Abstract
Substantial genetic variation exists in natural populations of Drosophila melanogaster. This segregating variation includes alleles at different loci that interact to cause lethality or sterility (synthetic incompatibilities). Fitness epistasis in natural populations has important implications for speciation and the rate of adaptive evolution. To assess the prevalence of epistatic fitness interactions, we placed naturally occurring X chromosomes into genetic backgrounds derived from different geographic locations. Considerable amounts of synthetic incompatibilities were observed between X chromosomes and autosomes: greater than 44% of all combinations were either lethal or sterile. Sex-specific lethality and sterility were also tested to determine whether Haldane's rule holds for within-species variation. Surprisingly, we observed an excess of female sterility in genotypes that were homozygous, but not heterozygous, for the X chromosome. The recessive nature of these incompatibilities is similar to that predicted for incompatibilities underlying Haldane’s rule. Our study also found higher levels of sterility and lethality for genomes that contain chromosomes from different geographical regions. These findings are consistent with the view that genomes are co-adapted gene complexes and that geography affects the likelihood of epistatic fitness interactions.
Keywords: Drosophila melanogaster, epistasis, Haldane’s rule, synthetic lethality, synthetic sterility, X-autosome interactions
Introduction
Genes act in a genomic context. Their epistatic interactions affect the evolution of natural populations (Wolf et al. 2000). Many different types of epistasis exist (Phillips 2008), but one unifying theme is that the effects of genes often depend upon genetic background. Some notable examples of epistatic interactions include olfactory behavior in Drosophila melanogaster (Anholt et al. 2003), plant growth in Arabidopsis thaliana (Alcazar et al. 2009), and blood and bone traits in Mus musculus and Rattus rattus (Shao et al. 2008). Regardless of the phenotype affected, epistasis must modify fitness to be evolutionarily relevant. Fitness epistasis lies at the core of evolutionary genetic dynamics, influencing both the rate of adaptation (Sanjuan et al. 2005; Kim 2007; Yukilevich et al. 2008; Griswold and Masel 2009) and the genetic architecture of speciation (Johnson 2000; Presgraves 2007).
Synthetic incompatibilities are an important type of fitness epistasis. These interactions take place between alleles at different loci causing fitness to be reduced when both alleles are present in the same individual. Synthetic incompatibilities can cause either lethality or sterility. Population genetics theory suggests that synthetic lethal alleles can segregate at relatively high frequencies at mutation-selection balance (Phillips and Johnson 1998). This is because single mutations are masked and only individuals containing multiple mutations are exposed to selection. The mechanistic basis of specific synthetic incompatibilities can be conserved over evolutionary time: >20% of gene combinations that lead to incompatibilities in Saccharomyces pombe also lead to incompatibilities in Saccharomyces cerevisiae (Dixon et al. 2008). Synthetic lethal alleles have been found segregating in common haplotypes of Caenorhabditis elegans (Seidel et al, 2008) and in populations of the copepod Tigriopus californicus (Harrison and Edmands 2006). Numerous studies of natural variation in the genus Drosophila have detected synthetic incompatibilities, particularly those involving interactions between alleles on different chromosomes (Krimbas 1960; Lucchesi 1968; Temin et al. 1969; Thompson 1986; Powell 1997). Most of the chromosome pairs tested in these studies did not result in large numbers of synthetic incompatibilities (on the order of zero to 5%). However, most of these studies only examined synthetic lethality. Much less is known about the synthetic sterility of naturally segregating alleles, especially in females.
Because synthetic incompatibilities can form the basis of Dobzhansky-Bateson-Muller (DBM) incompatibilities, they are relevant to speciation. DBM incompatibilities result in postzygotic reproductive isolation without either diverging population having to cross a fitness valley, and introgression studies indicate that many small genomic regions from one species cause DBM incompatibilities when placed in the genetic background of a sister species (Palopoli and Wu 1994; True et al. 1996; Tao et al. 2003; Masly and Presgraves 2007; Moyle and Nakazato 2009). Another example involves the Lethal hybrid rescue gene in D. simulans and the Hybrid male rescue gene in D. melanogaster. These genes interact to cause lethality in hybrid F1 males (Brideau et al. 2006). Within-species DBM incompatibilities have also been observed in the autoimmune response of Arabidopsis thaliana (Bomblies et al. 2007). Some types of synthetic incompatibilities do not result in DBM incompatibilities. For example, synergistic epistasis can occur between two deleterious alleles, resulting in synthetic lethality or sterility. Since each of these alleles is slightly deleterious by itself, this scenario is not an example of a DBM incompatibility.
Theoretical models such as Orr’s “snowball effect” suggest that the accumulation of DBM incompatibilities is contingent on the number of divergent epistatic loci (Orr 1995). In this model, substitutions that occur in each of two divergent populations can potentially result in deleterious interactions. There currently is some debate whether the “snowball effect” best describes the accumulation of reproductive incompatibilities in nature (Gourbiere and Mallet 2009). There is evidence that standing genetic variation can greatly affect time to speciation (Schluter and Conte 2009). It is therefore of great interest to assess whether natural populations harbor large amounts of synthetic lethal and synthetic sterile alleles.
The chromosomes involved in DBM incompatibilities are important to Haldane’s rule. This rule states that if only one sex of a hybrid is sterile or inviable, it tends to be the heterogametic sex (Haldane 1922). Four alternative hypotheses that explain Haldane’s rule are: faster-X, faster male, meiotic drive, and dominance (Coyne and Orr 2004). The faster-X hypothesis is sensitive to demography and it posits that rates of adaptive change differ between X-linked and autosomal loci (Mank et al. 2010). The faster male hypothesis posits that male traits may evolve faster due to sexual selection and the sensitivity of spermatogenesis to new mutations (Wu and Davis 1993). Divergence of meiotic drive suppression systems can cause hybrid sterility and lethality, and segregation distortion of sex chromosomes can distort sex ratios away from 50:50 (Frank 1991; Hurst and Pomiankowski 1991; Henikoff et al. 2001; Tao et al. 2001). Finally, the dominance hypothesis states that if alleles causing interspecific incompatibilities behave recessively in hybrids, then the heterogametic sex will be more likely to be affected (Turelli and Orr 1995). Data from a wide variety of taxa support the dominance hypothesis (Coyne and Orr 2004; Bierne et al. 2006; Carling and Brumfield 2008), as do theoretical models with a firm grounding in DBM incompatibilities (Turelli and Orr 2000). Drosophila have an XY system in which males are the heterogametic sex. Because recessive X-linked alleles cannot be masked in Drosophila males, sex-specific patterns can arise when epistasis involves the X chromosome. While Haldane’s rule is known to apply to between-species incompatibilities, it is unknown whether it applies to within-species sterility and lethality. Note, however, that the dominance theory does not make any predictions about the dominance of incompatible alleles within species. Recent work in Tribolium castaneum suggests that Haldane’s rule may apply to within-species incompatibilities between different populations (Demuth and Wade 2007). However, sterility in other heterogametic-male species (such as Homo sapiens) seems to be female-biased (Thonneau et al. 1991).
An additional consideration is that genomes can be viewed as locally co-adapted gene complexes (Dobzhansky 1970). If this is the case, then we would expect genes derived from different geographic regions to be more likely to exhibit synthetic incompatibilities. Natural selection may be unable to remove deleterious allelic combinations if populations are spatially structured. This is consistent with the phenomenon of outbreeding depression (Edmands 2007). Drosophila melanogaster is a human commensal that has a worldwide distribution, having recently expanded out of Africa (Keller 2007). Clines exist for multiple traits and isolation by distance has led to genetic differentiation among populations of D. melanogaster (Sezgin et al. 2004; Umina et al. 2005; Pool and Aquadro 2006). In addition, flies derived from Zimbabwe are sexually isolated from cosmopolitan populations (Wu et al. 1995). There is also evidence of phenotypic differentiation and partial prezygotic reproductive isolation between United States and Caribbean populations of D. melanogaster (Yukilevich and True 2008a, b). While previous studies detected only low levels of synthetic lethality in D. melanogaster, these studies did not test chromosomes derived from multiple geographic locations (Thompson 1986).
In this study we investigated synthetic incompatibilities in D. melanogaster on a chromosomal scale. Extracted-X lines were constructed by placing naturally segregating X chromosomes into multiple genetic backgrounds. Levels and types of synthetic incompatibilities were assessed (including lethality vs. sterility, recessive vs. dominant, and male vs. female incompatibilities). This allowed us to address the following questions: 1) How common are synthetic incompatibilities among naturally segregating chromosomes? 2) Does Haldane’s rule hold for within-species synthetic lethality and sterility? 3) Are synthetic incompatibilities more common when chromosomes are derived from different geographic locations?
Materials and Methods
Stocks and construction of lines
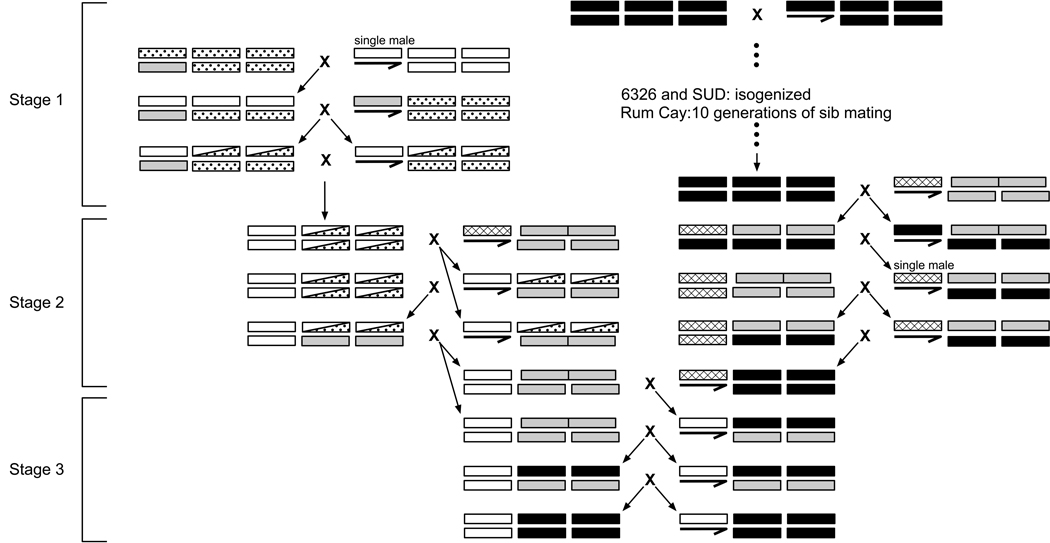
We constructed two sets of extracted-X lines using balancer chromosomes. Set 1 contains 118 different X chromosomes bred into a single autosomal background. Set 2 contains 52 different X chromosomes bred into three different autosomal backgrounds. Two of the lines in Set 2 were unable to be maintained, resulting in a total of 154 lines. X chromosomes tested in Set 2 overlap with X chromosomes tested in Set 1. Extracted-X lines were constructed via three stages (Figure 1). First, wild-caught isofemale lines were isogenized (either via balancer stocks or ten generations of sib-mating). This resulted in lines with genotypes of +x or +a (superscripts indicate whether a line serves as a source of extracted-X chromosomes or autosomes). Secondly, intermediate stocks containing markers and either an extracted X chromosome or an autosomal background were constructed. These crosses involved w1118;T(2;3)apXa/CyO:TM3 flies (Bloomington stock 2475), and resulted in +x;T(2;3)apXa/CyO:TM3 and w1118;+a;+a lines. Third, homozygous extracted-X lines were constructed. The final cross of this scheme involved crossing +x;+a/CyO;+a/TM3 flies and selecting wild-type offspring. Each of the resulting extracted-X lines has a genotype of +x;+a;+a. The fourth chromosome (∼2% of the genome) was not monitored.
Figure 1.
Construction of extracted-X lines. Color scheme is as follows: balancer chromosomes are labeled gray, w1118 X chromosomes are cross-hatched, genetic backgrounds of balancer stocks used in Stage 1 are labeled with dots, source lines of extracted-X chromosomes are labeled white (+x), and source lines of autosomal chromosomes are labeled black (+a). The series of crosses depicted here result in extracted-X lines that have a genotype of +x;+a;+a. Different types of synthetic incompatibilities are distinguishable by the offspring generated by each of the crosses in Stage 3.
X chromosomes used in these lines were derived from wild-caught isofemale lines, while autosomal backgrounds were derived from both wild caught and laboratory lines. Geographic origins of X chromosomes were: Sudbury, Ontario, Canada and Long Island, New York (collected by T. Merritt in 2005), Southern United States and the Caribbean (collected by R. Yukilevich in 2004–5), and Cameroon and Zimbabwe (collected by J. Pool and C. Aquadro in 1990, 1994 and 2004). See supplemental information for a full list of X chromosomes. The autosomal background used in Set 1 was derived from a mapping stock (Bloomington stock 6326). The three autosomal backgrounds used in Set 2 were: 6326, Sudbury (latitude: 46.49, longitude: −81.01), and Rum Cay, Bahamas (latitude: 23.38, longitude: −74.5). Note that Sudbury, Ontario is near the northern range limit of D. melanogaster, and Rum Cay is a southern location in the Bahamas. By contrast, the geographic origins of 6326 are unknown (R. Hoskins and A. Phan, personal communication). Each source line (X and autosomal) could be maintained as an isofemale line, indicating the absence of single-locus lethality or sterility. The 6326 and Sudbury lines used to provide autosomal backgrounds are isogenic, whereas the Rum Cay line was produced by ten generations of sib-mating.
Flies were cultured on standard corn meal/molasses/agar medium supplemented with antibiotics (either penicillin at 40 µg/ml or a mix of tetracycline and streptomycin at 63 µg/ml and 19 µg/ml, respectively). All crosses were performed at 25°C with a 12 hour light:dark cycle.
Lethality assays
The crossing scheme used to generate extracted-X lines allowed different types of synthetic lethality to be distinguished. In particular, when we were unable to construct homozygous lines the stage at which crosses were unable to proceed was recorded. This allowed us to determine the dominance of the fitness interactions, which chromosomes were involved, and whether synthetic lethality was male or female-specific (see bottom part of Figure 1). Set 1 of the extracted-X lines was tested for recessive synthetic lethality, and Set 2 was tested for dominant and recessive synthetic lethality. Dominant male lethality was assessed by crossing +x;T(2;3)apXa/CyO:TM3 females and w1118;+a;+a males. When this cross did not result in male offspring, dominant X-autosome interactions were implicated. Dominant female lethality was assessed by crossing +x;T(2;3)apXa/CyO:TM3 females and +x;+a;+a males. When this cross did not result in female offspring with curly wings and stubble bristles (i.e. flies with a +x;+a/CyO;+a/TM3 genotype), dominant X-autosome interactions were implicated. Note that dominance and recessivity in this case refers to the number of autosomal copies required for synthetic lethality.
Recessive lethal interactions were assessed by intercrossing +x;+a/CyO;+a/TM3 flies. Wild-type offspring from this cross can only appear in the absence of recessive lethal interactions. Recessive X-2nd lethal interactions cause all offspring from this cross to have curly wings (i.e. flies with +x;+a/CyO;+a/+a or +x;+a/CyO;+a/TM3 genotypes). Recessive X-3rd lethal interactions cause all offspring from this cross to have stubble bristles (i.e. flies with +x;+a/+a;+a/TM3 or +x;+a/CyO;+a/TM3 genotypes). Each X-autosome combination was replicated twice, and at least 60 offspring were genotyped for each combination of X chromosome and autosomal background.
Sterility assays
X-autosome synthetic sterility was detected by crossing synthetic genotypes to wild-type flies and looking for the presence of offspring. For each X-autosome combination, flies were mass mated with other flies belonging to the same extracted-X line. Newly emerged flies were aged three to five days prior to each cross. Three virgin +x;+a;+a females were then placed into a vial with three +x;+a;+a males. After seven days, adults were removed and vials were inspected for developing offspring. If larvae or pupae were observed, genotypes were considered to be fertile. Each of these crosses was replicated three times. Note that Y chromosomes in these sterility assays were derived from +x;T(2;3)apXa/CyO:TM3 balancer stocks.
The absence of viable offspring can be due to either male or female sterility. To determine if sterility was male or female-specific, extracted-X flies were outcrossed to SBU1, a wild-caught isofemale line from Stony Brook, NY (collected by J. R. True in 2005). Three males and females from each extracted-X line were mated with SBU1 flies of the opposite sex. Each of these crosses was replicated three times. If no offspring resulted from crossing female SBU1 flies with extracted-X males (+x;+a;+a), the line was considered male sterile. If no offspring resulted from crossing extracted-X females (+x;+a;+a) with SBU1 males, the line was considered female sterile. This scheme also allowed us to infer whether sterility was an organismal property or the property of a pair of mating individuals. With organismal sterility, flies of a particular sex and genotype are unable to produce offspring regardless of the genotype of their mating partner. With mating pair sterility, flies are only sterile when they are paired with flies of a particular genotype. If a particular genotype was unable to produce offspring when mated with either its own genotype or SBU1 flies, then the synthetic incompatibility was classified as organismal sterility.
Sperm motility was assayed for lines that contained males that were sterile when crossed to females of two different genotypes (organismal male sterile lines). After developing at 25°C, newly emerging males were separated by genotype and aged 4–5 days without access to females. Testes of individual males were dissected in a drop of Ringer’s solution, gently squashed under a coverslip, and examined under a stereomicroscope. If a single motile sperm was observed, males were classified as possessing motile sperm.
Dominance tests for female sterility
Because the lines used in the above sterility tests were homozygous for the X chromosome, it was unclear whether female sterility was due to dominant or recessive X-autosome interactions (i.e. is a single copy of an X chromosome sufficient to confer synthetic sterility?). To assess this, we controlled for genetic background and generated females heterozygous for a putatively sterile X chromosome. +x1;+a/CyO;+a/TM3 females were crossed with +x2;+a/CyO;+a/TM3 males (and vice versa) to generate +x1/+x2;+a;+a females (where +x1 and +x2 are two different X chromosomes). As per the above sterility assays, the resulting wild-type females were mass mated with +x1;+a;+a, +x2;+a;+a, and SBU1 males. Each putatively sterile X chromosome was tested with four other X chromosomes (two putatively sterile and two fertile X chromosomes). If all four combinations failed to generate offspring, female-specific synthetic sterility was classified as dominant. Otherwise, synthetic sterility was classified as (partially) recessive. A total of 14 different synthetic sterile X chromosomes were tested, and each cross was replicated twice. Note that these crosses are also complementation tests for X-linked sterility factors.
Sex ratio assays
Sex ratio data were recorded for 49 extracted-X lines from Set 1. Each of these lines was able to be maintained as a homozygous stock. For each X-autosome combination, three replicate vials were checked. Numbers of newly emerging males and females were recorded five different days for each vial. Emerging flies were counted no later than 17 days after the original cross was set up. Samples sizes were too small for 13 of the lines (less than 30 flies emerged), leaving a total of 36 lines. For each of the 36 remaining lines, a mean number of 127 flies were counted.
Non-parametric test of geographical patterns
A non-parametric test was used to determine whether synthetic incompatibilities were more likely when X chromosomes and autosomes were derived from different geographical locations. First, X chromosomes were classified as either northern or southern. Northern X chromosomes were derived from populations found at latitudes above 40 (approximately the Mason-Dixon line, see Figure 5A), and southern X chromosomes were derived from populations found at latitudes below 40. Under this formulation, Set 2 contains 18 northern X lines and 34 southern X lines. Note that the set of southern X chromosomes contains some lines with African X chromosomes. The Sudbury autosomal background has a northern origin, and the Rum Cay autosomal background has a southern origin. The following test statistic (γ) was calculated:
| (1) |
In this equation x refers to the proportion of X chromosomes that are incompatible with a particular autosomal background. Subscripts indicate the geographic origins of the X and autosomal chromosomes, with X chromosomal origin listed first. For example, xSN refers to the proportion of southern X chromosomes that are incompatible with a northern autosomal background. γ is negative if synthetic incompatibilities occur more often when X chromosomes and autosomes are mismatched (i.e. chromosomes are derived from different regions).
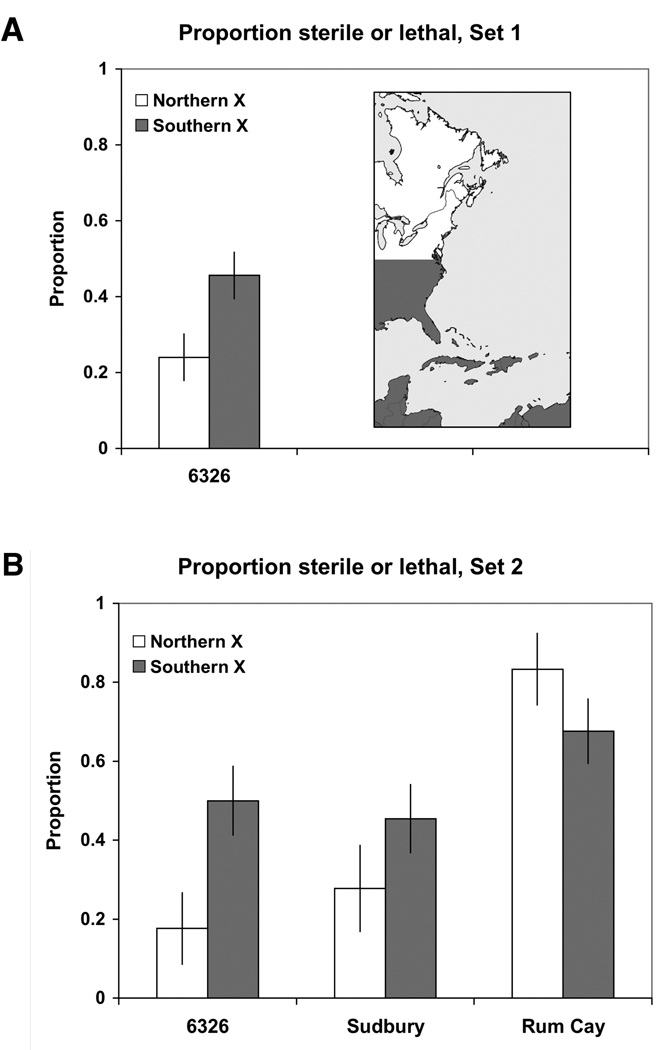
Figure 5.
Geographic patterns of sterility and lethality for different autosomal backgrounds. Lines are classified as northern or southern according to the geographic origins of X chromosomes, and the proportion of northern or southern X-autosome combinations that result in sterility or lethality are depicted. 118 different X chromosomes were tested in Set 1, and 52 different X chromosomes were tested for each autosomal background in Set 2. Northern X chromosomes are labeled white and southern X chromosomes are labeled gray. Note that the Sudbury autosomal background is derived from a northern population and the Rum Cay autosomal background is derived from a southern population. Error bars are +/− one standard error.
Given a null hypothesis that synthetic incompatibilities are independent of the geographic origin of chromosomes, Monte-Carlo simulations were run to determine the distribution of γ. The probability of synthetic incompatibility varies by autosomal background (38.5% of tested X chromosomes were incompatible with a northern autosomal background, and 73.1% were incompatible with a southern autosomal background). Using these probabilities, simulated datasets of 52 X chromosomes (18 northern and 34 southern) were generated for each autosomal background. Thus, the number of incompatible X-autosome combinations for each of the four possibilities (north-north, south-north, south-south, and north-south) follows a binomial distribution, and null expectations are equal proportions of incompatible northern and southern X chromosomes. γ was calculated for each simulation run. Monte Carlo simulations were run 100,000 times and the distribution of the test statistic γ was compared to the observed data.
Results
Overall levels of synthetic incompatibility
Substantial levels of synthetic incompatibilities were observed for both sets of extracted-X lines (Table 1). In Set 1, 43 of 118 X chromosomes (36.4%) resulted in synthetic sterility or lethality when placed on a 6326 genetic background. Similarly, 50.0% of the X-autosome combinations tested in Set 2 resulted in synthetic sterility or lethality. The numbers of synthetic incompatibilities varied by genetic background in Set 2: 20 of 51 X chromosomes (39.2%) were incompatible with a 6326 background, 19 of 52 X chromosomes (36.5 %) were incompatible with a Sudbury background, and 38 of 51 X chromosomes (74.5%) were incompatible with a Rum Cay background. The excess proportion of deleterious interactions on a Rum Cay background relative to other backgrounds was highly significant (two-tailed p-value < 0.001 for each comparison; Fisher’s exact test). Note that two lines were unable to be tested in this second data set, and levels of incompatibilities on a 6326 background were similar for Set 1 and Set 2. The proportion of incompatibilities that involved lethality or sterility also varied by genetic background. A larger percentage of incompatible X-autosome combinations involving the 6326 background were sterile (as opposed to lethal) compared to Sudbury and Rum Cay backgrounds (35.0% vs. 15.0% and 13.2%, single-tailed p-values of 0.137 and 0.056 for each comparison; Fisher’s exact test). As the presence of only a few larvae or pupae was sufficient to classify a line as fertile, many of the X-autosome combinations labeled as lacking synthetic incompatibilities actually exhibited semi-sterility and were difficult to maintain as a homozygous stocks. This detail was corroborated by the observation that 13 of the 49 lines assayed in the sex ratio experiment had insufficient statistical power due to low numbers of offspring. The relatively low level of synthetic lethality observed in Set 1 as opposed to Set 2 is due to the fact that Set 1 lines were only tested for recessive lethal interactions. In addition, Set 1 involved X chromosomes over a 6326 background (the background that was more likely to result in synthetic sterility). The net result of the data in Table 1 was that many otherwise viable and fertile X chromosomes interacted deleteriously with novel autosomal backgrounds.
Table 1.
Levels of synthetic incompatibility. Values indicate the number of lines that exhibit a particular type of incompatibility. All lines in Set 1 have a 6326 autosomal background. Data from multiple autosomal backgrounds (6326, Sudbury, and Rum Cay) are pooled for Set 2.
| Incompatible sex | Females only | Males only | Both | Neither |
|---|---|---|---|---|
| Lethality | ||||
| Set 1 | 1 | 3 | 1 | 113 |
| Set 2 | 7 | 7 | 48 | 92 |
| Sterility | ||||
| Set 1* | 13 | 3 | 22 | 80 |
| Set 2 | 3 | 1 | 11 | 139 |
| Lethality or sterility | ||||
| Set 1 | 14 | 6 | 23 | 75 |
| Set 2 | 10 | 8 | 59 | 77 |
indicates a significant excess of female-specific sterility (one-tailed p-value < 0.05; Fisher’s exact test).
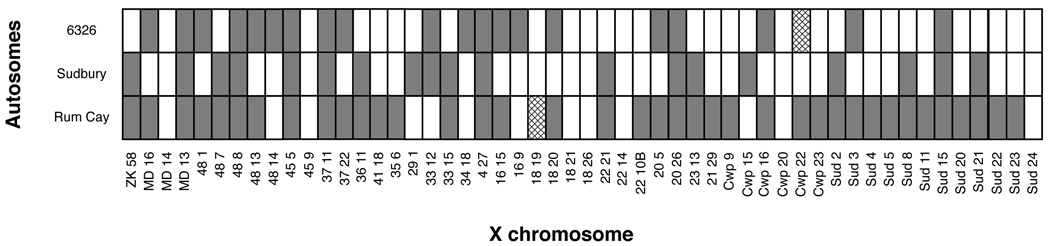
Does knowledge that a particular X-autosome combination is incompatible tell us anything about the lethality or sterility of another X-autosome combination? Set 2 of the extracted X lines was used for a test of independence, as Set 1 involved only a single autosomal background. Inspection of Figure 2 suggests the absence of any pattern: X chromosomes that were incompatible with a 6326 background (alternatively Sudbury or Rum Cay) were not any more or less likely to be incompatible with another autosomal background. Indeed, independence of the incompatibilities found in each background could not be rejected when the data in Figure 2 were converted into a 2×2×2 contingency table and a log-linear model was tested (p-value = 0.2742, χ2 = 5.13, d.f. = 4). Seven X chromosomes were incompatible with all three genetic backgrounds and eight X chromosomes were compatible with every tested genetic background. However, this was consistent with what one would expect from multiplying background-specific probabilities of synthetic lethality and sterility.
Figure 2.
X-autosome incompatibilities for Set 2. Each row corresponds to a different autosomal background, and each column corresponds to a different X chromosome. X-chromosomes are ordered by increasing latitude (i.e. the rightmost lines have X chromosomes that are derived from northern populations). Incompatible combinations are labeled gray and fertile combinations are labeled white. Two combinations were unable to be tested, and are labeled with cross-hatching. A log-linear test of independence yields a p-value of 0.2742 (χ2 = 5.13, d.f. = 4).
Chromosome-specific lethality patterns
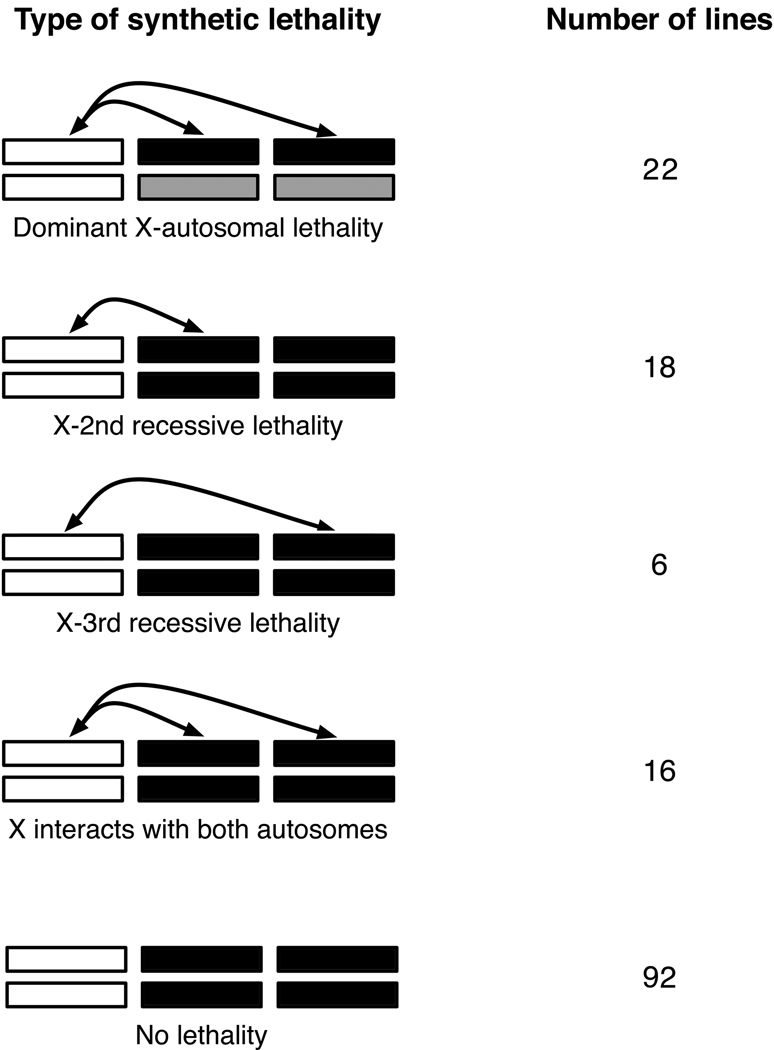
A majority of X-autosome interactions were recessive (requiring homozygous autosomes) and both X-2nd and X-3rd interactions were observed. Of the 154 lines tested in Set 2, 92 did not exhibit any synthetic lethality (Figure 3). Of the remaining synthetic lethal lines, 22 (35.5%) exhibited dominant synthetic lethality and 40 (64.5%) exhibited recessive lethality. Here, dominance and recessivity refers to the number of autosomal copies required for synthetic lethality (all lines tested were homozygous for an extracted X chromosome). One caveat is that our methodology may overestimate the frequency of synthetic lethals. This is because Mendelian segregation alone can cause a genotype to be absent (even if 60 flies were assayed per cross). Three times as many X-autosome combinations involving the 2nd chromosome resulted in recessive synthetic lethality relative to combinations involving the 3rd chromosome (two-tailed p-value < 0.001; Fisher’s exact test). While the Drosophila melanogaster second chromosome is slightly larger than the third chromosome (and thus a larger target for epistatic interactions), this alone is an insufficient explanation for the observed differences in X-2nd vs. X-3rd synthetic lethality. Finally, an appreciable number of lines (16) exhibited both X-2nd and X-3rd interactions, suggesting that complex epistasis may underlie synthetic lethality in these lines.
Figure 3.
Chromosomal patterns of synthetic lethality. Data are from Set 2 and involve pooled autosomal backgrounds (6326, Sudbury, Rum Cay). The color scheme is the same as Figure 1. There is a significant excess of X-2nd lethality relative to or X-3rd lethality (two-tailed p-value < 0.001; Fisher’s exact test).
Sterility tests
Sex-specific patterns of synthetic incompatibilities were observed. These patterns involved a very slight excess of male lethality over female lethality and a greater than three-fold excess of female sterility over male sterility (Table 1). Comparisons between female-specific and male-specific sterility revealed a statistically significant difference for Set 1, but not Set 2 (one-tailed p-value = 0.0086 for Set 1, one-tailed p-value = 0.3112 for Set 2, Fisher’s exact test). The excess of female sterility was less striking when sex-specific sterility and both-sexes-sterile data were pooled (one-tailed p-value = 0.0891 for Set 1, one-tailed p-value = 0.4190 for Set 2; Fisher’s exact test). Regardless of statistical significance, these patterns would not be expected if Haldane’s rule holds within species. While slight differences were observed for different autosomal backgrounds (greater levels of female sterility for lines containing a 6326 background), sample sizes were too small to say anything definitive about background-specific female sterility.
There was evidence for both organismal and mating pair specific sterility. Of 33 synthetic sterile lines tested in Set 1, 11 were unable produce offspring when mated with flies containing either the same combination of X and autosomes or SBU1 flies (i.e. sterility was an organismal property). The other 22 lines were able to produce offspring when mated with one, but not the other, genotype (i.e. sterility was a property of a mating pair). Of 15 synthetic sterile lines tested in Set 2, six exhibited organismal sterility and nine exhibited mating pair sterility. Note that the proportion of synthetic sterile lines exhibiting organismal sterility might be an overestimate. This is because flies were only tested against two genotypes of the opposite sex, and we cannot formally rule out the possibility that they might be able to produce viable offspring when mated with flies that contain a third, untested genotype. Males that were unable to sire offspring with females of multiple genotypes were assayed for sperm motility. Motile sperm were not observed for any of the six lines tested, suggesting that these incidences of male-specific organismal sterility involved defective spermatogenesis.
Tests of recessivity of female-sterile lines
Lines that exhibited female sterility were tested for recessivity of X effects. Hybrid flies used in these tests were homozygous for their respective autosomal backgrounds. When hybrid females containing a single putatively sterile X chromosome over a fertile X chromosome were tested for sterility, viable offspring resulted for all 14 of the lines tested. This indicates that X-autosome interactions causing female sterility in our study required flies to be homozygous for the same X chromosome (i.e. they were recessive). When hybrid females containing X chromosomes derived from two different female-sterile lines were tested for sterility, 13 of 14 cases resulted in viable offspring. The exception involved a line with an X chromosome derived from Cameroon, Africa over a 6326 background (MD 16). The MD 16 X chromosome was incompatible with Sud 24 (Sudbury, Ontario) and 18 26 (Montgomery, Alabama) X chromosomes. As this line was able to produce viable offspring when mated with fertile lines, there was still evidence that MD 16 contains a recessive X-linked female sterility factor. The failure of MD 16 to complement two other female sterile X chromosomes suggests that the same loci may be implicated in multiple cases of synthetic sterility.
Skewed sex ratios
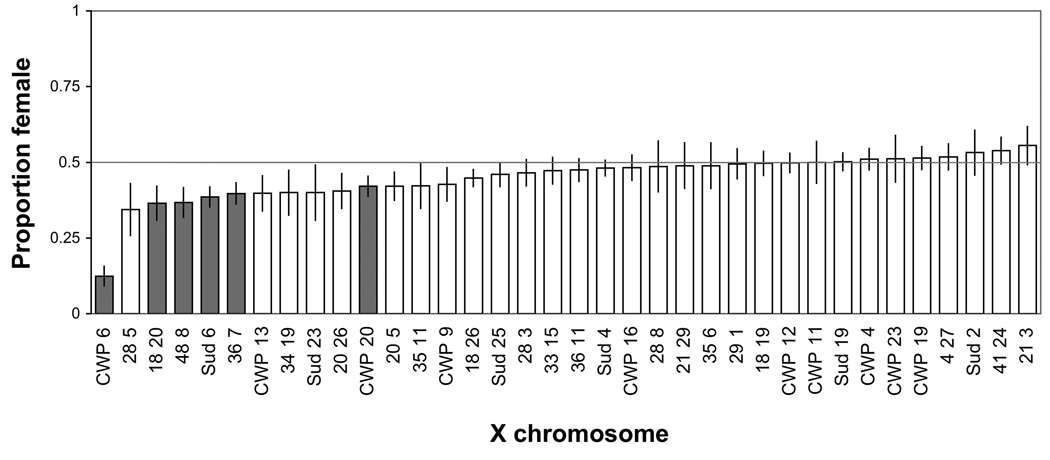
Additional evidence of sex-specific effects was observed in the form of skewed sex ratios. While 1:1 sex ratios were expected, we observed an excess of males for a number of extracted-X lines (Figure 4). Six of 36 lines tested from Set 1 had a significant excess of males (p-value < 0.05 after correcting for multiple tests using the Benjamini and Hochberg false discovery rate). These male-biased sex ratios are inconsistent with Haldane’s rule expectations. Lines were successfully maintained and sex ratios were reasonably stable over time, suggesting that Y-linked meiotic drive was not a cause of the observed patterns. As we observed an excess of males (as opposed to females) and our media contained antibiotics, Wolbachia can also be ruled out as a cause of unequal sex ratios. It is possible that female inviability could explain the male-biased sex ratios, but this was not assayed in our sex ratio tests.
Figure 4.
Biased sex ratios. Error bars are +/− one standard error. Lines are force ranked by % female. Lines with a significant excess of males are labeled in gray (p-value < 0.05 after correcting for multiple tests using the Benjamini and Hochberg false discovery rate). Autosomal background for all lines is 6326 (Set 1).
Geographic patterns
Interesting patterns arose when the geographic origins of chromosomes were considered (Figure 5). A general trend was that southern X chromosomes were more likely to result in synthetic incompatibilities than northern X chromosomes (two-tailed p-value = 0.013 for Set 1, two-tailed p-value = 0.259 for pooled data from Set 2; Fisher’s exact test). In addition, synthetic incompatibilities were much more common when X chromosomes were placed into the southern autosomal background (Rum Cay). More importantly, levels of synthetic incompatibilities depended on the combination of X chromosomes and autosomes. Southern X chromosomes were two and a half times more likely than northern X chromosomes to result in synthetic lethality or sterility when placed on a 6326 autosomal background (Set 2, single-tailed p-value = 0.081; Fisher’s exact test). When X chromosomes were placed into a northern autosomal background, southern X chromosomes were more likely to result in synthetic incompatibilities than northern X chromosomes (44.1% vs. 27.8%, single-tailed p-value = 0.198; Fisher’s exact test). When X chromosomes were placed into a southern autosomal background, northern X chromosomes were more likely to result in synthetic incompatibilities than southern X chromosomes (83.3% vs. 67.6%, single-tailed p-value = 0.190; Fisher’s exact test). While each of these geographical trends was not significant by itself, the data were what one would expect if local populations contain coadapted gene complexes. Sudbury and Rum Cay background data were combined to calculate the test statistic described in Equation 1, yielding γ = −0.320. This value of γ was statistically significant (p-value = 0.0445; only 4.45% of all Monte Carlo runs resulted in γ < −0.320). When African X chromosomes were omitted from Set 2, γ = −0.337 (p-value = 0.0397). Correlations between synthetic incompatibility and the great-circle distance between the geographic origin of X chromosomes and autosomes were weakly positive (ρ = 0.1461 for the Sudbury autosomal background and ρ = 0.0723 for the Rum Cay autosomal background). When African X chromosomes were omitted from Set 2, the correlations were slightly different (0.1366 for Sudbury and 0.1217 for Rum Cay). X-autosome combinations were more likely to result in incompatibilities when chromosomes were derived from different geographical regions.
Discussion
We observed substantial levels of synthetic incompatibility. These findings are consistent with theoretical models that predict recessive synthetic incompatibilities segregating at moderately high frequencies (Phillips and Johnson 1998; Proulx and Phillips 2005). Because each of the X chromosomes and autosomal backgrounds tested can be maintained indefinitely as an isofemale line, we were able to infer that observed incompatibilities are due to epistatic interactions involving X chromosomes and autosomes rather than single locus effects. Note that cytonuclear incompatibilities could be ruled out by our crossing scheme. The independence of X-autosome combinations and the failure of most female sterile X chromosomes to complement each other suggest that multiple genetic factors may be involved. Assuming that the incompatibilities observed in our study are the “stuff” of speciation, this perspective is consistent with the view that speciation can be due to genes at many different loci (Wu and Ting 2004). In addition, our data support past findings that the genetic basis of hybrid incompatibility is complex even at early stages of divergence (Good et al. 2008).
There are three likely reasons why our study detected much higher levels of synthetic incompatibilities than classic studies (Thompson 1986; Powell 1997). First, we assayed levels of synthetic sterility and lethality, as opposed to just synthetic lethality. Second, chromosomes assayed in our study differed in their geographic origins. Third, larger regions of the genome were tested in our study (instead of detecting incompatibilities between a single pair of chromosomes we were able to detect incompatibilities between X chromosomes and either autosome). Many of the lines constructed in this study are double handicapped: they contain homozygous variants for each chromosome (inbreeding depression) and X-autosome combinations from different geographic locations (outbreeding depression).
We did not observe increased levels of synthetic incompatibilities for the heterogametic sex, suggesting that Haldane’s rule may not apply for within-species variation of our study species. However, when we examined the genetic basis of female sterility we found evidence consistent with the dominance hypothesis of Haldane’s rule. This is because female-specific synthetic sterility involved recessive alleles in each case. As there is evidence of female-biased expression patterns on the X chromosome (Ranz et al. 2003), our findings are also consistent with the faster-X hypothesis. Had the faster male hypothesis held, we would have expected to observe greater levels of male sterility. Note that while our study reveals the recessivity of standing epistatic fitness variation, it does not directly explain what causes Haldane’s rule.
What can cause within species patterns of sterility and lethality to differ from between species patterns? One key difference between these two situations is that natural selection is able to eliminate deleterious combinations within species, but it is unable to eliminate deleterious combinations between species (barring reinforcement). Because X-linked sterility factors are unable to be masked in the heterogametic sex, natural selection is more effective at eliminating X-linked male sterility factors than recessive X-linked female sterility factors (Vicoso and Charlesworth 2006). Thus, within a single species the frequencies of naturally segregating X-linked alleles are likely to be greater for female synthetic sterile alleles than male synthetic sterile alleles. By contrast, sex-linked DBM incompatibilities between species have not had the chance to be filtered by natural selection. Note that a comprehensive mathematical treatment of synthetic incompatibilities at mutation-selection equilibrium only exists for autosomal loci at present (Phillips and Johnson 1998).
Another possible explanation for the absence of Haldane’s rule in our study is that we assayed homozygous flies. These are genotypes that can occur in the F2 generation of hybridizing populations. However, the original formulation and subsequent discussion of Haldane’s rule has largely focused on the F1 generation (Wu et al. 1996; Laurie 1997). It is unknown whether sex-biased patterns of synthetic incompatibilities are expected to differ between the F1 and F2 generations. For Set 2, 7.8% of X-autosome combinations resulted in male sterility and 9.1% of X-autosome combinations resulted in female sterility (Table 1, pooled sex-specific and both-sterile data). In natural populations this would actually result in higher frequencies of hemizygous males compared to homozygous females (assuming Hardy-Weinberg proportions).
Our data contained both sex-specific lethality and sex-specific sterility. While it is likely that genes affecting viability will have the same effect in both sexes, different physiological processes underlie female and male sterility. The D. melanogaster X chromosome is enriched for genes with female-biased expression and deficient for genes with male-biased expression (Ranz et al. 2003). Misregulation of X-linked genes (due to trans effects from the autosomal background) may affect each sex differently. Female-biased expression patterns of X-linked genes may also explain why we observed greater levels of female sterility for homozygous flies. However, mutation studies suggest that the number of X-linked genes involved in male and female fertility are approximately the same (Kaplan et al. 1970; Watanabe and Lee 1977).
Our data are consistent with the idea that genomes are locally coadapted gene complexes. X chromosomes and autosomes derived from the same geographic region were usually compatible. Conversely, northern X chromosomes were more likely to be incompatible with southern autosomes, and vice versa. In addition, there were weak positive correlations between synthetic incompatibility and the geographic distance between the origin of X chromosomes and autosomes. The existence of these incompatibilities may be due to the demographic history of this species. The spread of D. melanogaster into the New World likely arose via two separate routes: a northern route from Africa via Europe and a southern route involving direct immigration from Africa. North America and the Caribbean thus appear to be zones of secondary contact where potentially incompatible alleles can interact. Additional support for this hypothesis comes from the fact that Caribbean populations have phenotypes that are more similar to African than United States populations (Caracristi and Schlotterer 2003; Yukilevich and True 2008a). Synthetic incompatibilities can also be a byproduct of local selection pressures if locally adaptive alleles have pleiotropic effects. Despite the intriguing geographic patterns in our study, generalizations should be taken with caution. This is because only three autosomal backgrounds were tested and it is possible that the observed patterns are due to the specific backgrounds tested rather than geography. More data are needed, as are additional theoretical models of synthetic incompatibility that incorporate spatial population structure. Our findings indicate that levels of synthetic incompatibility may be underestimated if chromosomes from only a single location are tested. Natural populations may already contain the genetic potential for speciation in the form of cryptic DBM incompatibilities.
Supplementary Material
Acknowledgements
We thank Kelly Dyer, E. Hill-Burns, N. Johnson, R. Ng, R. Yukilevich, and three anonymous reviewers for helpful comments on this manuscript. Additional thanks are directed towards J. Lenci and J. Hyder for assistance in the genotyping of flies. We are grateful to T. Merritt, J. Pool, and R. Yukilevich for providing Drosophila strains. This work was supported in part by Stony Brook University, a National Institutes of Health grant (1RO1GM07171001) to J.R.T., and a National Institutes of Health predoctoral training grant (5 T32 GM007964–24).
References
- Alcazar R, Garcia AV, Parker JE, Reymond M. Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc Natl Acad Sci U S A. 2009;106:334–339. doi: 10.1073/pnas.0811734106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anholt RR, Dilda CL, Chang S, Fanara JJ, Kulkarni NH, Ganguly I, Rollmann SM, Kamdar KP, Mackay TF. The genetic architecture of odor-guided behavior in Drosophila: epistasis and the transcriptome. Nat Genet. 2003;35:180–184. doi: 10.1038/ng1240. [DOI] [PubMed] [Google Scholar]
- Bierne N, Bonhomme F, Boudry P, Szulkin M, David P. Fitness landscapes support the dominance theory of post-zygotic isolation in the mussels Mytilus edulis and M. galloprovincialis. Proc Biol Sci. 2006;273:1253–1260. doi: 10.1098/rspb.2005.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, Dangl JL, Weigel D. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 2007;5:e236. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science. 2006;314:1292–1295. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- Caracristi G, Schlotterer C. Genetic differentiation between American and European Drosophila melanogaster populations could be attributed to admixture of African alleles. Mol Biol Evol. 2003;20:792–799. doi: 10.1093/molbev/msg091. [DOI] [PubMed] [Google Scholar]
- Carling MD, Brumfield RT. Haldane's rule in an avian system: using cline theory and divergence population genetics to test for differential introgression of mitochondrial, autosomal, and sex-linked loci across the Passerina bunting hybrid zone. Evolution. 2008;62:2600–2615. doi: 10.1111/j.1558-5646.2008.00477.x. [DOI] [PubMed] [Google Scholar]
- Coyne J, Orr HA. Speciation. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- Demuth JP, Wade MJ. Population differentiation in the beetle Tribolium castaneum. II. Haldane'S rule and incipient speciation. Evolution. 2007;61:694–699. doi: 10.1111/j.1558-5646.2007.00049.x. [DOI] [PubMed] [Google Scholar]
- Dixon SJ, Fedyshyn Y, Koh JL, Prasad TS, Chahwan C, Chua G, Toufighi K, Baryshnikova A, Hayles J, Hoe KL, Kim DU, Park HO, Myers CL, Pandey A, Durocher D, Andrews BJ, Boone C. Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proc Natl Acad Sci U S A. 2008;105:16653–16658. doi: 10.1073/pnas.0806261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics of the Evolutionary Process. New York: Columbia University Press; 1970. [Google Scholar]
- Edmands S. Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol Ecol. 2007;16:463–475. doi: 10.1111/j.1365-294X.2006.03148.x. [DOI] [PubMed] [Google Scholar]
- Frank SA. Divergence of meiotic drive-suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution. 1991;45:262–267. doi: 10.1111/j.1558-5646.1991.tb04401.x. [DOI] [PubMed] [Google Scholar]
- Good JM, Handel MA, Nachman MW. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution. 2008;62:50–65. doi: 10.1111/j.1558-5646.2007.00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourbiere S, Mallet J. Are Species Real? The Shape of the Species Boundary with Exponential Failure, Reinforcement, and the 'Missing Snowball'. Evolution. 2009 doi: 10.1111/j.1558-5646.2009.00844.x. [DOI] [PubMed] [Google Scholar]
- Griswold CK, Masel J. Complex adaptations can drive the evolution of the capacitor [PSI], even with realistic rates of yeast sex. PLoS Genet. 2009;5:e1000517. doi: 10.1371/journal.pgen.1000517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 1922;12:101–109. [Google Scholar]
- Harrison JS, Edmands S. Chromosomal basis of viability differences in Tigriopus californicus interpopulation hybrids. J Evol Biol. 2006;19:2040–2051. doi: 10.1111/j.1420-9101.2006.01145.x. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Pomiankowski A. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane's rule and related phenomena. Genetics. 1991;128:841–858. doi: 10.1093/genetics/128.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N. In: Gene Interactions and the Origin of Species. Wolf J, Brodie E, Wade M, editors. New York: Epistasis and the Evolutionary Process. Oxford University Press; 2000. [Google Scholar]
- Kaplan WD, Seecof RL, Trout WE, Pasternack ME. Production and Relative Frequency of Maternally Influenced Lethals in Drosophila melanogaster. American Naturalist. 1970;104:261–271. [Google Scholar]
- Keller A. Drosophila melanogaster's history as a human commensal. Curr Biol. 2007;17:R77–R81. doi: 10.1016/j.cub.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Kim Y. Rate of adaptive peak shifts with partial genetic robustness. Evolution. 2007;61:1847–1856. doi: 10.1111/j.1558-5646.2007.00166.x. [DOI] [PubMed] [Google Scholar]
- Krimbas CB. Synthetic Sterility in Drosophila Willistoni. Proc Natl Acad Sci U S A. 1960;46:832–833. doi: 10.1073/pnas.46.6.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie CC. The weaker sex is heterogametic: 75 years of Haldane's rule. Genetics. 1997;147:937–951. doi: 10.1093/genetics/147.3.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JC. Synthetic lethality and semi-lethality among functionally related mutants of Drosophila melanfgaster. Genetics. 1968;59:37–44. doi: 10.1093/genetics/59.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Vicoso B, Berlin S, Charlesworth B. Effective population size and the faster-X effect: empirical results and their interpretation. Evolution. 2010;64:663–674. doi: 10.1111/j.1558-5646.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- Masly JP, Presgraves DC. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 2007;5:e243. doi: 10.1371/journal.pbio.0050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle LC, Nakazato T. Complex epistasis for Dobzhansky-Muller hybrid incompatibility in solanum. Genetics. 2009;181:347–351. doi: 10.1534/genetics.108.095679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics. 1995;139:1805–1813. doi: 10.1093/genetics/139.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palopoli MF, Wu CI. Genetics of hybrid male sterility between drosophila sibling species: a complex web of epistasis is revealed in interspecific studies. Genetics. 1994;138:329–341. doi: 10.1093/genetics/138.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC. Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet. 2008;9:855–867. doi: 10.1038/nrg2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips PC, Johnson NA. The population genetics of synthetic lethals. Genetics. 1998;150:449–458. doi: 10.1093/genetics/150.1.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Aquadro CF. History and structure of sub-Saharan populations of Drosophila melanogaster. Genetics. 2006;174:915–929. doi: 10.1534/genetics.106.058693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell J. Progress and Prospects in Evolutionary Biology: The Drosophila Model. New York: Oxford University Press; 1997. [Google Scholar]
- Presgraves DC. Speciation genetics: epistasis, conflict and the origin of species. Curr Biol. 2007;17:R125–R127. doi: 10.1016/j.cub.2006.12.030. [DOI] [PubMed] [Google Scholar]
- Proulx SR, Phillips PC. The Opportunity for Canalization and the Evolution of Genetic Networks. American Naturalist. 2005;165:147–162. doi: 10.1086/426873. [DOI] [PubMed] [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- Sanjuan R, Cuevas JM, Moya A, Elena SF. Epistasis and the adaptability of an RNA virus. Genetics. 2005;170:1001–1008. doi: 10.1534/genetics.105.040741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter D, Conte GL. Genetics and ecological speciation. Proc Natl Acad Sci U S A. 2009;106 Suppl 1:9955–9962. doi: 10.1073/pnas.0901264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezgin E, Duvernell DD, Matzkin LM, Duan Y, Zhu CT, Verrelli BC, Eanes WF. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics. 2004;168:923–931. doi: 10.1534/genetics.104.027649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Burrage LC, Sinasac DS, Hill AE, Ernest SR, O'Brien W, Courtland HW, Jepsen KJ, Kirby A, Kulbokas EJ, Daly MJ, Broman KW, Lander ES, Nadeau JH. Genetic architecture of complex traits: large phenotypic effects and pervasive epistasis. Proc Natl Acad Sci U S A. 2008;105:19910–19914. doi: 10.1073/pnas.0810388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Chen S, Hartl DL, Laurie CC. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics. 2003;164:1383–1397. doi: 10.1093/genetics/164.4.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Hartl DL, Laurie CC. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc Natl Acad Sci U S A. 2001;98:13183–13188. doi: 10.1073/pnas.231478798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temin RG, Meyer HU, Dawson PS, Crow JF. The influence of epistasis on homozygous viability depression in Drosophila melanogaster. Genetics. 1969;61:497–519. doi: 10.1093/genetics/61.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson V. Synthetic lethals: a critical review. Evolutionary Theory. 1986;8:1–13. [Google Scholar]
- Thonneau P, Marchand S, Tallec A, Ferial ML, Ducot B, Lansac J, Lopes P, Tabaste JM, Spira A. Incidence and main causes of infertility in a resident population (1,850,000) of three French regions (1988–1989) Hum Reprod. 1991;6:811–816. doi: 10.1093/oxfordjournals.humrep.a137433. [DOI] [PubMed] [Google Scholar]
- True JR, Weir BS, Laurie CC. A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics. 1996;142:819–837. doi: 10.1093/genetics/142.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Orr HA. The dominance theory of Haldane's rule. Genetics. 1995;140:389–402. doi: 10.1093/genetics/140.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Orr HA. Dominance, epistasis and the genetics of postzygotic isolation. Genetics. 2000;154:1663–1679. doi: 10.1093/genetics/154.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umina PA, Weeks AR, Kearney MR, McKechnie SW, Hoffmann AA. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science. 2005;308:691–693. doi: 10.1126/science.1109523. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Watanabe TK, Lee WH. Sterile mutation in Drosophila melanogaster. Genetical Research. 1977;30:107–113. [Google Scholar]
- Wolf J, Brodie E, Wade MJ, editors. Epistasis and the Evolutionary Process. New York: Oxford University Press; 2000. [Google Scholar]
- Wu CI, Davis AW. Evolution of postmating reproductive isolation: the composite nature of Haldane's rule and its genetic bases. Am Nat. 1993;142:187–212. doi: 10.1086/285534. [DOI] [PubMed] [Google Scholar]
- Wu CI, Hollocher H, Begun DJ, Aquadro CF, Xu Y, Wu ML. Sexual isolation in Drosophila melanogaster: a possible case of incipient speciation. Proc Natl Acad Sci U S A. 1995;92:2519–2523. doi: 10.1073/pnas.92.7.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Johnson NA, Palopoli MF. Haldane's rule and its legacy: Why are there so many sterile males? TREE. 1996;11:281–284. doi: 10.1016/0169-5347(96)10033-1. [DOI] [PubMed] [Google Scholar]
- Wu CI, Ting CT. Genes and speciation. Nat Rev Genet. 2004;5:114–122. doi: 10.1038/nrg1269. [DOI] [PubMed] [Google Scholar]
- Yukilevich R, Lachance J, Aoki F, True JR. Long-term adaptation of epistatic genetic networks. Evolution. 2008;62:2215–2235. doi: 10.1111/j.1558-5646.2008.00445.x. [DOI] [PubMed] [Google Scholar]
- Yukilevich R, True JR. African morphology, behavior and phermones underlie incipient sexual isolation between us and Caribbean Drosophila melanogaster. Evolution. 2008a;62:2807–2828. doi: 10.1111/j.1558-5646.2008.00488.x. [DOI] [PubMed] [Google Scholar]
- Yukilevich R, True JR. Incipient sexual isolation among cosmopolitan Drosophila melanogaster populations. Evolution. 2008b;62:2112–2121. doi: 10.1111/j.1558-5646.2008.00427.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.