Abstract
Acid-sensing ion channels, or ASICs, are members of the amiloride-sensitive cationic channel superfamily that are predicted to have intracellular amino and carboxyl termini and two transmembrane domains connected by a large extracellular loop. This prediction comes from biochemical studies of the mammalian epithelial sodium channels where glycosylation mutants identified the extracellular regions of the channel and a combination of antibody sensitivity and protease action substantiated the intracellular nature of the amino and carboxyl termini. However, although there are highly conserved regions within the different cation channel family members, membrane topology prediction programs provide several alternative structures for the ASICs. Thus, we used glycosylation studies to define the actual membrane topology of the ASIC2a subtype. We deleted the five predicted endogenous asparagine-linked glycosylation sites (Asn-Xaa-(Ser/Thr)) at Asn-22, Asn-365, Asn-392, Asn-478, and Asn-487 to map the extracellular topology. We then introduced exogenous asparagine-linked glycosylation sites at Lys-4, Pro-37, Arg-63, Tyr-67, His-72, Ala-81, Tyr-414, Tyr-423, and Tyr-453 to define the transmembrane domain borders. Finally, we used cell permeabilization studies to confirm the intracellular amino termini of ASIC2a. The data show that Asn-365 and Asn-392 are extracellular and that the introduction of asparagine-linked glycosylation sites at His-72, Ala-81, Tyr-414, and Tyr-423 leads to an increase in molecular mass consistent with an extracellular apposition. In addition, heterologous expression of ASIC2a requires membrane permeabilization for antibody staining. These data confirm the membrane topology prediction that the ASIC2a subtype consists of intracellular amino and carboxyl termini and two transmembrane domains connected by a large extracellular loop.
Acid-sensing ion channels (ASICs)1 are amiloride-sensitive cation channels that are widely expressed throughout the nervous system (for review see Ref. 1). There are eight ASIC subtypes encoded by four genes: ASIC1α, ASIC1b, ASIC-β, ASIC-β2, ASIC2a, ASIC2b, ASIC3, and ASIC4 (2, 3). In the peripheral nervous system ASICs act as nociceptors in sensory neurons (4, 5) and are implicated in pain perception during tissue acidosis (1, 6), particularly in the ischemic myocardium where ASICs induce anginal pain (7). In the central nervous system, ASICs act analogous to neurotransmitter receptors that respond to marked, local fluctuations in the extracellular pH that accompanies normal synaptic activity (8). ASICs also contribute to learning and memory (9) as well as acidosis-induced injury (10), where they may act as mediators of stimuli that respond to the acidosis that accompanies stroke, epilepsy, and trauma under pathological conditions. Although studies suggest that degenerins/epithelial Na+ channels act as mechanosensors (11), recent studies suggest that ASIC2 and ASIC3 do not contribute to mechanosensation (12, 13). However, ASICs are implicated in taste (14) and retinal function (15).
ASICs are members of the amiloride-sensitive cationic channel superfamily that includes the Caenorhabditis elegans degenerins (DEGs), the Helix aspersa Phe-Met-Arg-Phe-amidegated sodium (Na+) channel, and the mammalian epithelial Na+ channels (ENaC) (16). The amino acid identity between the different ENaC/DEG subfamilies is ~15–20%, whereas the primary amino acid identity between the four ASIC genes is ~45–60% (3). The membrane topology of these channel proteins is predicted to consist of intracellular amino and carboxyl termini with two transmembrane domains connected by a large extracellular loop (Fig. 1a). This prediction comes from hydrophobicity analysis, correlation with in vivo structure-function analyses of the C. elegans DEG mutant MEC-4 (17), and correlation with biochemical studies of the α-rENaC (rat) subunit where glycosylation mutants identified the extracellular regions of the channel; a combination of antibody sensitivity and protease action substantiated the intracellular nature of the amino and carboxyl termini (18). However, although there are highly conserved regions within the different cation channel families, to depict the membrane topology of the ASIC family based on homology with ENaCs or hydrophobicity analysis alone may prove erroneous. For example, analysis of each ASIC subtype using four server-based membrane topology prediction programs (PredictProtein (19, 20), TopPred (21, 22), ConPred II (23–25), and HMMTOP (26, 27)) resulted in several possible structural outcomes. For seven of the eight ASIC subtypes analyzed, PredictProtein predicted one structure consistent with the DEG/ENaC topology of an intracellular amino and carboxyl termini with two transmembrane domains connected by a large extracellular loop (Fig. 1a). However, the other topology prediction programs did not consistently predict this structure. For the ASIC2a subtype, TopPred analysis predicted four possible outcomes (Fig. 1b), ConPred analysis predicted two structures, each with three transmembrane domains (Fig. 1c), and HMMTOP analysis predicted one structure with a single transmembrane domain (Fig. 1d). Thus, we used biochemical studies and antibody permeabilization studies to unambiguously define the membrane topology of the ASIC2a protein. These data definitively show the membrane topology of ASIC2a to consist of intracellular amino and carboxyl termini with two transmembrane domains and a large extracellular domain.
Fig. 1. Depiction of the membrane topology of ASIC2a as analyzed by four membrane topology prediction programs.
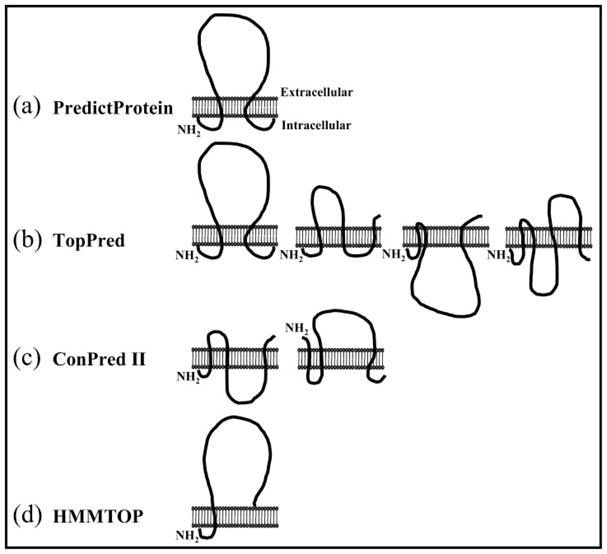
The DEG/ENaCs consists of intracellular amino and carboxyl termini with two transmembrane domains connected by a large extracellular loop. a, PredictProtein depicts one structure for ASIC2a that is identical to the DEG/ENaC structures. b, TopPred predicts four possible structure outcomes for ASIC2a, one of which is identical to the DEG/ENaC structures. c and d, ConPred II (c) and HMMTOP (d) depict possible alternative structures for ASIC2a.
MATERIALS AND METHODS
Site-directed Mutagenesis
The ASIC2a cDNA clone was a gift from Professor Michel Lazdunski, Institut de Pharmacologie Moleculaire et Cellulaire, Centre National de la Recherche Scientifique, Institut Paul Hamel, Sophia Antipolis, Valbonne, France. The ASIC2a cDNA was subcloned into the pCI vector (Promega Corporation, Madison, WI). For the glycosylation studies, five endogenous consensus sites for N-linked glycosylation (NX(S/T)) were deleted individually (N22S, N365S, N392S, N478S, and N487S) whereas nine separate glycosylation sites were introduced (K4N, P37N, R63N, Y67N, H72N, A81N, Y414N, Y423N/V425S, and Y453N/Y455S). Each mutant was generated with the QuikChange® mutagenesis kit (Stratagene, La Jolla, CA) in accordance with the manufacturer’s protocol using high pressure liquid chromatography-purified or PAGE-purified oligonucleotide primers (Sigma-Genosys, The Woodlands, TX). Individual mutants were verified by DNA sequence analysis, and the predicted amino acid sequences were determined by computer analysis.
ASIC2a Transfection and Immunoblot Analysis
The wild type and mutant ASIC2a cDNAs were transiently transfected into human embryonic kidney HEK293 cells using Lipofectamine 2000 (Invitrogen). The cells were collected after 48 h and lysed for immunoblotting using lysis buffer (100 mM NaCl, 20 mM Tris·HCl, pH 7.4, 1% Triton X-100, and 10 μl/ml protease inhibitor mixture). All samples were resuspended in SDS loading buffer (1×0.0625 M Tris-HCl (pH 6.8), 2% (w/v) SDS, 5% (v/v) β-mercaptoethanol, 10% (v/v) glycerol, and 0.002% bromphenol blue) and heated to 95 °C for 2 min. The proteins were separated by one-dimensional gel electrophoresis on an 8% SDS-polyacrylamide gel and transferred to polyvinylidene difluoride membrane (Immobilon-P; Millipore, Billerica, MA). The resultant protein blot was incubated with 5% (w/v) milk in Tris-buffered saline (TBS) containing 0.1% Tween (TBST) for 1.5 h at room temperature followed by incubation with the anti-ASIC2 antibody (Alomone; Jerusalem, Israel) diluted in 5% (w/v) milk in TBST for 1.5 h at room temperature. The blot was rinsed four times for 5 min each in TBST without milk and incubated with a horseradish peroxidase-conjugated secondary antibody (Bio-Rad Laboratories) for 1 h at room temperature. The blot was rinsed one time in TBST and four times in TBS, and the proteins were detected using an enhanced chemiluminescent Western blotting Kit (Amersham Biosciences) and Kodak BioMax film (Eastman Kodak Co.).
ASIC2a Transfection and Immunocytochemistry Analysis
The wild type ASIC2a cDNA was transiently transfected into HEK293 cells using FuGENE 6 transfection reagent (Roche Molecular Systems). The cells were plated onto poly-D-lysine-coated coverslips and fixed after 48 h using 2% paraformaldehyde. Permeabilized cells were incubated with 3% Triton X-100 in phosphate-buffered saline (PBS) for 20 min. All cells were blocked with 2% goat serum and 1% bovine serum albumin in PBS for 1 h and then incubated with 1.2 μg/ml anti-ASIC2a antibody (Alomone) and 15 μg/ml anti-α-tubulin antibodies overnight at 4 °C (Oncogene Research Products; Boston, MA). Cells were rinsed three times in PBS and then incubated with 1:200 goat anti-rabbit fluorescein isothiocyanate-conjugated second antibody and goat anti-mouse Cy3-conjugated second antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) in PBS containing 2% goat serum and 1% bovine serum albumin for 1 h. Cells were washed three times in PBS, mounted on glass slides with 4′,6-diamidino-2-phenylindole mounting medium (Vector Laboratories, Burlingame, CA), and viewed on a Leica microscope.
Electrophysiological Recording of ASIC2a Current
Mutant and wild type ASIC2a cDNAs were transcribed to produce sense strand cRNA using the mMessage mMachine® kit (Ambion; Austin, TX). Manually defoliculated stage V Xenopus laevis oocytes were injected with 50 nl (50 ng) of cRNA using an INJECT+MATIC system (INJECT+MATIC; Genéva, Switzerland) and stored at 18 °C in ND96 buffer (96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 5 mM sodium pyruvate, and 5 mM HEPES, pH 7.6) for 3–7 days with fresh solution changes daily. Two-electrode voltage clamp recordings were made from the oocytes using a GeneClamp 500B amplifier with a Digidata 1322 analog-to-digital converter and pClamp 8.2 acquisition software (Axon Instruments; Union City, CA). ND96 adjusted to varying pH concentrations was applied to the oocytes via a U-tube perfusion system, and reproducible responses were recorded with a 5-min interval between applications. All data are shown as means ± S.E. of the mean with significant differences calculated using the Student’s t test where n is the number of oocytes. Using Origin 6.0 (MicroCal; LLC, Northampton, MA) concentration-response curves for pH sensitivity were fitted with the Hill equation Y = [(X)nH × M]/[(X)nH × (pH50)nH] where Y is response, X is pH, nH is the Hill coefficient, M is maximum response, and pH50 is the acidity that evokes 50% of the maximum response.
RESULTS
ASIC2a Is a Glycoprotein
The ASIC2a cDNA encodes a protein of 512 amino acids with a predicted molecular mass of 58 kDa. Immunoblot analysis of HEK293 cells transiently expressing ASIC2a identified a protein of ~63 kDa that was absent from non-transfected HEK293 cells (Fig. 2a). This difference in molecular mass suggests that the protein is post-translationally modified by mechanisms such as phosphorylation, acetylation, farnesylation, and glycosylation. Proteins that are at least in part located in the extracellular space can be glycosylated on asparagine (N-linked), serine/threonine (O-linked), or tryptophan (C-linked) residues as they pass through the endoplasmic reticulum (ER) and Golgi during their biosynthesis. The enzymes responsible for this process are restricted to the lumen of these organelles and, thus, only parts of the protein that ultimately will have an extracellular disposition will be glycosylated. There are five conserved N-linked glycosylation sites (NX(S/T)) in the ASIC2a sequence (Asn-22, Asn-365, Asn-392, Asn-478, and Asn-487). We used enzymatic deglycosylation to determine the extent of the post-translational modification of ASIC2a by glycosylation. Endoglycosidase H removes N-linked chains that have been added in the ER, whereas N-glycosidase F (PNGase) will remove N-glycans that have been added both in the ER and the Golgi. Treatment of HEK293 cells expressing ASIC2a with endoglycosidase H and PNGase reduced the apparent molecular mass of the receptor by ~5 kDa (Fig. 2a). As one individual glycosylation adds ~2–3 kDa of mass to a protein, these results suggest that the ASIC2a channel has two natural N-linked glycosylation sites.
Fig. 2. Immunoblot analysis of endogenous ASIC2a glycosylation sites.
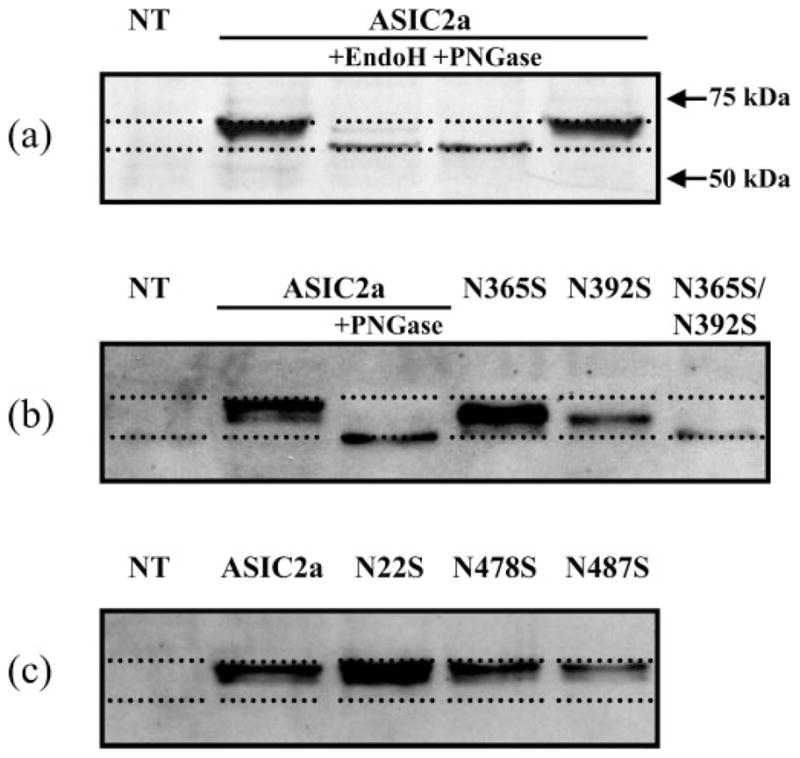
a, the effect of Endo H and PNGase on ASIC2a receptor protein. HEK293 cells transiently expressing the ASIC2a protein were prepared and treated with either Endo H or PNGase. The proteins were separated by SDS-PAGE, transferred to nylon membrane, and the resultant blot was probed with ASIC2a primary antibody (Alomone). The immunoblot shows that both Endo H and PNGase reduced the molecular mass of ASIC2a by ~5 kDa, implying that two glycosylation sites may exist for ASIC2a. b, ASIC2a contains two N-linked glycosylation sites. Mutations N365S and N392S produced ASIC2a channels with a reduced molecular mass, indicating they are both sites for N-linked glycosylation. A double mutant of N365S/N392S produced a reduced molecular mass equal in size to that of PNGase-treated ASIC2a wild type, indicating that only these two sites are required for full glycosylation of ASIC2a. c, the three remaining predicted N-linked glycosylation sites are not glycosylated. Immunoblot analysis of N22S, N478S, and N487S showed no reduction in molecular mass; thus, these ASIC2a proteins are not glycosylated in HEK293 cells. NT, non-transfected.
To determine the identity of the sites for N-linked glycosylation and the functional consequences of this glycosylation, we generated individual point mutants (N22S, N365S, N392S, N478S, and N487S) to remove endogenous asparagine residues in the consensus sequence (NX(S/T)). As glycosylation only occurs on the extracellular face of the protein, we predict, based on a hydrophobicity analysis and a putative membrane topology similar to that of the ENaCs, that residues Asn-365 and Asn-392 are glycosylated. If an asparagine residue is involved in glycosylation, the respective mutant should result in a reduction of the molecular mass of the protein detected by the anti-ASIC2a antibody. Of the mutants, only N365S and N392S resulted in a decrease in mass of the ASIC2a protein, whereas the double mutant N365S/N392S gave a further decrease in size, and the band identified by the anti-ASIC2a antibody was indistinguishable from WT ASIC2a treated with PNGase (Fig. 2b). The double mutant N365S/N392S also had a reduced level of receptor expression. These results confirm that amino acid residues Asn-365 and Asn-392 are in the extracellular domain of the protein and are glycosylated. In contrast, the mutants N22S, N478S, and N487S did not show any change in mass compared with WT ASIC2a, consistent with an intracellular localization for these amino acids (Fig. 2c).
The effects of glycosylation on the properties of ASIC2a were determined in electrophysiological studies. For WT ASIC2a expressed in Xenopus oocytes, acidification of the ND96 buffer resulted in an inward current that was dependent on the concentration of protons and had a pH50 of 3.58 ± 0.07 (n = 3) (Fig. 3, a and b). The individual mutants N365S and N392S that removed natural glycosylation sites on the protein had no effect on the proton sensitivity of the channel (pH50 of 3.50 ± 0.25 (n = 4) and pH50 of 3.96 ± 0.25 (n = 4), respectively). However, although mutant N365S had no effect on current amplitude relative to control WT ASIC2a, mutant N392S had reduced amplitude responses (49.5% ± 12, n = 6, p < 0.05). In addition, removal of both glycosylation sites (N365S/N392S) altered pH sensitivity (pH50 of 2.90 ± 0.14, n = 7, p < 0.01) and resulted in a significant reduction in the peak current amplitude (70.1% ± 5, n = 5, p < 0.01), consistent with a reduction in protein expression. An accurate determination of the pH sensitivity curve was not possible, as the current did not saturate at pH 2, and treatment with pH 1 resulted in cell death.
Fig. 3. Effect of removing the glycosylation site on pH sensitivity.
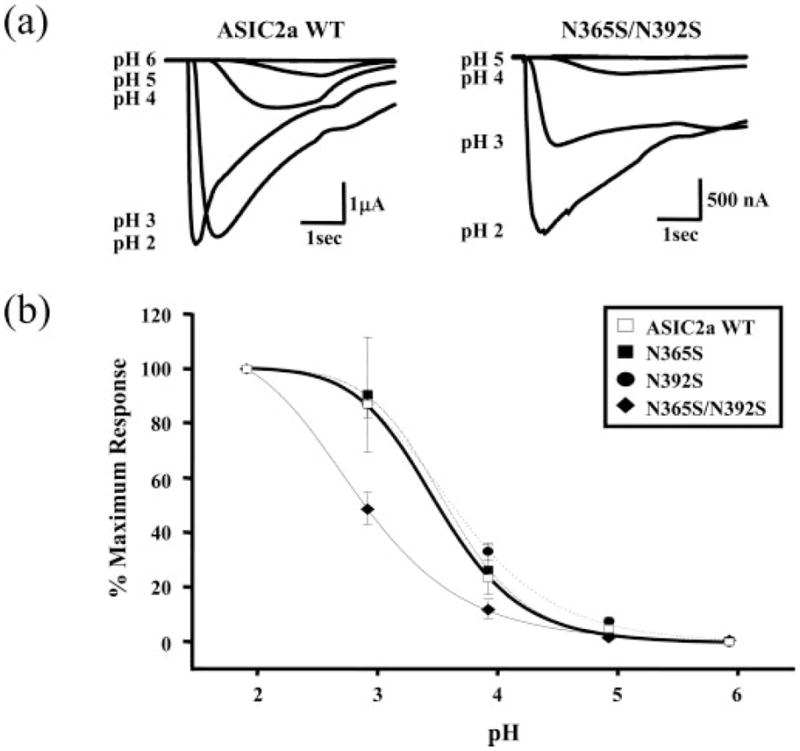
a, pH has a decreased effect at double mutant N365S/N392S. Wild type ASIC2a and double mutant N365S/N392S receptors were expressed in Xenopus oocytes, and two-electrode voltage clamp analysis (−60mV) was carried out by comparing current with decreasing pH. Trace data shows that an increasing level of acidity is required for double mutant activation as compared with wild type ASIC2a. b, only the double mutant N365S/N392S has altered affinity for increased acidity. Predicted glycosylation site mutants N365S (closed box, dash-dot line) and N392S (closed circle, dotted line) are similar to wild type ASIC2a (open box, solid bold line), exhibiting increased inward currents with increasing pH. N365S/N392S (closed diamond, solid line) shows a significant difference (pH50, 2.90 ± 0.14) compared with wild type (pH50, 3.58 ± 0.07).
Introduction of Potential N-Linked Glycosylation Sites
To investigate further the membrane topology of ASIC2a, we generated a series of mutant channels to introduce consensus sequences for glycosylation. We generated mutants first to investigate the regions that are predicted to be in the intracellular domains (K4N and P37N of the amino terminus compared with N478S and N487S of the carboxyl terminus) and second, to define the regions post-TM1 (R63N, Y67N, H72N, and A81N) and pre-TM2 (Y414N and Y423N/V425S) that can be glycosylated (Fig. 4). The mutants K4N and P37N did not result in any change in the molecular mass of the ASIC2a protein, consistent with the effect of mutating N478S and N487S and supporting the prediction that these amino acids are located intracellularly (Fig. 4a). The post-TM1 mutants H72N and A81N showed an increase in molecular mass compared with WT channels, demonstrating that these residues are glycosylated, whereas R63N and Y67N had no effect on the molecular mass of the ASIC channel (Fig. 4b). Similarly, the pre-TM2 mutants Y414N and Y423N/V425S both resulted in a doublet of bands corresponding to the WT channel and an additional extra-glycosylated form, whereas the post-TM2 mutant Y453N/Y455S did not result in any change in molecular mass of the ASIC2a protein (Fig. 4c). These results help to define the boundaries of the extracellular domain of the channel, as depicted in Fig. 6.
Fig. 4. Immunoblot analysis of exogenously introduced N-linked glycosylation sites.
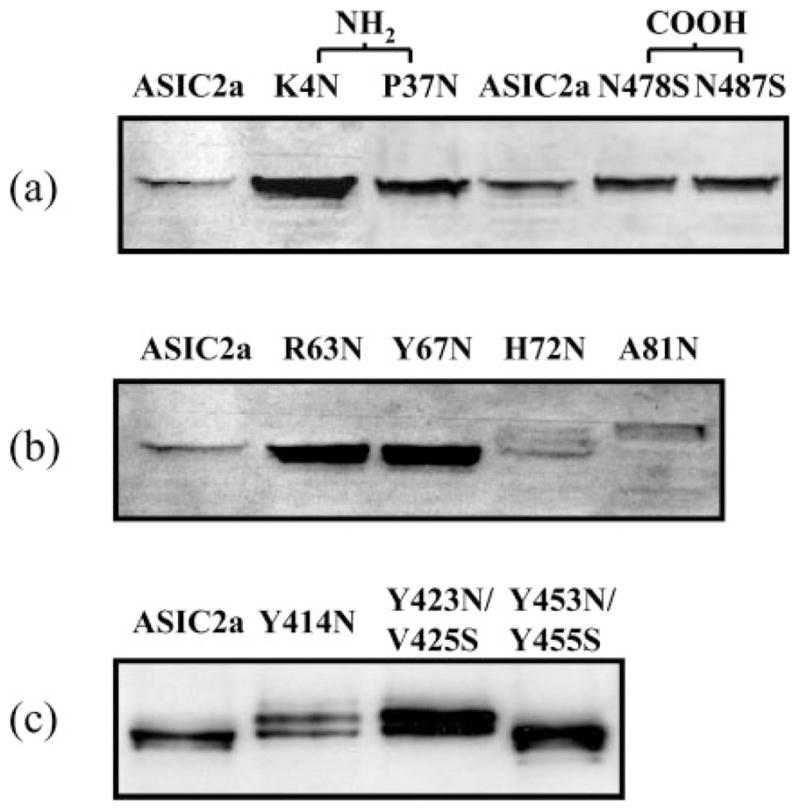
a, amino terminus Lys-4 and Pro-37 are not glycosylated. Carboxyl terminus Asn-478 and Asn-487 are not glycosylated. Also, Tyr-453 is not glycosylated (see panel c). b, extracellular loop-predicted start region is localized around residue Ala-81; the introduced NQS at Ala-81 is glycosylated. c, the extracellular loop end region is localized around residue Tyr-423; the introduced NES at Tyr-423 is glycosylated.
Fig. 6. Summary of glycosylation studies.
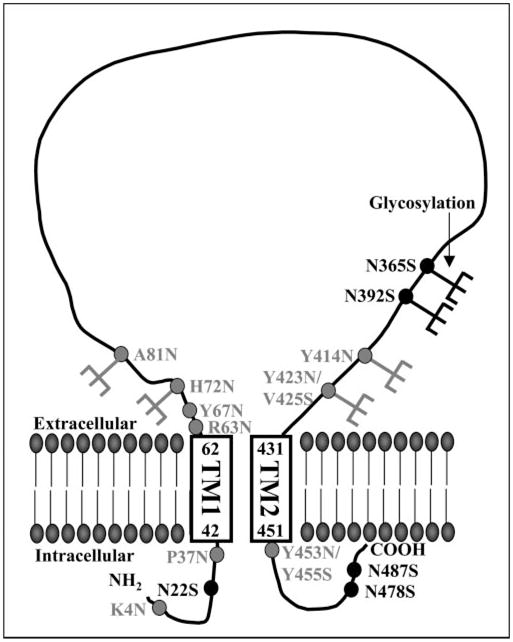
The model depicts the membrane topology of ASIC2a based on glycosylation and antibody permeabilization studies. The ASIC2a consists of two TM domains (TM1 and TM2, with amino acid numbers corresponding to the boundaries of the TM domains), intracellular amino and carboxyl termini, and a large extracellular loop. Consensus sites for N-linked glycosylation are shown in black, mutants to introduce consensus N-linked glycosylation sites are shown in gray, and glycosylated residues (H72N, A81N, N365, N392, Y414N, and Y423N/V425S) are shown with the addition of a sugar group.
To gain additional independent evidence for the intracellular location of the amino-terminal domain, we performed permeabilization studies on ASIC2a-transfected cells. The epitope of the ASIC2a antibody (Alomone) is directed against amino acids 1–20 of the intracellular amino terminus; thus, we compared ASIC2a expression in permeabilized versus non-permeabilized cells. The data show that ASIC2a expression is detected in cells permeabilized with 3% Triton X-100 prior to staining, whereas there is no ASIC2a expression in non-permeabilized cells (Fig. 5). These data provide additional support for an intracellular localization of the amino-terminal domain.
Fig. 5. Permeabilization studies support an intracellular amino terminus.
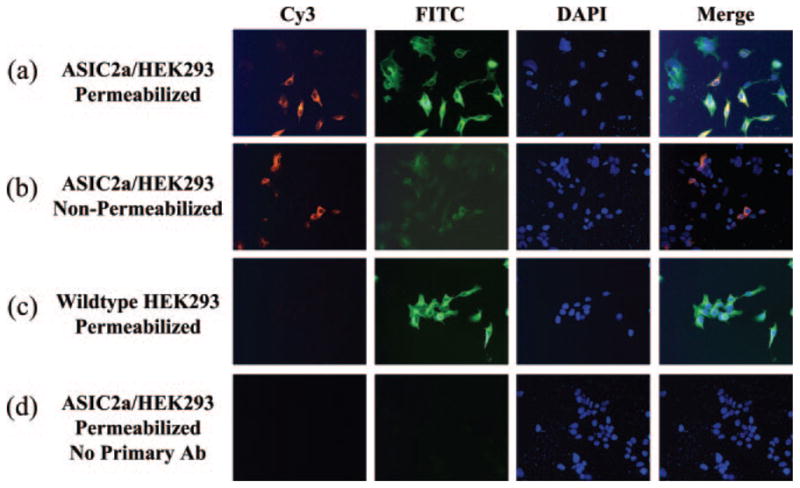
The anti-ASIC2a epitope is directed against amino acids 1–20. a, ASIC2a-expressing HEK293 cells permeabilized with 3% Triton X-100 are positive for ASIC2a and tubulin. b, ASIC2a-expressing HEK293 cells that are not permeabilized with Triton X-100 show a low level of ASIC2a staining that overlaps with tubulin; thus, these cells are compromised and are permeable to the antibodies. c, wild type HEK293 cells permeabilized with 3% Triton X-100 show no ASIC2a staining but are positive for tubulin. d, ASIC2a-expressing HEK293 cells permeabilized with 3% Triton X-100 but not exposed to the ASIC2a or tubulin antibodies show no ASIC2a or tubulin staining. FITC, fluorescein isothiocyanate; DAPI, 4′,6-diamidino-2-phenylindole; Ab, antibody.
DISCUSSION
The ASIC2a receptor protein is glycosylated at two asparagines residues (Asn-365 and Asn-392). These sugar modifications of the channel were removed by treatment with Endo H that cleaves high mannose oligosaccharides added in the ER from glycoproteins (28). Glycosylation can also occur in the Golgi, and this modification is sensitive to PNGase that removes N-glycans added in both the ER and the Golgi. Compared with Endo H treatment, the lack of an additional effect of PNGase on the molecular mass of ASIC2a shows that glycosylation is complete upon exit from the ER before transit to the Golgi. Removal of the Asn-365 glycosylation site had no measurable effect on the pH sensitivity or expression of the ASIC2a channels, whereas removal of Asn-392 resulted in ~50% reduction in expression relative to WT ASIC2a. Removal of both glycosylation sites resulted in a marked reduction in channel expression and pH sensitivity. These results suggest that the ASIC2a site requires glycosylation at either Asn-365 or Asn-392 for normal channel expression and trafficking. The change in decreased sensitivity to pH upon the removal of both glycosylation sites also indicates that glycosylation contributes to binding or gating of the ASIC2a receptor channel. This is in contrast to studies on ENaC channels (18) that have six glycosylation sites, and these sites can be removed with no obvious effect on channel function.
Examination of the exogenously added N-linked glycosylation sites (H72N, A81N, Y414N, and Y423N/V425S) demonstrated that these residues are on the extracellular face of the receptor (Fig. 4), as expected from the predicted membrane topology according to hydrophobicity analysis and homology with ENaC channels (3). The predicted end of TM1 is at residue 62. It is therefore perhaps not surprising that the introduced glycosylation site mutants R63N and Y67N had no effect on the molecular mass of the ASIC channel, as they may be within the transmembrane domain or because folding at this region of the protein renders these residues inaccessible at the extracellular face of the protein. Renard et al. (28) suggested that for the ENaC channel there may be an H5-like P-loop just before TM2 that contributes to the pore of the channel (28). Our studies showed the mutant Y423N/V425S acquires additional glycosylation, and this site is close to the predicted start of the second transmembrane region (residue 431). This finding suggests that for ASIC2a channels a P-loop that dives into the membrane and forms part of the channel pore is unlikely. These studies of the glycosylation of introduced asparagine residues indicate that the extracellular domain contains at least the residues between His-72 and Tyr-423 as depicted in Fig. 6 and support a membrane topology for ASIC2a as depicted in Fig. 1a.
The lack of effect on molecular mass following mutation to remove the consensus sites for glycosylation at positions Asn-22, Asn-478, and Asn-487 support an intracellular localization for these amino acids. These data also support the configuration depicted in Fig. 1a and are not consistent with alternative membrane topology predictions. The intracellular location of the amino terminus is supported by studies on chimeric ASIC2/2b constructs that identified a nine-amino acid region preceding the predicted first transmembrane domain of ASIC2 that is involved in ion permeation and is critical for Na+ selectivity (29). In particular, three amino acids in this region (Ile-19, Phe-20, and Thr-25) appear important, as channels mutated at these residues discriminate poorly between Na+ and K+. Our results confirm that this region is intracellular, and the immunocytochemistry studies showing that ASIC2a staining requires cell permeabilization are consistent with an intracellular amino terminus. In summary, although there are highly conserved regions within the different cation channel families, to depict the membrane topology of the ASIC family based on homology with ENaCs or hydrophobicity analysis alone may prove erroneous. Thus, we used a combination of glycosylation and antibody permeabilization studies on the ASIC2a subtype to confirm that the membrane topology of the ASIC2a subtype consists of two transmembrane domains, intracellular amino and carboxyl termini, and a large extracellular loop.
Footnotes
The abbreviations used are: ASIC, acid-sensing ion channel; DEG, degenerin; ENaC, epithelial Na+ channel; ER, endoplasmic reticulum; PBS, phosphate-buffered saline; pH50, acidity evoking 50% of the maximum response; PNGase, peptide:N-glycosidase; TBST, Tris-buffered saline with Tween 20; TM, transmembrane; WT, wild type.
This work was supported by the Legacy Research Foundation (to J. A. S.), the Oregon Medical Research Foundation (to J. A. S.), and the Wellcome Trust (to R. J. E.).
References
- 1.Krishtal O. Trends Neurosci. 2003;26:477–483. doi: 10.1016/S0166-2236(03)00210-8. [DOI] [PubMed] [Google Scholar]
- 2.Waldmann R, Champigny G, Lingueglia E, De Weille JR, Heurteaux C, Lazdunski M. Ann N Y Acad Sci. 1999;868:67–76. doi: 10.1111/j.1749-6632.1999.tb11274.x. [DOI] [PubMed] [Google Scholar]
- 3.Kellenberger S, Schild L. Physiol Rev. 2002;82:735–767. doi: 10.1152/physrev.00007.2002. [DOI] [PubMed] [Google Scholar]
- 4.Krishtal OA, Pidoplichko VI. Neurosci Lett. 1981;24:243–246. doi: 10.1016/0304-3940(81)90164-6. [DOI] [PubMed] [Google Scholar]
- 5.Krishtal OA, Pidoplichko VI. Brain Res. 1981;214:150–154. doi: 10.1016/0006-8993(81)90446-7. [DOI] [PubMed] [Google Scholar]
- 6.Bevan S, Yeats J. J Physiol. 1991;433:145–161. doi: 10.1113/jphysiol.1991.sp018419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sutherland SP, Benson CJ, Adelman JP, McCleskey EW. Proc Natl Acad Sci U S A. 2001;98:711–716. doi: 10.1073/pnas.011404498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chesler M, Kaila K. Trends Neurosci. 1992;15:396–402. doi: 10.1016/0166-2236(92)90191-a. [DOI] [PubMed] [Google Scholar]
- 9.Wemmie JA, Chen J, Askwith CC, Hruska-Hageman AM, Price MP, Nolan BC, Yoder PG, Lamani E, Hoshi T, Freeman JH, Jr, Welsh MJ. Neuron. 2002;34:463–477. doi: 10.1016/s0896-6273(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 10.Xiong ZG, Zhu XM, Chu XP, Minami M, Hey J, Wei WL, Mac-Donald JF, Wemmie JA, Price MP, Welsh MJ, Simon RP. Cell. 2004;118:687–698. doi: 10.1016/j.cell.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 11.Welsh MJ, Price MP, Xie J. J Biol Chem. 2002;277:2369–2372. doi: 10.1074/jbc.R100060200. [DOI] [PubMed] [Google Scholar]
- 12.Drew LJ, Rohrer DK, Price MP, Blaver KE, Cockayne DA, Cesare P, Wood JN. J Physiol. 2004;556:691–710. doi: 10.1113/jphysiol.2003.058693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roza C, Puel JL, Kress M, Baron A, Diochot S, Lazdunski M, Waldmann R. J Physiol. 2004;558:659–669. doi: 10.1113/jphysiol.2004.066001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ugawa S, Yamamoto T, Ueda T, Ishida Y, Inagaki A, Nishigaki M, Shimada S. J Neurosci. 2003;23:3616–3622. doi: 10.1523/JNEUROSCI.23-09-03616.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ettaiche M, Guy N, Hofman P, Lazdunski M, Waldmann R. J Neurosci. 2004;24:1005–1012. doi: 10.1523/JNEUROSCI.4698-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darboux I, Lingueglia E, Champigny G, Coscoy S, Barbry P, Lazdunski M. J Biol Chem. 1998;273:9424–9429. doi: 10.1074/jbc.273.16.9424. [DOI] [PubMed] [Google Scholar]
- 17.Hong K, Mano I, Driscoll M. J Neurosci. 2000;20:2575–2588. doi: 10.1523/JNEUROSCI.20-07-02575.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snyder PM, McDonald FJ, Stokes JB, Welsh MJ. J Biol Chem. 1994;269:24379–24383. [PubMed] [Google Scholar]
- 19.Rost B, Fariselli P, Casadio R. Protein Sci. 1996;5:1704–1718. doi: 10.1002/pro.5560050824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rost B, Yachdav G, Liu J. Nucleic Acids Res. 2004;32:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Claros MG, von Heijne G. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- 22.von Heijne G. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 23.Arai M, Mitsuke H, Ikeda M, Xia JX, Kikuchi T, Satake M, Shimizu T. Nucleic Acids Res. 2004;32:W390–W393. doi: 10.1093/nar/gkh380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikeda M, Arai M, Lao DM, Shimizu T. In Silico Biol. 2002;2:19–33. [PubMed] [Google Scholar]
- 25.Ikeda M, Arai M, Okuno T, Shimizu T. Nucleic Acids Res. 2003;31:406–409. doi: 10.1093/nar/gkg020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tusnady GE, Simon I. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 27.Tusnady GE, Simon I. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 28.Renard S, Lingueglia E, Voilley N, Lazdunski M, Barbry P. J Biol Chem. 1994;269:12981–12986. [PubMed] [Google Scholar]
- 29.Coscoy S, de Weille JR, Lingueglia E, Lazdunski M. J Biol Chem. 1999;274:10129–10132. doi: 10.1074/jbc.274.15.10129. [DOI] [PubMed] [Google Scholar]


