Abstract
Plumbagin (5-hydroxy-2-methyl-1,4 naphthoquinone) is a naturally occurring low molecular weight lipophilic phytochemical derived from roots of plants of the Plumbago genus. Plumbagin has been reported to have several clinically relevant biological activities in non-neural cells including antiatherosclerotic, anticoagulant, anticarcinogenic, antitumor and bactericidal effects. In a recent screen of a panel of botanical pesticides we identified plumbagin as having neuroprotective activity. In the present study we determined if plumbagin could modify the developmental fate of rat E14.5 embryonic neural progenitor cells (NPC). Plumbagin exhibited no cytotoxicity when applied to cultured NPC at concentrations below 1 µM. At a concentration of 0.1 µM, plumbagin significantly enhanced the proliferation of NPC as indicated by a 17% increase in the percentage of cells incorporating bromo-deoxyuridine. plumbagin at a concentration of 0.1 pM (but not 0.1 µM), stimulated the production of astrocytes as indicated by increased GFAP expression. Plumbagin selectively induced the proliferation and differentiation of glial progenitor cells without affecting the proliferation or differentiation of neuron-restricted progenitors. Plumbagin (0.1 pM) rapidly activated the transcription factor Stat3 in NPC, and a Stat3 inhibitor peptide prevented both plumbagin-induced astrocyte formation and proliferation. These findings demonstrate the ability of a low molecular weight naturally occurring phytochemical to control the fate of glial progenitor cells by a mechanism involving the Stat3 signaling pathway.
Keywords: Neural progenitors, neurogenesis, Stat3, GFAP, CNTF
Introduction
Phytochemicals, especially those containing electrophilic structures, have recently received considerable attention because of their ability to induce a battery of cell survival genes including those involved in detoxification, and resistance to oxidative and metabolic stress (Mattson and Cheng, 2006). One adaptive stress response pathway involves the transcription factor Nrf-2 (nuclear factor E2-related factor 2) pathway which binds to the antioxidant response element (ARE), a DNA sequence located upstream of several different genes that encode antioxidant and phase-2 enzymes; examples include NAD(P)H quinone oxidoreductase-1 (NQO1), glutamate cystine ligase, glutathione peroxidase, thioredoxin, thioredoxin reductase, peroxiredoxin, heme oxygenase-1, and glutathione S-transferase (Dinkova-Kostova et al. 2005; Mattson and Cheng, 2006; Satoh and Lipton 2007; Calabrese et al., 2008; Johnson, 2008). Plumbagin (5-hydroxy-2-methyl-1,4 naphthoquinone) is a naturally occurring product present in relatively high amounts in the roots of plants in the Plumbago genus (Van der Vijver, 1972). It has been shown to have cardiotonic, hypolipidemic, antiatherosclerotic, anticoagulant, anticarcinogenic, antitumor, antimutagenic, wound healing, anti-fertility, antimalarial antifungal and antibacterial properties (Bhargava, 1984; Igoigawa et al., 1991; de Paiva et al., 2003; Hsu et al., 2006). These different biological actions of plumbagin may result from its ability to alter cellular redox state in different cell types. In human cancer cell lines, plumbagin induces reactive oxygen species (ROS) resulting in apoptosis and cell cycle arrest (Srinivas et al., 2004; Wang et al., 2008). In a recent screen of botanical pesticides, plumbagin was shown to activate the Nrf2 – ARE pathway and promote neuronal survival (Son et al., 2010).
In addition to affecting cell survival, some electrophilic phytochemicals also alter the fate of stem cells. One example is curcumin, a yellow pigment in the spice turmeric, which affect several signaling pathways known to regulate stem cell proliferation and differentiation including the PI3 kinase – Akt pathway, MAP kinases and Stat3 (signal transducer and activator of transcription 3) (Aggarwal and Sung, 2009). Stat3 signaling maintains mouse embryonic stem cells (ESC) in a self-renewing state (Okita and Yamanaka, 2006) and promotes gliogenesis in the mouse cortex (Yoshimatsu et al., 2006). Recently, curcumin was reported to stimulate the proliferation of embryonic neural progenitor cells and neurogenesis in the adult hippocampus (Kim et al., 2008).
Rat spinal cord neural tube provides a useful model for studying neural development. At E10.5 rat, the neural tube consists of a homogenous population of neuroepithelial cells (NEPs), which are neural stem cells capable of self-renewal, and these cells can generate differentiated cells by first producing intermediate precursor cells (Kalyani et al., 1997; Mayer-Proschel et al., 1997). Neuron-restricted precursors (NRPs) and glia-restricted precursors (GRPs) are two types of neural progenitor cells with limited developmental differentiation potential (Kalyani et al., 1998). At E14.5 rat, epithelial progenitor cells decrease to less than 10% of the neural tube population, while NRPs and GRPs comprise about 80% of the cells (Kalyani et al., 1997; Mujtaba et al., 1999; Cai et al., 2002). The remaining population consists of sparse postmitotic neurons and small numbers of endothelial cells and connective tissue elements (Cai et al., 2002). Thus, E14.5 is a useful stage at which NRP and GRP can be isolated and studied in culture (Kalyani et al., 1997). NRP and GRP have been identified and isolated using the cell surface markers PSA-NCAM and A2B5, respectively, and their distinct transcriptomes have been characterized previously (Luo et al., 2002). In the present study we evaluated the biological effects of plumbagin on NRP and GRP proliferation, differentiation and survival. We report that plumbagin selectively stimulates the proliferation and differentiation of GRP into astrocytes, without affecting the fate of NRP. The gliogenic action of plumbagin is mediated by the transcription factor Stat3.
Materials and Methods
Isolation and culture of neural progenitors, and experimental treatments
The procedures for preparation of rat E14.5 neural progenitor cells are described in detail previously (Luo et al. 2002). Briefly, experimentally naive, pregnant Sprague-Dawley rats at day 8 gestation were purchased from Harlan and housed individually in standard cages in the National Institute on Aging (NIA) Intramural Research Program facility. All procedures were approved by the NIA Animal Care and Use Committee. On gestational day E14.5 the pregnant rats were euthanized by anesthesia (isoflurane) overdose, and embryos were removed and caudal neural tubes were dissected in fresh cold PBS. To obtain neural progenitors, the acutely dissected spinal cords were incubated with 0.05% trypsin-EDTA (Invitrogen) for 10 min at 37 °C and then gently triturated with a Pasteur pipette in NEP-basal medium (DMEM/F12 medium with 1x N2 and B27 supplements, Invitrogen) with bFGF (20 ng/ml, Peprotech). To isolate GRP- or NRP- enriched cells from this mixture of neuronal progenitor cells, magnetic activated cell sorting (MACS) was performed using anti-A2B5 or anti-PSA-NCAM microbeads (Miltenyi Biotec Inc.). Briefly, freshly dissociated E14.5 progenitor cells (107) were labeled with either anti-A2B5 or anti-PSA-NCAM microbeads in a buffer containing PBS (pH 7.2), 0.5% bovine serum albumin and 2 mM EDTA for 15 min at 4 °C. The labeled cells were washed 3 times with 1 ml of buffer and then separated using a MACS Separator. Cells were plated in culture dishes that had been coated sequentially with poly-L-lysine (15 µg/mL, Sigma) and laminin (15 µg/mL, Invitrogen). The culture medium was consisted of NEP basal medium containing bFGF (20 ng/ml) and the cultures were maintained at 37°C in a 5% CO2/95% room air atmosphere. Experiments were performed on culture day 4. The cell purities in cultures of sorted cells were 75 ± 4% (n=4) and 80 ± 5 % (n=4) for A2B5+ and PSA-NCAM+ cells, respectively (determined by immunostaining).
For differentiation and treatment experiments, the cells were cultured triplicates in NEP basal medium containing a very low level of bFGF (1 ng/ml) in 96 well plates. For positive controls, astrocyte differentiation was stimulated using either 10% fetal calf serum (FCS) or 4 nM CNTF, and neuron differentiation was promoted by treating the cells with BDNF 10 ng/ml, NT-3 10 ng/ml and retinoic acid 10 µM. The chemicals and above inducers were added freshly every two days. After four days (for neuronal differentiation conditions) or six days (for glial differentiation conditions) the cells were used to perform RT-PCR, immunoblot or immunostaining analyses. For Stat3 peptide inhibitor treatment, the cells were preincubated with the inhibitor for 1 h and then exposed to 0.1 pM plumbagin, 0.4 nM CNTF or vehicle for 4 days. Plumbagin was purchased from Sigma and further purified by high-performance liquid chromatography to greater than 99.5% purity. Plumbagin was prepared as a stock solution in dimethylsulfoxide at a concentration of 10 mM. MTT, (3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyl tetrasodium bromide) was purchased from Chemicon International.
MTT assay
The MTT assay was performed using the manufacture’s (Chemicon International) protocol with slight modifications. Cells (105) were plated into flat bottom 96-well plates that had been precoated with poly-L-lysine and laminin, and were maintained in 100 µl of NEP basal medium containing bFGF (20 ng/ml) for 2 days. The medium was then replaced with NEP basal medium containing 1 ng/ml bFGF with or without additional treatments, and the cultures were incubated an additional 24 h. The cell cultures were then exposed to MTT (final concentration 0.5 mg/ml) for 4 h at 37°C in an atmosphere of 5%CO2/95% room air. The MTT formazan product was dissolved in 100 µl of isopropanol containing 0.04 N HCL and the absorbance of 570 nm light was measured.
BrdU(bromodeoxyuridine) incorporation assay
Cells at ~60% confluence were treated with experimental chemicals or vehicle (control) for 24 h, and were then pulsed with 10 µM BrdU in NEP basal medium containing 1 ng/ml bFGF for designated time periods. For Stat3 peptide inhibitor treatment, the cells were preincubated with the inhibitor for 1 h, and then exposed to 0.1 µM plumbagin or vehicle for 24 h followed by BrdU labeling. The cells were fixed in a solution of 2% paraformaldehyde in PBS for 30 min and the DNA was denatured by incubating in a solution of 2N HCl for 10 min. The cells were then rinsed with PBS and immunostaining with BrdU antibody was performed as described below. For quantification, three microscope fields of the cells in each well were randomly taken and both BrdU- and DAPI-positive cells were counted. The results show average of percentage of BrdU positive cells to total DAPI stained cells.
RNA preparation, and reverse transcriptase PCR amplification
Total RNA from the cell cultures was isolated using RNA STAT-60™ (Tel-Test, Inc). The cDNAs were synthesized using 1 µg total RNA in the presence of Superscript II and oligo(dT)12–18 (both from Invitrogen). The PCR was performed in a 20 µl reaction solution containing 2 µl 10x PCR buffer, 150 nmol MgCl2, 10 nmol dNTP, 20 pmol primer, 1 µl 10x diluted cDNA, and 1 U RedTag DNA polymerase (Sigma, St. Louis, MO). Primer sequences for GFAP and β-actin were described previously (Cai et al., 2002, Szabo et al., 2000). The PCR reaction cycle parameters were: 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and a final extension for 10 min at 72°C.
Immunoblot analysis
After treatment, the cells were washed with ice-cold PBS and lysed by adding 100 µl of ice-cold lysis buffer (25 mM Hepes, pH 7.5, 300 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.1% Triton X 100, 20 mM β-glycerophosphate, 1x ptrotease inhibitor cocktail (Sigma), 1x phosphatase inhibitor cocktail I and II (Sigma) and 0.5 mM DTT) followed by sonication for 10 seconds. The cellular extracts were then centrifuged for 5 minutes at 13,000 rpm to remove debris. The supernatant was collected, aliquoted and stored at −20°C. Protein concentration was determined using a BCA protein assay kit (Pierce). For immunoblot analysis, equal amounts of supernatant protein (80 µg/lane) were run on a 4 – 12% Bis-Tris NuPAGE® gradient gel and electrophoretically transferred to PVDF membrane (Invitrogen). The membrane blots were first blocked with 5% nonfat dry milk in PBST buffer (10 mM sodium phosphate, 500 mM NaCl and 0.01% Tween-20, pH 7.4) and then incubated overnight with different primary antibodies in PBST containing 1% BSA at 4 °C. The primary antibodies included: p-Stat3 (1:2000, Cell Signaling), GFAP (1:1000, Cell Signaling), β III tubulin (1:400, Sigma), Nrf2 (1:1000, Santa Cruz Biotechnology, Inc), HO-1 (1:1000, Santa Cruz Biotechnology, Inc) and β-actin (1:5000, Sigma.). Immunoreactivity was detected by sequential incubation with horseradish peroxidase-conjugated secondary antibody (1:10,000, Jackson Immuno Research) and SuperSignal® West Femto Maximum Sensitivity Substrate (Pierce) following image acquisition using ChemoFlure™ 8900 (Alpha Innotech Corp.). To evaluate protein loading in the gel, the bound antibodies were stripped off by incubation in BlotFresh™ Western Blot Stripping Reagent (Ver. II) (SignlGen® Laboratories) for 10 minutes at room temperature, and the same membranes were then immunoblotted with β-actin antibody. For quantification, optical density for each band was measured using ImageQuant 5.2 software. A relative intensity value for each immunoreactive band of interest was normalized to the intensity of the β-actin band in the same lane. Results were presented as fold change compared to the value of the vehicle-treated control group.
Immunocytochemistry and quantification of stained cells
Immunostaining procedures for cells were described previously (Mayer-Proschel et al. 1997). Briefly, the cells were fixed with 2% paraformaldehyde in PBS for 20 min. For nuclear p-Stat3 staining, the cells were exposed to ice-cold 100% methanol for 5 min and then incubated in the methanol for 10 min at −20°C. The cells were then incubated with blocking buffer (5% normal goat serum, 1% BSA and 0.1% Triton X-100 in PBS; pH 7.4) for 1 h. Cells were then incubated with a primary antibody in blocking buffer overnight at 4°C followed by incubation with Alexa Fluor® 568 anti-goat IgG (1:500, Invitrogen) at room temperature for 0.5 h. The cells were examined and images were acquired using a fluorescence microscope. The procedures for immunostaining with A2B5 and PSA-NCAM antibodies were the same as those described above except that the cell membranes were not permeabilixed with Triton X-100. The sources and dilutions of primary antibodies were as follows: A2B5 (1:500, Chemicon), PSA-NCAM (1:400, Chemicon), β III tubulin (1:1000, Sigma), MAP2 (1:1000, Sigma), GFAP (1:1000, Cell Signaling), p-Stat3 (1:100, Cell Signaling), and BrdU (1:500, Sigma). For quantification, images of three microscope fields of cells in each well were acquired and both immunostained cells and DAPI-positive cells were counted.
Statistics analysis
The data were analyzed by ANOVA using JUMP software. Pairwise comparisons were performed using either Dunnett’s or Tukey-Kramer HSD tests.
Results
Plumbagin stimulates neural progenitor proliferation
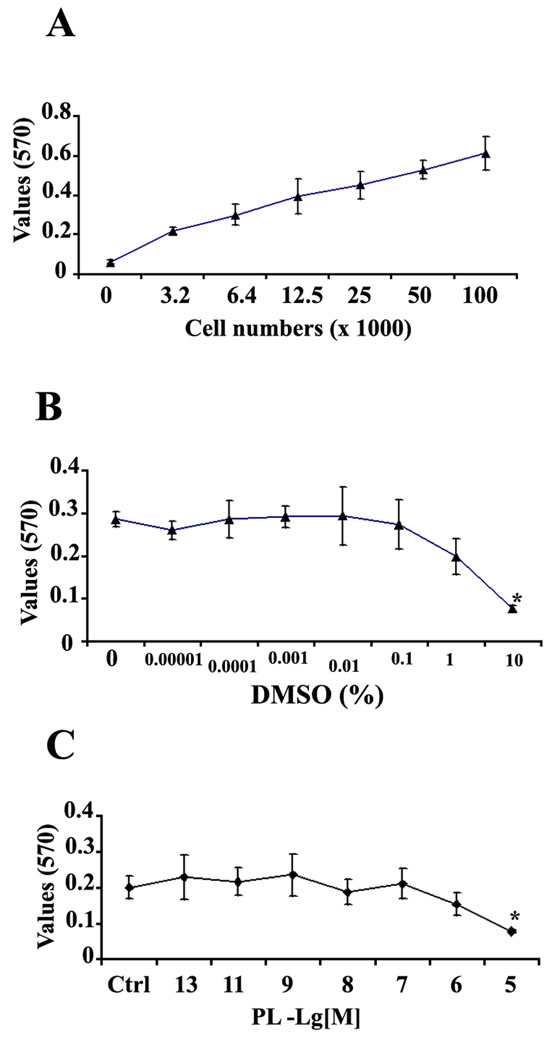
To investigate whether plumbagin can affect the developmental fate of rat E14.5 neural progenitor cells (NPC), we exposed NPC to increasing concentrations of plumbagin and then quantified cell viability using the MTT assay (Morgan 1998). We first established the validity of the MTT assay by evaluating MTT reduction levels in cultures with increasing numbers of cells. There was a linear relationship between the cell number and formation of formazan (Fig. 1A). Under our culture conditions the correlation coefficient for cell number versus MTT reduction was 0.9. We next tested cytotoxicity of dimethylsulfoxide (DMSO), the vehicle used to dissolve plumbagin; at concentrations from 0.0001% to 1%, DMSO did not show significant cellular toxicity during a 24 hour exposure period compared to untreated control cultures (Fig. 1B). Based on above result, we chose 0.1% DMSO as the final concentration of vehicle to which cells were exposed in both plumbagin-treated and control cultures. As indicated in Fig. 1C, treatment of the cells with plumbagin at concentrations from 0.1 pM to 1 µM did not cause significant cell death during a 24 hour exposure period relative to vehicle-treated control cultures. Plumbagin at a higher concentration (10 µM) did show cellular toxicity.
Figure 1. Concentrations of plumbagin below 1 micromolar are not toxic to cultured neural progenitor cells.
A. E14.5 rat neural progenitor cells (NPC) were plated at increasing cell densities and 2 days later the MTT cell viability assay was performed. There was a linear relationship between the cell numbers and MTT reaction product absorbance at 570 nm; Y (570 nm absorbance value) = 0.004 X (cell #) + 0.176 with R2 = 0.9. Values are the mean and SEM (n = 3). B. NPC were exposed to different concentrations of dimethylsulfoxide (DMSO) for 24 hr and cell viability was assessed using the MTT assay. Based on this cytotoxicity curve, a concentration at 0.1% DMSO was chosen as a vehicle for plumbagin (PL) in all subsequent experiments. C. The cells were treated with different concentrations of PL for 24 hrs. PL at 10 µM shows a significant decrease in MTT reduction compared to vehicle (0.1% DMSO) control group. * P < 0.05, comparison with a vehicle control in panel C or a 0% DMSO group in panel B using Dunnett’s Method.
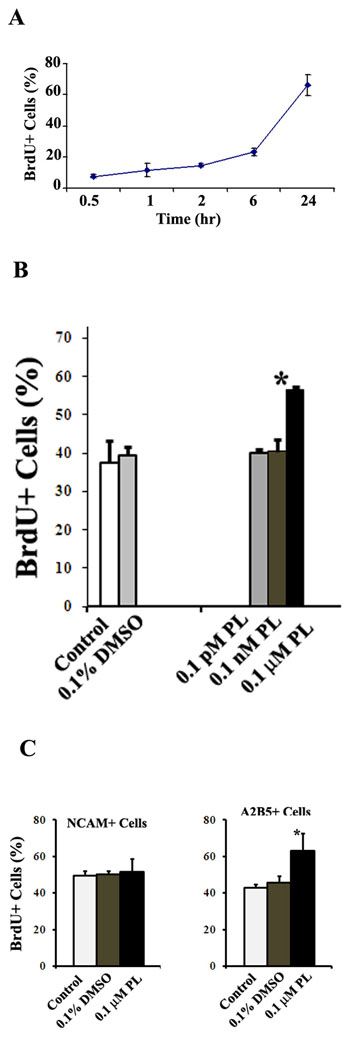
Based on the results from MTT cytotoxicity assay, we next determined whether non-toxic concentrations of plumbagin affected NPC proliferation using a BrdU incorporation cell proliferation assay (BrdU is thymidine analog that is incorporated into newly synthesized DNA strands of actively proliferating cells). We first determined the optimal time required for BrdU incorporation in the cultured NPC using pulse tests. As shown in Fig 2A, pulses of BrdU less than 6 hours gave a low incorporation (23.1 ± 2.5 %, n = 3). Between 6 and 24 hours after a pulse of BrdU, incorporation increased from 23.1% to 66.2%. We therefore chose 22 hours as the time period following a BrdU pulse at which BrdU incorporation into cells was quantified. Plumbagin at concentrations of 0.1 pM and 0.1 nM had no significant effect on BrdU incorporation (Fig. 2B). However, plumbagin at a concentration of 0.1 µM did significantly increase BrdU incorporation by 17.2 ± 0.8% (p < 0.05) compared to vehicle-treated cells. At this time point, the total cell numbers counted by DAPI staining was also increased 32.3 ± 11.1 % (p < 0.05) by 0.1 µM plumbagin compared to vehicle-treated cells. Thus, 0.1 µM plumbagin promoted progenitor proliferation. Since the NPC cultures include a mixture of NRP and GRP (Kalyani et al., 1997, 1998; Luo et al., 2002), we next examined which subtypes of the progenitors was affected by application of plumbagin. We used magnetic activated cell sorting (MACS) to enrich PSA-NCAM+ NRP and A2B5+ GRP. Rat E14.5 neural cells were freshly isolated and MACS was performed using anti-A2B5 and anti-PSA-NCAM microbeads as described in Method section. This technique enriched the A2B5+ GRP from an original level of ~30% to a post-MACS level of ~75%, and enriched the PSA-NCAM+ NRP from an original level of ~60% to a post-MACS level of ~85%. Using these enriched subtypes of progenitors, we performed BrdU assays in cultures exposed to 0.1 µM plumbagin or vehicle. plumbagin stimulated BrdU incorporation in A2B5+ GRP, but not in PSA-NCAM+ NRP (Fig. 2C). These results suggest that plumbagin can selectively stimulate the proliferation of GRP.
Figure 2. Neural progenitor cell proliferation is stimulated with a relatively high concentration of plumbagin.
A. NPC were plated and grown ~60% confluence. The cells were pulsed with 10 µM concentration of BrdU at the indicated time points. The data suggest that the amount of BrdU incorporation is greatest during the time window between 6 to 24 hours after a BrdU pulse. B. NPC were treated for 24 hours with different concentrations of PL, vehicle alone (0.1% DMSO) or no treatment (control), and were then pulsed 10 µM BrdU for 22 hours. NPC treated with 0.1 µM PL exhibited a significant increase in BrdU incorporation compared to vehicle-treated and untreated cells. C. NCAM+ and A2B5+ progenitors were enriched using MACS and then planted to perform BrdU assay as above. The results showed that only A2B5+ cells had an increasing BrdU incorporation in the presence of 0.1 µM PL. *p< 0.05 (n = 3 separate experiments; ANOVA with Dunnett’s post hoc test compared to 0.1% DMSO group).
Plumbagin induces gliogenesis from glial cell-restricted precursor cells
To determine the effects of plumbagin on differentiation of GRPs we evaluated the expression of the astrocyte marker GFAP. Mixed progenitor cell cultures were prepared and exposed to increasing concentrations of plumbagin (picomolar to 0.1 micromolar) in differentiation medium. Every two days the culture medium was replaced with fresh medium with or without plumbagin. After six days, the cells were harvested and levels of GFAP mRNA and protein were measured by RT-PCR and immunoblot analysis, respectively. As shown in Figure 3, plumbagin treatment affected GFAP expression in a concentration-dependent manner. At 100 nM, a concentration that stimulated BrdU incorporation (Fig. 2B), plumbagin had no significant effect on GFAP expression at the mRNA or protein levels (Fig. 3). Plumbagin at 0.1 nM, also had no significant effect on GFAP expression. In contrast, plumbagin at 0.1 pM significantly increased GFAP mRNA levels (>100%) measured using either quantitative PCR (at 35 cycles) or semi-quantitative PCR using 23 to 31 cycles (Fig. 3A). Moreover, plumbagin at 0.1 pM also significantly increased GFAP protein levels (50.2 ± 7.8 %) compared to vehicle control (Fig. 3B). Immunocytochemistry evaluation indicated that plumbagin at 0.1 pM significantly increased the production of GFAP-positive cells by 19 ± 3.3 % (Fig. 6D). This plumbagin-induced GFAP expression was specific since application of menadione, a chemical with a structure very similar to plumbagin (Supplemental Fig. 1A) did not increase GFAP protein levels (Fig. 3B).
Figure 3. Low dose plumbagin promotes astrocytic differentiation of NPC.
A. After 6 days of treatments of the cells with the indicated concentrations of PL, or vehicle, total RNA was isolated and used to perform RT-PCR analysis to quantify GFAP mRNA levels. Top panel shows a RT-PCR result performed in 35 cycles. The bottom is a semi-quantitative PCR result run within 31 cycles. PL at 0.1 pM increased levels of GFAP mRNA compared to vehicle-treated cells. NPC treated with 10% FCS served as a positive control. B. Levels of GFAP protein were also increased in response to treatment with 0.1 pM PL but not with menadione for 6 days. Shown is a representative immunoblot and quantification results from 4 separate experiments. *p <0.05 compared to vehicle-treated cells (ANOVA with Dunnett’s post hoc test).
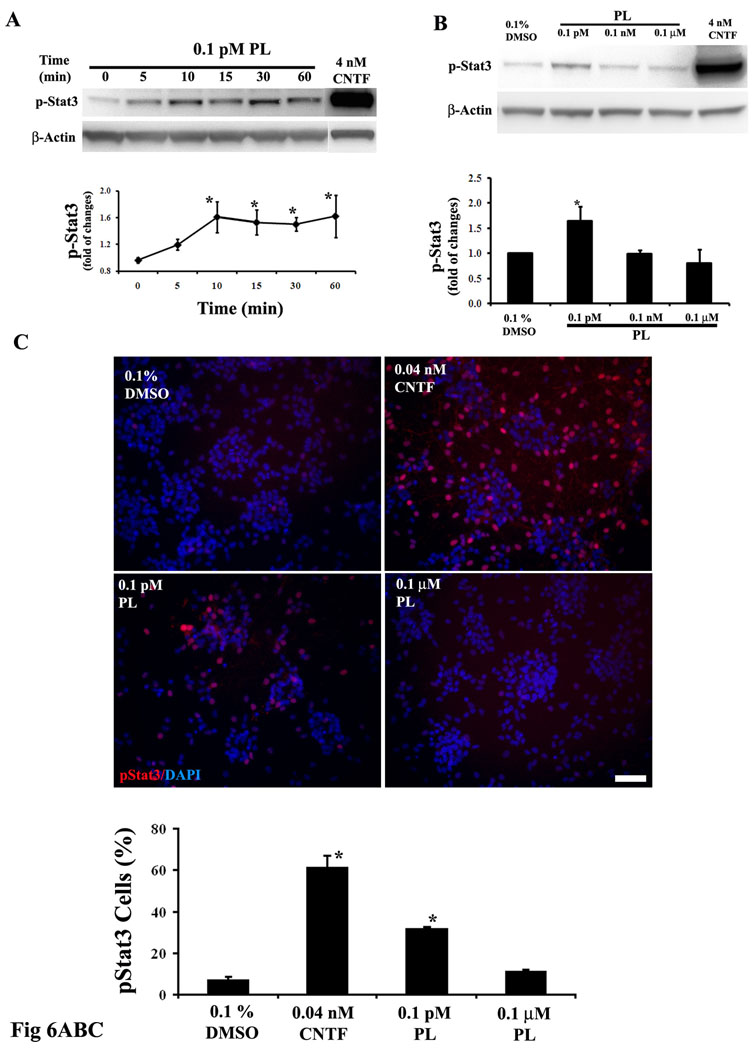
Figure 6. Activation of Stat3 is required for the proliferation and the astrocyte-inducing action of plumbagin in neural progenitor cells.
A. Enriched A2B5+ glial progenitors were stimulated with 0.1 pM PL for increasing time periods. Activation of Stat3 was examined by immunoblot analysis using an antibody that selectively recognizes Stat3 only when it is phosphorylated on Tyr705. PL at 0.1 pM significantly enhanced phosphorylation of Stat3 within 10 min of exposure and the elevated phosphorylation level was maintained for at least 60 min. Application of 4 nM CNTF, a positive control, resulted in a much stronger stimulation of phospho-Tyr705 Stat3 compared to that induced by PL. *p<0.05 compared to the 0 time point value. B. Enriched A2B5+ cells were stimulated treated for 15 min with PL at the indicated concentrations. Only the cells treated with 0.1 pM PL group exhibited a significant increase in phospho-Stat3. Quantification of data in both panel A and B were performed in four separate experiments. *p<0.05 compared to the vehicle value. C. Enriched A2B5+ cells were treated for 15 min with PL (0.1 pM) or CNTF (0.04 nM) and then used to perform phospho-Stat3 (Tyr705) immunocytochemistry. Quantification results (n = 3): values represent the percentage of total cells (DAPI-positive) that were also pStat3 immunoreactive. In agreement with the immunoblot analysis, both PL and CNTF enhanced nuclear phospho-Stat3 immunostaining. bar = 50 µm. *p<0.05 compared to the vehicle value. D. Rat E14.5 mixture of neural progenitors were pretreated for 1 hour with 5 to 50 µM Stat3 inhibitor peptide or its vehicle (water), and were then treated with the indicated agents for 4 days. Astrocyte generation was evaluated using GFAP immunostaining. Shown only are representative GFAP immunostaining images in the presence or absence (0) of 10 µM Stat3 inhibitor peptide. Quantitative data were present as percentages of GFAP immunostained astrocytes response to different concentration of Stat3 inhibitor peptides (n = 3). The Stat3 inhibitor peptide, even at 5 µM, prevented PL-induced astrocyte formation. CNTF (0.4 nM) was served as a positive control. E. Rat E14.5 mixture of neural progenitors were pretreated for 1 hour with 5 µM Stat3 inhibitor peptide or its vehicle (water), and were then treated with the indicated agents for 24 hours. The cells were pulsed 10 µM BrdU for 22 hours and used for performing BrdU immunocytochemistry. Application of 5 µM Stat3 inhibitor peptide prevented PL-induced BrdU incorporation. Stat3 inhibitor peptide at 5 µM did not appear to inhibitory effect on BrdU incorporation compared to vehicle control group. *p<0.05 in both panel D and E. (ANOVA with Dunnett’s post hoc test compared with 0.1% DMSO group, n=3).
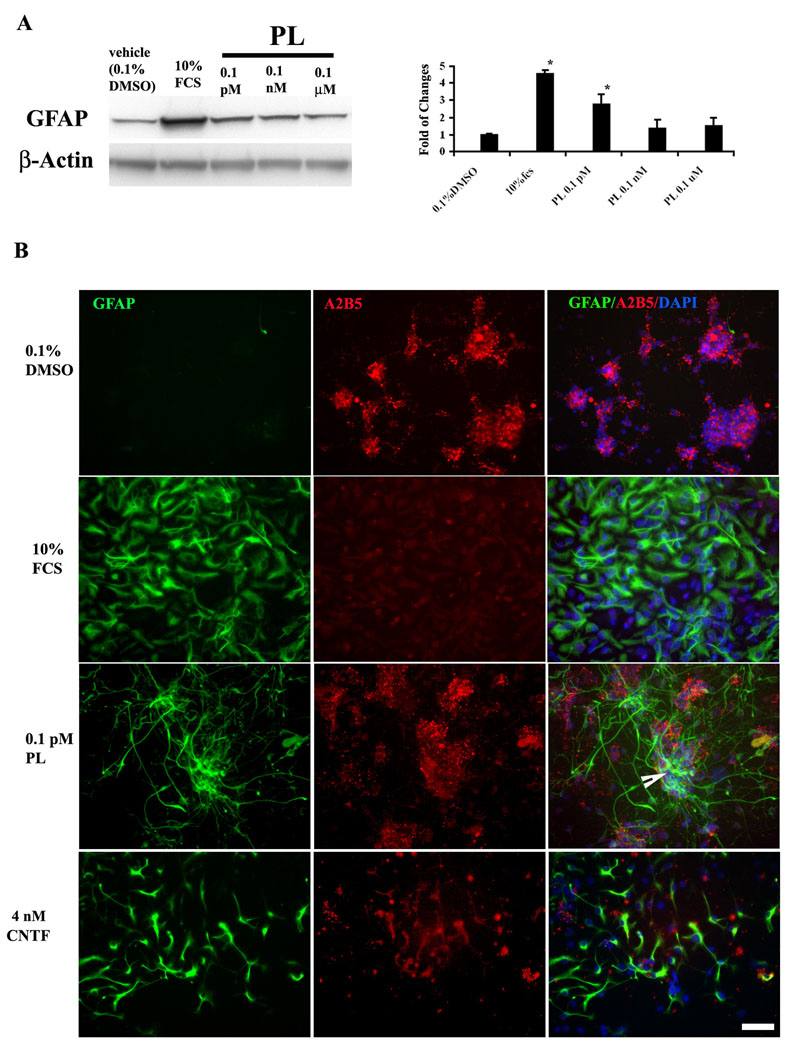
The relatively modest effect of 0.1 pM plumbagin on GFAP expression might be due to the fact that primary cultures of E14.5 neural tube cells contain about 60% NRP and 30% GRP (Cai et al. 2002). Astrocytes in the spinal cord neural tube are derived from GRP that can be identified by their expression of the surface marker A2B5 (Rao et al., 1998). We therefore used MACS to enrich A2B5+ glial progenitors. We then used GRP-enriched cultures to evaluate the ability of plumbagin to promote astrocyte differentiation using same paradigm as above. In agreement with the results in mixed cultures of neuronal and glial precursors, plumbagin at 0.1 µM and 0.1 nM did not stimulate GFAP expression (Fig. 4A). Plumbagin at 0.1 pM induced a significant (177 ± 22%) increase in GFAP expression (Fig. 4A). A greater increase in GFAP expression was also observed in response to exposure to 10% FCS in A2B5+-enriched cultures (Fig. 4A) compared to mixed cell cultures (Fig. 3B). These findings suggest that plumbagin at 0.1 pM selectively induces the production of astrocyte-like cells from GRP.
Figure 4. Plumbagin stimulates the formation of type 2 astrocytes.
A. Glial progenitor cells, enriched using anti-A2B5 microbeads, were treated with different concentrations of PL for 6 days as described in the Methods. PL at 0.1 pM significantly increased levels of the glial marker protein GFAP. Shown is a representative immunoblot image and quantification results for three separate experiments. *p <0.05 compared to the value for vehicle-treated cultures (ANOVA with Dunnett’s posthoc test). B. The A2B5-enriched glial progenitors were treated with the indicated agents for 6 days and were then immunostained with antibodies against A2B5 and GFAP. Type 1 astrocytes (induced by 10% FCS) were only immunoreactive with the GFAP antibody, and not with the A2B5 antibody. Type 2 astrocytes (induced by 4 nM CNTF) were immunostained by both GFAP and A2B5. The morphology of the cells treated with PL was similar to type 2 astrocytes with long extensions, and the cells also expressed both GFAP and A2B5 immunoreactivities (arrow). Scale bar = 50 µm.
We next examined and compared the phenotypes of cells differentiated from GRP in response to by plumbagin, 10% FCS, and ciliary neurotrophic factor (CNTF). GRP-enriched cultures were treated for 6 days with vehicle, 0.1 pM plumbagin, 10% FCS or 4 nM CNTF, and the cells were then fixed and immunostained with GFAP antibody. FCS induced type 1 astrocytes characterized by a flat polygonal morphology, robust GFAP immunoreactivity, and little or no A2B5 immunoreactivity (Fig. 4B). Three different subpopulations of cells were observed in CNTF-treated cultures: polygonal GFAP-positive presumptive type 1 astrocytes; ramified GFAP-positive presumptive type 2 astroglial cells; and cells that expressed both GFAP and A2B5, which may be cells that remained glial progenitors or were in the process of differentiating into oligodendrocytes (Mujtaba et al. 1999). In cultures treated with 0.1 pM plumbagin, many cells expressed GFAP with little or no A2B5 immunoreactivity and exhibited a ramified morphology with long processes (Fig. 4B). Some of the GFAP-positive cells also exhibited A2B5 immunoreactivity.
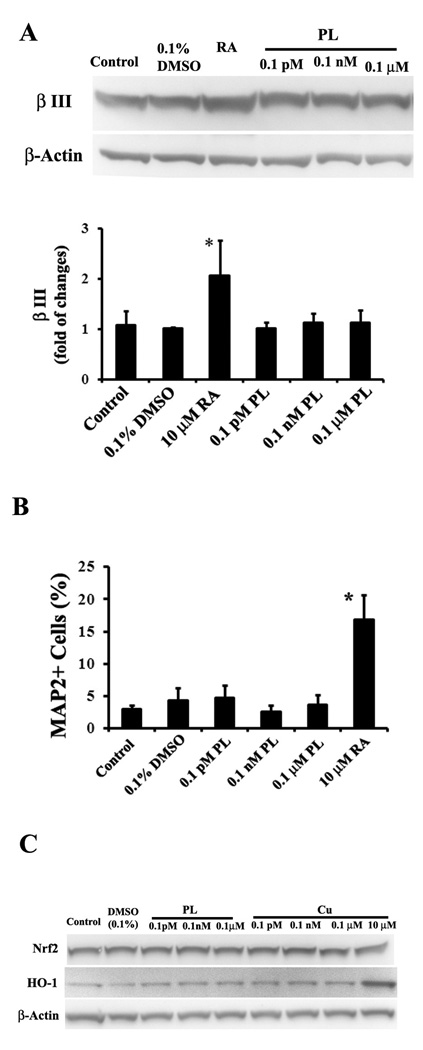
Plumbagin does not affect neurogenesis or Nrf2/HO-1 protein expression
Because the results above suggested that plumbagin might selectively enhance gliogenesis, we examined the effects of plumbagin on neurogenesis in rat E14.5 NPC cultures. The progenitors were stimulated with plumbagin for 4 days and then the expression of the neuronal marker βIII tubulin was first determined by immunoblot analysis. Plumbagin treatment had no discernable effect on the level of βIII tubulin (Fig. 5A). In contrast, when NPC were treated with a differentiation factors (10 µM retinoic acid, 10 ng/ml BDNF and 10 ng/ml NT-3) the levels of βIII tubulin protein were significantly greater than in vehicle-treated control cultures. Similarly, plumbagin did not affect βIII tubulin levels in PSA-NCAM+ (NRP)-enriched cultures (data not shown). To confirm these results, we evaluated expression of another neuronal marker MAP2. The E14.5 progenitors were induced for 4 days with designed chemicals. Plumbagin at concentrations ranging from 0.1 pM to 0.1 µM did not increase the number of MAP2-positive cells, whereas retinoic acid did increase the number of MAP2-postive cells (Fig. 5B). Collectively, the findings above suggest that very low doses of plumbagin selectively stimulate gliogenesis from GRP.
Figure 5. Plumbagin does not increase the expression of β III tubulin and MAP2 or HO-1 proteins.
A. E14.5 neural progenitors were exposed to the indicated treatments for 4 days and immunoblots were performed on cell lysates using a β III tubulin antibody. PL treatment did not affect β III tubulin levels in E14.5 embryonic NPC, whereas RA (10 µM retinoic acid plus 10 ng/ml BDNF and 10 ng/ml NT-3) did increase β III tubulin levels. Bar graph shows a quantitative result from four separated experiments. B. E14.5 progenitors were plated in 96 wells and treated with the indicated chemicals for 4 days and then used to perform immunocytochemistry using anti-MAP2 antibody, another neuronal marker. Shown are percentages of MAP2 immunostained cells from three experiments. Only positive control group RA (10 µM retinoic acid plus 10 ng/ml BDNF and 10 ng/ml NT-3) shows a significant increase in MAP2 positive neurons. C. To determine if PL could activate the Nrf2/HO-1 pathway, Rat E14.5 neural cells were stimulated for 6 hours by indicated reagents. Curcumin (Cu) was used as a positive control. PL from 0.1 pM to 0.1 µM had no detectable effect on Nrf2 or HO-1 protein levels. Cu at 10 µM, but not from 0.1 pM to 0.1 mM, did induce HO-1 expression. All data shown are representative of three separate experiments. *p <0.05 compared to the value for vehicle-treated cultures (ANOVA with Dunnett’s posthoc test).
Plumbagin is a naphthoquinone with a para-quinone structure and may therefore be capable of generating reactive oxygen species (ROS). We previously reported that 100 nM – 1 µM plumbagin can activate the ROS-responsive transcription factor Nrf-2 resulting in increased expression of the antioxidant enzyme heme oxygenase-1 (HO-1) in cultured human neuroblastoma cells (Son et al. 2010). To determine if concentrations of plumbagin that induce gliogenesis act via the Nrf-2 pathway we measured levels of Nrf-2 and HO-1 in primary rat E14.5 NPC that had been exposed for 6 hours to plumbagin at concentrations from 0.1 pM to 0.1 µM. Immunoblots of protein samples from the NPC indicated that plumbagin, at each concentration tested, had little or no effect on the levels of either Nrf-2 or HO-1 (Fig. 5C). In contrast, HO-1 levels were increased in response to curcumin, an agent previously reported to activate Nrf-2 (Balogun et al., 2003).
Activation of the transcription factor Stat3 is required for plumbagin-induced gliogenesis and BrdU incorporation
Activation of the transcription factor signal transducer and activator of transcription 3 (Stat3) plays an important role in astrocyte differentiation from NPC (Bonni et al. 1997; Nakashima and Taga 2002). Stat3 is activated by phosphorylation at Tyr705, which results in dimerization, nuclear translocation, and DNA binding (Darnell et al. 1994). In the promoter region of the GFAP gene there is a CpG dinucleotide binding site for Stat3; this binding site is methylated during the process of neuronal differentiation, and becomes demethylated during astrocyte differentiation (Takizawa et al. 2001). To determine whether Stat3 is involved in plumbagin-induced gliogenesis, we first examined the effects of plumbagin on phosphorylation at Try705 of Stat3 using a specific anti-phospho(Tyr705) Stat3 antibody. Enriched A2B5+ glial progenitors were stimulated with 0.1 pM plumbagin for increasing time periods. Levels of pTyr705 Stat3 were rapidly increased within 10 min of exposure to plumbagin and remained elevated through 60 min of exposure (Fig. 6A). However, the magnitude of the Stat3 response to plumbagin was small compared to the large increase in pTyr705 Stat3 levels in cells treated with 4 nM CNTF (Fig. 6A), a positive control for stimulation of Stat3 (Boulton et al. 1995). Using a time point of 15 min, we performed a plumbagin concentration – Stat3 response study and found that only the low concentration (0.1 pM) increased pTyr705 Stat3 levels (Fig. 6B). Treatment of enriched A2B5+ glial progenitors with 0.1 pM plumbagin also induced an increase in the nuclear localization and staining intensity of pTyr705 immunoreactive cells, albeit to a lesser extent than occurred in CNTF-treated cells (Fig. 6C). Thus, the data from both the immunoblot and the immunocytochemistry studies indicate that 0.1 pM plumbagin stimulates the activation and nuclear localization of Stat3 in NPC.
We next determined if activated Stat3 is a critical mediator of the gliogenesis-inducing action of plumbagin. To this end, we employed a cell-permeable Stat3 peptide inhibitor PY*LKTK-mts (mts, membrane translocating sequence). This Stat3 inhibitor is capable of binding the Stat3 SH2 domain, thus disrupting Stat3 dimerization, DNA binding and target gene regulation (Turkson et al. 2001). This Stat3 peptide inhibitor was previously reported to block gliogenesis induced by interleukin-6 and leukaemia inhibitory factor in cultured embryonic rat brain NPC (Nakanishi et al. 2007). We preincubated mixed neural progenitor cells with 5 to 50 µM of the Stat3 inhibitor peptide for 1 hour or its vehicle (water), and then treated the cells with plumbagin or CNTF for 4 days. Astrocyte generation was evaluated using GFAP immunostaining. Treatment with the Stat3 inhibitor peptide from 5 to 50 µM alone did not significantly affect the basal level of GFAP-expressing astrocytes (Fig. 6D). However, even at 5 µM the Stat3 inhibitor prevented plumbagin-induced astrocyte formation. The ability of CNTF (0.4 nM) to induce astrocyte formation was also blocked by the Stat3 inhibitor (Fig. 6D). The Stat3 also prevented plumbagin-induced gliogenesis in GRP-enriched cultures (data not shown). Together, these data suggest that activation of Stat3 mediates plumbagin-induced generation of astrocytes from GRP.
Finally, we determined whether Stat3 mediated plumbagin-induced proliferation of neural progenitors. To this end, rat E14.5 mixture of neural progenitors were pretreated for 1 hour with 5 µM Stat3 inhibitor peptide or its vehicle (water), and were then treated with 0.1 µM plumbagin or its vehicle (0.1% DMSO) for 24 hours. The cells were then pulsed 10 µM BrdU for 22 hours and BrdU incorporation was analyzed. As expected, plumbagin (0.1 µM) significantly increased BrdU incorporation by 16.6 ± 3.5 % (Fig. 6E). Pretreatment of the cells with 5 µM Stat3 inhibitor peptide prevented plumbagin-stimulated BrdU incorporation without affecting the basal level of BrdU incorporation (Fig. 6E).
Discussion
Several endogenous signals that regulate the fate of neural stem and progenitor cells have recently been identified, including bFGF which promotes self-renewal of NPC (Maric et al. 2003; Cheng et al. 2004), and BDNF which promotes neuronal differentiation (Pencea et al. 2001; Cheng et al. 2003). Yet other signals, including CNTF, bone morphogenic protein (BMP) and leukemia inhibitory factor (LIF) direct NPC to differentiate into astrocytes (Hughes et al. 1988; Bonaguidi et al. 2005). We found that the low molecular weight phytochemical plumbagin directs embryonic rat glial cell-restricted NPC to differentiate into astrocytes. In contrast, plumbagin did not affect the fate of neuron-restricted progenitor cells. Plumbagin induced a rapid activation of the transcription factor Stat3 in GRP, and treatment of GRP with a Stat3 inhibitor blocked plumbagin-induced astrocyte differentiation, demonstrating a pivotal role for Stat3 as a mediator of the differentiation-promoting effect of plumbagin on GRP. These findings suggest that plumbagin exerts its effects on GRP fate in a manner similar to that of CNTF, BMPs and LIF which bind to receptors coupled to Stat3 activation and astrocyte differentiation (Bonni et al. 1997; Rajan et al. 2003). It remains to be determined whether the activation of Stat3 in response to plumbagin is due to a direct action of plumbagin on Stat3 or an action upstream of Stat3.
The effects of plumbagin on GRP were biphasic with a very low concentration (0.1 pM) promoting astrocyte differentiation, intermediate concentrations having no detectable effects and a high concentration 100 nM enhancing GRP proliferation. Because 0.1 pM plumbagin is a very low concentration, similar to or lower than concentrations of endogenous molecules that regulate NPC fate (CNTF, BMP, BDNF and others), it is unlikely that the effect on GRP fate was the result of a non-specific action. Consistent with the notion that plumbagin stimulates a specific signaling pathway, we found that exposure of GRP to 0.1 pM resulted in the rapid activation of Stat3 and that a Stat3 inhibitor blocked the gliogenesis-promoting effect of plumbagin. Although we did not establish the mechanism underlying the different (proliferation-promoting) effect of the higher concentration (100 nM) of plumbagin on GRP, it could result from induction of mild cellular stress. Because of its quinone structure and redox-cycling properties, high concentrations of plumbagin can induce oxidative stress and toxicity in a range of organisms and cell types including nematodes and cultured mammalian cells (Farr et al. 1985; Darr and Fridovich 1995; Inbaraj and Chignell 2004; Kawiak et al. 2007). Preclinical studies have focused on the cytotoxicity of micromolar concentrations of plumbagin in various types of cultured cancer cells, with multiple mechanisms being proposed including inhibition of topoisomerases (Kawiak et al. 2007) and NF-κB (Sandur et al. 2006). We previously showed that low micromolar concentrations of plumbagin activate the Nrf2-antioxidant response element pathway resulting in increased expression of the antioxidant proteins HO-1 and NQO1 in neuroblastoma cells (Son et al. 2010). Other studies have shown that activation of Nrf2 can stimulate cell proliferation (Reddy et al., 2008; Homma et al., 2009), consistent with the possibility that Nrf2 also mediates the stimulatory effect of high concentrations of plumbagin on GRP proliferation.
Although the present studies were performed on embryonic NPC in culture, under well-controlled experimental conditions, it is reasonable to consider that plumbagin might also influence the fate of NPC in vivo. It was recently reported that plumbagin inhibits p300 histone acetyltransferase activity in a noncompetitive manner in liver cells when administered intraperitoneally to mice (Ravindra et al. 2009). Other studies have revealed roles for p300 in the cell fate determination of NPC (Vojtek et al. 2003; Lee et al. 2009), suggesting a possible role for p300 in the actions of plumbagin on NPC demonstrated in the present study. Indeed, we demonstrated a key role for Stat3 in the astrocyte differentiation-promoting effects of low dose plumbagin, and others have shown that Stat3 forms a complex with p300 to promote astrocyte production from neuroepithelial cells (Fukuda et al. 2007) and p300 mediates some transcription-regulating actions of Stat3 (Hou et al. 2008). Plumbagin readily enters the central nervous system when administered peripherally (Son et al. 2010) and so would be expected to affect Stat3 activity and NPC fate in vivo. It will therefore be of considerable interest to determine the consequences of enhanced astrocyte generation in response to plumbagin in physiological and pathological settings. The ability to selectively affect the fate of GRP with lipophilic low molecular weight chemicals, without reducing neurogenesis, might be particularly valuable in conditions where trophic support of neurons is desirable.
Supplementary Material
Acknowledgements
This research was supported by the Intramural Research Program of the National Institute on Aging.
References
- Aggarwal BB, Sung B. Pharmacological basis for the role of curcumin in chronic diseases: an age-old spice with modern targets. Trends Pharmacol Sci. 2009;30:85–94. doi: 10.1016/j.tips.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Bhargava SK. Effects of plumbagin on reproductive function of male dog. Indian J Exp Biol. 1984;22:153–156. [PubMed] [Google Scholar]
- Bonaguidi MA, McGuire T, Hu M, Kan L, Samanta J, Kessler JA. LIF and BMP signaling generate separate and discrete types of GFAP-expressing cells. Development. 2005;132:5503–5514. doi: 10.1242/dev.02166. [DOI] [PubMed] [Google Scholar]
- Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME. Regulation of gliogenesis in the central nervous system by the JAK-STAT signaling pathway. Science. 1997;278:477–483. doi: 10.1126/science.278.5337.477. [DOI] [PubMed] [Google Scholar]
- Boulton TG, Zhong Z, Wen Z, Darnell JE, Jr, Stahl N, Yancopoulos GD. STAT3 activation by cytokines utilizing gp130 and related transducers involves a secondary modification requiring an H7-sensitive kinase. Proc Natl Acad Sci U S A. 1995;92:6915–6919. doi: 10.1073/pnas.92.15.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Wu Y, Mirua T, Pierce JL, Lucero MT, Albertine KH, Spangrude GJ, Rao MS. Properties of a fetal multipotent neural stem cell (NEP cell) Dev Biol. 2002;251:221–240. doi: 10.1006/dbio.2002.0828. [DOI] [PubMed] [Google Scholar]
- Calabrese V, Cornelius C, Mancuso C, Pennisi G, Calafato S, Bellia F, Bates TE, Giuffrida Stella AM, Schapira T, Dinkova Kostova AT, Rizzarelli E. Cellular stress response: a novel target for chemoprevention and nutritional neuroprotection in aging, neurodegenerative disorders and longevity. Neurochem Res. 2008;33:2444–2471. doi: 10.1007/s11064-008-9775-9. [DOI] [PubMed] [Google Scholar]
- Cheng A, Wang S, Cai J, Rao MS, Mattson MP. Nitric oxide acts in a positive feedback loop with BDNF to regulate neural progenitor cell proliferation and differentiation in the mammalian brain. Dev Biol. 2003;258:319–333. doi: 10.1016/s0012-1606(03)00120-9. [DOI] [PubMed] [Google Scholar]
- Cheng A, Tang H, Cai J, Zhu M, Zhang X, Rao M, Mattson MP. Gap junctional communication is required to maintain mouse cortical neural progenitor cells in a proliferative state. Dev Biol. 2004;272:203–216. doi: 10.1016/j.ydbio.2004.04.031. [DOI] [PubMed] [Google Scholar]
- Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Darr D, Fridovich I. Adaptation to oxidative stress in young, but not in mature or old, Caenorhabditis elegans. Free Radic Biol Med. 1995;18:195–201. doi: 10.1016/0891-5849(94)00118-4. [DOI] [PubMed] [Google Scholar]
- de Paiva SR, Figueiredo MR, Aragao TV, Kaplan MA. Antimicrobial activity in vitro of plumbagin isolated from Plumbago species. Mem Inst Oswaldo Cruz. 2003;98:959–961. doi: 10.1590/s0074-02762003000700017. [DOI] [PubMed] [Google Scholar]
- Dinkova-Kostova AT, Holtzclaw WD, Wakabayashi N. Keap1, the sensor for electrophiles and oxidants that regulates the phase 2 response, is a zinc metalloprotein. Biochemistry. 2005;44:6889–6899. doi: 10.1021/bi047434h. [DOI] [PubMed] [Google Scholar]
- Farr SB, Natvig DO, Kogoma T. Toxicity and mutagenicity of plumbagin and the induction of a possible new DNA repair pathway in Escherichia coli. J Bacteriol. 1985;164:1309–1316. doi: 10.1128/jb.164.3.1309-1316.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda S, Abematsu M, Mori H, Yanagisawa M, Kagawa T, Nakashima K, Yoshimura A, Taga T. Potentiation of astrogliogenesis by STAT3-mediated activation of bone morphogenetic protein-Smad signaling in neural stem cells. Mol Cell Biol. 2007;27:4931–4937. doi: 10.1128/MCB.02435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou T, Ray S, Lee C, Brasier AR. The STAT3 NH2-terminal domain stabilizes enhanceosome assembly by interacting with the p300 bromodomain. J Biol Chem. 2008;283:30725–30734. doi: 10.1074/jbc.M805941200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YL, Cho CY, Kuo PL, Huang YT, Lin CC. Plumbagin (5-hydroxy-2-methyl-1,4-naphthoquinone) induces apoptosis and cell cycle arrest in A549 cells through p53 accumulation via c-Jun NH2-terminal kinase-mediated phosphorylation at serine 15 in vitro and in vivo. J Pharmacol Exp Ther. 2006;318:484–494. doi: 10.1124/jpet.105.098863. [DOI] [PubMed] [Google Scholar]
- Hughes SM, Lillien LE, Raff MC, Rohrer H, Sendtner M. Ciliary neurotrophic factor induces type-2 astrocyte differentiation in culture. Nature. 1988;335:70–73. doi: 10.1038/335070a0. [DOI] [PubMed] [Google Scholar]
- Inbaraj JJ, Chignell CF. Cytotoxic action of juglone and plumbagin: a mechanistic study using HaCaT keratinocytes. Chem Res Toxicol. 2004;17:55–62. doi: 10.1021/tx034132s. [DOI] [PubMed] [Google Scholar]
- Itoigawa M, Takeya K, Furukawa H. Cardiotonic action of plumbagin on guinea-pig papillary muscle. Planta Med. 1991;57:317–319. doi: 10.1055/s-2006-960106. [DOI] [PubMed] [Google Scholar]
- Kalyani A, Hobson K, Rao MS. Neuroepithelial stem cells from the embryonic spinal cord: isolation, characterization, and clonal analysis. Dev Biol. 1997;186:202–223. doi: 10.1006/dbio.1997.8592. [DOI] [PubMed] [Google Scholar]
- Kalyani AJ, Rao MS. Cell lineage in the developing neural tube. Biochem Cell Biol. 1998;76:1051–1068. [PubMed] [Google Scholar]
- Kalyani AJ, Piper D, Mujtaba T, Lucero MT, Rao MS. Spinal cord neuronal precursors generate multiple neuronal phenotypes in culture. J Neurosci. 1998;18:7856–7868. doi: 10.1523/JNEUROSCI.18-19-07856.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawiak A, Piosik J, Stasilojc G, Gwizdek-Wisniewska A, Marczak L, Stobiecki M, Bigda J, Lojkowska E. Induction of apoptosis by plumbagin through reactive oxygen species-mediated inhibition of topoisomerase II. Toxicol Appl Pharmacol. 2007;223:267–276. doi: 10.1016/j.taap.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Son TG, Park HR, Park M, Kim MS, Kim HS, Chung HY, Mattson MP, Lee J. Curcumin stimulates proliferation of embryonic neural progenitor cells and neurogenesis in the adult hippocampus. J Biol Chem. 2008;283:14497–14505. doi: 10.1074/jbc.M708373200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee B, Lee JW, Lee SK. Retinoid signaling and neurogenin2 function are coupled for the specification of spinal motor neurons through a chromatin modifier CBP. Neuron. 2009;62:641–654. doi: 10.1016/j.neuron.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Rao MS. Glial progenitors in the CNS and possible lineage relationships among them. Biol Cell. 2004;96:279–290. doi: 10.1016/j.biolcel.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Luo Y, Cai J, Liu Y, Xue H, Chrest FJ, Wersto RP, Rao M. Microarray analysis of selected genes in neural stem and progenitor cells. J Neurochem. 2002;83:1481–1497. doi: 10.1046/j.1471-4159.2002.01260.x. [DOI] [PubMed] [Google Scholar]
- Maric D, Maric I, Chang YH, Barker JL. Prospective cell sorting of embryonic rat neural stem cells and neuronal and glial progenitors reveals selective effects of basic fibroblast growth factor and epidermal growth factor on self-renewal and differentiation. J Neurosci. 2003;23:240–251. doi: 10.1523/JNEUROSCI.23-01-00240.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Cheng A. Neurohormetic phytochemicals: Low-dose toxins that induce adaptive neuronal stress responses. Trends Neurosci. 2006;29:632–639. doi: 10.1016/j.tins.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Mayer-Proschel M, Kalyani AJ, Mujtaba T, Rao MS. Isolation of lineage-restricted neuronal precursors from multipotent neuroepithelial stem cells. Neuron. 1997;19:773–785. doi: 10.1016/s0896-6273(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Morgan DM. Tetrazolium (MTT) assay for cellular viability and activity. Methods Mol Biol. 1998;79:179–183. doi: 10.1385/0-89603-448-8:179. [DOI] [PubMed] [Google Scholar]
- Mujtaba T, Piper DR, Kalyani A, Groves AK, Lucero MT, Rao MS. Lineage-restricted neural precursors can be isolated from both the mouse neural tube and cultured ES cells. Dev Biol. 1999;214:113–127. doi: 10.1006/dbio.1999.9418. [DOI] [PubMed] [Google Scholar]
- Nakanishi M, Niidome T, Matsuda S, Akaike A, Kihara T, Sugimoto H. Microglia-derived interleukin-6 and leukaemia inhibitory factor promote astrocytic differentiation of neural stem/progenitor cells. Eur J Neurosci. 2007;25:649–658. doi: 10.1111/j.1460-9568.2007.05309.x. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Taga T. Mechanisms underlying cytokine-mediated cell-fate regulation in the nervous system. Mol Neurobiol. 2002;25:233–244. doi: 10.1385/MN:25:3:233. [DOI] [PubMed] [Google Scholar]
- Okita K, Yamanaka S. Intracellular signaling pathways regulating pluripotency of embryonic stem cells. Curr Stem Cell Res Ther. 2006;1:103–111. doi: 10.2174/157488806775269061. [DOI] [PubMed] [Google Scholar]
- Pencea V, Bingaman KD, Wiegand SJ, Luskin MB. Infusion of brain-derived neurotrophic factor into the lateral ventricle of the adult rat leads to new neurons in the parenchyma of the striatum, septum, thalamus, and hypothalamus. J Neurosci. 2001;21:6706–6717. doi: 10.1523/JNEUROSCI.21-17-06706.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan P, Panchision DM, Newell LF, McKay RD. BMPs signal alternately through a SMAD or FRAP-STAT pathway to regulate fate choice in CNS stem cells. J Cell Biol. 2003;161:911–921. doi: 10.1083/jcb.200211021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MS, Noble M, Mayer-Proschel M. A tripotential glial precursor cell is present in the developing spinal cord. Proc Natl Acad Sci U S A. 1998;95:3996–4001. doi: 10.1073/pnas.95.7.3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindra KC, Selvi BR, Arif M, Reddy BA, Thanuja GR, Agrawal S, Pradhan SK, Nagashayana N, Dasgupta D, Kundu TK. Inhibition of lysine acetyltransferase KAT3B/p300 activity by a naturally occurring hydroxynaphthoquinone, plumbagin. J Biol Chem. 2009;284:24453–24464. doi: 10.1074/jbc.M109.023861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandur SK, Ichikawa H, Sethi G, Ahn KS, Aggarwal BB. Plumbagin (5-hydroxy-2-methyl-1,4-naphthoquinone) suppresses NF-kappaB activation and NF-kappaB-regulated gene products through modulation of p65 and IkappaBalpha kinase activation, leading to potentiation of apoptosis induced by cytokine and chemotherapeutic agents. J Biol Chem. 2006;281:17023–17033. doi: 10.1074/jbc.M601595200. [DOI] [PubMed] [Google Scholar]
- Satoh T, Lipton SA. Redox regulation of neuronal survival mediated by electrophilic compounds. Trends Neurosci. 2007;30:37–45. doi: 10.1016/j.tins.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Son TG, Camandola S, Arumugam TV, Cutler RG, Telljohann RS, Mughal MR, Moore TA, Luo W, Yu QS, Johnson DA, Johnson JA, Greig NH, Mattson MP. Plumbagin, a novel Nrf2/ARE activator, protects against cerebral ischemia. J Neurochem. 2010;112:1316–1326. doi: 10.1111/j.1471-4159.2009.06552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Turkson J, Ryan D, Kim JS, Zhang Y, Chen Z, Haura E, Laudano A, Sebti S, Hamilton AD, Jove R. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem. 2001;276:45443–45455. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- Van der Vijver LM. Distribution of plumbagin in the Plumbagin aceae. Phytochemistry. 1972;11:3247–3248. [Google Scholar]
- Vojtek AB, Taylor J, DeRuiter SL, Yu JY, Figueroa C, Kwok RP, Turner DL. Akt regulates basic helix-loop-helix transcription factor-coactivator complex formation and activity during neuronal differentiation. Mol Cell Biol. 2003;23:4417–4427. doi: 10.1128/MCB.23.13.4417-4427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liu Y, Chesnut JD, Rao MS. Isolation of neural stem and precursor cells from rodent tissue. Methods Mol Biol. 2008;438:39–53. doi: 10.1007/978-1-59745-133-8_5. [DOI] [PubMed] [Google Scholar]
- Yoshimatsu T, Kawaguchi D, Oishi K, Takeda K, Akira S, Masuyama N, Gotoh Y. Non-cell-autonomous action of STAT3 in maintenance of neural precursor cells in the mouse neocortex. Development. 2006;133:2553–2563. doi: 10.1242/dev.02419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.