Abstract
It has been recently hypothesized that many of the signals detected in genome-wide association studies (GWAS) to T2D and other diseases, despite being observed to common variants, might in fact result from causal mutations that are rare. One prediction of this hypothesis is that the allelic associations should be population-specific, as the causal mutations arose after the migrations that established different populations around the world. We selected 19 common variants found to be reproducibly associated to T2D risk in European populations and studied them in a large multiethnic case-control study (6,142 cases and 7,403 controls) among men and women from 5 racial/ethnic groups (European Americans, African Americans, Latinos, Japanese Americans, and Native Hawaiians). In analysis pooled across ethnic groups, the allelic associations were in the same direction as the original report for all 19 variants, and 14 of the 19 were significantly associated with risk. In summing the number of risk alleles for each individual, the per-allele associations were highly statistically significant (P<10−4) and similar in all populations (odds ratios 1.09–1.12) except in Japanese Americans the estimated effect per allele was larger than in the other populations (1.20; Phet = 3.8×10−4). We did not observe ethnic differences in the distribution of risk that would explain the increased prevalence of type 2 diabetes in these groups as compared to European Americans. The consistency of allelic associations in diverse racial/ethnic groups is not predicted under the hypothesis of Goldstein regarding “synthetic associations” of rare mutations in T2D.
Author Summary
Single rare causal alleles and/or collections of multiple rare alleles have been suggested to create “synthetic associations” with common variants in genome-wide association studies (GWAS). This model predicts that associations with common variants will not be consistent across populations. In this study, we examined 19 T2D variants for association with T2D risk in 6,142 cases and 7,403 controls from five racial/ethnic populations in the Multiethnic Cohort (European Americans, African Americans, Latinos, Japanese Americans, and Native Hawaiians). In racial/ethnic pooled analysis, all 19 variants were associated with T2D risk in the same direction as previous reports in Europeans, and the sum total of risk variants was significantly associated with T2D risk in each racial/ethnic group. The consistent associations across populations do not support the Goldstein hypothesis that rare causal alleles underlie GWAS signals. We also did not find evidence that these markers underlie racial/ethnic disparities in T2D prevalence. Large-scale GWAS and sequencing studies in these populations are necessary in order to both improve the current set of markers at these risk loci and identify new risk variants for T2D that may be difficult, or impossible, to detect in European populations.
Introduction
Multiple common risk alleles have been identified as reproducibly associated with risk of type 2 diabetes (T2D) [1]–[13]. With the exception of the KCNQ1 locus which was identified in the Japanese population [1], [2], all of the well-replicated risk variants were first identified in populations of Northern European ancestry [3]–[13]. T2D morbidity varies widely across racial/ethnic groups; the prevalence is more than twice as high among African Americans, Japanese Americans, Latinos and Native Hawaiians as European Americans [14], [15]. It is important to evaluate whether and how genetic variation may contribute to health disparities between populations. For example, genetic variation at 8q24 may contribute to population differences in risk of prostate cancer [16], [17], and genetic variation at MYH9 contributes substantially to the higher rates of kidney disease in African Americans [18].
It has recently been argued that single rare causal variants and/or collections of multiple different rare variants on unrelated haplotypes may create “synthetic associations” of common variants with disease risk [19]–[21]. One prediction of this model is that the associations with common variants will not be consistent across populations (since many of the mutations will be young in age, and post-date the migrations that led to the founding of modern continental populations). Type 2 diabetes has been specifically discussed as a possible case in which synthetic associations might be operative, based on the lack of statistical significance in very small studies that examined allelic associations for T2D in multi-ethnic samples.
Testing the association of each validated risk allele for T2D in multiple populations is an important step to determine (a) whether these genetic markers can be used to better understand population risk in non-European populations, (b) to measure their association with racial/ethnic variation in disease risk, and (c) to test a prediction of the Goldstein “common SNP, rare mutation” hypothesis [19]–[21].
To allow for comparability of estimates of genetic risk among racial/ethnic groups requires large studies comprised of cases and controls defined using identical criteria and sampled ideally from the same study population. In the present study, we, as part of the Population Architecture using Genomics and Epidemiology (PAGE) Study, examined genetic associations with 19 validated risk alleles for T2D in European American, African American, Latino, Japanese American, and Native Hawaiian T2D cases (n = 6,142) and controls (n = 7,403) from the population-based Multiethnic Cohort study (MEC). We also evaluated whether these variants can be utilized to model the genetic risk of T2D in each population and their association to disparities in risk.
Results
The age of the cases and controls ranged from 45 to 77 at cohort entry, with the mean age of cases (mean 59.0 years) being essentially the same as the controls (mean 58.8 years), and African Americans being on average the oldest (mean 60.2 years) and Native Hawaiians the youngest (mean 55.6 years). Compared to controls, cases were heavier, more likely to be a current or former smoker, less physically active and had fewer years of education (Table 1). Compared to the other groups, the Japanese were leaner (for cases and controls, men and women).
Table 1. The descriptive characteristics of type 2 diabetes cases and controls in the MEC at baseline by racial/ethnic group and sex.
| Characteristic | European Americans | African Americans | Latinos | Japanese Americans | Native Hawaiians | |||||
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |
| Men | ||||||||||
| N | 288 | 504 | 408 | 634 | 1,063 | 1,051 | 978 | 986 | 258 | 493 |
| Age (yr, mean ± SD) | 57.6±8.1 | 57.5±8.0 | 62.6±7.9 | 61.9±7.9 | 59.7±6.9 | 59.7±6.9 | 59.0±8.2 | 59.1±8.2 | 55.5±7.0 | 55.6±7.0 |
| BMI (kg/m2, mean ± SD) | 29.4±4.7 | 25.1±3.1 | 28.9±4.5 | 26.2±3.8 | 28.4±4.2 | 26.2±3.3 | 26.9±4.0 | 24.5±3.1 | 31.0±4.8 | 28.0±5.2 |
| Weight (lbs, mean ± SD) | 209±38 | 180±25 | 206±37 | 187±30 | 188±31 | 174±25 | 172±30 | 156±23 | 215±38 | 194±40 |
| Former/current smoker (%) | 77.7 | 60.1 | 75.7 | 73.5 | 72.4 | 63.3 | 72.3 | 67.8 | 70.9 | 66.1 |
| Education (≤12 years, %) | 27.9 | 11.1 | 39.6 | 39.7 | 59.3 | 55.9 | 29.1 | 26.0 | 48.8 | 40.6 |
| Physical activity (0 hrs / wk, %)a | 30.9 | 21.0 | 45.8 | 38.3 | 34.6 | 29.5 | 35.3 | 30.9 | 24.4 | 19.3 |
| Women | ||||||||||
| N | 245 | 502 | 669 | 835 | 1,157 | 1,133 | 758 | 775 | 318 | 490 |
| Age (yr, mean ± SD) | 58.1±8.3 | 58.3±8.3 | 59.1±8.6 | 58.6±8.6 | 59.0±7.1 | 59.1±7.0 | 59.4±8.5 | 59.5±8.4 | 55.7±7.6 | 55.7±7.6 |
| BMI (kg/m2, mean ± SD) | 30.2±6.2 | 24.4±4.5 | 31.8±6.3 | 27.6±5.7 | 30.0±5.9 | 26.2±4.4 | 26.6±4.6 | 22.8±3.5 | 31.4±6.7 | 26.4±5.2 |
| Weight (lbs, mean ± SD) | 179±39 | 147±28 | 191±39 | 166±35 | 170±34 | 148±26 | 145±27 | 124±21 | 186±42 | 155±33 |
| Former or current smoker (%) | 45.7 | 48.6 | 51.9 | 51.3 | 36.3 | 31.0 | 35.6 | 30.2 | 51.3 | 47.1 |
| Education (≤12 years, %) | 30.2 | 20.9 | 37.1 | 30.5 | 74.9 | 68.5 | 34.7 | 31.9 | 51.4 | 46.0 |
| Physical activity (0 hrs / wk, %)a | 55.1 | 44.2 | 64.6 | 58.6 | 66.8 | 59.7 | 64.3 | 61.9 | 50.3 | 37.1 |
Hours per week of vigorous activity of both work and sports.
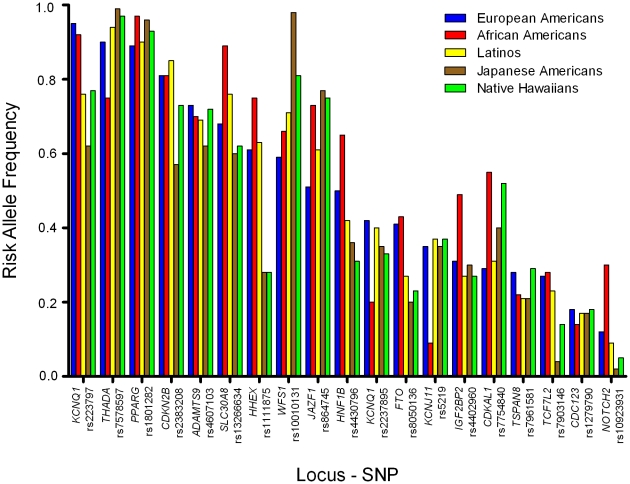
The established T2D risk SNPs were polymorphic (frequency>0.05) in all racial/ethnic groups (Figure 1), except for rs10923931 (NOTCH2) in Japanese and Native Hawaiians and rs7903146 (TCF7L2) in Japanese (Table 2). In European populations these 19 SNPs have very modest odds ratios (1.1–1.3 per copy of the risk allele), and required studies of more than ten thousand cases and controls to reach genome-wide significance [3]–[13], [22]. Our sample sizes, although substantial, provided limited power to detect these modest effects (Table S1; power to achieve nominal significance (P = 0.05) of 34%, 47%, 67%, 54%, and 33%, in European Americans, African Americans, Latinos, Japanese Americans, and Native Hawaiians, respectively).
Figure 1. Risk allele frequencies by racial/ethnic group.
Risk allele frequencies for each variant in European Americans (Blue), African Americans (Yellow), Latinos (Purple), Japanese Americans (Red), and Native Hawaiians (Green). The order of the variants is based on the frequency of the risk allele in European Americans (high to low).
Table 2. The association of known risk alleles for T2D by race/ethnicity.a b .
| SNP | Risk Allelec | Chr. | NearestGene | European Americans533 cases1,006 controls | African Americans1,077 cases1,469 controls | Latinos2,220 cases2,184 controls | Japanese Americans1,736 cases1,761 controls | Native Hawaiians576 cases983 controls | Pooled6,142 cases7,403 controls | P value | Phet d | |
| rs10923931 | T | 1 | NOTCH2 | OR(95%CI) | 0.84 (0.64–1.10) | 1.10 (0.97–1.25) | 1.17 (1.01–1.35) | 1.00 (0.70–1.44) | 0.76 (0.50–1.15) | 1.06 (0.98–1.16) | 0.16 | 0.086 |
| RAF | 0.12 | 0.29 | 0.09 | 0.02 | 0.05 | 0.11 | ||||||
| rs7578597 | T | 2 | THADA | OR(95%CI) | 1.42 (1.06–1.91) | 1.04 (0.90–1.19) | 1.12 (0.93–1.35) | 1.10 (0.68–1.78) | 1.65 (1.01–2.70) | 1.13 (1.02–1.25) | 0.016 | 0.21 |
| RAF | 0.90 | 0.75 | 0.94 | 0.99 | 0.97 | 0.91 | ||||||
| rs1801282 | C | 3 | PPARG | OR(95%CI) | 1.26 (0.95–1.68) | 1.95 (1.30–2.92) | 1.03 (0.89–1.19) | 1.05 (0.82–1.36) | 1.14 (0.84–1.55) | 1.13 (1.02–1.26) | 0.018 | 0.048 |
| RAF | 0. 89 | 0.97 | 0.90 | 0.96 | 0.93 | 0.93 | ||||||
| rs4607103 | C | 3 | ADAMTS9 | OR(95%CI) | 1.11 (0.92–1.34) | 1.03 (0.91–1.18) | 0.99 (0.89–1.08) | 1.05 (0.94–1.17) | 0.98 (0.82–1.16) | 1.02 (0.97–1.08) | 0.45 | 0.78 |
| RAF | 0.73 | 0.70 | 0.69 | 0.61 | 0.72 | 0.68 | ||||||
| rs4402960 | T | 3 | IGF2BP2 | OR(95%CI) | 1.02 (0.85–1.23) | 1.14 (1.01–1.28) | 1.05 (0.95–1.16) | 1.24 (1.11–1.38) | 1.17 (0.99–1.39) | 1.13 (1.07–1.19) | 2.2×10−5 | 0.26 |
| RAF | 0.31 | 0.49 | 0.27 | 0.30 | 0.27 | 0.33 | ||||||
| rs10010131 | G | 4 | WFS1 | OR(95%CI) | 1.18 (0.99–1.40) | 0.94 (0.83–1.07) | 1.15 (1.04–1.27) | 1.45 (0.98–2.13) | 1.27 (1.03–1.56) | 1.11 (1.04–1.19) | 1.8×10−3 | 0.032 |
| RAF | 0.59 | 0.66 | 0.71 | 0.98 | 0. 81 | 0.76 | ||||||
| rs7754840 | C | 6 | CDKAL1 | OR(95%CI) | 1.26 (1.05–1.50) | 1.03 (0.92–1.16) | 1.09 (0.99–1.19) | 1.37 (1.24–1.52) | 1.39 (1.19–1.62) | 1.20 (1.13–1.26) | 4.1×10−11 | 6.2×10−4 |
| RAF | 0.29 | 0.55 | 0.31 | 0.40 | 0.52 | 0.40 | ||||||
| rs864745 | T | 7 | JAZF1 | OR(95%CI) | 0.98 (0.83–1.16) | 1.16 (1.01–1.32) | 1.30 (1.19–1.43) | 1.19 (1.05–1.35) | 1.13 (0.93–1.36) | 1.20 (1.13–1.27) | 1.3×10−9 | 0.054 |
| RAF | 0.51 | 0.73 | 0.61 | 0.77 | 0.75 | 0.68 | ||||||
| rs13266634 | C | 8 | SLC30A8 | OR(95%CI) | 1.28 (1.06–1.54) | 1.21 (0.99–1.48) | 1.11 (1.00–1.23) | 1.18 (1.06–1.31) | 1.04 (0.88–1.22) | 1.15 (1.08–1.22) | 9.7×10−6 | 0.47 |
| RAF | 0.68 | 0. 89 | 0.75 | 0.60 | 0.62 | 0.72 | ||||||
| rs2383208 | A | 9 | CDKN2B | OR(95%CI) | 1.35 (1.07–1.69) | 1.14 (0.98–1.33) | 1.15 (1.01–1.31) | 1.26 (1.13–1.40) | 1.02 (0.85–1.22) | 1.18 (1.11–1.26) | 2.1×10−7 | 0.27 |
| RAF | 0. 81 | 0. 81 | 0. 85 | 0.56 | 0.74 | 0.75 | ||||||
| rs1111875 | C | 10 | HHEX | OR(95%CI) | 0.93 (0.78–1.11) | 1.10 (0.95–1.26) | 1.03 (0.94–1.13) | 1.21 (1.08–1.36) | 0.93 (0.78–1.11) | 1.07 (1.01–1.13) | 0.028 | 0.037 |
| RAF | 0.61 | 0.74 | 0.63 | 0.28 | 0.28 | 0.52 | ||||||
| rs7903146 | T | 10 | TCF7L2 | OR(95%CI) | 1.55 (1.29–1.87) | 1.32 (1.16–1.51) | 1.31 (1.19–1.45) | 1.74 (1.38–2.20) | 1.12 (0.90–1.40) | 1.36 (1.27–1.45) | 1.1×10−19 | 0.068 |
| RAF | 0.27 | 0.28 | 0.23 | 0.04 | 0.14 | 0.19 | ||||||
| rs12779790 | G | 10 | CDC123 | OR(95%CI) | 1.02 (0.82–1.28) | 1.09 (0.92–1.29) | 1.19 (1.06–1.33) | 1.01 (0.88–1.15) | 1.16 (0.95–1.41) | 1.10 (1.03–1.18) | 5.9×10−3 | 0.36 |
| RAF | 0.17 | 0.14 | 0.17 | 0.17 | 0.18 | 0.17 | ||||||
| rs2237895e | C | 11 | KCNQ1 | OR(95%CI) | 0.98 (0.82–1.16) | 1.04 (0.90–1.21) | 1.15 (1.05–1.28) | 1.12 (0.98–1.27) | 1.16 (0.97–1.39) | 1.11 (1.04–1.18) | 7.8×10−4 | 0.17 |
| RAF | 0.42 | 0.20 | 0.40 | 0.35 | 0.33 | 0.34 | ||||||
| rs2237897e | C | 11 | KCNQ1 | OR(95%CI) | 0.86 (0.59–1.26) | 1.13 (0.90–1.42) | 1.23 (1.09–1.39) | 1.26 (1.11–1.44) | 1.06 (0.86–1.31) | 1.21 (1.13–1.30) | 3.2×10−7 | 0.10 |
| RAF | 0.95 | 0.92 | 0.76 | 0.62 | 0.78 | 0.79 | ||||||
| rs5219 | T | 11 | KCNJ11 | OR(95%CI) | 1.24 (1.05–1.47) | 1.03 (0.84–1.26) | 1.09 (1.00–1.20) | 1.26 (1.13–1.40) | 1.03 (0.88–1.21) | 1.15 (1.08–1.21) | 3.3×10−6 | 0.13 |
| RAF | 0.35 | 0.09 | 0.37 | 0.35 | 0.37 | 0.31 | ||||||
| rs7961581 | C | 12 | TSPAN8 | OR(95%CI) | 1.03 (0.86–1.24) | 0.92 (0.80–1.06) | 1.03 (0.93–1.15) | 1.00 (0.88–1.14) | 1.12 (0.94–1.33) | 1.01 (0.95–1.08) | 0.71 | 0.51 |
| RAF | 0.29 | 0.23 | 0.21 | 0.21 | 0.29 | 0.23 | ||||||
| rs8050136 | A | 16 | FTO | OR(95%CI) | 0.89 (0.75–1.06) | 1.07 (0.95–1.20) | 1.02 (0.92–1.12) | 1.04 (0.92–1.18) | 1.01 (0.84–1.21) | 1.02 (0.96–1.08) | 0.48 | 0.68 |
| RAF | 0.41 | 0.43 | 0.27 | 0.20 | 0.23 | 0.30 | ||||||
| rs4430796 | G | 17 | HNF1B | OR(95%CI) | 0.96 (0.82–1.14) | 1.10 (0.97–1.25) | 0.96 (0.88–1.05) | 1.18 (1.06–1.32) | 1.09 (0.92–1.30) | 1.05 (1.00–1.11) | 0.058 | 0.043 |
| RAF | 0.50 | 0.65 | 0.42 | 0.36 | 0.31 | 0.45 |
Odds ratios (and 95% confidence intervals) for allele dosage effects are adjusted for age (quartiles), BMI (quartiles), sex, and ethnicity (in pooled analysis).
Ethnic specific risk allele frequencies (RAF) calculated for each SNP.
NCBI build 36 (forward strand).
Phet = P value for heterogeneity of allelic effects across ethnic groups (4 df test).
rs2237895 and rs2237897 adjusted for each other.
We first assessed whether the “risk allele” of each SNP was associated in the same direction (odds ratios>1) in each ethnic group. Whereas the null hypothesis is that 50% of “risk” alleles would trend in the same direction, we observed from 12 (63%; P = 0.18; binomial probability) in European Americans to 19 (100%; P = 1.9×10−6) in Japanese Americans. The number of these associations that reached nominal significance (P<0.05) ranged from 3 (P = 0.067; binomial probability) in Native Hawaiians to 10 (P = 5.9×10−9) in Japanese (Table 2). For the majority of alleles with positive associations, odds ratios for homozygous carriers were greater than for heterozygous carriers in each population, which provides support for their associations and allele dosage effects (Table S2). In African Americans, results were similar after adjustment for percent European ancestry (Table S3). Adjustment for education, a proxy for socio-economic status (SES) and European ancestry, did not influence the results (Table S4) [23].
We next performed analyses that combined evidence for association across the five ethnic groups. In this analysis the power to achieve nominal significance for the allelic effects reported previously was >80% for 18 out of 19 alleles (average 94%; Table S1). In this analysis all 19 (100%; P = 1.9×10−6, binomial probability) variants were associated with risk in the same direction as the initial report (odds ratios>1) and 14 (P = 5.7×10−15; binomial probability) with nominal statistical significance (P<0.05). All 19 associations remained in the same direction as previous reports (OR>1) and 13 of the variants were significantly associated with T2D risk when the European American subjects were excluded from the analysis. The association of rs8050136 in FTO was attenuated by adjustment for BMI (odds ratio (95% confidence interval), 1.06(1.00–1.11) prior to adjustment; 1.02(0.96–1.08) after adjustment). Only 5 of the 19 risk variants showed nominal evidence for heterogeneity in the odds ratio across ethnic groups, and only one of these (CDKAL1) was significant after correction for having performed 19 tests (PPARG, rs1801282, Phet = 0.048; WFS1, rs10010131, Phet = 0.032; CDKAL1, rs7754840, Phet = 6.2×10−4; HHEX, rs1111875, Phet = 0.037; and, HNF1B, rs4430796, Phet = 0.043; Table 2).
Summary Measures of Risk
We next calculated a summary risk score comprised of an unweighted count of the 19 risk-associated alleles. The average increment in risk per allele was generally similar in all populations, except Japanese Americans, where the effect of each allele was nearly double that observed in Europeans ((odds ratio, 95% confidence interval): African Americans, 1.09, 1.05–1.12; (P = 3.0×10−6); Native Hawaiians, 1.10, 1.06–1.15 (P = 1.2×10−5); European Americans, 1.11, 1.06–1.17 (P = 1.2×10−5); Latinos, 1.12, 1.09–1.14 (P = 7.5×10−19); and, Japanese, 1.20, 1.17–1.24; (P = 7.0×10−32); Phet = 3.8×10−4). Results were similar when limiting the analysis to individuals with complete genotype data for all variants and when including only those markers associated with risk (at P<0.10) (Table S5). Individuals in the top quartile of the risk allele distribution were at 1.6 (African Americans, P = 5.3×10−4) to 3.1-fold (Japanese Americans, P = 7.9×10−26) greater risk of diabetes compared to those in the lowest quartile (Table 3).
Table 3. The association of the total risk score with T2D risk by racial/ethnic population.a .
| European Americans | African Americans | Latinos | Japanese Americans | Native Hawaiians | Pooled | ||
| Total Risk alleles, Mean (range) | 18.8 (10–27) | 20.2 (11–27) | 18.6 (10–28) | 17.2 (10–26) | 18.1 (10–26) | 18.5 (10–28) | |
| Per allele additive model | |||||||
| n (cases/controls) | 533/1,006 | 1,077/1,469 | 2,220/2,184 | 1,736/1,761 | 576/983 | 6,142/7,403 | |
| OR(95% CI)b c | 1.11 (1.06–1.17) | 1.09 (1.05–1.12) | 1.12 (1.09–1.14) | 1.20 (1.17–1.24) | 1.10 (1.06–1.15) | 1.13 (1.11–1.15) | |
| P-value | 1.2×10−5 | 3.0×10−6 | 7.5×10−19 | 7.0×10−32 | 1.2×10−5 | 4.7×10−59 | |
| P-value vs. European Americans | Ref. | 0.43 | 0.76 | 0.007 | 0.86 | ||
| Quartiles of Risk Alleles d | |||||||
| Q1 | n (cases/controls) | 69/196 | 206/360 | 362/464 | 267/441 | 119/251 | 1,023/1,712 |
| OR(95% CI) | 1.00(ref) | 1.00(ref) | 1.00(ref) | 1.00(ref) | 1.00(ref) | 1.00(ref) | |
| Q2 | n (cases/controls) | 112/273 | 273/437 | 520/605 | 424/518 | 150/307 | 1,479/2,140 |
| OR(95% CI) | 1.17 (0.79–1.73) | 1.10 (0.86–1.40) | 1.17 (0.97–1.42) | 1.39 (1.12–1.72) | 1.11 (0.81–1.52) | 1.21 (1.08–1.35) | |
| P-value | 0.44 | 0.45 | 0.10 | 3.2×10−3 | 0.51 | 8.4×10−4 | |
| Q3 | n (cases/controls) | 170/281 | 353/391 | 626/587 | 297/306 | 167/259 | 1,613/1,824 |
| OR(95% CI) | 1.52 (1.05–2.21) | 1.59 (1.25–2.01) | 1.50 (1.25–1.81) | 1.85 (1.46–2.36) | 1.39 (1.02–1.90) | 1.61 (1.44–1.80) | |
| P-value | 0.027 | 1.3×10−4 | 2.1×10−5 | 5.4×10−7 | 0.039 | 4..2×10−17 | |
| Q4 | n (cases/controls) | 182/256 | 245/281 | 712/528 | 748/496 | 140/166 | 2,027/1,727 |
| OR(95% CI) | 1.88 (1.29–2.74) | 1.58 (1.22–2.04) | 1.99 (1.65–2.40) | 3.06 (2.48–3.77) | 2.09 (1.49–2.94) | 2.17 (1.95–2.42) | |
| P-value | 9.3×10−4 | 5.3×10−4 | 7.6×10−13 | 7.9×10−26 | 2.0×10−5 | 4.6×10−44 | |
Odds ratios (and 95% confidence intervals) adjusted for age (quartiles), BMI (quartiles), sex, and ethnicity (in pooled analysis).
P = 3.8×10−4 for heterogeneity of allelic effects across ethnic groups (4 df test).
P≤0.02 for each ethnic group for the comparison with Japanese Americans.
Quartiles are defined separately for each population.
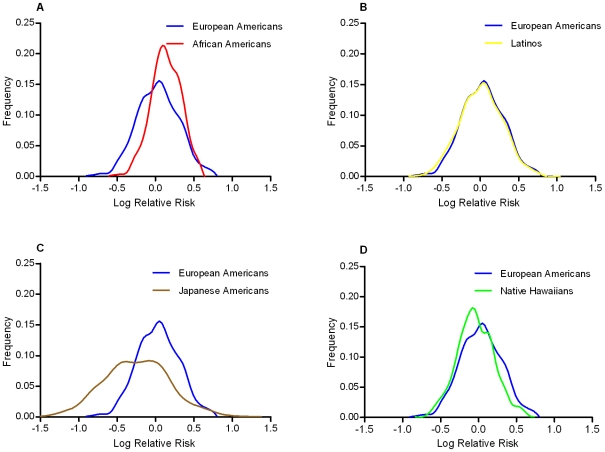
Using these ethnic-specific per allele odds ratio estimates and the aggregate risk allele counts, we built a quantitative risk model to compare the distribution of genetic risks between populations associated with these marker alleles. The higher average number of risk alleles in African Americans caused their distribution to be slightly right shifted (towards higher log ORs) compared to European Americans, however their relatively smaller per allele odds ratio resulted in wide overlap with the European American distribution (Figure 2). The Japanese Americans had a wider distribution of risk because of the large per allele odds ratio, but the low average risk allele counts caused the Japanese distribution to be left-shifted (towards lower log ORs) compared to European Americans. The distributions for Latinos and Native Hawaiians were very similar to the European Americans.
Figure 2. Predicted distribution of T2D risk from common variants by racial/ethnic group compared to European Americans.
Comparison of the predicted risk distributions conveyed by the risk alleles relative to European Americans (Blue in Panels A–D): Panel A, African Americans (Red); Panel B, Latinos (Yellow); Panel C, Japanese Americans (Brown); Panel D, Native Hawaiians (Green). The x-axis is the log relative risk for each population centered (at log RR = 0) around the average total risk allele count in the multiethnic sample (18.5). The y-axis is the relative frequency of the population with that level of risk.
Discussion
We tested 19 common genetic risk markers that were discovered in European populations. We found that association with all 19 of these SNPs trended in the same direction in this large multiethnic study, and the majority of these variants were nominally significant in their association with diabetes risk. A risk score comprised of these alleles was significantly associated with diabetes risk in all five racial/ethnic groups, with the only significant heterogeneity being larger effect sizes in Japanese Americans. However, in comparing the distribution of risk conferred by these alleles between populations we found that they explain little, if any, of known differences in the prevalence of diabetes between these populations.
These observations indicate that most, if not all, of these alleles show directionally similar association to T2D across many populations. Such a pattern indicates that the causal alleles at these validated risk loci (which have yet to be found) likely predate the migrations that separated these populations now residing in Europe, Africa, East Asia, the Pacific Islands and the Americas. We note that this pattern is unexpected under the recently described “common SNP, rare mutation” model of Goldstein that suggests that GWAS signals with common alleles for T2D and other diseases may be “synthetic associations” created by one or more rare alleles [19]–[21]. Under the Goldstein Hypothesis the consistent associations that we noted at these loci across populations would only be observed if, in each population, one or more distinct rare alleles arose at each locus, and they happened to arise each time on the same haplotype background. Although possible, this scenario seems unlikely, and a more parsimonious explanation would be the “synthetic association” hypothesis of Goldstein does not apply to a majority of these T2D SNPs.
The modest number of cases and controls in this study (as compared to the initial discovery studies) likely underlies the lack of statistically significant associations in some groups. Weaker associations in some racial/ethnic groups may also be due to differences in allele frequencies, linkage disequilibrium, and environmental and genetic modifiers. In two cases (WFS1 and CDKAL1), significant heterogeneity by race/ethnicity reflected a lack of association in African Americans, perhaps because of lower linkage disequilibrium between the marker and the biologically relevant allele.
It is interesting that the odds ratios observed for these marker SNPs were larger in Japanese Americans than in the original discovery cohorts, and in the other ethnic groups in our study. A meta-analysis of 7 association studies in Japanese populations replicated associations from studies in European populations for 7 loci under study (TCF7L2, CDKAL1, CDKN2B, IGF2BP2, SLC30A8, KCNJ11, and HHEX) [24]. A recent GWAS in Japanese observed significant associations in KCNQ1 as well as these same 7 loci and, similar to our observations, noted magnitudes of effect that were generally stronger than previously observed in European populations [25]. Additional studies in other Asian populations have replicated associations with many of these loci as well [24], [26]–[28].
In the Multiethnic Cohort, we have found the prevalence of T2D to be at least 2-fold higher in African Americans, Latinos, Japanese and Native Hawaiians compared to European Americans, with these differences being independent of body weight [14]. We examined the extent to which the known genetic risk alleles for diabetes could explain these disparities by quantifying and comparing the relative risk distributions between populations. Compared to European Americans, we did not observe evidence of greater genetic risk in any population. Our findings therefore indicate that these risk markers explain little, if any, of racial/ethnic disparities in T2D prevalence. It remains possible that the actual causal alleles in these regions may be more common in frequency and/or have larger effects than the index signals in non-European populations. As seen with KCNQ1 [1], [2], GWAS in non-European populations are effective in discovering risk loci that are important in multiple populations but difficult to identify in European populations where the alleles are rare.
This study had a number of limitations. First, a self-report of diabetes and use of medication for diabetes was used to define cases and controls. We observed that approximately 1% of a random sample of the controls in this study had HbA1C levels above 7.0%, which suggests that only a small portion of controls had undiagnosed diabetes (see Materials and Methods). Also, our case definition did not differentiate between T1D and T2D, however we expect this misclassification to be minor as <3% of T2D cases had a previous diagnosis of T1D based on other sources (see Materials and Methods). The highly consistent findings of this study, as compared to the discovery GWAS reports, argue that our phenotypic characterization is adequate to observe the association to T2D.
Some caution should also be given to the interpretation of the risk modeling conducted in each ethnic group, as the genetic markers included are unlikely to be the causal alleles. Future fine-mapping and sequencing studies to identify the functional variants (common and/or rare) and large-scale testing of each allele will be required to more precisely model risk as well as assess differences in the distribution of genetic risk across populations.
Another limitation is that we did not account for the potential confounding effects of population stratification. However, odds ratios were essentially unchanged after adjusting for global European ancestry in a subset of African Americans (336 cases 397 controls) for whom ancestry markers were available, suggesting that effects due to population substructure were not substantial, at least in this group. We also noted that controlling for education, a proxy for SES which has been shown to be significantly associated with Native American ancestry in Latinos [23], had little effect on the associations with these risk alleles. Furthermore, the risk alleles were not generally more frequent in Latinos than in European Americans which would be likely if these alleles were proxies for more general ancestry differences. While population stratification is unlikely to fully explain these findings, it remains possible that at some loci, the causal alleles may be more correlated with ancestry than the index SNPs.
In summary, our data provide strong support for common genetic variation contributing to T2D risk in multiple populations. Our findings in T2D do not support the theory that GWAS signals are due to rare alleles. Nonetheless, GWAS and sequencing studies in these and other racial/ethnic populations are needed to reveal a more complete spectrum of risk alleles that are important globally as well as those that may contribute to risk disparities.
Materials and Methods
Ethics Statement
The Institutional Review Boards at the University of Southern California and University of Hawaii approved the study protocol.
Study Population
The MEC consists of 215,251 men and women, and comprises mainly five self-reported racial/ethnic populations: European Americans, African Americans, Latinos, Japanese Americans and Native Hawaiians [29]. Between 1993 and 1996, adults between 45 and 75 years old were enrolled by completing a 26-page, self-administered questionnaire asking detailed information about dietary habits, demographic factors, level of education, personal behaviors, and history of prior medical conditions (e.g. diabetes). Potential cohort members were identified through Department of Motor Vehicles drivers' license files, voter registration files and Health Care Financing Administration data files. In 2001, a short follow-up questionnaire was sent to update information on dietary habits, as well as to obtain information about new diagnoses of medical conditions since recruitment. Between 2003 and 2007, we re-administered a modified version of the baseline questionnaire. All questionnaires inquired about history of diabetes, without specification as to type (1 vs. 2). Between 1995 and 2004, blood specimens were collected from ∼67,000 MEC participants at which time a short questionnaire was administered to update certain exposures, and collect current information about medication use.
Cohort members in California are linked each year to the California Office of Statewide Health Planning and Development (OSHPD) hospitalization discharge database which consists of mandatory records of all in-patient hospitalizations at most acute-care facilities in California. Records include information on the principal diagnosis plus up to 24 other diagnoses (coded according to ICD-9), including T1D and T2D. In Hawaii cohort members have been linked with the diabetes care registries for subjects with Hawaii Medical Service Association (HMSA) and Kaiser Permanente Hawaii (KPH) health plans (∼90% of the Hawaiian population has one of these two plans) [15]. Information from these additional databases have been utilized to assess the percentage of T2D controls (as defined below) with undiagnosed T2D, as well as the percentage of identified diabetes cases with T1D rather than T2D. Based on the OSHPD database <3% of T2D cases had a previous diagnosis of T1D. We did not use these sources to identify T2D cases because they did not include information on diabetes medications, one of our inclusion criteria for cases (see below).
In this study, diabetic cases were defined using the following criteria: (a) a self-report of diabetes on the baseline questionnaire, 2nd questionnaire or 3rd questionnaire; and (b) self-report of taking medication for T2D at the time of blood draw; and (c) no diagnosis of T1D in the absence of a T2D diagnosis from the OSHPD (California Residents). Controls were defined as: (a) no self-report of diabetes on any of the questionnaires while having completed a minimum of 2 of the 3 (79% of controls returned all 3 questionnaires); and (b) no use of medications for T2D at the time of blood draw; and (c) no diabetes diagnosis (type 1 or 2) from the OSHPD, HMSA or KPH registries. To preserve DNA for genetic studies of cancer in the MEC, subjects with an incident cancer diagnosis at time of selection for this study were excluded. Controls were frequency matched to cases on age at entry into the cohort (5-year age groups) and for Latinos, place of birth (U.S. vs. Mexico, South or Central America), oversampling African American, Native Hawaiian and European American controls to increase statistical power.
Fasting glucose (FG) and HbA1C measurements were used to validate the case-control selection criteria. Among 185 T2D cases and 1,048 controls who met the T2D case-control definitions above and with FG measurements available from ongoing studies in the MEC, 57% of cases (ranging from 43% in European Americans to 63% in Japanese Americans) and 3% of controls (ranging from 1% in African Americans to 6% in Latinos) had a FG value >125 mg/dl. We also measured HbA1C (ARUP Laboratories, Salt Lake City, Utah) in 50 cases and 50 controls per each sex-ethnic group. Just over 1% (6/500) of controls were likely to have unreported T2D (HbA1C value ≥7%). In contrast, ∼47% (234/500) of T2D cases had HbA1C ≥7% (ranging from 41% in European Americans to 57% of Native Hawaiians). Since hypoglycemic medication use was part of the case selection criteria, some cases were expected to have FG and HbA1C levels in the normal range.
Altogether, this study included 6,142 T2D cases and 7,403 controls (European American (533/1,006), African American (1,077/1,469), Latino (2,220/2,184), Japanese American (1,736/1,761) and Native Hawaiian (576/983)). Genotyping was conducted by the TaqMan allelic discrimination assay (Applied Biosystems, Foster City, CA) [30]. For all SNPs, genotype call rates were >95% among case and control groups in each population and HWE p-values among controls were >0.05 in at least 4 of the 5 ethnic groups and none of the values were <0.01 (Table S6). Subjects missing data for >5 SNPs (n = 82) were removed from the analysis.
Statistical Analysis
Odds ratios and 95% confidence intervals were calculated for each allele in unconditional logistic regression models while adjusting for age at cohort entry (quartiles), body mass index (BMI, kg/m2, quartiles), sex, and race/ethnicity (pooled analysis) in ethnic-stratified and pooled analyses. Associations with the two variants at KCNQ1 were examined adjusting for the other allele. Potential confounding factors including, smoking history, education, physical activity, and history of hypertension were evaluated but did not influence the results. Potential confounding by percent European ancestry was examined in a subset of African American men (336 cases, 397 controls) with available genetic ancestry information [16], [31], [32].
We also modeled the cumulative genetic risk of T2D using these markers. We summed the number of risk alleles for each individual and estimated the odds ratio per allele for this aggregate unweighted allele count variable as an approximate risk score appropriate for unlinked variants with independent effects of approximately the same magnitude for each allele. We also examined a second model where each allele was weighted and multiplied by the log of the published odds ratio prior to summing all alleles. The results of the more parsimonious unweighted risk score is presented as the two risk scores were highly correlated in each ethnic group (Pearson r≥0.92) and similar associations with T2D risk were observed for each score. For individuals missing genotypes for a given SNP, we assigned the average number of risk alleles within each ethnic group (2× risk allele frequency) to replace the missing value for that SNP. We used these ethnic-specific per allele summary odds ratios and the total number of risk alleles among control subjects to estimate the distribution of relative risks conveyed by all risk alleles. To avoid making the reference group carriers of zero risk alleles (a group which does not exist) we centered the distribution on the mean number of risk alleles observed in the control population (18.5). The log relative risk for each subject was calculated as logRR = (RA−18.5)×log(ORi) (where RA is equal to the subject's total risk alleles and log(ORi) is the log of the ethnic specific per allele odds ratio. A spline function was used to capture the shape of the distributions of log OR for display purposes. Two variants in KCNQ1 were included in the risk modeling because both were significantly associated with T2D when co-modeled (results were similar when only the most significant of the two, rs2237897, was included). The variant in FTO was excluded from risk modeling procedures, as we found (as have others) that it is not a risk factor for diabetes independent of its effect on obesity.
Supporting Information
Power estimates (α = 0.05) to detect relative risks in previous studies.
(0.10 MB DOC)
Association with T2D risk by genotype.
(0.13 MB DOC)
Effects of European ancestry adjustment in African Americans.
(0.07 MB DOC)
Effects of adjustment for education.
(0.12 MB DOC)
Associations with risk score among subjects with and without complete genotype data.
(0.05 MB DOC)
Genotyping efficiency: genotype call rates, Hardy-Weinberg Equilibrium testing.
(0.08 MB DOC)
Acknowledgments
We thank Dr. Jose C. Florez for his help in selecting the diabetes medications in this study.
Footnotes
The authors have declared that no competing interests exist.
The Multiethnic Cohort Study was supported by National Cancer Institute grants CA63464, CA54281, and 1U01HG004802. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40:1098–1102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 2.Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40:1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 3.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 4.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 5.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 9.Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 10.Gloyn AL, Weedon MN, Owen KR, Turner MJ, Knight BA, et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52:568–572. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- 11.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39:770–775. doi: 10.1038/ng2043. [DOI] [PubMed] [Google Scholar]
- 12.Sandhu MS, Weedon MN, Fawcett KA, Wasson J, Debenham SL, et al. Common variants in WFS1 confer risk of type 2 diabetes. Nat Genet. 2007;39:951–953. doi: 10.1038/ng2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 14.Maskarinec G, Grandinetti A, Matsuura G, Sharma S, Mau M, et al. Diabetes prevalence and body mass index differ by ethnicity: the Multiethnic Cohort. Ethn Dis. 2009;19:49–55. [PMC free article] [PubMed] [Google Scholar]
- 15.Maskarinec G, Erber E, Grandinetti A, Verheus M, Oum R, et al. Diabetes incidence based on linkages with health plans: the multiethnic cohort. Diabetes. 2009;58:1732–1738. doi: 10.2337/db08-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proc Natl Acad Sci U S A. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 20.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare Variants Create Synthetic Genome-Wide Associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang K, Dickson SP, Stolle CA, Krantz ID, Goldstein DB, et al. Interpretation of association signals and identification of causal variants from genome-wide association studies. Am J Hum Genet. 2010;86:730–742. doi: 10.1016/j.ajhg.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonsson A, Isomaa B, Tuomi T, Taneera J, Salehi A, et al. A variant in the KCNQ1 gene predicts future type 2 diabetes and mediates impaired insulin secretion. Diabetes. 2009;58:2409–2413. doi: 10.2337/db09-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Florez JC, Price AL, Campbell D, Riba L, Parra MV, et al. Strong association of socioeconomic status with genetic ancestry in Latinos: implications for admixture studies of type 2 diabetes. Diabetologia. 2009;52:1528–1536. doi: 10.1007/s00125-009-1412-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabara Y, Osawa H, Kawamoto R, Onuma H, Shimizu I, et al. Replication study of candidate genes associated with type 2 diabetes based on genome-wide screening. Diabetes. 2009;58:493–498. doi: 10.2337/db07-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeuchi F, Serizawa M, Yamamoto K, Fujisawa T, Nakashima E, et al. Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes. 2009;58:1690–1699. doi: 10.2337/db08-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee YH, Kang ES, Kim SH, Han SJ, Kim CH, et al. Association between polymorphisms in SLC30A8, HHEX, CDKN2A/B, IGF2BP2, FTO, WFS1, CDKAL1, KCNQ1 and type 2 diabetes in the Korean population. J Hum Genet. 2008;53:991–998. doi: 10.1007/s10038-008-0341-8. [DOI] [PubMed] [Google Scholar]
- 27.Tan JT, Ng DP, Nurbaya S, Ye S, Lim XL, et al. Polymorphisms identified through genome-wide association studies and their associations with type 2 diabetes in Chinese, Malays, and Asian-Indians in Singapore. J Clin Endocrinol Metab. 2010;95:390–397. doi: 10.1210/jc.2009-0688. [DOI] [PubMed] [Google Scholar]
- 28.Wu Y, Li H, Loos RJ, Yu Z, Ye X, et al. Common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX/IDE genes are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Diabetes. 2008;57:2834–2842. doi: 10.2337/db08-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolonel LN, Henderson BE, Hankin JH, Nomura AM, Wilkens LR, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. Am J Epidemiol. 2000;151:346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee LG, Connell CR, Bloch W. Allelic discrimination by nick-translation PCR with fluorogenic probes. Nucleic Acids Res. 1993;21:3761–3766. doi: 10.1093/nar/21.16.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng CY, Kao WH, Patterson N, Tandon A, Haiman CA, et al. Admixture mapping of 15,280 African Americans identifies obesity susceptibility loci on chromosomes 5 and X. PLoS Genet. 2009;5:e1000490. doi: 10.1371/journal.pgen.1000490. doi: 10.1371/journal.pgen.1000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deo RC, Patterson N, Tandon A, McDonald GJ, Haiman CA, et al. A high-density admixture scan in 1,670 African Americans with hypertension. PLoS Genet. 2007;3:e196. doi: 10.1371/journal.pgen.0030196. doi: 10.1371/journal.pgen.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Power estimates (α = 0.05) to detect relative risks in previous studies.
(0.10 MB DOC)
Association with T2D risk by genotype.
(0.13 MB DOC)
Effects of European ancestry adjustment in African Americans.
(0.07 MB DOC)
Effects of adjustment for education.
(0.12 MB DOC)
Associations with risk score among subjects with and without complete genotype data.
(0.05 MB DOC)
Genotyping efficiency: genotype call rates, Hardy-Weinberg Equilibrium testing.
(0.08 MB DOC)