Abstract
Hundreds of proteins are post-translationally inserted into the endoplasmic reticulum (ER) membrane by a single carboxy-terminal transmembrane domain (TMD)1. During targeting through the cytosol, the hydrophobic TMD of these tail-anchored (TA) proteins requires constant chaperoning to prevent aggregation or inappropriate interactions. A central component of this targeting system is TRC40, a conserved cytosolic factor that recognizes the TMD of TA proteins and delivers them to the ER for insertion2-4. The mechanism that permits TRC40 to effectively find and capture its TA protein cargos in a highly crowded cytosol is unknown. Here, we identify a conserved three-protein complex composed of Bat3, TRC35, and Ubl4A that facilitates TA protein capture by TRC40. This Bat3 complex is recruited to ribosomes synthesizing membrane proteins, interacts with the TMDs of newly released TA proteins, and transfers them to TRC40 for targeting. Depletion of the Bat3 complex allows non-TRC40 factors to compete for TA proteins, explaining their mislocalization in the analogous yeast deletion strains5-7. Thus, the Bat3 complex acts as a TMD selective chaperone that effectively channels TA proteins to the TRC40 insertion pathway.
The final step in TA protein targeting to the mammalian ER is mediated by TRC40 (also called Asna1)2,4. This factor interacts directly with TA substrates via their TMD, and upon targeting to the ER membrane via putative receptor(s), releases the TA protein in an ATPase-dependent manner for insertion into the lipid bilayer. The yeast homolog of TRC40, termed Get3, plays an analogous function in conjunction with its ER receptors Get1 and Get2 (3), and still poorly understood cytosolic factors Get4 and Get5 (5-10). In both cases, TRC40/Get3 must selectively and efficiently capture TA proteins via recognition of a hydrophobic TMD that is prone to aggregation, inappropriate interactions, or sequestration by other chaperones.
To investigate how TRC40 efficiently captures its TA protein cargo, we reconstituted this event in vitro and analyzed its requirements (Sup. Fig. S1). Sucrose gradient purified ribosome-nascent chains (RNCs) containing a tRNA-tethered TA protein (Sec61β) were released with puromycin in the presence of cytosolic fractions. Capture of the radiolabeled Sec61β by TRC40 was assayed by crosslinking. As expected, Sec61β released into a complete cytosol was captured by TRC40 in a TMD-dependent and energy-stimulated manner. Surprisingly, TRC40 in size fractionated cytosol was unable to capture Sec61β, which instead crosslinked to an unidentified protein we provisionally term p38 (Sup. Fig. S2). TRC40 capture could be restored by adding back one of the missing fractions. Further fractionation confirmed this ‘capture-stimulating’ factor is distinct from TRC40 (Sup. Fig. S3). Thus, efficient substrate capture by TRC40 in a complex environment requires an additional protein factor, the absence of which leads to substrate interactions with non-TRC40 proteins (e.g., p38) despite the availability of TRC40.
We postulated the capture-stimulating factor could be an intermediary that delivers substrates to TRC40. Such a factor should recognize TA substrates in a TMD-dependent manner, be found in fractions that contain ‘capture-stimulating’ activity, and be more easily observed upon TRC40 saturation. Crosslinking analyses of Sec61β overproduced in complete cytosol revealed two interacting partners of ~120 kD and 35 kD that met these criteria (Sup. Fig. S4). Our earlier affinity purification of Sec61β from large-scale translation reactions2 contained an associated product of ~120 kD (in addition to the major TRC40 band) that we identified by mass spectrometry as Bat3 (also known as Scythe/Bag6) (Sup. Fig. S5). Bat3 affinity purified from reticulocyte lysate co-purified with proteins of ~18 kD and 35 kD (Fig. 1A) that we identified by mass spectrometry as Ubl4A and C7ORF20, for which we propose the name TRC35 (Sup. Fig. S5). Antibodies against either Bat3, Ubl4A or TRC35 could co-immunoprecipitate the other two components, and affinity-depletion of any one of these components resulted in substantial depletion of the other two proteins (Sup. Fig. S6). Thus, Bat3 is part of a stable heterotrimeric complex in the cytosol.
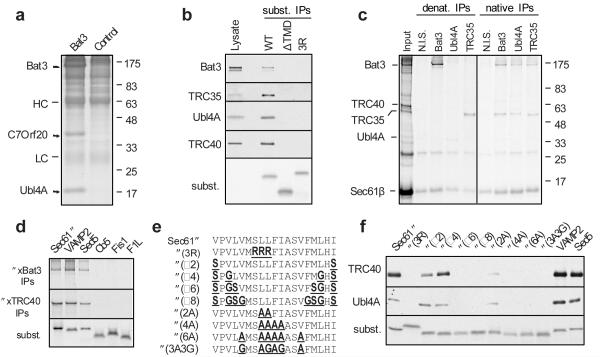
Fig. 1. Identification of a TMD-interacting protein complex.
a, Cytosolic proteins bound and eluted from anti-Bat3 or anti-GFP (control) affinity columns are shown. HC and LC are IgG heavy and light chain. b, Sec61β (WT), a deletion construct lacking its TMD (ΔTMD), or the insertion-deficient 3R mutant were translated in reticulocyte lysate, affinity purified on an anti-Sec61β column, and immunoblotted for the indicated products. Total lysate was included for comparison. An autoradiograph of the blot revealed equal recovery of the three translated substrates. c, Crosslinking products of Sec61β (from pooled sucrose gradient fractions 6-9 in Sup. Fig. S4) were immunoprecipitated under denaturing or native conditions. Non-immune serum (N.I.S) was included as a control. d, Versions of Sec61β containing the TMD from the indicated proteins were analyzed for interaction with Bat3 and TRC40 by in vitro translation, crosslinking, and immunoprecipitation. An aliquot of the total translation reaction is shown for each substrate (‘subst.’), as well as the immunoprecipitation products of the crosslinking reactions. e, The TMD of Sec61β was mutated to change its hydrophobicity as indicated. f, Each construct was analyzed for its interactions with Bat3 complex and TRC40 as in panel b. An aliquot of the total translation product was analyzed by autoradiography to visualize the substrates.
The TMD-dependent interaction of the Bat3 complex with TA proteins was verified by immunoblotting of Sec61β affinity purified from in vitro translation reactions (Fig. 1B). Furthermore, the TMD-dependent ~120 kD and 35 kD crosslinks to Sec61β (Sup. Fig. S4) could be specifically immunoprecipitated under denaturing conditions with anti-Bat3 and anti-TRC35 antibodies, respectively, or under non-denaturing conditions by anti-Ubl4A antibodies (Fig. 1C). Analysis of additional TA protein TMDs by crosslinking showed that both TRC40 and the Bat3 complex interacted with the ER-targeted VAMP2 and Sed5 TMDs, while neither interacted with the TMDs from spontaneously inserting cytochrome b5 (Cb5) or mitochondrially-targeted Fis1 or F1L (Fig. 1D). Differences in TMD hydrophobicity (either length or maximal hydrophobicity) among these substrates appeared to be a key determinant, since altering Sec61β TMD hydrophobicity to match that of Cb5, Fis1, or F1L markedly reduced interaction with both Bat3 complex and TRC40 (Fig. 1E,F; Sup. Fig. S7). Thus, the Bat3 complex interacts directly with the TMDs of several ER-targeted TA proteins with similar substrate-specificity as TRC40.
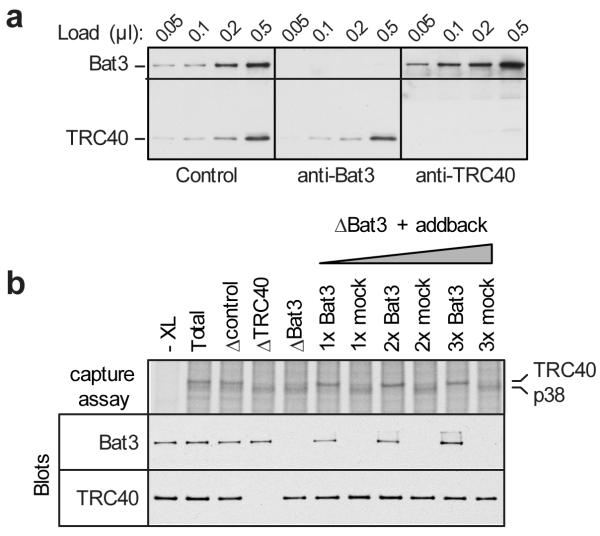
We surmised that the Bat3 complex was responsible for the stimulatory activity needed by TRC40 for efficient substrate capture. In cytosolic fractions immunodepleted (by ~95%) of Bat3 (which also depletes Ubl4A and TRC35 without affecting TRC40 levels; Fig. 2A and Sup. Fig. S6), Sec61β capture by TRC40 is diminished, with a concomitant increase in capture by p38 (Fig. 2B); a similar effect is seen after immunodepletion of TRC40, where p38 was the primary soluble interaction partner. Replenishment of Bat3 depleted extracts to the original level with affinity purified Bat3 complex [prepared using an anti-TRC35 antibody (Sup. Fig. S8)] fully restored TRC40 capture activity (Fig. 2B). The effect of Bat3 depletion on TRC40 substrate capture was not simply a consequence of the RNC-release assay since translation of full-length TA substrate in Bat3-depleted reticulocyte lysate also showed diminished TRC40 interactions with increased p38 interaction (Sup. Fig. S9). Thus, maximally efficient capture by TRC40 of TA proteins upon their release from the ribosome requires the Bat3 complex. These findings are consistent with recent observations suggesting that depletion of Bat3 can impair insertion of a TRC40-dependent TA protein11.
Fig. 2. The Bat3 complex mediates substrate capture by TRC40.
a, Translation extracts were passed over anti-GFP (control), anti-Bat3, or anti-TRC40 affinity resins and different amounts of each depleted lysate analyzed by immunoblotting. b, Substrate capture assay using either total cytosol, or cytosol immunodepleted (‘Δ’) with the indicated affinity resins. In this assay (see Sup. Fig. S1), radiolabeled Sec61β RNCs are released with puromycin and capture by TRC40 is assessed by crosslinking. The portion of gel showing the TRC40-Sec61β crosslink is shown. Failure of TRC40 to capture substrate typically results in capture by a 38 kD protein (p38). The ‘addback’ lanes are Bat3-depleted cytosol replenished with affinity purified Bat3 complex (prepared using an anti-TRC35 resin; Sup. Fig. S8) at three concentrations spanning that present in the original cytosol. The ‘mock’ addback sample was prepared in parallel, but employed an irrelevant affinity resin (anti-GFP) in lieu of TRC35 affinity resin (Sup. Fig. S8). A reaction lacking crosslinker (XL) is shown in the first lane. Aliquots of each reaction (prior to crosslinking) were also analyzed by immunoblot against Bat3 and TRC40 to document their relative amounts in the reactions.
Although the biochemical functions of Bat3, Ubl4A, and TRC35 are poorly understood, the latter two proteins have recognizable homologues in yeast (Mdy2/TMA24/Get5 and Yor164C/Get4, respectively) that have been analysed by recent biochemical, structural, and genetic studies5-7,12,13. Get4 and Get5 form a stable complex that can interact with Get3, the yeast TRC40 homolog5,7-10,12,13. Deletion strains of Get4 and Get5 appear to phenocopy Get1, Get2, or Get3 deletions5-7, and all five genes cluster together in synthetic genetic interaction arrays5,7. These observations have implicated Get4 and Get5 in TA insertion at a step prior to Get3, but their functions have been unclear.
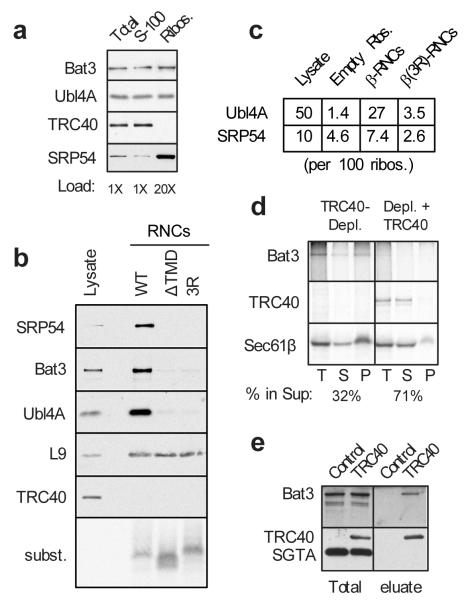
Given that Bat3 complex facilitates TRC40-substrate interactions, we hypothesized that Bat3 complex might capture TA substrates at the ribosome and transfer them to TRC40. Such a model requires the Bat3 complex to be at or near the ribosome. Indeed, affinity-purified RNCs containing a TA protein were markedly enriched in the Bat3 complex relative to empty ribosomes (Fig. 3A,B). Quantification showed that Bat3 complex and SRP occupy ~27% and ~7%, respectively, of TMD-containing RNCs (Fig. 3C). For comparison, SRP occupancy on a genuine SRP substrate isolated by the same method was ~30% (ref. 14; Sup. Fig. S10). Notably, Bat3 complex occupancies of <5% were observed for empty ribosomes and RNCs containing a mutant TMD, illustrating substrate selectivity for Bat3 complex recruitment. We confirmed that our affinity purified RNC preparations were tRNA-associated, as judged by their sensitivity to alkaline hydrolysis and selective precipitation with cetyl-trimethyl-ammonium-bromide (CTAB) (Sup. Fig. S11). Selective enrichment of Bat3 complex on TA protein RNCs was also seen using a magnetic bead ‘pull-up’ strategy that minimizes non-specific recovery of aggregates (Sup. Fig. S12). Thus, as suggested for SRP14, the Bat3 complex appears to be recruited to ribosomes when a functional TMD is inside the ribosome.
Fig. 3. Bat3 complex captures substrates on ribosomes for transfer to TRC40.
a, Ribosomes were purified under native conditions from reticulocyte lysate and analyzed for TRC40, Bat3, Ubl4A, and SRP54 by immunoblotting. Approximately 30-50% of SRP, 2-5% of Bat3 complex, and undetectable (<1%) amounts of TRC40 are ribosome-bound. b, Affinity and size purified RNCs (from 60 ul translation reactions) of Sec61β, a TMD-lacking version (ΔTMD), and the 3R mutant were analyzed by immunoblotting for the indicated antigens. For comparison, 0.5 ul translation lysate was analyzed. L9 is a ribosomal protein. Autoradiography confirmed equal recovery of each substrate. c, The amounts of ribosomes, Ubl4A, and SRP54 were quantified in total lysate, purified empty ribosomes, or purified RNC preparations. Shown are the amounts of SRP54 and Ubl4A, normalized to 100 ribosomes, averaged from multiple independent purifications (n=3 for empty ribosomes, n=7 for Sec61β-RNCs, and n=6 from β(3R)-RNCs). d, Translations of Sec61β in TRC40-depleted lysates lacking or replenished with recombinant zebrafish TRC40 were rapidly diluted, crosslinked, and separated by centrifugation into soluble and ribosome fractions. Sec61β, Bat3 crosslinks (immunopreciptiated with anti-Bat3), and TRC40 crosslinks are shown in the total (T), soluble (S), and ribosome (P) fractions. e, Cytosolic fractions from HT1080 cells lacking or stably over-expressing HA-tagged human TRC40 were bound to anti-HA resin and selectively eluted with TEV protease (which cleaves between the HA tag and TRC40). The eluted products, along with starting lysates, were analyzed by immunoblotting for Bat3, TRC40, or SGTA (a control protein). Note that endogenous TRC40 is present, but not visible at this exposure of the blot. Identical results were obtained when elution of the affinity column was with HA peptide instead of TEV protease (data not shown).
Analysis of various TMDs for their capacity to recruit Bat3 complex and SRP to the ribosome from inside the tunnel showed a general correlation with hydrophobicity, with bona fide ER-targeted TMDs (from Sec61β, VAMP2, and Sed5) showing the highest recruitment (Sup. Fig. S10). However, a strict concordance seems unlikely since a moderately hydrophobic Sec61β mutant (3A3G) that fails to interact with Bat3 complex post-translationally (Fig. 1F) nonetheless mediated its recruitment from inside the ribosome (Sup. Fig. S10). Interestingly, Bat3 complex did not crosslink to ribosome-tethered substrate (Sup. Fig. S13), but only upon release with puromycin or translational termination. This is opposite to SRP, which contacts substrates on the ribosome, but not after release. Thus, ribosomes synthesizing TMD-containing proteins can recruit both SRP and Bat3 complex at a stage before TMD emergence into the cytosol. Both of these complexes stay on the ribosome during further translation (Sup. Fig. S10), but upon substrate release, only the Bat3 complex remains associated in a TMD-dependent manner. Because changes in SRP recruitment do not influence Bat3 recovery on RNCs, the two factors may not compete with each other. However, this remains to be determined, as the binding site for Bat3 complex is not known.
Selective recruitment of the Bat3 complex to ribosomes before TA protein release suggested this may be the site of initial substrate capture. We therefore looked for a substrate-Bat3-ribosome intermediate upon TRC40 depletion. Sec61β translated in a TRC40-depleted lysate was ~70% ribosome-associated, where it could be crosslinked to Bat3 (Fig. 3D). When recombinant TRC40 was included in the depleted translation extract, the proportion of Sec61β in the ribosome fraction was decreased to ~30%, crosslinking to Bat3 was lost, and Sec61β in the soluble fraction was crosslinked to TRC40. Non-ribosomal Sec61β-Bat3 complexes can also transfer substrate to TRC40, since incubation of this fraction with TRC40-containing fractions resulted in a decrease in Bat3 crosslinks and concomitant increase in TRC40 crosslinks (data not shown). Thus, Bat3 complex is recruited to the ribosome, where it can interact with TA substrates upon their translational termination. This putative Bat3-substrate intermediate, whether on the ribosome or free in solution, is then converted into the productive Sec61β-TRC40 targeting complex. Consistent with this model, Bat3 and TRC40 can interact as determined by their co-immunoprecipitation under detergent free conditions (Fig. 3E).
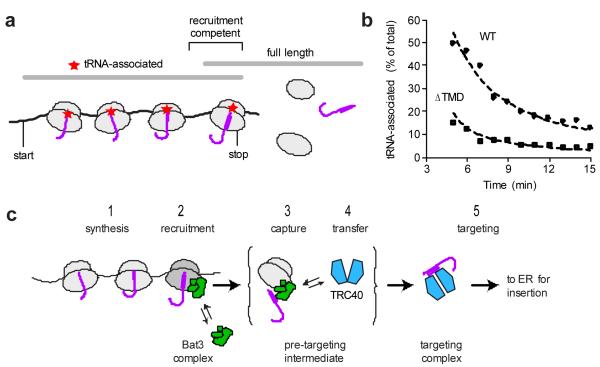
Bat3 recruitment to RNCs is restricted to the period between synthesis of the TMD and release from the ribosome (Fig. 4A), a time window that seems exceedingly brief. To explain this conundrum, we measured the recruitment window by performing a time course of TA protein synthesis combined with selective precipitation of tRNA-associated polypeptides with CTAB. Nascent chains available for Bat3 complex recruitment would be nearly full-length, but still contain a covalent tRNA. After validating that such tRNA-associated full-length species can be selectively recovered (Sup. Fig. S14), we quantified the amount of this population during a time course of Sec61β synthesis. Five minutes into this time course (~2 min after full-length Sec61β chains are first detected), a remarkable 50% of full-length Sec61β was precipitated by CTAB (Fig. 4B). This proportion diminished over time as translational termination ensued and completed Sec61β chains accumulated. Comparison of experimental data to theoretical expectations (Sup. Fig. S15) estimate a t1/2 for Sec61β termination of ~ 1 min.
Fig. 4. Translation termination is delayed for a TA protein.
a, Schematic of Sec61β synthesis illustrating that nascent chains competent for recruitment of Bat3 complex would be near full-length and contain a covalently associated tRNA (red star). Such recruitment-competent polypeptides should migrate on gels close to the full-length size, but be precipitated by CTAB, which selectively precipitates tRNA-associated proteins. b, Time course of Sec61β and Sec61β(ΔTMD) (both tagged at the C-terminus with the same 12-residue epitope tag) synthesized at 25°C in reticulocyte lysate. At each time point, samples were analyzed directly or after CTAB precipitation. The proportion of total full-length polypeptide that was CTAB precipitated is plotted. The dashed lines indicate theoretical expectations (Sup. Fig. S15) for a t1/2 of translational termination of 15 and 60 sec. c, Model for shuttling of TA proteins from the ribosome to TRC40 via a pre-targeting intermediate involving recruitment of the Bat3 complex to the ribosome.
Remarkably, Sec61β-ΔTMD (lacking the TMD) showed substantially less tRNA-associated polypeptide at each time point (Fig. 4B) corresponding to a termination t1/2 of ~ 15 sec. Importantly, the last 12 codons (containing an epitope tag) of both constructs are identical, arguing against differences in residues close to the peptidyl transferase centre as the basis for differences in termination rate. The translation elongation rate (~1.5-1.8 sec per residue) and the ~10-fold slower termination rate (relative to elongation) we measured for our control substrate (Sec61β-ΔTMD) are consistent with earlier estimates using this same in vitro system15. However, the unexpectedly long termination rate of Sec61β suggests that the TMD slows this reaction. The magnitude of termination delay seen for Sec61β (~1 min) is comparable to the elongation ‘arrest’ mediated by SRP measured under similar conditions in this same system16. This provides a plausible explanation for how Bat3 complex could have sufficient time to be recruited to RNCs containing a TMD inside the tunnel. The sequence parameters that modulate termination remain to be established, but is unlikely to be simple hydrophobicity given earlier studies showing similar termination delays by relatively hydrophilic sequences17.
Our findings explain how the hydrophobic TMDs of TA proteins are safely shielded from inappropriate interactions and aggregation during their delivery to the ER membrane (Fig. 4C). Our working model posits that the TMD is shielded throughout targeting, first by the ribosome, then by the Bat3 complex, and finally by TRC40. The sequential handoffs of substrate between these complexes are likely to be tightly regulated to prevent exposure of the TMD to aqueous solution. Perhaps the Bat3 complex regulates the nucleotide cycle and/or conformation of TRC40 to facilitate its loading with substrate. Based on recent crystal structures of Get3 (18-22), efficient substrate loading likely requires a nucleotide-dependent transition from an ‘open’ to ‘closed’ conformation18,19 that is perhaps aided by the Bat3 complex. In the absence of this activity, capture by TRC40 would be slower, explaining why it cannot operate efficiently in the presence of competing cytosolic factors, but does manage in a purified system (data not shown).
A role for the Bat3 complex in TA protein targeting may therefore not be absolutely essential23, but would increase fidelity and efficiency. Such a function would seem to be highly conserved, since yeast have a homologous complex (Get4 and Get5) that shows genetic, physical, and functional interactions with Get35-10,12,13, and whose absence can lead to aggregation and partial mislocalization of TA proteins5-7. Thus, the Bat3 complex appears to represent a conserved TMD-selective chaperone that acts at the ribosome. Where on the ribosome the Bat3 complex binds, how it is recruited when a TMD is inside the ribosomal tunnel, and how its function is coordinated with several other ribosome-associating factors including SRP24, NAC25, and RAC26, remain important questions for future studies27.
Methods Summary
Reagents and standard methods
Constructs, antibodies, and recombinant proteins derived from earlier studies2,18 are fully described in Supplementary Methods. Antibodies to Bat3 and Ubl4A were produced in rabbits immunized with recombinant Bat3 fragment (residues 1-250) or full-length Ubl4A, respectively. TRC35 antibodies were against a synthetic peptide (NH2-CGSPIELD-COOH) conjugated to KLH. In vitro transcription, translation in reticulocyte lysate, sucrose gradient fractionation, chemical crosslinking, and immunoprecipitation was as before2. RNCs were generated by transcription and translation of PCR products lacking a stop codon and sucrose gradient purified for use in the capture assay. CTAB precipitation of tRNA-associated polypeptides has been described28.
RNC release assay
[35S]-labeled RNCs were incubated with the indicated additions for 15 min at 32°C. Standard assay conditions were 100 mM KAc, 20 mM Hepes, pH 7.4, 7 mM MgAc2. Energy regenerating system consisted of 1 mM ATP, 1 mM GTP, 10 mM Creatine phosphate, 40 ug/ml creatine kinase. The base cytosolic fraction was prepared from reticulocyte lysate that was bound and eluted from DEAE-sepharose to remove hemoglobin and concentrated to half the original volume. Immunodepletions from this lysate employed immobilized antibodies. Following RNC release, crosslinking was with 250 uM bismaleimidohexane (BMH, a homo-bifunctional cysteine-reactive crosslinker) on ice for 30 min and quenched with 25 mM 2-mercaptoethanol.
Bat3 complex purification
Reticulocyte lysate was bound to phenyl-sepharose, washed in 50 mM Hepes pH 7.4, and eluted with 1% Triton X-100, 50 mM Hepes, pH 7.4. The eluate was bound and acid-eluted from anti-Bat3 affinity resin to identify interacting proteins by mass spectrometry. Functional Bat3 complex was isolated from the phenyl sepharose fraction using anti-TRC35 resin, eluted with immunizing peptide, and concentrated by binding and elution from S-sepharose.
RNC purification
Affinity purification of size-fractionated RNCs was essentially as described24, but with an antibody against the N-terminus of Sec61β and elution with immunizing peptide. The final RNCs were sedimented to remove any spontaneously released polypeptides.
Supplementary Material
Acknowledgements
We are grateful to S. Appathurai, and Maureen Downing for technical assistance, Hegde lab members for advice, and J. Weissman and W. Clemons for useful discussions and sharing results prior to publication. This work was supported by the Intramural Research Program of the National Institutes of Health (RSH) and Edward Mallinckrodt, Jr. Foundation (RJK).
Methods
DNA constructs
The SP64 vector-based constructs encoding wild type and ΔTMD Sec61β appended at the C-terminus with an epitope recognized by the 3F4 antibody have been described2. Mutants of Sec61β (including those where the entire TMD was replaced with other native TMDs) were generated by either PCR-based site-directed mutagenesis of Sec61β-3F4 (to generate the 3R mutant) or by inserting synthetic oligos into Sec61β(ΔTMD)-3F4 at a unique Age1 site between the Sec61β cytosolic domain and 3F4 tag. Sec61β-CFP and CFP-Sec61β have been described2. Sec61β-TR was generated by inserting synthetic oligos encoding the TMD of human transferrin receptor (IAVIVFFLIGFMIGYLGY) into the Bsg1 site at codon 50 in the cytosolic domain of Sec61β-3F4. This positions the TMD outside the ribosomal tunnel when the Sec61β TMD is inside the tunnel. Note that the TMD from TR has been experimentally verified to bind SRP as a co-translational substrate29. Sec61β-TR* was generated by inserting the same oligos in the reverse orientation, thereby coding for an irrelevant hydrophilic sequence (YPKYPIMNPIKKKTITAI). Translations of full-length products was as before2. To make RNCs, the above constructs were used to PCR amplify the coding region with an SP6 5′ primer and a 3′ primer that lacks a stop codon. For Sec61 β and the 3R mutant, the primer anneals at codons 91 to 96 of Sec61 β, after the TMD, but before the epitope tag. For the substrate where the TMD of Sec61 β was replaced with an irrelevant but equal length hydrophilic spacer sequence, codons encoding the spacer sequence were part of the primer used for PCR amplification. The final product codes for residues 1-66 of Sec61 β plus an irrelevant sequence (see Sup. Fig. S1). These three RNCs are therefore the same length, and differ only in the C-terminal region that resides within the ribosomal tunnel. For other RNCs (e.g., the various mutants and other TMD constructs), the reverse primer annealed in the 3F4 tag, or in the case of the F1L construct, at the end of the TMD. For the Sec61β-CFP RNCs, the reverse primer annealed within the CFP such that the Sec61β TMD was 46 residues away, and therefore outside the ribosomal tunnel.
Antibodies and proteins
All custom antibodies were raised in rabbits by Lampire Biological Laboratories. Rabbit polyclonal antibodies against N- and C-terminal peptides within mammalian TRC40 have been described2. Antibodies against TRC40, Bat3 and Ubl4A were raised in rabbits against full-length human TRC40, residues 1-250 of human Bat3, and full-length human Ubl4, respectively. These antigens were produced as His-tagged recombinant proteins expressed from the pRSETA vector and purified by Co+2 affinity chromatography. Antibodies against TRC35 were raised in rabbits against a synthetic peptide (CGSPIELD) conjugated to KLH. Recombinant zebrafish TRC40 was expressed as a His-tagged protein and purified as described previously18. The Bat3 complex used for functional reconstitution of depleted lysates was purified by a three-step procedure. First, crude ribosome-free reticulocyte lysate was passed over phenyl-sepharose (column volume was 0.4X the volume of lysate). After extensive washing in column buffer (10 mM Hepes, pH 7.4, 40 mM KAc, 1 mM MgAc2), the bound proteins were eluted with 1% Triton X-100 in column buffer. This eluate was adjusted to 150 mM KAc and 2 mM MgAc2, passed over a protein A resin coupled to anti-TRC35 antibodies (or a control anti-GFP column for the ‘mock’ addback). After washing with the same buffer, followed by a low-detergent buffer (25 mM Hepes, pH 7.0, 50 mM KAc, 2 mM MgAc2, 0.1% Triton X-100), selective elution was with 1 mg/ml immunizing peptide in the low-detergent buffer. This eluate was bound to S-sepharose, washed with detergent-free buffer, and step-eluted in 50 mM Hepes, pH 7.4, 500 mM KAc, 2 mM MgAc2. The concentration relative to starting lysate was quantified by comparative immunoblotting of serial dilutions. Bat3 complex purification in Fig. 1A was from the phenyl-sepharose elution using anti-Bat3 affinity resin. Control resin contained anti-GFP. Elution was with 0.1 M Glycine, pH 2.3, 1% Triton X-100.
Capture assay
In vitro transcription and translation in reticulocyte lysate to generate [35S]-labelled substrate was as before2. Translation time for RNC preparation was for 15 min at 32°C, after which the samples (typically 200 ul volume) were chilled on ice and immediately layered onto 2 ml 10-50% sucrose gradients in physiologic salt buffer (PSB: 100 mM KAc, 50 mM Hepes, pH 7.4, 2 mM MgAc2). Centrifugation was for 1 h at 55,000 rpm at 4°C in the TLS-55 rotor (Beckman), after which 200 ul fractions were removed from the top. The peak ribosomal fractions (6 and 7) were pooled, and used as the RNCs. Standard assay conditions were 100 mM KAc, 20 mM Hepes, pH 7.4, 7 mM MgAc2. Energy regenerating system consisted of 1 mM ATP, 1 mM GTP, 10 mM Creatine phosphate, 40 ug/ml creatine kinase. The base cytosolic fraction was prepared from reticulocyte lysate that was bound and eluted from DEAE-sepharose to remove hemoglobin and concentrated to half the original volume. Release was induced by addition of puromycin to 1 mM and incubation at 32°C for 15 min. Crosslinking was with 250 uM BMH on ice for 30 min and quenched with 25 mM 2-mercaptoethanol.
RNC affinity purification
RNCs were affinity purified using an antibody against the nascent chain similar to previous methods24. In brief, 300 ul translation reactions were subjected to a three-step procedure. Immediately after a 15 min translation reaction, samples were chilled on ice, adjusted to 1 uM emetine, and run by gravity over a 1 ml Sephacryl S-300 resin equilibrated in PSB. The void fraction, containing the RNCs but lacking molecules less than ~1500 kD, was then passed over an anti-Sec61 β affinity resin that recognizes the very N-terminus of Sec61β30. After washing with PSB, the bound RNCs were eluted with 1.5 mM of the immunizing peptide in PSB. Finally, the eluate was sedimented at 70,000 rpm in the TL100.2 rotor for 1 h, and the RNC pellet was analyzed by immunoblotting. As an alternate strategy in some experiments, the affinity column step above was replaced by magnetic bead isolation. Protein A-bound magnetic beads (Dyanbeads; Invitrogen) were first pre-bound with the anti-Sec61β antibody, washed, and added to the sample (the void fraction from the gel filtration step). After incubation at 4°C for 1 h, the beads were washed five times with PSB using a magnet to secure the beads on the side of the tube between washes. Elution and subsequent processing was as above.
Miscellaneous biochemistry
Analysis of crosslinking partners for full-length Sec61 β (or its mutant) in individual sucrose gradients and immunoprecipitation to confirm crosslinking adducts was as before2. For denaturing immunoprecipitations, samples after crosslinking were adjusted to 1% SDS, heated, and diluted 10-fold with IP buffer (1% Triton X-100, 50 mM Hepes, pH 7.4, and 100 mM NaCl). Native immunoprecipitations were performed on samples diluted directly into ice-cold IP buffer. SDS-PAGE was on either 8.5% or 12% Tris-tricine gels. The former resulted in separation of p38 crosslinks from TRC40 crosslinks, while the latter allowed visualization of the ~10 kD substrate. Typically, samples were run on both types of gels in parallel to both resolve the crosslinks and confirm equal sample recovery and loading. Preparation and use of antibody affinity columns was by standard methods. Depletions were performed on columns run by gravity flow. Preliminary experiments were used to determine the minimum amount of resin required to effect at least 90% depletion (as estimated by comparative blotting using serial dilutions). CTAB precipitation was as before28. In brief, samples to be precipitated were adjusted to 2% CTAB, mixed with an equal volume of 0.5M NaAc containing 0.2 mg/ml yeast or bovine liver tRNA, and incubated at 32°C for 10 min. The precipitate was collected by centrifugation for 10 min in a microcentrifuge, washed once in 1% CTAB, 0.25M NaAc at room temperature, and dissolved in sample buffer for subsequent analysis. Note that CTAB is a denaturing detergent, so translation (and other biochemical reactions) are instantly stopped upon addition to CTAB. Quantification of Bat3 complex levels in reticulocyte lysate was by semi-quantitative immunoblotting of serial dilutions relative to known quantities of purified recombinant Ubl4A. Ribosome levels in lysate were determined by their isolation by sedimentation and measurement of absorbance at 260 nm. Levels of proteins in RNCs were quantified by immunoblotting relative to serial dilutions of either lysate (whose abundances of individual proteins was known), purified ribosomes, and/or purified Ubl4A. Multiple independent determinations were made and averaged to generate the values in Fig. 3. Figures were prepared using Adobe Photoshop and Illustrator.
- 29.High S, Gorlich D, Wiedmann M, Rapoport TA, Dobberstein B. The identification of proteins in the proximity of signal-anchor sequences during their targeting to and insertion into the membrane of the ER. J Cell Biol. 1991;113:35–44. doi: 10.1083/jcb.113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fons RD, Bogert BA, Hegde RS. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003;160:529–39. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Footnotes
Supplementary Information consisting of 15 figures accompanies the paper.
References
- 1.Rabu C, Schmid V, Schwappach B, High S. Biogenesis of tail-anchored proteins: the beginning for the end? J Cell Sci. 2009;122:3605–12. doi: 10.1242/jcs.041210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–59. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 3.Schuldiner M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–45. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Favaloro V, Spasic M, Schwappach B, Dobberstein B. Distinct targeting pathways for the membrane insertion of tail-anchored (TA) proteins. J Cell Sci. 2008;121:1832–40. doi: 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jonikas MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–7. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copic A, et al. Genomewide analysis reveals novel pathways affecting endoplasmic reticulum homeostasis, protein modification and quality control. Genetics. 2009;182:757–69. doi: 10.1534/genetics.109.101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costanzo M, et al. The genetic landscape of a cell. Science. 2010;327:425–31. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang YW, et al. Crystal structure of Get4-Get5 complex and its interactions with Sgt2, Get3, and Ydj1. J Biol Chem. 2010;285:9962–70. doi: 10.1074/jbc.M109.087098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bozkurt G, et al. The structure of Get4 reveals an alpha-solenoid fold adapted for multiple interactions in tail-anchored protein biogenesis. FEBS Lett. 2010;584:1509–14. doi: 10.1016/j.febslet.2010.02.070. [DOI] [PubMed] [Google Scholar]
- 10.Chartron JW, Suloway CJM, Zaslaver M, Clemons WM., Jr. Structural characterization of the Get4/5 complex and its interaction with Get3. Proc Natl Acad Sci. 2010 doi: 10.1073/pnas.1006036107. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leznicki P, Clancy A, Schwappach B, High S. Bat3 promotes the membrane integration of tail-anchored proteins. J. Cell Sci. 2010 doi: 10.1242/jcs.066738. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito T, et al. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci. 2001;98:4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krogan NJ, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 14.Berndt U, Oellerer S, Zhang Y, Johnson AE, Rospert S. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc Natl Acad Sci. 2009;106:1398–403. doi: 10.1073/pnas.0808584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lodish HF, Jacobsen M. Regulation of hemoglobin synthesis. Equal rates of translation and termination of - and -globin chains. J Biol Chem. 1972;247:3622. [PubMed] [Google Scholar]
- 16.Wolin SL, Walter P. Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J Cell Biol. 1989;109:2617. doi: 10.1083/jcb.109.6.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao J, Geballe AP. Coding sequence-dependent ribosomal arrest at termination of translation. Mol Cell Biol. 1996;16:603. doi: 10.1128/mcb.16.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mateja A, et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461:361–6. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bozkurt G, et al. Structural insights into tail-anchored protein binding and membrane insertion by Get3. Proc Natl Acad Sci. 2009;106:21131–6. doi: 10.1073/pnas.0910223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suloway CJ, Chartron JW, Zaslaver M, Clemons WM., Jr. Model for eukaryotic tail-anchored protein binding based on the structure of Get3. Proc Natl Acad Sci. 2009;106:14849–54. doi: 10.1073/pnas.0907522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamagata A, et al. Structural insight into the membrane insertion of tail-anchored proteins by Get3. Genes Cells. 2010;15:29–41. doi: 10.1111/j.1365-2443.2009.01362.x. [DOI] [PubMed] [Google Scholar]
- 22.Hu J, Li J, Qian X, Denic V, Sha B. The crystal structures of yeast Get3 suggest a mechanism for tail-anchored protein membrane insertion. PLoS One. 2009;4:e8061. doi: 10.1371/journal.pone.0008061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Desmots F, Russell HR, Lee Y, Boyd K, McKinnon PJ. The reaper-binding protein scythe modulates apoptosis and proliferation during mammalian development. Mol Cell Biol. 2005;25:10329–37. doi: 10.1128/MCB.25.23.10329-10337.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halic M, et al. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–14. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 25.Wang S, Sakai H, Wiedmann M. NAC covers ribosome-associated nascent chains thereby forming a protective environment for regions of nascent chains just emerging from the peptidyl transferase center. J Cell Biol. 1995;130:519–28. doi: 10.1083/jcb.130.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gautschi M, et al. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc Natl Acad Sci. 2001;98:3762–7. doi: 10.1073/pnas.071057198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol. 2009;16:589–97. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 28.Hobden AN, Cundliffe E. The mode of action of alpha sarcin and a novel assay of the puromycin reaction. Biochem J. 1978;170:57–61. doi: 10.1042/bj1700057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.