 0 <ti ≤ tn, and that the two-locus model is always proportional at the different ti. Hence, applying the product-limit estimation, we obtain:
where Kti is the time-point prevalence at ti and; ti ≤ tn.
From Kti we can calculate (Genotype)ti from the cumulative multilocus genotype penetrances [Genotype(tn)] previously used to compute K(tn) and H2(tn):
Similarly, time-point cumulative estimated multilocus genotype penetrances [Genotype(ti)] are proportional to K(ti). These penetrances can be used to derive the time-point cumulative estimated H2 or H2(ti).
Once (Genotype)ti values had been calculated, a population of 65000 individuals was built considering all the time-intervals 0 < ti ≤ tn. Herein, from 5 survival distributions with 3 different H2(tn) and two tn we obtained 40 populations where the outcome was related to the epistatic interaction between SNP A and SNP B (see Additional file 1). To each of these populations 13 unrelated SNPs in HWE, with MAF ranging from 0.1 to 0.5 were added. An additional 5% censoring/year was also added to account for hypothetical non-event related causes of withdrawal from observation.
From the simulated populations we finally randomly draw 100 samples of 200 cases and 200 controls (e.g. 50% censorship) or 120 cases and 280 controls (e.g. 70% censorship). A total of 4,000 datasets were then generated for simulation. Power was estimated as the number of times SDR correctly identified the two functional SNPs out of 100 datasets/model/degree-of-censorship. Datasets can be obtained upon request from the authors.
Application of the SDR algorithm to the RA dataset
The SDR algorithm was then tested in a real-world dataset which consists of previously unpublished data about 386 Dutch patients with (1) a diagnosis of RA according to ACR criteria [26], (2) a disease activity score (DAS28) >3.2 [27] and (3) previous treatment with at least two other anti-rheumatics including methotrexate (MTX) at an optimal dose (maximum dose of 25 mg/week) or intolerance for MTX, that underwent treatment with anti-TNFα agents. These patients were extracted from the Dutch Rheumatoid Arthritis Monitoring (DREAM) registry [28] and genotyped for 39 candidate SNPs and evaluated every 3 months to ascertain whether they had reached a clinical remission, defined as a DAS28 ≤ 2.6 [27]. Genotyping details can be found in: Pavy et al [29], Coenen et al [30], Toonen et al [31] and Alizadeh et al [32]. The choice of the studied SNPs was motivated by results from previous association and/or pharmacogenomics studies in RA [18-20,33]. A detailed list of the analysed genetic variants is provided in Additional file 2, Table S1. The k-nearest-neighbour method was used to impute genotypes with missing data <10% [34]. The open-source Orange data mining software (available at: http://www.ailab.si/orange) was used for imputation. Overall the right-censorship of this dataset was 68%; 5-fold cross-validation was used for the SDR analysis. An empirical P-value for the SDR results was calculated by performing 100-fold permutation testing [35]. A whole SDR analysis, up to the 3rd dimension was conducted in the permutated datasets during the permutation procedure.
For all the analyses a modified version of the freely available SDR algorithm written in Python http://sourceforge.net/projects/sdrproject/ was used.
Results
Simulated datasets
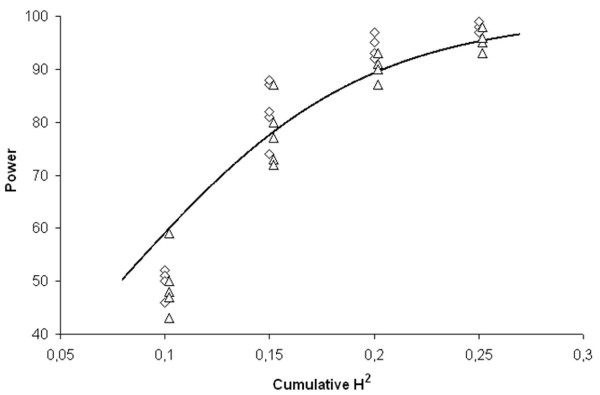
The penetrance functions for the simulated datasets are reported in Additional file 1; as it can be observed, time-point H2 across the different models were consistently low, with a median of 0.017 (interquartile range [IQR]: 0,011 - 0.025). Power for the SDR algorithm to correctly identify the causative pair of SNPs in the 20 simulated survival models with 2 different degrees of censorship is reported in Table 1. Overall, the median power across all the datasets was 87.5% (IQR, 62.25% - 95%). The relationship between H2(tn) and power followed a direct logistic distribution as shown in Figure 2, (overall, R2 = 0.846; 50% censoring, R2 = 0.886; 70% censoring, R2 = 0.870). Moreover, the power resulted to be independent from the survival distribution the sample was withdrawn from whilst it was inversely related to the degree of censorship. The H2(tn) values we employed correspond to mild-to-moderate hazard ratios (HR) for high-risk vs low-risk combinations in the different epistatic models: HR = 1.38 for H2(tn) = 0.10, HR = 1.5 for H2(tn) = 0.15, HR = 1.58 for H2(tn) = 0.20 and HR = 1.69 for H2(tn) = 0.25. Altogether these results suggest that SDR has a satisfactory power to identify a pair of causative genes in purely epistatic lifetime models with mild-to-moderate effect size and for which the proportionality of hazards holds.
Table 1.
Power for the Survival Dimensionality Reduction (SDR) algorithm in models with cumulative prevalence K(tn) = 0.750
| H2(t5) | |||||
|---|---|---|---|---|---|
| Model | RCR | 0.10 | 0.15 | 0.20 | 0.25 |
| UBT | 50% | 52 | 81 | 95 | 97 |
| 70% | 59 | 73 | 93 | 93 | |
| DFR | 50% | 52 | 87 | 97 | 98 |
| 70% | 48 | 77 | 87 | 96 | |
| IFR | 50% | 51 | 88 | 92 | 99 |
| 70% | 50 | 87 | 91 | 98 | |
| BT | 50% | 46 | 74 | 93 | 97 |
| 70% | 43 | 72 | 91 | 95 | |
| EXP | 50% | 50 | 82 | 95 | 99 |
| 70% | 47 | 80 | 90 | 96 | |
Power of the SDR algorithm in simulated datasets modelled using the 5 classes of survival distribution described by the logistic-exponential equation. Different degrees of right-censoring rate (RCR), and cumulative heritability at the survival time [H2(t5)] are considered. UBT, upside-down bathtub-shaped failure rate; DFR, decreasing failure rate; IFR, increasing failure rate; BT bathtub-shaped failure rate; EXP, exponential.
Figure 2.
Trend of the power for the survival dimensionality reduction algorithm in the different models. Relationship between power and cumulative broad-sense heritability (H2) according to the different degrees of censorship of the sample datasets (squares = 70%; triangles = 50%). The line represents the power estimated by the logistic function (R2 = 0.846).
RA dataset
The RA datasets comprises 386 anti-TNFα-treated patients, aging 45 ± 13.7 (mean ± standard deviation) at the onset of disease; 78.3% of patients tested positive for the rheumatoid factor and 262 (67.9%) were females. One-hundred-forty-five patients (37.6%) were treated with adalimumab, 201 (52.1%) with infliximab and 40 (10.4%) with etanercept; overall 346 patients (89.6%) were treated with anti-TNF antibodies (e.g. adalimumab and infliximab). DAS28 at the beginning of therapy was 5.64 ± 1.09. Clinical remission, based on DAS28, was observed in 123 cases (31.8%).
Details about the genotyped SNPs along with their frequencies in the studied population are reported in Additional file 2, Table S1; all the SNPs were in HWE. None of the single SNPs showed a statistically significant association with response to anti-TNF agents, either under a dominant or recessive model, as illustrated in Additional file 2, Table S2 (log-rank-associated p values with 1 degree of freedom, corrected for the number of comparison by Bonferroni adjustment >0.05).
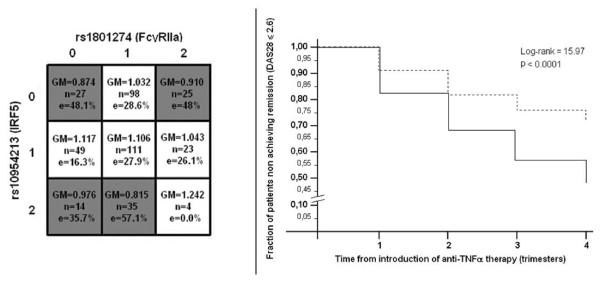
The SDR algorithm sorted out two-way interaction model, involving the rs1801274 (Fc gamma receptor 2a, FcγRIIa) and the rs10954213 (interferon regulatory factor 5, IRF5) SNPs, as the most predictive for responses to anti-TNF therapy in patients with active RA. Table 2 shows the full analysis conducted by the SDR algorithm in the RA datasets. As expected, the model overfits in the training population as the number of SNPs included in the model increases. Yet, cross-validation prevented over-fitting as the minimum meta-IBS was observed for the 2-way interaction, that was thus chosen as the best epistatic model. This model was significant at the 0.05 threshold after 100-fold permutation testing. Figure 3a summarizes the multilocus cells for the rs1801274 × rs10954213 interaction along with SDR "high-" and "low-risk" assignments (e.g. "responders" and "not responders" to therapy). This interaction had the typical non-linear behaviour of epistatic model. Plotting this SDR assignments we can observe that patients labelled as "responders" achieved earlier and higher rates of clinical remission after anti-TNF therapy compared to patients labelled as "non-responders" (figure 3b) [36].
Table 2.
Survival dimensionality reduction (SDR) model for the rheumatoid arthritis (RA) dataset.
| IBS | ||||
|---|---|---|---|---|
| n-way | SNPs (genes) in each dimension | Training | Testing | p |
| 1 | rs2327832 (TNFAIP3, OLIG3) | 0.263 | 0.2366 | - |
| 2 | rs1801274 (FcγRIIa), rs10954213 (IRF5) | 0.2339 | 0.2354 | <0.05 |
| 3 | rs1801274 (FcγRIIa), rs10954213 (IRF5), rs3761847 (TRAF1) | 0.2219 | 0.2393 | - |
Selection of the best combination of attributes by the SDR method. The model with the minimum testing IBS value in the cross-validated testing sets is indicated in boldface type. p values associated with the log-rank test statistic calculated by the 100-fold permutation test. SNP, single nucleotide polymorphism.
Figure 3.
Survival dimensionality reduction (SDR) model for the rheumatoid arthritis dataset. a. Multidimensional matrix for the single nucleotide polymorphisms (SNPs) interaction; GM, geometric mean of the differences; n, number of cases; e, percentage of events. b. Kaplan-Meier estimated survival patterns associated with "high-" and "low-risk" assignments. P values are those associated with a chi-square distribution with 1 degree of freedom.
Discussion
In the present paper we introduce SDR, an algorithm specifically conceived to detect non-linear gene-gene interactions in presence of right-censored data. The need for such a bioinformatics tool comes from the observation that several studies in the medical field deal with the loss of data during the period of study, and that methods that do not take into account censored data give upwardly biased estimates of failure and/or suffer from information loss due to reduced sample size. Cox regression [37], the most popular statistical technique used to analyse time-to-event multivariate model, is not adequate to detect non-linearities either. Indeed, to properly model interaction in Cox regression, the user should have a priori knowledge of the variable relationships and may need to enter nonlinear transforms of the predictors, but this is often a trial and error approach. Also, the number of polynomial terms needed to model complex interactions is inflated as the number of predictors grows, increasing standard errors and, thus, type I error [6,7].
Using simulated epistatic lifetime datasets, we demonstrated that the SDR algorithm retains a fully satisfactory power to sort out a set of causative genes with mild-to-moderate epistatic effect size from a pool of candidate genes. These results have been accomplished by the intrinsic properties of the SDR methodology. Firstly, SDR is non-parametric, in the sense that it is not necessary to make a priori assumptions about the underlying interaction model. Also, SDR requires no assumptions concerning the nature or shape of the underlying survival distribution. Secondly, SDR performs well even in datasets where the right-censorship rate is high (up to 70% of cases in our simulation). Notably, when we run MDR ignoring censorship on the same simulated datasets we observed a dramatic reduction in the power to detect the causative pair of genes, that ranged from 5% to more than 90% (results not shown). Beside power, an additional advantage of SDR is that, combining cross-validation with permutation testing, the chance of false-positive findings is minimized [14]. Yet, to generate the simulated datasets we required the hazards among multilocus cells to be proportional along the lifetime distribution. Hence, further simulations are needed to ascertain whether SDR is suitable for situations where this assumption does not hold, as for instance in case of additive hazard models. Similarly, further simulations are needed to establish SDR performance in larger datasets, in presence of genetic heterogeneity, linkage disequilibrium, or different ranges of MAF.
SDR is, de facto, an extension of the MDR algorithm optimized to analyse lifetime distributions, hence it suffers from similar limitations [14] and it shares some peculiarities with the latter. Namely, the power of SDR is influenced by the epistatic effect size, which is strictly related to the (cumulative) heritability of the model [38]. Moreover, as with MDR, the biological significance of SDR models may be difficult to interpret due to the non-linear distribution of high-risk and low-risk cells across the multidimensional space [39]. Finally, SDR cannot make predictions when multilocus cells contain no data and GM estimates may be upwardly or downwardly inflated when multilocus cells contain few data.
Having demonstrated the capability of SDR to detect gene-gene interactions in lifetime datasets, we applied the algorithm to a population of patients with active RA to identify epistatic interactions that may affect time-related responses to anti-TNF biological agents. We did show that among a set of 39 candidate-gene loci, none of which had a detectable marginal effect on the outcome variable, the non-linear interaction between the rs1801274 (FcγRIIa) and the rs10954213 (IRF5) SNPs significantly predicted the responses to anti-TNF therapy.
Whilst it is difficult to dissect the biological meaning of statistical epistatic models [39], it should be noted that several lines of evidence support a role for Fcγ receptors and IRF5 in rheumatoid arthritis and TNF driven processes. Associations between IRF5 polymorphisms and RA have been described in different populations [40] and a recent genome-wide association study (GWAS) showed that the rs4728142 variant in the IRF5 gene, in tight linkage disequilibrium with rs10954213, is strongly associated with RA susceptibility [41]. Of interest, functional experiments demonstrated that the rs10954213 SNP significantly alters IRF5 mRNA expression [42]. The association between IRF5 gene and RA may be linked, at least in part, to the ability of IRF5 to regulate the secretion of pro-inflammatory cytokines. Indeed, Takaoka et al [43] using mouse models deficient in the IRF5 gene, showed that IRF5 is generally involved downstream of the toll-like receptor (TLR)-MyD88 signalling pathway for gene induction of TNFα and other cytokines relevant to the pathogenesis of RA. Of interest, the use of anti-TNF agents was shown to decrease TLRs expression on different cellular types [44,45]. Similarly to IRF5, the FcγRIIa is involved in TNFα production in the rheumatoid synovia, as observed by Clavel and co-workers [46]. The interaction between the genetic variants of IRF5 and Fcγ receptors could thus influence TNFα production and/or availability, affecting the clinical response to anti-TNF agents. Additionally, as postulated by Cañete at al [47], polymorphisms of the FcγRIIa may alter the clearance rate of anti-TNF antibodies modulating plasma concentrations and consequently their biological effect in subjects with active RA. Theoretically, this effect should not restricted only to anti-TNF antibodies, as also anti-TNFα receptors (e.g. etanercept) contain a Fc portion of IgG1 capable of binding to FCγ receptors to produce biological effects, such as antibody-dependent cell-mediated cytotoxicity [48].
Conclusions
Herein we introduced SDR, an innovative algorithm to detect epistasis in lifetime datasets. Simulation studies and application in a real-world setting, demonstrate the capability of SDR to detect non linear gene-gene interactions in studies aimed at evaluating the effect of candidate genes on time-dependent outcomes. Further studies are necessary to evaluate its applicability in large-scale datasets as well.
Supplementary Material
Epistatic models and simulation specifics. The file contains the settings used to generate the five survival distributions upon which epistatic models were modelled. For each epistatic model, ttime-point and cumulative multilocus genotype penetrances are reported, along with the time-point and the cumulative broad-sense heritability and prevalence of the event.
Genotype frequencies and univariate analysis in the rheumatoid arthritis (RA) dataset. The file lists in tabular form the single nucleotide polymorphisms (SNPs) included in the RA case-control study. It also reports the associations by the log-rank test statistics under the dominant and recessive model between the studied SNPs and the occurrence of clinical remission after therapy with anti-TNF agents.
Contributor Information
Lorenzo Beretta, Email: lorberimm@hotmail.com.
Alessandro Santaniello, Email: sandraniello@gmail.com.
Piet LCM van Riel, Email: P.vanRiel@reuma.umcn.nl.
Marieke JH Coenen, Email: M.Coenen@antrg.umcn.nl.
Raffaella Scorza, Email: raffaella.scorza@gmail.com.
References
- Thornton-Wells TA, Moore JH, Haines JL. Dissecting trait heterogeneity: a comparison of three clustering methods applied to genotypic data. BMC Bioinformatics. 2006;7:204–21. doi: 10.1186/1471-2105-7-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum. Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- Bateson W. Mendel's Principles of Heredity. Cambridge, UK: Cambridge University Press; 1909. [Google Scholar]
- Fisher RA. The correlations between relatives on the supposition of Mendelian inheritance. Transactions of the Royal Society of Edinburgh. 1918;52:399–433. [Google Scholar]
- Bellman R. Adaptive Control Processes. Princeton NJ: Princeton University Press; 1961. [Google Scholar]
- Concato J, Feinstein AR, Holford TR. The risk of determining risk with multivariable models. Ann. Int. Med. 1996;118:201–210. doi: 10.7326/0003-4819-118-3-199302010-00009. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav. Brain. Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 2008, 40: 695-701. [DOI] [PubMed] [Google Scholar]
- Moore JH, Williams SM. New strategies for identifying gene-gene interactions in hypertension. Ann. Med. 2002;34:88–95. doi: 10.1080/07853890252953473. [DOI] [PubMed] [Google Scholar]
- Heidema AG, Boer JM, Nagelkerke N, Mariman EC, van der ADL, Feskens EJ. The challenge for genetic epidemiologists: how to analyze large numbers of SNPs in relation to complex diseases. BMC Genet. 2006;21:7–23. doi: 10.1186/1471-2156-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronek LP, Reddy A. Logical analysis of survival data: prognostic survival models by detecting high-degree interactions in right-censored data. Bioinformatics. 2008;24:248–53. doi: 10.1093/bioinformatics/btn265. [DOI] [PubMed] [Google Scholar]
- Ishwaran H, Kogalur UB. Random survival forests for R. Rnews. 2006;7:25–31. [Google Scholar]
- Ripley BD, Ripley RM. In: Artificial Neural Networks: Prospects for Medicine. Dybowski R, Gant V, editor. Landes Bio-sciences Publishers; 1998. Neural networks as statistical methods in survival analysis. [Google Scholar]
- Kalbfleisch JD, Prentice RL. The Statistical Analysis of Failure Time Data. New York: Wiley; 1980. [Google Scholar]
- Ritchie MD, Hahn LW, Roodi N, Bailey LR, Dupont WD, Parl FF, Moore JH. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am. J. Hum. Genet. 2001;69:138–47. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–82. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- Kaplan E, Meier P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958;53:457–81. doi: 10.2307/2281868. [DOI] [Google Scholar]
- Graf E, Schmoor C, Sauerbrei W, Schumacher M. Assessment and comparison of prognostic classification schemes for survival data. Stat Med. 1999;18:2529–45. doi: 10.1002/(SICI)1097-0258(19990915/30)18:17/18<2529::AID-SIM274>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Bansard C, Lequerré T, Daveau M, Boyer O, Tron F, Salierm JP, Vittecoqm O, Le-Loët X. Can rheumatoid arthritis responsiveness to methotrexate and biologics be predicted? Rheumatology (Oxford) 2009;11:1021–8. doi: 10.1093/rheumatology/kep112. [DOI] [PubMed] [Google Scholar]
- Ranganathan P. An update on pharmacogenomics in rheumatoid arthritis with a focus on TNF-blocking agents. Curr. Opin. Mol. Ther. 2008;10:562–7. [PubMed] [Google Scholar]
- Rego-Pérez I, Fernández-Moreno M, Blanco FJ. Gene polymorphisms and pharmacogenetics in rheumatoid arthritis. Curr. Genomics. 2008;9:381–93. doi: 10.2174/138920208785699553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn KA. A comprehensive review of genetic association studies. Genet. Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Greene CS, Penrod NM, Williams SM, Moore JH. Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS One. 2009;4:e5639. doi: 10.1371/journal.pone.0005639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds MC, Higgins JP, Stewart LA, Tierney JF, Clarke MJ, Thompson SG. Meta-analysis of individual patient data from randomized trials: a review of methods used in practice. Clin. Trials. 2005;2:209–17. doi: 10.1191/1740774505cn087oa. [DOI] [PubMed] [Google Scholar]
- Leemis LM, Lam Y. The Logistic-Exponential Survival Distribution. Naval Research Logistics. 2008;55:252–264. doi: 10.1002/nav.20279. [DOI] [Google Scholar]
- Culverhouse R, Suarez BK, Lin J, Reich T. A perspective on epistasis: limits of models displaying no main effect. Am J Hum Genet. 2002;70:461–71. doi: 10.1086/338759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnett FC, Edworthy SM, Bloch DA, Mcshane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, Medsger TA, Mitchell DM, Neustadt DH, Pinals RS, Schaller JG, Sharp JT, Wilder RL, Hunder GG. The American Rheumatism Association revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1987;31:315–24. doi: 10.1002/art.1780310302. 1988. [DOI] [PubMed] [Google Scholar]
- Prevoo ML, van't Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995;38:44–8. doi: 10.1002/art.1780380107. [DOI] [PubMed] [Google Scholar]
- Kievit W, Fransen J, Oerlemans AJ, Kuper HH, van der Laar MA, de Rooij DJ, De Gendt CM, Ronday KH, Jansen TL, van Oijen PC, Brus HL, Adang EM, van Riel PL. The efficacy of anti-TNF in rheumatoid arthritis, a comparison between randomised controlled trials and clinical practice. Ann. Rheum. Dis. 2007;66:1473–8. doi: 10.1136/ard.2007.072447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavy S, Toonen EJ, Miceli-Richard C, Barrera P, van Riel PL, Criswell LA, Mariette X, Coenen M. TNF alpha -308 G > A polymorphism is not associated with response to TNF-alpha-blockers in Caucasian patients with rheumatoid arthritis: systematic review and meta-analysis. Ann. Rheum. Dis. 2009. in press . [DOI] [PubMed]
- Coenen MJ, Trynka G, Heskamp S, Franke B, van Diemen CC, Smolonska J, van Leeuwen M, Brouwer E, Boezen MH, Postma DS, Platteel M, Zanen P, Lammers JW, Groen HJ, Mali WP, Mulder CJ, Tack GJ, Verbeek WH, Wolters VM, Houwen RH, Mearin ML, van Heel DA, Radstake TR, van Riel PL, Wijmenga C, Barrera P, Zhernakova A. Common and different genetic background for rheumatoid arthritis and celiac disease. Hum. Mol. Genet. 2009;18:4195–203. doi: 10.1093/hmg/ddp365. [DOI] [PubMed] [Google Scholar]
- Toonen EJ, Coenen MJ, Kievit W, Fransen J, Eijsbouts AM, Scheffer H, Radstake TR, Creemers MC, de Rooij DJ, van Riel PL, Franke B, Barrera P. The tumour necrosis factor receptor superfamily member 1b 676T > G polymorphism in relation to response to infliximab and adalimumab treatment and disease severity in rheumatoid arthritis. Ann. Rheum. Dis. 2008;67:1174–7. doi: 10.1136/ard.2008.088138. [DOI] [PubMed] [Google Scholar]
- Alizadeh BZ, Valdigem G, Coenen MJ, Zhernakova A, Franke B, Monsuur A, van Riel PL, Barrera P, Radstake TR, Roep BO, Wijmenga C, Koeleman BP. Association analysis of functional variants of the FcgRIIa and FcgRIIIa genes with type 1 diabetes, celiac disease and rheumatoid arthritis. Hum. Mol. Genet. 2007;16:2552–9. doi: 10.1093/hmg/ddm194. Altman DG, Royston P: What do we mean by validating a prognostic model? Stat. Med. 2000, 19:453-473. [DOI] [PubMed] [Google Scholar]
- Coenen MJ, Gregersen PK. Rheumatoid arthritis: a view of the current genetic landscape. Genes Immun. 2009;10:101–11. doi: 10.1038/gene.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Good P. Permutation Tests: a Practical Guide to Resampling Methods for Testing Hypotheses. Springer, New York; 2000. [Google Scholar]
- Peto R, Peto J. Asymptotically Efficient Rank Invariant Test Procedures. Journal of the Royal Statistical Society. Series A (General) 1972;135:185–207. doi: 10.2307/2344317. [DOI] [Google Scholar]
- Cox DR. Regression Models and Life-tables (with discussion) Journal of the Royal Statistical Society B. 1972;24:187–220. [Google Scholar]
- Edwards TL, Lewis K, Velez DR, Dudek S, Ritchie MD. Exploring the performance of Multifactor Dimensionality Reduction in large scale SNP studies and in the presence of genetic heterogeneity among epistatic disease models. Hum Hered. 2009;67:183–92. doi: 10.1159/000181157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH, Williams SM. Traversing the conceptual divide between biological and statistical epistasis: systems biology and a more modern synthesis. BioEssays. 2005;27:637–646. doi: 10.1002/bies.20236. [DOI] [PubMed] [Google Scholar]
- Han SW, Lee WK, Kwon KT, Lee BK, Nam EJ, Kim GW. Association of polymorphisms in interferon regulatory factor 5 gene with rheumatoid arthritis: a metaanalysis. J. Rheumatol. 2009;36:693–7. doi: 10.3899/jrheum.081054. [DOI] [PubMed] [Google Scholar]
- Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, Li Y, Kurreeman FA, Zhernakova A, Hinks A, Guiducci C, Chen R, Alfredsson L, Amos CI, Ardlie KG. BIRAC Consortium; Barton A, Bowes J, Brouwer E, Burtt NP, Catanese JJ, Coblyn J, Coenen MJ, Costenbader KH, Criswell LA, Crusius JB, Cui J, de Bakker PI, De Jager PL, Ding B, Emery P, Flynn E, Harrison P, Hocking LJ, Huizinga TW, Kastner DL, Ke X, Lee AT, Liu X, Martin P, Morgan AW, Padyukov L, Posthumus MD, Radstake TR, Reid DM, Seielstad M, Seldin MF, Shadick NA, Steer S, Tak PP, Thomson W, van der Helm-van Mil AH, van der Horst-Bruinsma IE, van der Schoot CE, van Riel PL, Weinblatt ME, Wilson AG, Wolbink GJ, Wordsworth BP. YEAR Consortium. Wijmenga C, Karlson EW, Toes RE, de Vries N, Begovich AB, Worthington J, Siminovitch KA, Gregersen PK, Klareskog L, Plenge RM. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–14. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LR, Baechler EC, Plenge RM, Koeuth T, Ortmann WA, Hom G, Bauer JW, Gillett C, Burtt N, Cunninghame Graham DS, Onofrio R, Petri M, Gunnarsson I, Svenungsson E, Rönnblom L, Nordmark G, Gregersen PK, Moser K, Gaffney PM, Criswell LA, Vyse TJ, Syvänen AC, Bohjanen PR, Daly MJ, Behrens TW, Altshuler D. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci USA. 2007;104:6758–63. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW, Taniguchi T. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;10:243–9. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- De Rycke L, Vandooren B, Kruithof E, De Keyser F, Veys EM, Baeten D. Tumor necrosis factor alpha blockade treatment down-modulates the increased systemic and local expression of Toll-like receptor 2 and Toll-like receptor 4 in spondylarthropathy. Arthritis Rheum. 2005;52:2146–58. doi: 10.1002/art.21155. [DOI] [PubMed] [Google Scholar]
- Netea MG, Radstake T, Joosten LA, van der Meer JW, Barrera P, Kullberg BJ. Salmonella septicemia in rheumatoid arthritis patients receiving anti-tumor necrosis factor therapy: association with decreased interferon-gamma production and Toll-like receptor 4 expression. Arthritis Rheum. 2003;48:1853–7. doi: 10.1002/art.11151. [DOI] [PubMed] [Google Scholar]
- Clavel C, Nogueira L, Laurent L, Iobagiu C, Vincent C, Sebbag M, Serre G. Induction of macrophage secretion of tumor necrosis factor alpha through Fcgamma receptor IIa engagement by rheumatoid arthritis-specific autoantibodies to citrullinated proteins complexed with fibrinogen. Arthritis Rheum. 2008;58:678–88. doi: 10.1002/art.23284. [DOI] [PubMed] [Google Scholar]
- Cañete JD, Suárez B, Hernández MV, Sanmartí R, Rego I, Celis R, Moll C, Pinto JA, Blanco FJ, Lozano F. Influence of variants of Fc gamma receptors IIA and IIIA on the American College of Rheumatology and European League Against Rheumatism responses to anti-tumour necrosis factor alpha therapy in rheumatoid arthritis. Ann. Rheum. Dis. 2009;68:1547–52. doi: 10.1136/ard.2008.096982. [DOI] [PubMed] [Google Scholar]
- Mitoma H, Horiuchi T, Tsukamoto H, Tamimoto Y, Kimoto Y, Uchino A, To K, Harashima S, Hatta N, Harada M. Mechanisms for cytotoxic effects of anti-tumor necrosis factor agents on transmembrane tumor necrosis factor alpha-expressing cells: comparison among infliximab, etanercept, and adalimumab. Arthritis Rheum. 2008;58:1248–57. doi: 10.1002/art.23447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Epistatic models and simulation specifics. The file contains the settings used to generate the five survival distributions upon which epistatic models were modelled. For each epistatic model, ttime-point and cumulative multilocus genotype penetrances are reported, along with the time-point and the cumulative broad-sense heritability and prevalence of the event.
Genotype frequencies and univariate analysis in the rheumatoid arthritis (RA) dataset. The file lists in tabular form the single nucleotide polymorphisms (SNPs) included in the RA case-control study. It also reports the associations by the log-rank test statistics under the dominant and recessive model between the studied SNPs and the occurrence of clinical remission after therapy with anti-TNF agents.