Abstract
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces apoptosis through its cognate receptors death receptor 4 (DR4) and death receptor 5 (DR5), preferentially in malignant cells. However, many malignant cells remain resistant to TRAIL cytotoxicity by poorly characterized mechanisms. Here, using cholangiocarcinoma cells, as a model for TRAIL resistance, we identified a role for the oncogenic Hedgehog (Hh)-GLI pathway in the regulation of TRAIL cytotoxicity. Blockade of Hh using pharmacological and genetic tools sensitizes the cells to TRAIL cytotoxicity. Restoration of apoptosis sensitivity coincided with upregulation of DR4 expression, while expression of other death effector proteins remained unaltered. Knockdown of DR4 mimics Hh-mediated resistance to TRAIL cytotoxicity. Hh regulates the expression of DR4 by modulating the activity of its promoter. Luciferase, chromatin immunoprecipitation and expression assays show that the transcription factor GLI3 binds to the DR4 promoter and Hh requires an intact GLI3-repression activity to silence DR4 expression. Finally, small interfering RNA (siRNA)-targeted knockdown of GLI3, but not GLI1 or GLI2, restores DR4 expression and TRAIL sensitivity, indicating that the Hh effect is exclusively mediated by this transcription factor. In conclusion, these data provide evidence of a regulatory mechanism, which modulates TRAIL signaling in cancer cells and suggest new therapeutic approaches for TRAIL-resistant neoplasms.
Keywords: apoptosis, cholangiocarcinoma, GLI transcription factors, TRAIL
Introduction
Tumor necrosis factor–related apoptosis-inducing ligand or TRAIL (TNSF10) is a ligand that initiates signaling cascades by binding to two cognate receptors termed death receptor 4, DR4 (TNSF10A, also referred to as TRAIL receptor 1), and death receptor 5, DR5 (TNSF10B, also referred to as TRAIL receptor 2/KILLER/TRICK-2) (Johnstone et al., 2008). TRAIL can signal both apoptosis, especially in transformed cells (Johnstone et al., 2008), and non-apoptotic signaling cascades (Ishimura et al., 2006). Pro-apoptotic signaling by TRAIL and its receptors has been best characterized. Clustering and oligomerization of DR4 and DR5 by TRAIL results in conformational changes of the death domains within their cytoplasmic tails, facilitating recruitment and activation of caspases 8 and 10 within a death-inducing signaling complex (Kischkel et al., 1995; Ashkenazi and Dixit, 1999; Falschlehner et al., 2007). If activation of these initiator caspases is sufficiently robust, they directly activate caspase 3, which in turn results in cellular demise (Daniel et al., 2001; Eggert et al., 2001). This is referred to as type I death receptor signaling. However, if the magnitude of caspase 8 and 10 activation is not sufficient to directly activate caspase 3, TRAIL may still induce apoptosis by a pathway involving mitochondrial dysfunction, type II death receptor signaling (Daniel et al., 2001). In this pathway, caspase 8 and 10 result in the cleavage of the pro-apoptotic BH3-only protein Bid to generate a truncated Bid (tBID). This pro-apoptotic protein triggers the mitochondrial death pathway, a pathway of apoptosis, which is regulated by members of the mitochondrial B-cell leukemia 2 protein family (Liu et al., 1996; Kluck et al., 1997). Although these aspects of the cascade including its mitochondrial inhibitory mechanism are well characterized, the non-mitochondrial regulatory events modulating the response to TRAIL are still poorly understood.
Here, we used cholangiocarcinoma cells as a model to study TRAIL resistance. Given the emerging data implicating Hedgehog (Hh) signaling in gastrointestinal tumor biology (Lees et al., 2005; Parkin and Ingham, 2008), we explored Hh signaling as a mechanism contributing to TRAIL resistance by cancer cells. The data support a regulatory mechanism independent of mitochondria modulating cell-death response to TRAIL. We show that activation of the oncogenic Hh-GLI pathway represses the expression of DR4 receptor through regulation of its promoter. Mechanistically, we have shown that silencing of DR4 by Hh requires an intact GLI3 activity. Consistent with these data, the inhibition of Hh pathway sensitizes cholangiocarcinoma cells to TRAIL-induced apoptosis. Thus, our data define a mechanism by which oncogenic signaling could prevent TRAIL-induced cell death in cancer cells.
Results
Blockade of the Hh pathway sensitizes cholangiocarcinoma cells to TRAIL-induced apoptosis by upregulating DR4
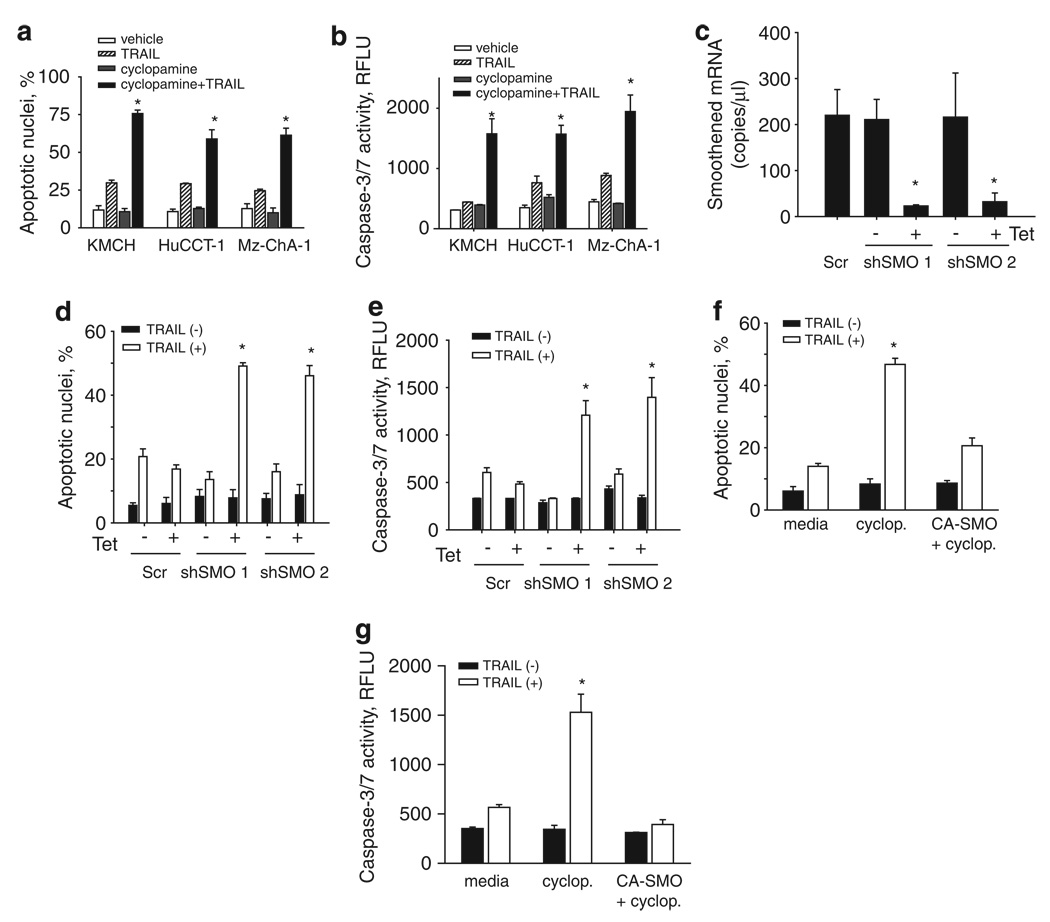
We first ascertained whether inhibiting Smoothened, the signaling component of the Hh receptor complex, using cyclopamine (Taipale et al., 2000; Chen et al., 2002), sensitizes cholangiocarcinoma cells to TRAIL-induced cytotoxicity. Neither TRAIL nor cyclopamine alone induced significant apoptosis in cholangiocarcinoma cells such as KMCH, HuCCT-1 and Mz-ChA-1. However, cyclopamine sensitized all the three human cholangiocarcinoma cell lines to TRAIL cytotoxicity (Figures 1a and b). This effect of cyclopamine was concentration- and time-dependent with sensitization as low as 500 nm, a maximal effect at 5 µm and utmost toxicity by 8 h of incubating KMCH cells with cyclopamine plus TRAIL (Supplementary Figure 1). In contrast, cyclopamine did not sensitize the TRAIL-resistant human hepatoblastoma cell line, human hepatocellular liver carcinoma cell line or the non-malignant normal rat cholangiocyte cell line to TRAIL cyctotoxicity (Supplementary Figure 2). Therefore, sensitization to TRAIL-mediated apoptosis by cyclopamine was concentration- and cell-specific.
Figure 1.
Cyclopamine sensitizes cholangiocarcinoma cells to Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated cytotoxicity. (a) Cells were pretreated with vehicle (ethanol, 0.01% v/v) or cyclopamine (5 µm) for 24 h. After pretreatment, human recombinant TRAIL was added where indicated at 5 ng/ml in fresh medium and the cells were incubated for an additional 5 h. Cells were then stained with 4′,6-diamino-2-phenylindole dihydrochloride (DAPI) and cells with apoptotic morphology were counted. Mean ± s.e.m., *P<0.001. (b) In parallel, cells were pretreated with vehicle or cyclopamine and treated with TRAIL as in panel (a) but after 6 h caspase-3/7 activity was measured. Mean ± s.e.m., *P<0.001. (c) KMCH cells were transfected with either of two independent short hairpin RNA (shRNA) expression vectors targeting Smoothened (shSMO 1 and shSMO 2) or a scrambled shRNA vector, followed by quantitative RT–PCR for Smoothened using total RNA. Results expressed as copies of each complementary DNA. Mean ± s.e.m., *P<0.001. (d) Cells transfected as in panel (c) were then treated with TRAIL (5 ng/ml) for 6 h, then stained with DAPI and cells with apoptotic morphology were counted. (e) Caspase-3/7 activity was measured 6 h after TRAIL treatment of cells transfected as in panel (c). Mean ± s.e.m., *P<0.001. (f, g) KMCH cells were pretreated with vehicle or cyclopamine as above, or transfected with a constitutively active Smoothened vector (CA-SMO). Cells were then stained with DAPI and cells with apoptotic morphology were counted (f) or caspase-3/7 activity was measured 6 h after TRAIL treatment (g). Mean ± s.e.m., *P<0.001.
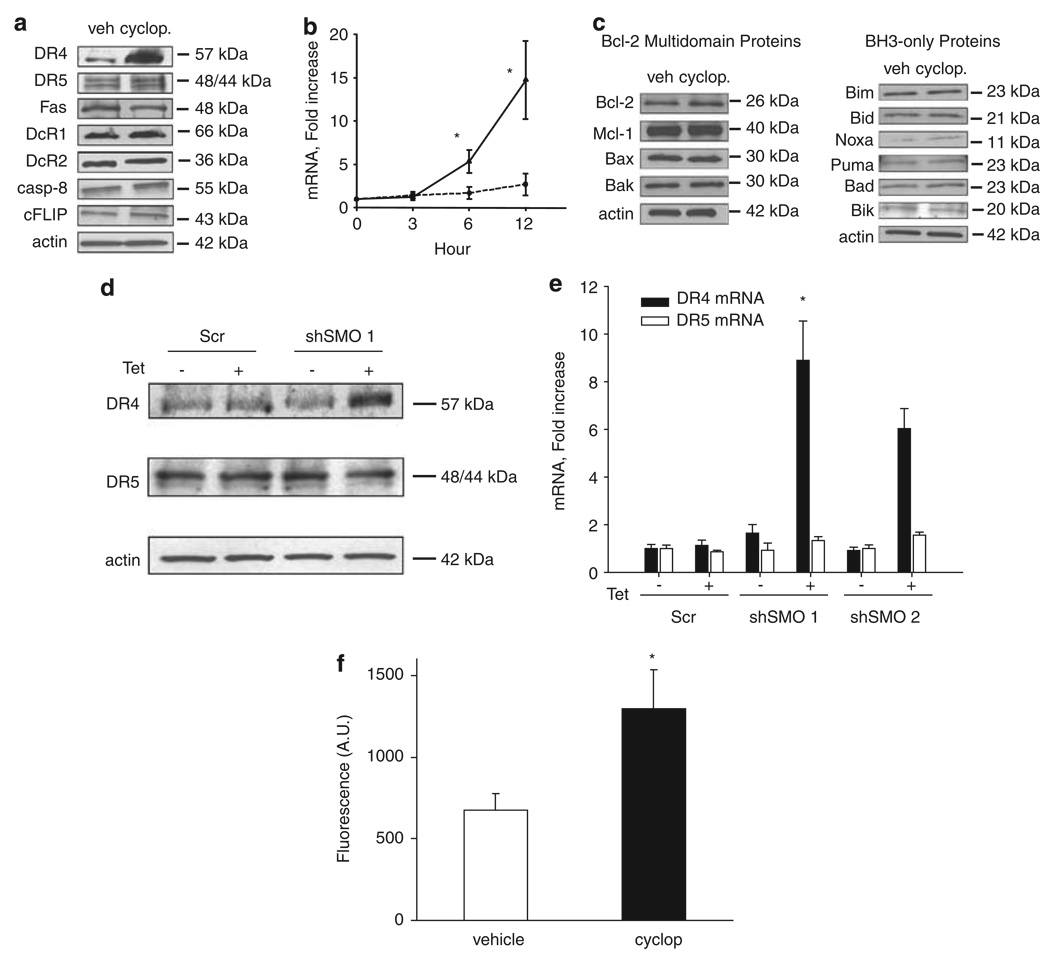
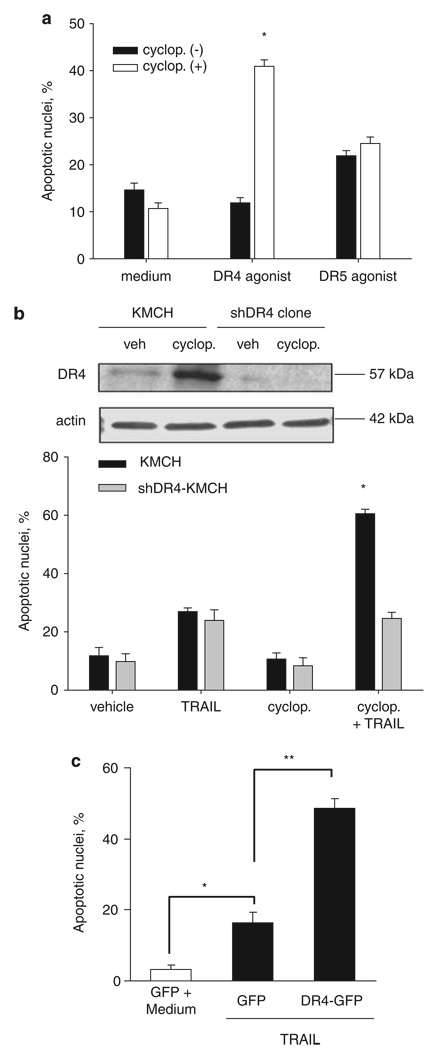
Validation using short hairpin RNA (shRNA)-targeted knockdown of Smoothened also sensitizes KMCH cells to TRAIL cytotoxicity (Figures 1c and e), showing that the relevant effect of cyclopamine is on Smoothened inhibition. Similarly, overexpression of a constitutively active, cyclopamine-insensitive, Smoothened was able to rescue the cyclopamine effect and restore the resistance to TRAIL cytotoxicity in KMCH cells (Figures 1f and g). Next, we sought to identify potential changes in expression of apoptotic mediators in cyclopamine treated cells, which could explain this sensitization to TRAIL-mediated apoptosis. After 24 h of cyclopamine treatment, DR4, but not DR5 protein and mRNA were upregulated in KMCH cells, whereas protein levels of other apoptotic mediators including Fas, TRAIL decoy receptors 1 and 2 (DcR1, DcR2), cellular FLICE-inhibitory protein, caspase 8 and members of the B-cell leukemia 2 protein family were unaltered (Figures 2a and c). In addition, validation using shRNA-targeted knockdown of Smoothened also upregulated DR4 expression at both mRNA and protein level (Figures 2d and e). This increase in total cellular DR4 protein levels was also associated with an increase in cell surface expression of the death receptor (Figure 2f). Increased DR4 mRNA and protein expression on cyclopamine treatment or knockdown of Smoothened suggested that sensitization to TRAIL cytotoxicity could be mediated specifically through DR4. Consistent with this interpretation, a DR4-specific agonistic antibody, but not a DR5-specific agonistic antibody, triggered apoptosis in cyclopamine-treated cells (Figure 3a). In addition, cells stably expressing shRNA-targeting DR4 were not sensitized to TRAIL cytotoxicity by cyclopamine (Figure 3b). Finally, DR4 overexpression sensitized the cells to a TRAIL-mediated cell death (Figure 3c). Taken together, these observations implicate DR4 upregulation as a likely mechanism responsible for TRAIL cytotoxicity in cyclopamine-treated cholangiocarcinoma cells.
Figure 2.
Cyclopamine upregulates death receptor 4 (DR4) expression. (a) Immunoblotting of cell lysates from KMCH cells treated with vehicle or cyclopamine (5 µm) for Fas, TRAIL receptors DR4, DR5, DcR1 and DcR2, and proximal signaling proteins caspase 8 and cellular FLICE-inhibitory protein. Actin was used as a loading control. (b) Quantitative RT–PCR for DR4 and DR5 using total RNA from KMCH cells after treatment with cyclopamine for the indicated times. Expression was normalized to 18S rRNA levels and presented as fold increase over time. Mean ± s.e.m., *P<0.01. (c) Whole-cell lysates of KMCH cells treated with vehicle or cyclopamine (5 µm) were prepared for immunoblotting for B-cell leukemia 2family proteins, including BH3-only proteins. (d) KMCH cells transfected with scrambled control short hairpin RNA (shRNA) or two shRNA sequences targeting Smoothened (SMO shRNA 1 and SMO shRNA 2) were treated with tetracycline (1 mg/ml) to induce shRNA expression and whole-cell lysates prepared for immunoblotting. (e) In parallel, total RNA was prepared from KMCH cells transfected as indicated and RT–PCR for DR4 or DR5 was performed. Mean ± s.e.m., presented as fold increase over scrambled. *P<0.01. (f) DR4 surface expression was performed by using confocal microscopy. KMCH cells treated with cyclopamine (5 µm) for 24 h. Quantification of DR4-green fluorescent protein (GFP) localization was accomplished using the LSM210 imaging software. Fluorescence intensity for the green channel was quantified. Mean ± s.e.m. *P<0.05.
Figure 3.
Death receptor 4 (DR4) receptor activation is necessary and sufficient for apoptosis in cyclopamine-sensitized cells. (a) Cells were pretreated with vehicle or cyclopamine (5 µm) for 24 h. DR4 agonist (0.1 µg/ml) or DR5 agonist (1 µg/ml) was then added in fresh medium and the cells were further incubated for the indicated time-intervals. Cells were then stained with 4′,6-diamino-2-phenylindole dihydrochloride (DAPI) and cells with apoptotic morphology were counted. Mean ± s.e.m., *P<0.001. (b) KMCH cells stably transfectred with DR4-shRNA and parent KMCH cells were incubated with vehicle or 5 µm cyclopamine for 24 h. Upper, Immunoblotting of whole cell lysates was performed using antibody against DR4. Actin was used as a loading control. Lower, Cells were pretreated where indicated with cyclopamine then treated with TRAIL (5 ng/ml) for 5 h followed by DAPI staining. Cells with apoptotic morphology were counted. Mean ± s.e.m., *P<0.001 compared with parent cells treated with TRAIL and cyclopamine. (c) KMCH cells transiently transfected with green fluroscent protein (GFP) (control) or DR4-GFP were treated where indicated with TRAIL (5 ng/ml) for 6 h. The number of GFP-positive cells with apoptotic nuclear morphology is presented as a percent of total GFP-positive cells. Data are mean ± standard error, *P<0.05; **P<0.01.
Cholangiocarcinoma cells have canonical Hh pathway
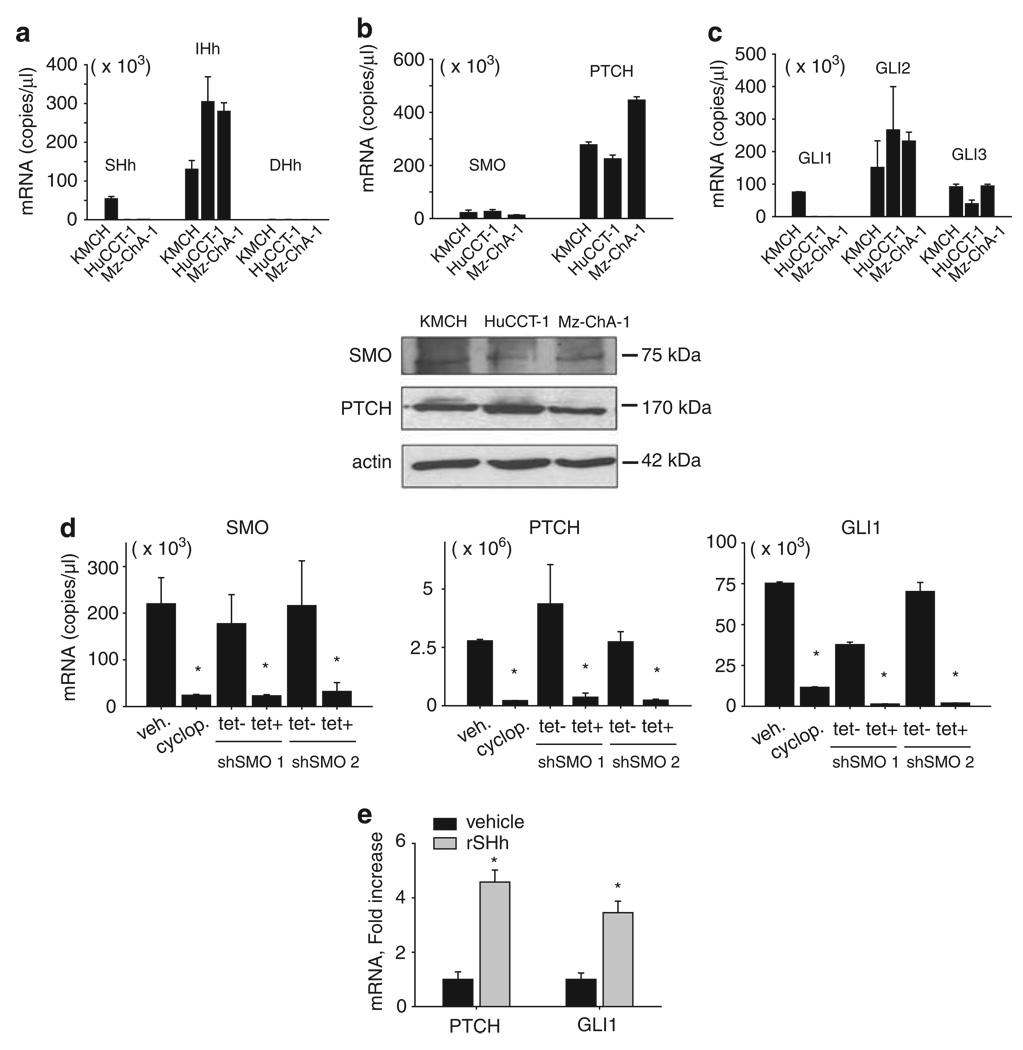
Sensitization to TRAIL-mediated apoptosis by cyclopamine suggests cholangiocarcinoma cells express and respond to Hh signaling ligands. To confirm this hypothesis, expression of the mediators of Hh-GLI signaling pathway was examined by quantitative RT–PCR (Figure 4). All three Hh ligands (Sonic, Indian and Desert) could be identified at the mRNA level in the three human cholangiocarcinoma cell lines. However, both Sonic Hedgehog (Shh) and Indian Hedgehog (Ihh) were predominantly expressed by KMCH cells whereas Indian Hedgehog was predominantly expressed only by HuCCT-1 and Mz-ChA-1 cells (Figure 4a). As acceptable commercial anti-sera for Hh ligands at the protein level are not available, protein expression for the ligands could not be confirmed. Expression for the cell surface proteins, Smoothened and Patched1, was confirmed by both RT–PCR and immunoblot analysis (Figure 4b); the specificity of the anti-sera for Smoothened was confirmed by using appropriate positive and negative controls (Supplementary Figure 3). The cell lines expressed the transcription factors regulated by Hh, GLI1, GLI2 and GLI3. (Figure 4c). Finally, in all the 47 human cholangiocarcinoma samples (containing cancer plus admixed stromal cells), mRNA transcripts were also identified for all the Hh pathway mediators (Supplementary Figure 4), thus supporting the biological relevance of our findings. To address whether the components of the Hh cascade are signaling through the canonical pathway, the cholangiocarcinoma cells were treated with cyclopamine, and the mRNA expression for GLI1 and Patched1, established transcriptional targets of canonical Hh pathway, were assayed by RT–PCR. GLI1 and Patched1 were downregulated by cyclopamine treatment as well as shRNA targeting the knockdown of Smoothened (Figure 4d). Conversely, both GLI1 and Patched1 were upregulated in KMCH cell treated with recombinant Shh ligand (Figure 4e). Collectively, these data suggest that the Hh signaling is constitutively active and acts through the canonical ligand activated pathway in human cholangiocarcinoma cells. Interestingly, cyclopamine alone did not alter cell proliferation or survival over 96 h (data not shown), suggesting the major effect of Hh inhibition is sensitization to TRAIL cytotoxicity.
Figure 4.
Cholangiocarcinoma cell lines expressed Hedgehog pathway mediators and respond to Hedgehog pathway activation and inhibition. (a) Quantitative RT–PCR for Sonic Hedgehog (Shh), Indian Hedgehog (Ihh), and Desert Hedgehog (Dhh) using total RNA from KMCH, HuCCT-1 and Mz-ChA-1 cells. Results are expressed as copies of each complementary DNA (cDNA) (Mean ± s.e.m.). (b) Upper, Quantitative RT–PCR for Smoothened and Patched-1 using total RNA from KMCH, HuCCT-1 and Mz-ChA-1 cells. Results are expressed as copies of each cDNA (Mean ± s.e.m.). Lower, Whole cell lysates from KMCH, HuCCT-1 and Mz-ChA-1 cells were probed using a Smoothened or a Patched-1 antibody. Actin was used as a loading control. (c) Quantitative RT–PCR for Hedgehog transcription factors, GLI1, -2 and -3, using total RNA from KMCH, HuCCT-1 and Mz-ChA-1 cells. Results expressed as copies of each cDNA (Mean ± s.e.m.). (d) Quantitative RT–PCR for Smoothened, Patched-1 and GLI1 using total RNA from KMCH cells after treatment with cyclopamine or gene silencing of Smoothened by a tetracycline-inducible shRNA technique. Results are expressed as copies of each cDNA (Mean ± s.e.m.). Cells were treated with cyclopamine 5 µm for 24 h before isolating mRNA or were transfected with Smoothened shRNA for 5 days before isolating mRNA. *P<0.001. (e) Quantitative RT–PCR for Patched-1, and GLI1 using total RNA from KMCH cells after treatment with recombinant sonic hedgehog ligand (rSHh). Results were normalized to 18S and expressed as fold increase over vehicle. Mean ± s.e.m., *P<0.01.
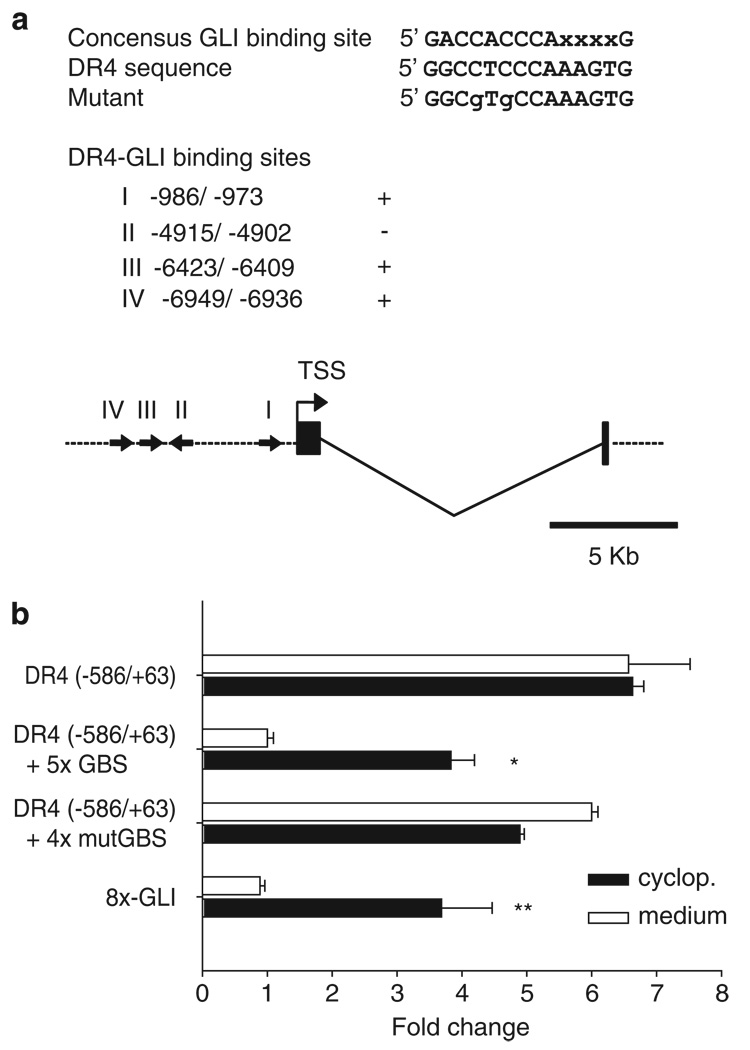
Hh silencing of the DR4 promoter requires intact GLI-binding sites
To determine if DR4 is transcriptionally regulated by GLI, we searched the 8 kb 5′-flanking region of the human and mouse DR4 gene for the 5′-GACCACCCAXXXXG-3′ GLI consensus-binding motif using the Enhancer Element Locator software (http://www.cs.helsinki.fi/u/kpalin/EEL/). In all, four candidate-binding sites (Figure 5a) were identified; all with the same sequence 5′-GGCCTCCCAAAGTG-3′, each had two mismatches (underlined) as compared with the consensus sequence. Using an earlier described luciferase reporter driven by the DR4-promoter fragment (−586/+63; Guan et al., 2002), we inserted a tandem repeat (five copies) of the DR4 candidate GLI-binding sites. To confirm whether GLI sites are required to modulate the DR4-promoter activity, we performed a luciferase-based assay using either the DR4-promoter construct alone (parental) or the DR4 promoter plus GLI-enhancer sequences. The parental construct was not responsive to cyclopamine, consistent with a lack of identified GLI-binding sites. However, when GLI-binding sites were inserted, the construct was Hh-responsive, as luciferase activity was increased by cyclopamine treatment (Figure 5b). In contrast, when mutant GLI-binding sites (four tandem copies) were incorporated into the reporter gene construct instead, luciferase activity was not enhanced by cyclopamine treatment. Consistent with these data, reporter gene activity was actually enhanced in the DR4-promoter construct containing mutated GLI-enhancer sequences compared with that containing intact GLI-binding sites, presumably because of the release from tonic GLI-mediated suppression (Figure 5b). Additionally, we assayed promoter function using an unrelated Hh-responsive construct containing eight tandem GLI-binding sites (Chen et al., 2009). This additional promoter was also stimulated by the cyclopamine treatment (Figure 5b). These data suggest that Hh may regulate DR4 gene expression through GLI-binding sites in the DR4 promoter.
Figure 5.
GLI3 directly repressed death receptor 4 (DR4) promoter function. (a) Putative GLI-binding sites in the DR4 promoter region. Nucleotide positions (I, II, III and IV) were counted from the transcription start site (TSS). Potential binding sites were observed in both the forward and the reverse direction; (+, sense; −, anti-sense) as indicated also by arrow on the schematic. (b) A DR4 promoter fragment (−586/+63) in pGL3, the same promoter plus five copies of the GLI-binding sequence (DR4 (−586/+63) + 5 × GBS), and a reporter construct containing 8 × GLI-binding sites (8 × -GLI) were used to evaluate the promoter activity on cyclopamine treatment. KMCH cells were co-transfected with 25 ng of pRL-CMV and 0.5 µg of indicated pGL3-based DR4 promoter reporter plasmids or 8 × -GLI reporter plasmids. Twenty-four hours after transfection, medium or cyclopamine (5 µm) was added where indicated for 24 h. Both firefly and Renilla luciferase activities were quantified and data (firefly/Renilla luciferase activity) are expressed as fold increase over DR4 promoter-reporter constructs containing 5 × GLI-binding sites treated with medium. Mean ± s.e.m., *P<0.001, **P<0.05.
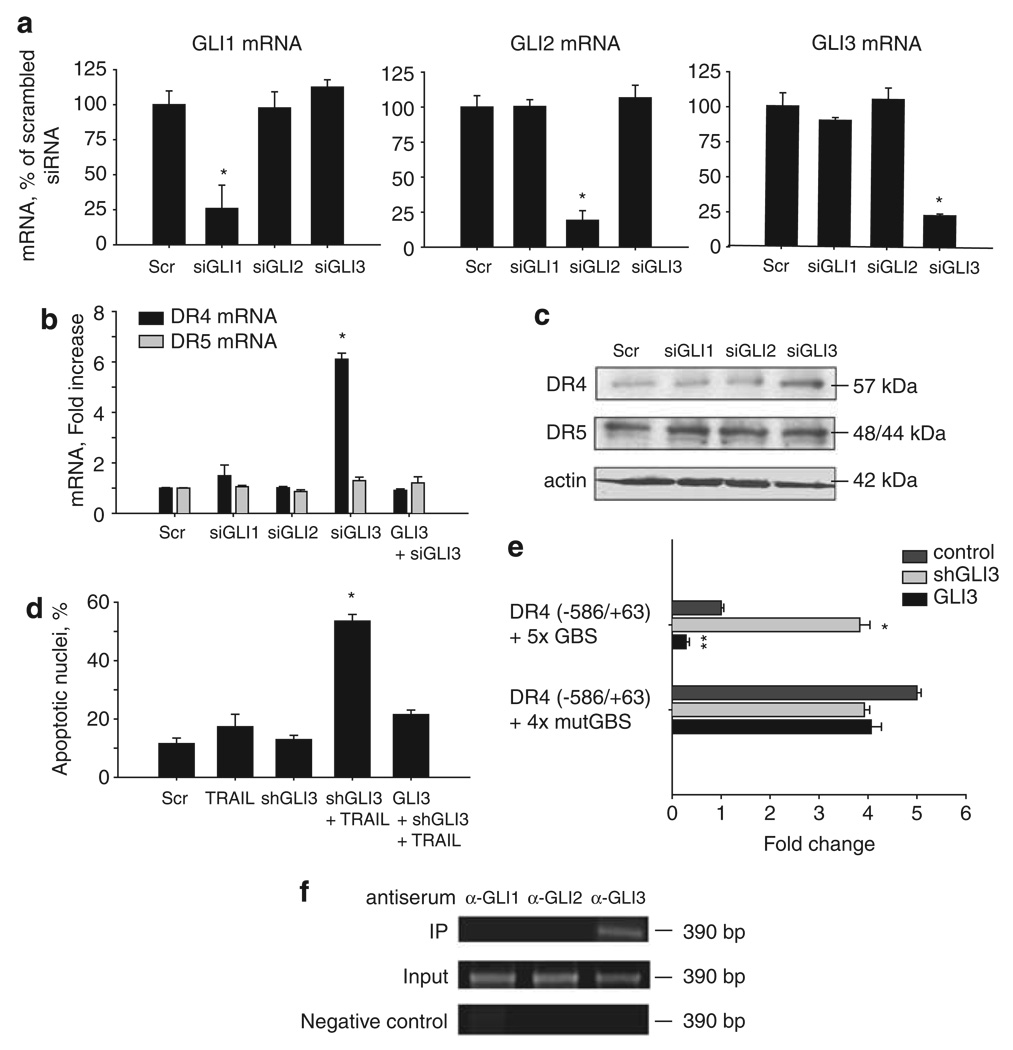
GLI3 knockdown restores DR4 levels and sensitizes cholangiocarcinoma cells to TRAIL-mediated apoptosis
Canonical Hh pathway signaling uses the GLI1, GLI2 and GLI3 proteins as transcriptional effectors of its cellular functions. To ascertain which of these GLI proteins likely suppresses the expression of DR4; GLI1, GLI2 and GLI3 were selectively knocked down by a small interfering RNA (siRNA) approach in KMCH cells (Figure 6a). DR4 expression was increased by gene silencing of GLI3, while gene silencing of GLI1 or GLI2 did not alter DR4 expression (Figures 6b and c). In contrast, gene silencing of GLI1, GLI2 or GLI3 did not alter the DR5 expression. These data suggest that the transcriptional factor, GLI3, downregulates DR4 expression in human cholangiocarcinoma cells by actively repressing its promoter. If this interpretation is correct, knockdown of GLI3 should sensitize cells to TRAIL cytotoxicity. Indeed, KMCH cells stably expressing shRNA-targeting GLI3 were sensitized to TRAIL-mediated apoptosis, while cells co-transfected with shRNA-targeting GLI3 plus an shRNA-resistant GLI3 were resistant to TRAIL cytotoxicity (Figure 6d). A parsimonious interpretation of these data is that GLI3 represses DR4 expression. Based on this assessment, we used the luciferase reporter assay to measure the effect of GLI3 silencing or overexpression on DR4-promoter function. Similar to cyclopamine, GLI3 gene silencing increased luciferase activity (Figure 6e). Consistent with repression by GLI3, DR4-reporter activity was decreased in GLI3 overexpressing cells (Figure 6e), as was the activity of an earlier described GLI-responsive promoter (Supplementary Figure 5). However, the reporter containing mutant GLI-binding sites did not have increased activity on GLI3 silencing, nor decreased function when GLI3 was overexpressed (Figure 6e). Finally, chromatin immunoprecipaitation assay showed that only GLI3 was found to bind consistently to the DR4 promoter (Figure 6f). We note that on occasion GLI2 binding was observed, suggesting that Gli dimers may participate in suppression of DR4 expression, but this observation could not be reliably replicated. Given that Gli3 siRNA alone relieved Hh repression of DR4, and the consistent results of the chromatin immunoprecipitation assay, we can only confidently implicate Gli3 in DR4 regulation.
Figure 6.
Gene silencing of GLI3 sensitizes cholangiocarcinoma cells to TRAIL-mediated apoptosis through death receptor 4 (DR4) upregulation. (a) Quantitative RT–PCR for GLI1, -2 and -3 using total RNA from KMCH cells transfected with the specific siRNA for GLI1, -2 or -3. Forty-eight hours after transfection, total RNA was isolated from cells. Cells transfected with scramble small interfering RNA (siRNA) were used as a control. Results normalized to 18S and expressed as percentage of scrambled. Mean ± s.e.m., *P<0.001. (b) Quantitative RT–PCR for DR4 and DR5 using total RNA from KMCH cells after the specific GLI-siRNA transfection. Forty-eight hours after transfection, total RNA was isolated from cells. Cells transfected with scramble siRNA were used as a control. Results normalized to 18S and expressed as fold increase over scramble. GLI3 silencing was rescued where indicated by co-transfection with a GLI3 expression plasmid resistant to the siRNA. Mean ± s.e.m., *P<0.001. (c) Immunoblotting of whole cell lysates obtained from KMCH cells transfected with the specific siRNA targeting GLI1, -2, or -3. Seventy-two hours after transfection, total cellular protein was isolated from the cells. Membranes were probed using a DR4 and a DR5 antibody. Actin was used as a loading control. (d) KMCH cells transfectred with GLI3 shRNA or co-transfected with shGLI3 plus a GLI3-expression-plasmid resistant to the shRNA were incubated with 5 ng/ml human recombinant TRAIL for 5 h. Cells were then stained with 4′,6-diamino-2-phenylindole dihydrochloride (DAPI) and cells with apoptotic morphology were counted. Mean ± s.e.m., *P<0.001 compared with vehicle. (e) DR4 promoter constructs (−586/+63) with a tandem repeat of the candidate GLI-binding site (DR4 (−586/+63) + 5 × GBS) or a tandem repeat of a mutated GLI-binding site (DR4 (−586/+63) + 4 × mutGBS) were assayed in KMCH cells. Cells were co-transfected with shGLI3 or a GLI3-expression construct as indicated. Mean ± s.e.m. *P<0.001 compared with medium; **P<0.005 compared with medium. (f) Chromatin immunoprecipitation was performed on KMCH cells using anti-GLI1, -GLI2 or –GLI3 anti-serum and the pull down used for DR4 promoter polymerase chain reaction using primers flanking the candidate GLI-binding site I. The polymerase chain reaction amplicon was visualized by ethidium bromide staining after agarose electrophoresis.
Discussion
Our study is consistent with previous reports that Hh signaling components are constitutively expressed by cholangiocarcinoma cells (Berman et al., 2003). However, this study extends these observations by showing that the Hh pathway imparts survival signals by inhibiting TRAIL cytotoxicity. This observation is quite relevant to human cholangiocarcinoma which paradoxically expresses TRAIL in vivo (Ishimura et al., 2006), and is, therefore, reliant on mechanisms to prevent TRAIL death signaling for survival. Given the role of immune surveillance in cancer biology by natural killer and natural killer T cells which eradicate target cells by TRAIL-mediated apoptosis (Johnstone et al., 2008), this function of Hh signaling in cancer biology could be pertinent to other cancers as well as cholangiocarcinoma.
The Hh signaling has been shown previously to regulate apoptotic pathways. For example, in basal cell carcinoma and central nervous system tumors, Hh regulates expression of the anti-apoptotic protein B-cell leukemia 2 and cellular FLICE-inhibitory protein (Bigelow et al., 2004; Regl et al., 2004; Kump et al., 2008). Despite its ability to promote apoptosis, cyclopamine treatment did not alter the B-cell leukemia 2or myeloid cell leukemia 1 protein expression in human cholangiocarcinoma cells. This was unexpected given that B-cell leukemia 2 family proteins, especially myeloid cell leukemia 1, are pivotal in inhibiting death ligand-induced apoptosis in cholangiocarcinoma cells (Taniai et al., 2004; Kobayashi et al., 2005; Mott et al., 2007). Cyclopamine did, however, result in enhanced DR4 expression. The upregulation of DR4 appears to be the mechanism for cyclopamine sensitization to TRAIL as cells in which DR4 expression was suppressed by targeted-shRNA, retained TRAIL resistance despite cyclopamine treatment. Inhibition of Hh signaling with enhanced DR expression also appears to overcome myeloid cell leukemia 1-mediated resistance to TRAIL cytotoxicity in cholangiocarcinoma cells. This observation suggests cyclopamine switches TRAIL signaling from a Type II pathway (dependent on mitochondrial dysfunction for apoptosis) to a Type I pathway (mitochondrial-independent pathway of apoptosis). Further studies will be required to examine this concept.
Why DR5, which is expressed by these cells, does not compensate for low-level DR4 expression in TRAIL killing is unclear and requires further investigation. TRAIL receptors undergo post-translational modification by O-glycosylation and perhaps other processes modulating their cytotoxicity (Wagner et al., 2007). Selective inactivation of DR5 through such modifications is a possible mechanism for this finding.
Previous studies reported that DNA-damaging agents, such as VP-16 and doxorubicin, induce DR4 expression by either p53-dependent or –independent mechanisms (Guan et al., 2001). There are few reports dealing with the issue of p53-independent regulation of DR4 expression. Nuclear factor-κB and activator protein-1 were recently reported to regulate DR4 expression at the transcriptional level (Guan et al., 2002; Mendoza et al., 2008). In this study, we have shown that gene silencing of GLI3 increased the expression of DR4, indicating that GLI3 suppresses DR4 expression. The inhibition of DR4 expression by GLI3 appears to be direct, as multiple GLI-binding elements are present in the promoter, and mutation of these elements increases the activity of the DR4 promoter. In addition, our data show a physical interaction of GLI3 protein with the endogenous DR4 promoter, which further supports regulation through direct binding. As our studies were performed in KMCH cells with mutant p53 (Wehbe et al., 2006), our observations provide mechanistic insight into the p53-independent regulation of this death receptor gene. Tonic suppression by GLI3 may be an important mechanism for its regulation.
In conclusion, our studies implicate Hh suppression of DR4 expression as a mechanism for TRAIL resistance in human cholangiocarcinoma cells. As these cancers express TRAIL in vivo, pharmacological inhibition of Hh signaling may promote autocrine and/or paracrine cholangiocarcinoma autonomous cytotoxicity. Collectively, these concepts suggest Hh pathway inhibitors, which are under clinical development (Rubin and de Sauvage, 2006; Rudin et al., 2009), could have therapeutic efficacy in the treatment of human cholangiocarcinoma.
Materials and methods
Cell culture
The human cholangiocarcinoma cell lines such as KMCH, HuCCT-1 and Mz-ChA-1 were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 000 U/l penicillin, 100 mg/l streptomycin and 100 mg/l gentamycin as previously described (Kobayashi et al., 2005; Isomoto et al., 2007).
Quantitative RT–PCR
Total RNA was isolated from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and complementary DNA was prepared using random primers and moloney murine leukemia virus-reverse transcriptase as previously described in detail (Higuchi et al., 2001). The complementary DNA product was amplified by polymerase chain reaction with Taq DNA polymerase using standard protocols. Polymerase chain reaction primers are depicted in Supplementary Table 1. Primers for 18S ribosomal RNA (rRNA) were purchased from Ambion (Austin, TX, USA). RT–PCR was performed with the Roche LightCycler using SYBR green as the fluorophore (Invitrogen) as previously described (Kobayashi et al., 2005). The copy number of the target mRNA in each sample was normalized as a ratio using the copy number for 18S rRNA in the denominator.
Immunoblot analysis
We obtained whole-cell lysates as previously described (Isomoto et al., 2007). SDS–PAGE resolved protein samples, transferred to nitrocellulose membranes and blotted with the indicated primary antibodies at a dilution of 1:500 to 1:1000. Peroxidase-conjugated secondary antibodies (Biosource International, Camarillo, CA, USA) were incubated at a dilution of 1:3000 to 1:5000. Bound antibodies were visualized using enhanced chemiluminescence reagents (Amersham, Arlington Heights, IL, USA) and Kodak X-OMAT film (Eastman Kodak, Rochester, NY, USA). Primary anti-sera taken, were those raised to Smoothened (Santa Cruz Biotechnology, Santa Cruz, CA, USA: H-300), patched-1 (Cell Signaling, Danvers, MA, USA), Bcl-2 and Mcl-1 (Santa Cruz Biotechnology: B19 for Bcl-2, S19 for Mcl-1), Bak (Upstate, Charrlotteville, VA, USA), Bax (Santa Cruz Biotechnology: C-12), Bid (R&D Systems, Minneapolis, MN, USA), Bim (BD Biosciences, San Jose, CA, USA), Bik and Bad (Santa Cruz Biotechnology: N-19), Puma (Abcam, Cambridge, MA, USA), Noxa (ProSci, Poway, CA, USA), caspase 8 (BD Biosciences), Fas (Santa Cruz Biotechnology: C-20), DR4 (Alexis, San Diego, CA, USA), DR5 and cFLIP (ProSci), DcR1 and DcR2 (Enzo Life Science, Plymouth Meeting, PA, USA), and actin (Santa Cruz Biotechnology: C-11).
Apoptosis
Apoptosis was quantified morphologically by staining the nuclei with 4′,6-diamino-2-phenylindole dihydrochloride and assessing the characteristic nuclear changes of apoptosis (that is, chromatin condensation and nuclear fragmentation) by fluorescence microscopy using excitation and emission wavelengths of 380 and 430 nm, respectively. Caspase-3/7 activity in cell cultures was used to assess apoptosis biochemically and was measured by quantifying rhodamine release from the caspase-3/7 substrate rhodamine-110, bis (N-CBZ-l-aspartyl-l-glutamyl-l-valyl-l-aspartic acid amide), using the Apo-ONE homogeneous caspase-3/7 kit (Promega, Madison, WI, USA) following the manufacturer’s instructions.
RNA interference
A siRNA was employed to silence GLI1, -2 or -3 expression. All siRNAs were designed and synthesized using the software available at http://www.ambion.com and the Silencer siRNA Construction kit from Ambion (Austin, TX, USA). The siRNA sequences used to inhibit GLI1, -2 and -3 expression were as follows: GLI1, 5′-AACTTTGATCCTTACCTCCCACCTGTCTC-3′; GLI2, 5′-AACTCTTGAGGTCTCTAACATCCTGTCTC-3′; GLI3, 5′-AAGGCATAATGTTGTCACAGACCTGTCTC-3′ (partial T7-promoter sequence is bold). As control, cells were transfected with a scrambled RNA duplex with the following sequence: 5′-AACGTGATTTATGTCACCAGA-3′. Inhibition of target mRNA expression was assessed after transient transfection of cells with siRNA by RT–PCR. Cells grown in six-well dishes were transiently transfected with 20 nm siRNA using 2.5 µl/ml LipofectAMINE 2000 (Invitrogen) in a total transfection volume of 2 ml of OPTI-MEM I (Gibco-Invitrogen, Carlsbad, CA, USA). After incubation at 37 °C and 5% CO2 for 4 h, the medium was exchanged with 2 ml of medium.
The shRNA for DR4 was from Sigma-Aldrich (St Louis, MO, USA) (MISSION short hairpin RNA lentiviral plasmid, targeting nucleotides 1499–1519, Genebank accession # NM 003844 for DR4-shRNA). To generate the GLI3-targeted shRNA construct, the pSSH1 plasmid containing the human H1 promoter for the expression of shRNA was used. A double-stranded DNA template (5′-GATCCCCGCAATAGGCTTTAGGAAAATTCAAGAGATTTTCCTAAAGCCTATTGCTTTTTGGAAA-3′) was inserted into the pSSH1 plasmid after the H1 RNA promoter. The DNA insert contains both sense and anti-sense sequences (bold type) for the GLI3 mRNA and a 9-nucleotide linker sequence, yielding transcription of an shRNA-targeting GLI3. For stable transfection, KMCH cells were transfected using OPTI-MEM I (Gibco-Invitrogen) containing 5 µl/ml LipofectAMINE 2000 (Invitrogen) and 3 µg/ml plasmid DNA. Forty-eight hours after transfection, fresh medium containing 1 µg/ml puromycin or 1.2 g/l G418 was added. Surviving clones were separated using cloning rings and individually cultured. lentiviral particles containing shRNA to Smoothened were constructed using the BLOCK-iT Inducible H1 Lentiviral RNAi System (Invitrogen), following the manufacturer’s protocol. Sequences (inserted into pLenti4/BLOCK-iT-DEST) are depicted in Supplementary Table 2. Cells were transduced with lentiviral particles containing pLenti6/TR 1-day before transduction with the inducible constructs as described above by following manufacturer’s protocol and the expression was induced with 1 mg/ml tetracycline (Sigma-Aldrich). After an additional 48 h, cells were used for the experiments. Target suppression was assessed by immunoblot analysis (DR4 and Smoothened) or by RT–PCR (GLI3).
DR4-surface expression
The KMCH cells were cultured on glass cover slip and allowed to grow to ~50% confluency. Following treatment with vehicle or cyclopamine, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline containing 0.1 M PIPES, 1 mm ethylene glycol tetra acetic acid and 3 mm magnesium sulfate. Cells were then blocked for 1 h at room temperature with phosphate-buffered saline containing 5% bovine serum albumin, 5% glycerol and 0.04% sodium azide. After incubation with polyclonal rabbit anti-DR4 (ProSci) in blocking buffer at 4 °C overnight, the cells were washed three times with phosphate-buffered saline , incubated with Alexa Flour 488-conjugated chicken anti-rabbit IgG (Molecular Probes, Carlsbad, CA, USA) at a dilution of 1:500 in blocking buffer for 1 h at 37 °C. Cells were then washed three times in phosphate-buffered saline and three times in H2O, mounted onto slides using ProLong Antifade Kit (Molecular Probes), and imaged by confocal microscopy, with excitation and emission wavelengths of 488 and 507 nm, respectively. Cell surface fluorescence was quantified using the LSM210 imaging software (Carl Zeiss Microimaging Inc., Thornwood, NJ, USA).
DR4 overexpression
The KMCH cells were transfected with green fluorescent protein (pEGFP-N1) or DR4-GFP (Akazawa et al., 2009) using FuGene HD. Twenty-four hours after transfection, fresh medium was added (untreated) or cells were treated with TRAIL (5 ng/ml) for 6 h, followed by 4′,6-diamino-2-phenylindole dihydrochloride staining. Green fluorescent protein - positive cells were then analyzed for apoptotic nuclear morphology.
Expression constructs for GLI3 and Smoothened
The GLI3 expression construct included the GLI3 cDNA in a pRK5 backbone, and was a kind gift from Dr Steven Yan Cheng (University of California, San Francisco, CA, USA). Cells were transfected using OPTI-MEM I (Gibco-Invitrogen) containing 5 µl/ml LipofectAMINE 2000 (Invitrogen) and 3 µg/ml plasmid DNA. Transfected cells were used for experiments 72 h after transfection. The coding sequence for Smoothened containing the M2 activating mutation and HSV glycoprotein d-amino terminal epitope tag (CA-SMO) was subcloned from pRK7 (Xie et al., 1998) into pCDNA 3.1 + by digestion at HindIII and EcoRI using standard recombinant DNA methodology. Cells were transfected using OPTI-MEM I (Gibco-Invitrogen) containing 5 µl/ml LipofectAMINE 2000 (Invitrogen), and 3 µg/ml plasmid DNA. Forty-eight hours after transfection, fresh medium containing 1.2 g/l G418 was added. Surviving clones were separated using cloning rings and individually cultured.
Reporter constructs and DR4 promoter-reporter assay
The luciferase reporter plasmid containing 0.65 kb DR4 5′-flanking region, pGL3 (−586/+63), has been described previously (Guan et al., 2002). Double-stranded oligonucleotides corresponding to the candidate GLI-binding sites in the DR4 promoter region were synthesized (Mayo DNA Synthesis Core Facility, Rochester, MN, USA) and ligated in tandem (five copies) at the BamHI site of the pGL3 (−586/+63) vector. As a negative control, double-stranded oligonucleotides with two C-to-G mutations in the DR4 GLI-binding site (Figure 5a) were synthesized and ligated (four tandem copies) at the BamHI site of the pGL3 (−586/+63) vector. The mutations were positioned within the consensus-binding region to disrupt GLI-binding. The vectors containing the correct and mutant inserts were sequenced to confirm that the GLI-binding site was as expected and that the luciferase gene did not contain cloning artifacts. As a positive control to determine GLI activity, a reporter containing eight consecutive GLI-binding sites downstream of the luciferase gene (8×-GLI) was included in the experimental paradigm (Sasaki et al., 1997). The 8×-GLI reporter was kindly provided by Dr Chi-chung Hui (Research Institute, Toronto, ON, Canada). The plasmid with the TRAIL-R1/DR4 (luciferase + DR4 promoter), the TRAIL-R1/DR4 GLI-binding site (luciferase + DR4 promoter + GLI-binding sites), the mutant binding site (luciferase + DR4 promoter + mutant GLI-binding sites) or 8×-GLI (luciferase + 8× GLI-binding sites) were transfected into KMCH cells using FuGene HD (Roche Diagnosis, Basel, Switzerland). Cells were co-transfected with 25 ng of a plasmid expressing Renilla luciferase (pRL-CMV) (Promega). Twenty-four hours after transfection, cell lysates were prepared, and both firefly and Renilla luciferase activities were quantified using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency and cell number.
Chromatin immunoprecipitation assay
The KMCH cells were treated with 1% formaldehyde for 15 min at room temperature, harvested in lysis buffer (R&D Systems) and sheared to fragment DNA of ~500 bp. Samples were then immunoprecipitated using GLI1, GLI2 or GLI3 anti-sera (R&D Systems) or beads alone at 4 °C overnight. Following immunoprecipitation, samples were washed and eluted using the chromatin immunoprecipitation kit (R&D systems) following manufacturer’s instructions. Cross-links were removed at 65 °C for 4 h and immunoprecipitated DNA was purified (Qiagen, Valencia, CA, USA) and subsequently amplified by polymerase chain reaction using a primer set for an area containing potential GLI-biding site I in DR4 promoter sequence: forward 5′-AGCGCAATGGCTCCATCTCGGCTC-3′, reverse 5′-AGTCACGGTCCTGCCTGCGAAGAA-3′. Polymerase chain reaction products were visualized by ethidium bromide staining after electrophoresis in a 2% agarose gel.
Supplementary Material
Acknowledgements
We thank Erin Nystuen-Bungum for her excellent secretarial assistance. This work was supported by NIH R01 Grants DK59427 (GJG), CA136526, (MEF-Z), CA100882 (LRR), the Clinical and Optical Microscopy Cores for P30 DK 84567, Mayo Clinic Pancreatic SPORE P50 CA102701 (MEF-Z), and the Mayo Foundation.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Akazawa Y, Mott JL, Bronk SF, Werneburg NW, Kahraman A, Guicciardi ME, et al. Death receptor 5 internalization is required for lysosomal permeabilization by TRAIL in malignant liver cell lines. Gastroenterology. 2009;136:2365–2376. e2361–e2367. doi: 10.1053/j.gastro.2009.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi A, Dixit VM. Apoptosis control by death and decoy receptors. Curr Opin Cell Biol. 1999;11:255–260. doi: 10.1016/s0955-0674(99)80034-9. [DOI] [PubMed] [Google Scholar]
- Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- Bigelow RL, Chari NS, Unden AB, Spurgers KB, Lee S, Roop DR, et al. Transcriptional regulation of bcl-2 mediated by the sonic hedgehog signaling pathway through gli-1. J Biol Chem. 2004;279:1197–1205. doi: 10.1074/jbc.M310589200. [DOI] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Wilson CW, Li YJ, Law KK, Lu CS, Gacayan R, et al. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel PT, Wieder T, Sturm I, Schulze-Osthoff K. The kiss of death: promises and failures of death receptors and ligands in cancer therapy. Leukemia. 2001;15:1022–1032. doi: 10.1038/sj.leu.2402169. [DOI] [PubMed] [Google Scholar]
- Eggert A, Grotzer MA, Zuzak TJ, Wiewrodt BR, Ho R, Ikegaki N, et al. Resistance to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in neuroblastoma cells correlates with a loss of caspase-8 expression. Cancer Res. 2001;61:1314–1319. [PubMed] [Google Scholar]
- Falschlehner C, Emmerich CH, Gerlach B, Walczak H. TRAIL signalling: decisions between life and death. Int J Biochem Cell Biol. 2007;39:1462–1475. doi: 10.1016/j.biocel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Guan B, Yue P, Clayman GL, Sun SY. Evidence that the death receptor DR4 is a DNA damage-inducible, p53-regulated gene. J Cell Physiol. 2001;188:98–105. doi: 10.1002/jcp.1101. [DOI] [PubMed] [Google Scholar]
- Guan B, Yue P, Lotan R, Sun SY. Evidence that the human death receptor 4 is regulated by activator protein 1. Oncogene. 2002;21:3121–3129. doi: 10.1038/sj.onc.1205430. [DOI] [PubMed] [Google Scholar]
- Higuchi H, Bronk SF, Takikawa Y, Werneburg N, Takimoto R, El-Deiry W, et al. The bile acid glycochenodeoxycholate induces trail-receptor 2/DR5 expression and apoptosis. J Biol Chem. 2001;276:38610–38618. doi: 10.1074/jbc.M105300200. [DOI] [PubMed] [Google Scholar]
- Ishimura N, Isomoto H, Bronk SF, Gores GJ. Trail induces cell migration and invasion in apoptosis-resistant cholangiocarcinoma cells. Am J Physiol Gastrointest Liver Physiol. 2006;290:G129–G136. doi: 10.1152/ajpgi.00242.2005. [DOI] [PubMed] [Google Scholar]
- Isomoto H, Mott JL, Kobayashi S, Werneburg NW, Bronk SF, Haan S, et al. Sustained IL-6/STAT-3 signaling in cholangiocarcinoma cells due to SOCS-3 epigenetic silencing. Gastroenterology. 2007;132:384–396. doi: 10.1053/j.gastro.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer. 2008;8:782–798. doi: 10.1038/nrc2465. [DOI] [PubMed] [Google Scholar]
- Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, et al. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD. The release of cytochrome c from mitochondria: a primary site for Bcl-2 regulation of apoptosis. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Werneburg NW, Bronk SF, Kaufmann SH, Gores GJ. Interleukin-6 contributes to Mcl-1 up-regulation and TRAIL resistance via an Akt-signaling pathway in cholangiocarcinoma cells. Gastroenterology. 2005;128:2054–2065. doi: 10.1053/j.gastro.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Kump E, Ji J, Wernli M, Hausermann P, Erb P. Gli2 upregulates cFlip and renders basal cell carcinoma cells resistant to death ligand-mediated apoptosis. Oncogene. 2008;27:3856–3864. doi: 10.1038/onc.2008.5. [DOI] [PubMed] [Google Scholar]
- Lees C, Howie S, Sartor RB, Satsangi J. The hedgehog signalling pathway in the gastrointestinal tract: implications for development, homeostasis, and disease. Gastroenterology. 2005;129:1696–1710. doi: 10.1053/j.gastro.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Mendoza FJ, Ishdorj G, Hu X, Gibson SB. Death receptor-4 (DR4) expression is regulated by transcription factor NF-kappaB in response to etoposide treatment. Apoptosis. 2008;13:756–770. doi: 10.1007/s10495-008-0210-0. [DOI] [PubMed] [Google Scholar]
- Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin CA, Ingham PW. The adventures of Sonic Hedgehog in development and repair. I. Hedgehog signaling in gastrointestinal development and disease. Am J Physiol Gastrointest Liver Physiol. 2008;294:G363–G367. doi: 10.1152/ajpgi.00457.2007. [DOI] [PubMed] [Google Scholar]
- Regl G, Kasper M, Schnidar H, Eichberger T, Neill GW, Philpott MP, et al. Activation of the BCL2 promoter in response to Hedgehog/GLI signal transduction is predominantly mediated by GLI2. Cancer Res. 2004;64:7724–7731. doi: 10.1158/0008-5472.CAN-04-1085. [DOI] [PubMed] [Google Scholar]
- Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov. 2006;5:1026–1033. doi: 10.1038/nrd2086. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, et al. Treatment of Medulloblastoma with Hedgehog Pathway Inhibitor GDC-0449. N Engl J Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- Taniai M, Grambihler A, Higuchi H, Werneburg N, Bronk SF, Farrugia DJ, et al. Mcl-1 mediates tumor necrosis factor-related apoptosis-inducing ligand resistance in human cholangiocarcinoma cells. Cancer Res. 2004;64:3517–3524. doi: 10.1158/0008-5472.CAN-03-2770. [DOI] [PubMed] [Google Scholar]
- Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K, et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med. 2007;13:1070–1077. doi: 10.1038/nm1627. [DOI] [PubMed] [Google Scholar]
- Wehbe H, Henson R, Lang M, Meng F, Patel T. Pifithrin-alpha enhances chemosensitivity by a p38 mitogen-activated protein kinase-dependent modulation of the eukaryotic initiation factor 4E in malignant cholangiocytes. J Pharmacol Exp Ther. 2006;319:1153–1161. doi: 10.1124/jpet.106.109835. [DOI] [PubMed] [Google Scholar]
- Xie J, Murone M, Luoh SM, Ryan A, Gu Q, Zhang C, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.