Abstract
The mammalian target of rapamycin (mTOR) has attracted substantial attention because of its involvement in a variety of diseases, such as cancer, cardiac hypertrophy, diabetes and obesity. Current knowledge indicates that mTOR functions as two distinct multiprotein complexes, mTORC1 and mTORC2. mTORC1 phosphorylates p70 S6 kinase (S6K1) and eukaryotic initiation factor 4E (eIF4E) binding protein 1 (4E-BP1), and regulates cell growth, proliferation, and survival by integrating hormones, growth factors, nutrients, stressors and energy signals. In contrast, mTORC2 is insensitive to nutrients or energy conditions. However, in response to hormones or growth factors, mTORC2 phosphorylates Akt, and regulates actin cytoskeleton and cell survival. These findings not only reveal the crucial role of mTOR in physiology and pathology, but also reflect the complexity of the mTOR signaling network. In this review, we discuss the advances in studies of the mTOR complexes, including the interacting proteins, the upstream regulators and the downstream effectors of mTOR complexes, as well as their implication in certain human diseases.
Keywords: Rapamycin, mTOR, S6K1, 4E-BP1, raptor, rictor, cancer, diabetes
1. Introduction
Rapamycin, a macrocyclic lactone produced by Streptomyces hygroscopicus, was first isolated from a soil sample of Easter Island (Rapa Nui) during a discovery program for anti-microbial agents in the early 1970s, and subsequently discovered to have equally potent immunosuppressive and anti-tumor properties [1-5]. In the early 1990s, during the yeast genetic screens for mutations that rescue the growth-inhibitory properties of rapamycin, two rapamycin target genes named TOR1 (the target of rapamycin 1) and TOR2 were identified [6, 7]. Further studies revealed that rapamycin requires an intracellular receptor, FKBP12 (FK506-binding protein 12 kDa). Upon entering the cells, rapamycin forms a complex with FKBP12 and then binds a region in the C terminus of TOR proteins termed FRB (FKB12-rapamycin binding) domain, thereby exerting its cell growth-inhibitory and cytotoxic effects by inhibiting the functions of TOR [8, 9]. Subsequent biochemical studies extended this model to mammalian cells, leading to the discovery of the mammalian target of rapamycin (mTOR) [10-13].
mTOR, also known as FRAP (FKBP12-rapamcyin-associated protein), RAFT1 (rapamycin and FKBP12 target), RAPT 1 (rapamycin target 1), or SEP (sirolimus effector protein), is a 289 kDa atypical serine/threonine (S/T) kinase [11-14]. mTOR is considered a member of the phosphatidylinositol 3-kinase (PI3K)-kinase-related kinase (PIKK) superfamily since its C-terminus shares strong homology to the catalytic domain of PI3K [15, 16]. A variety of members in this family, also including MEC1, ATM, ATR, DNA-PKcs, SMG-1 and TRRAP, are associated with diverse cellular functions, such as control of cell growth, cell cycle and DNA damage checkpoints, recombination and maintenance of telomere length [Fig. (1)] [17-19]. Cumulative evidence indicates that mTOR acts as a ‘master switch’ of cellular anabolic and catabolic processes, regulating the rate of cell growth and proliferation by virtue of its ability to sense mitogen, energy and nutrient levels [20, 21]. Deregulation of the mTOR pathway is frequently observed in various human diseases, such as cancer and diabetes. For example, activation of the mTOR pathway was noted in squamous cancers [22], adenocarcinomas [23], bronchioloalveolar carcinomas [24], colorectal cancers [25], astrocytomas [26] and glioblastomas [27]. A recent immunohistochemical study performed in tissue arrays containing 124 tumors from 8 common human tumor types showed that approximately 26% of tumors (32/124) are predicted to be sensitive to mTOR inhibition [28]. These findings indicate a crucial role of mTOR signaling in tumorigenesis.
Fig. (1).
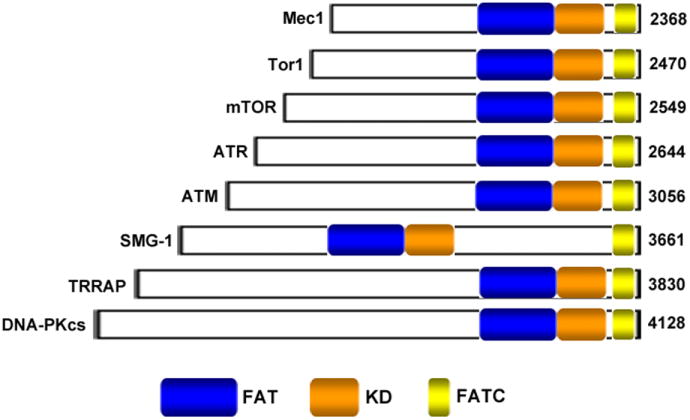
Domain structures of selected PIKKs. mTOR shown is of rat origin, and Mec1 and Tor1 are S. cerevisiae proteins. Other five proteins shown are of human origin. The number of residues for each protein is indicated in parentheses.
mTOR functions as two distinct signaling complexes, mTOR complex 1/2 (mTORC1/2), which are evolutionarily conserved from yeast to mammals. These two complexes consist of unique mTOR-interacting proteins which determine their substrate specificity. A rapamycin and nutrient-sensitive complex, mTORC1, consisting of mTOR, mLST8 (also termed G-protein β-subunit-like protein, GβL, a yeast homolog of LST8) and raptor (regulatory associated protein of mTOR) was first reported in 2002 [29, 30]. The main function of mTORC1 is to regulate cell growth, proliferation and survival by sensing mitogen, energy and nutrient signals [31]. In 2004, mTORC2, containing mTOR, mLST8 and rictor (rapamycin-insensitive companion of mTOR) was identified [32, 33]. mTORC2 was originally thought to be rapamycin-insensitive [32]. However, further studies demonstrated that prolonged rapamycin treatment inhibits the assembly and function of mTORC2 in some cell lines as well [34]. mTORC2 regulates the actin cytoskeleton by mediating phosphorylation state of protein kinase C α (PKCα) [33], and modulates cell survival in response to growth factors by phosphorylating its downstream effector Akt (protein kinase B, PKB) at the hydrophobic motif site, S473 [32, 35, 36]. These findings expand the role of mTOR in cells, and also reflect the complexity of the mTOR signaling network. In this review, we focus on the emerging studies of the mTOR complexes, including the interacting proteins, the upstream regulators and the downstream effectors of mTOR complexes, as well as their implication in some human diseases, such as cancer, cardiac hypertrophy, diabetes and obesity. Because of space limitation, we apologize for not being able to cite all related published studies. The readers who are particularly interested in learning more on how mTOR complexes are regulated are referred to the excellent review articles [37, 38].
2. mTORC1
Currently it is known that mTORC1 consists of mTOR, raptor, mLST8, and two negative regulators, PRAS40 (proline-rich Akt substrate 40 kDa) and DEPTOR (DEPDC6, DEP domain-containing protein 6) [Fig (2)] [30, 39-41]. mTOR is the core component of mTORC1/2 [11-14]. mTOR and yeast TOR proteins share >65% identity in their carboxy-terminal catalytic domains and >40% identity in overall sequence [42]. At the amino-acid level, human, mouse and rat TOR proteins share a 95% identity [43, 44]. Structurally, mTOR contains 2549 amino acids and the first 1200 amino acids comprise a tandemly repeated HEAT motifs including Huntingtin, elongation factor 3 (EF3), A subunit of protein phosphatase 2A (PP2A), and TOR [Fig. (3)] [45]. Tandem HEAT repeats are present in many proteins and may form an extended superhelical structure to create multiple large interfaces for protein-protein interaction. Immediately downstream of the HEAT repeat region lies a FAT (FRAP, ATM, and TRRAP) domain, an FKPB12-rapamycin binding (FRB) domain, a catalytic kinase domain, an auto-inhibitory (repressor domain or RD domain), and a FAT carboxy-terminal (FATC) domain, located at the C-terminus of the protein [Fig. (3)]. The FAT domain is always found in combination with the FATC region, so it has been hypothesized that the interactions between FAT and FATC might modulate catalytic kinase activity of mTOR [45, 46].
Fig. (2).
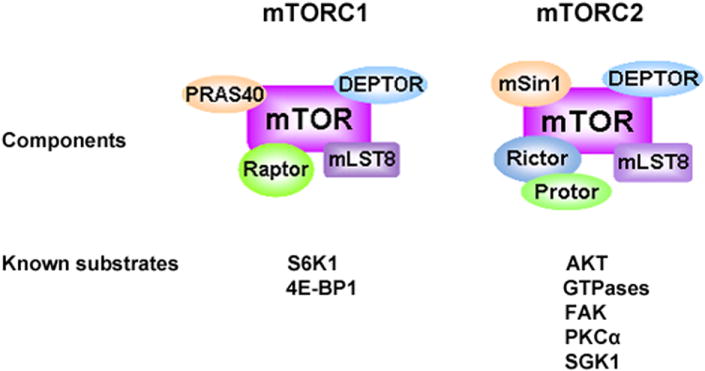
mTOR complexes and their known substrates. mTOR functions as two distinct complexes, mTORC1 and mTORC2. Besides mTOR, mTORC1 contains a positive regulatory subunit, raptor, two negative regulators, PRAS40 and DEPTOR, and a protein of unknown function called mLST8. mTORC2 also contains mTOR, mLST8 and DEPTOR, as well as other unique subunits, rictor, mSin1 and PROTOR.
Fig. (3).
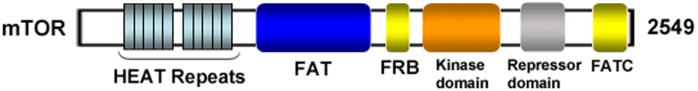
Schematic structure of mTOR. N-terminus of mTOR contains two tandemly repeated HEAT motifs. Following the HEAT repeat region, there exist a FAT domain, an FRB domain, a catalytic kinase domain, an auto-inhibitory (repressor domain or RD domain), and a FATC domain that is located at the C-terminus of the protein.
Raptor is an 150 kDa mTORC1 constitutively binding protein, which contains a highly conserved N-terminal domain followed by three HEAT repeats and seven WD40 repeats [29, 30]. Raptor is necessary for the regulation of mTORC1 activity in response to insulin, nutrient and energy level [29]. It has been described that raptor functions as an essential scaffold protein for mTOR to phosphorylate the downstream effectors, p70 S6 kinase (S6K1) and eukaryotic initiation factor 4E (eIF4E) binding protein 1 (4E-BP1), but whether raptor positively or negatively regulates mTOR kinase activity remains controversial [30]. It was speculated that at least two interactions exist between raptor and mTOR under nutrient-rich and -poor conditions. One mTOR-raptor complex that forms in the absence of nutrients is stable and leads to a repression of the mTOR catalytic activity. The other complex that forms under nutrients-rich conditions is unstable, but it is important for in vivo mTOR function [29]. Recent studies suggested that mTORC1 activity can be regulated by the phosphorylation status of raptor [47]. Phosphorylation of raptor on S722/792 mediated by AMPK is required for the inhibition of mTORC1 and cell cycle arrest induced by energy stress [47], whereas phosphorylation of raptor on S719/721/722 mediated by the p90 ribosomal S6 kinases (RSKs) is essential for the activation of mTORC1 by mitogen stimulation [47, 48]. Most recently, S863 in raptor was identified as mTOR-mediated phosphorylation site and proposed to be critical for the activation of mTORC1 in response to insulin [49].
mLST8, an additional subunit of mTORC1, was speculated to bind to the kinase domain of mTOR and positively regulate the mTOR kinase activity when first identified [39]. It was suggested that mLST8 is essential for a nutrient- and rapamycin-sensitive interaction between raptor and mTOR [39]. However, recent studies indicated that mLST8 is necessary for maintaining the rictor-mTOR interaction and the signaling of mTORC2, but not for that of mTORC1 [50]. Clearly, further studies are needed to elucidate the role of mLST8 in mTOR signaling.
PRAS40, another subunit of mTORC1, has been identified to be a direct negative regulator of mTORC1 function [40]. Initially, PRAS40 was identified as a novel substrate of Akt and a major 14-3-3 binding protein [51]. Akt activated by insulin directly phosphorylates PRAS40 at T246 [51]. Subsequent studies showed that PRAS40 associates with mTORC1 via raptor and inhibits mTORC1 activity [40]. Recently, PRAS40 has been implicated as a physiological substrate of mTORC1 [52, 53]. When binding to raptor, PRAS40 is phosphorylated on S183 by mTORC1 both in vivo and in vitro [52, 53]. Most recently, two novel sites in PRAS40 phosphorylated by mTORC1, S212 and S221, have been identified [54]. Rapamycin treatment reduced the phosphorylation of S183 and S221 but not S212, indicating that besides mTORC1, other kinases may regulate the phosphorylation of S212 in vivo [54]. The functional significance of phosphorylation of PRAS40 on S183, S212 and S221 remains to be determined.
2.1. Upstream regulators of mTORC1
As mentioned above, mTORC1 is rapamycin-sensitive and the main function of mTORC1 is to regulate cell growth, proliferation and survival by sensing mitogen, energy and nutrient signals [31]. The mTOR signaling can be activated by upstream signals, including growth factors, such as insulin and type I insulin-like growth factor (IGF-1), energy, stress and nutrients [31].
2.1.1. Growth factors
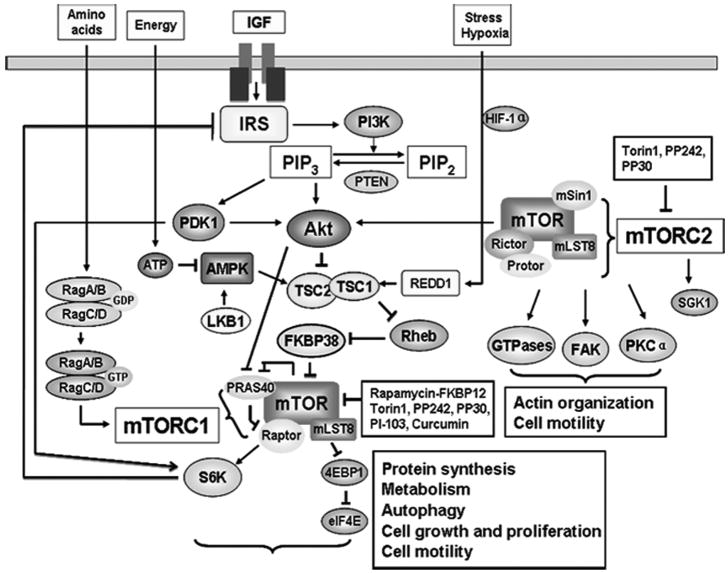
In response to ligand binding, the type I insulin-like growth factor receptor (IGFR), a transmembrane tyrosine kinase, is activated via auto-phosphorylation of multiple tyrosine residues. Activated IGFR in turn phosphorylates the insulin receptor substrates 1-4 (IRS1-4) and src- and collagen-homology (SHC) adaptor proteins, which can trigger multiple downstream signal transduction pathways, such as phosphatidylinositol 3′ kinase (PI3K) pathway [Fig. (4)] [55]. Phosphorylated IRS recruits the p85 subunit of PI3K and signals to the p110 catalytic subunit of PI3K, resulting in activation of PI3K. Activated PI3K catalyzes the conversion of phosphatidylinositol (4, 5)-bisphosphate (PIP2) to phosphatidylinositol-3, 4, 5-trisphosphate (PIP3). This pathway is negatively regulated by PTEN (phosphatase and tensin homolog on chromosome ten), also known as MMAC1 (mutated in multiple advanced cancers), a dual-specificity protein and lipid phosphatase. Increased PIP3 binds to the pleckstrin homology (PH) domain of Akt/PKB (protein kinase B) and, in combination with additional S/T phosphorylation of Akt by phosphoinositide-dependent kinase 1 (PDK1) and PDK2, results in full activation of Akt. Recent studies revealed that mTORC2 directly phosphorylates Akt on S473 and identified mTORC2 as the long-sought PDK2 [36]. Activated PI3K or Akt may positively regulate mTOR, leading to increased phosphorylation of S6K1 and 4E-BP1, the two best-characterized downstream effector molecules of mTORC1 [56]. Studies have placed tuberous sclerosis complex (TSC), a heterodimer that comprises TSC1 and TSC2 subunits, as a modulator between PI3K/Akt and mTOR [57-59]. The TSC1/2 complex acts as a repressor of mTOR function [57-59]. TSC2 has GTPase-activating protein (GAP) activity towards the Ras family small GTPase Rheb (Ras homolog enriched in brain), and TSC1/2 antagonizes the mTOR signaling pathway via stimulation of GTP hydrolysis of Rheb [58-63]. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38 [64], though this remains controversial [65].
Fig. (4).
mTOR signaling network. mTOR regulates multiple cellular processes by sensing energy and nutrient signals, and growth factors. Arrows represent activation, whereas bars represent inhibition.
2.1.2. Energy and stress
mTORC1 responds to cellular energy signals through AMP-activated protein kinase (AMPK), a highly conserved sensor of cellular energy status. AMPK functions as a S/T kinase and enhance TSC2 activity by direct phosphorylation of TSC2 [21]. Under conditions of low cellular energy (high AMP/ATP ratio), LKB1 phosphorylates AMPK on T172, and subsequently, activated AMPK phosphorylates TSC2 at T1227 and S1345, activating the Rheb-GAP activity of TSC, which catalyzes the conversion of Rheb-GTP to Rheb-GDP and thus inhibits mTORC1 activity [58-63]. Activation of AMPK by the AMP mimetic 5-amino-4-imidazolecarboxamide ribose (AICAR) represses mTOR signaling, leading to dephosphorylation of a number of targets of mTOR signaling including S6K1, eIF4G and 4E-BP1 [66]. By contrast, LKB1 deficient mouse-embryo fibroblasts (MEFs) exhibit hyperactivation of mTOR signaling [67]. In certain cells, such as neurons, endothelial cells, and lymphocytes, AMPK can also be activated by a Ca2+-dependent and AMP-independent process which is mediated by a calmodulin-dependent protein kinase kinase (CaMKK) [68, 69].
In addition to the direct phosphorylation of TSC2, Akt activates mTOR also by regulation of cellular energy [70]. In response to growth factors, Akt phosphorylates TSC2 on T1462 and inhibits the function of TSC1/2, thereby activating mTOR. However, in response to high level of ATP, Akt activates mTOR through inhibition of AMPK-mediated phosphorylation of TSC2 on T1227 and S1345 [70]. Thus, it was proposed that Akt lies upstream of two separate pathways that activate mTOR, the Akt-mediated phosphorylation of TSC2 on T1462 and the inhibition of AMPK-mediated phosphorylation of TSC2 on T1227 and S1345 [70].
mTOR signaling pathway is also regulated by hypoxia [71, 72]. Hypoxia induces rapid and reversible hypophosphorylation of mTOR at S2481 and its downstream targets 4E-BP1, S6K1, S6 and eIF4G [71]. Several studies identified REDD1 (Regulated in Development and DNA damage responses) as a gene that is induced at the mRNA level in response to stresses such as hypoxia or DNA damage [73-76]. In REDD1-/- MEFs, hypoxia failed to dephosphorylate of S6K and S6, indicating that REDD1 is required for the hypoxia-induced downregulation of mTOR signaling [75]. In addition, it was suggested that REDD1 acts upstream of TSC2. The inhibitory effect of REDD1 on mTOR signaling depends on the presence of the TSC1/2 complex, but independent on the LKB1-AMPK signaling [75]. However, most recent studies proposed that hypoxia and the LKB1-AMPK signaling are highly interrelated in head and neck squamous cell carcinoma (HNSCC) [72]. In response to prolonged hypoxia, REDD1 expression was enhanced by AMPK activation, leading to the inhibition of mTOR pathway. Indeed, it was demonstrated that prolonged hypoxia would induce ATP depletion and eventually activate AMPK [72]. Taken together, under hypoxic stress, the inhibition of mTOR activity by REDD1 activation may through AMPK-independent and -dependent mechanisms.
2.1.3. Amino acids
mTOR activity is also regulated by amino acid levels. Silencing expression of TSC1/2 confers resistance to amino acid deprivation, indicating that TSC1/2 is involved in the regulation of mTOR function by amino acids [57]. Inhibition of Rheb binding to mTOR is critical for the inhibitory effect of amino acid withdrawal on mTOR signaling [77]. However, other studies did not support this notion. For example, amino acid withdrawal still impairs mTOR signaling in TSC2-/- cells, suggesting that other than TSC2, additional mechanisms may also be involved in the regulation of mTOR by amino acids [78]. Recently, the class III PI3K, hVps34 (human vacuolar protein sorting 34), was proposed to be activated by amino acid and be involved in the transduction of signal from amino acids to mTORC1. However, currently this model remains controversial because similar data were not observed in Drosophila [79]. Most recent studies in mammalian cells and in Drosophila identified Rag GTPases as activators of mTORC1 by sensing amino acid signals [80, 81]. The proposal of these different hypotheses reflects the complexity of the signaling from amino acids to mTORC1. Therefore, more studies are required to understand how mTORC1 responds to amino acids.
2.2. Downstream targets of mTORC1
In mammals, S6Ks and 4E-BPs are the best characterized downstream targets of mTOR. In addition, mTOR is also involved in the regulation of some proteins including CLIP-170 (cytoplasmic linker protein-170) [82], eukaryotic elongation factor 2 (eEF2) kinase [83], ornithine decarboxylase (ODC) [84], glycogen synthase [85], hypoxia-inducible factor 1α (HIF-1α) [86, 87], lipin [88], PKCδ and PKCε [89], protein phosphatase 2A (PP2A) [90], p21Cip1 and p27Kip1 cyclin-dependent kinase inhibitors [91, 92], retinoblastoma protein (Rb) [93], and signal transducer and activator of transcription 3 (STAT3) [94].
Mammalian cells contain two similar S6 kinase proteins, S6K1 and S6K2 [95, 96]. S6K2, which has 70% overall amino acid identity with S6K1, was discovered much later than S6K1 [97]. Both the activation of S6K1 and S6K2 are regulated by mTOR [98, 99]. S6K1 is ubiquitously expressed and appears to be more critical in the control of cell growth. S6K1 can be activated by a wide variety of extracellular signals. Among the phosphorylation sites of S6K1, T229, S371 and T389 appear essential for S6K1 activation [100-102]. T229, which is located in the activation loop, can be phosphorylated by the “loop kinase” PDK1 [103, 104]. Moreover, for S6K1 activation, mTOR can directly phosphorylate S371 in vitro, and this event modulates T389 phosphorylation by mTOR [102, 105]. S6K1 may also be activated by TOR-insensitive signaling pathways, such as MAP kinases, which mediate phosphorylation of sites on the C-terminal autoinhititory domain [106-108]. S6K1 is known as the major ribosomal protein S6 (rpS6) kinase in mammalian cells and is pointed as a key player in the control of cell growth (cell size) and proliferation [109-111]. Early studies suggested that activated S6K1 regulates protein synthesis through phosphorylation of the 40S ribosomal protein S6, and this was thought to increase the translational efficiency of a class of mRNA transcripts with a 5’-terminal oligopolypirymidine (5’-TOP) [112, 113]. However, this model has been challenged by the recent findings that neither S6K1 activity nor rpS6 phosphorylation is required for the translational regulation of TOP mRNAs [114, 115]. New data indicate that mTOR and S6K1 control on and off the eukaryotic initiation factor 3 (eIF3) translation initiation complex in a growth factor- and rapamycin-sensitive manner [116]. S6K1 associates with the eIF3 complex when inactive, but dissociates from the eIF3 complex upon stimulation by insulin or amino acids. Activated S6K1 then phosphorylates its translational targets, including the 40S ribosomal protein S6 and eIF4B, promoting translation initiation.
4E-BP1 is the other well characterized downstream of mTORC1. In mammalian, 4E-BPs, a family of translational repressor proteins, consist of three low molecular weight proteins, 4E-BP1, 4E-BP2, and 4E-BP3 [117-119]. 4E-BP1 (also known as PHAS-I), a repressor of the translation initiation factor eIF4E, was first identified as an adipocyte protein that underwent phosphorylation by MAP kinase in response to insulin treatment [117, 118]. In addition, mTOR and ATM have also been identified to be involved in phosphorylation of 4E-BP1 [120-123]. It was shown that mTOR phosphorylated 4E-BP1 at serine and threonine residues in insulin-stimulated human embryonic kidney cells and these phosphorylations are inhibited by rapamycin [120]. Hypophosphorylated 4E-BP1 binds tightly to eIF4E, the mRNA cap-binding protein, and represses cap-dependent translation by blocking the binding of eIF4E to eIF4G. In response to sufficient growth factors and nutrients stimulation, six sites (T37, T46, S65, T70, S83 and S112) of 4E-BP1 can be phosphorylated [124]. The phosphorylation of 4E-BP1 at multiple site induce the dissociation of the 4E-BP1 from eIF4E, allowing eIF4E to engage eIF4G [118, 125]. eIF4G serves as a scaffold protein for the assembly of other initiation factors including eIF4A, which acts as an ATP-dependent RNA helicase, and further interacts with eIF3, which recruits the 40S ribosome to the 5’ end of the mRNA [126]. Thus, 4E-BP1 phosphorylation allows several important initiation factors, as well as the 40S ribosomal subunit, to be positioned at the 5’ end of the mRNA to begin the process of scanning.
3. mTORC2
Like mTORC1, mTORC2 also includes mTOR and mLST8, but instead of raptor, mTORC2 contains two special subunits, rictor and mSin1 (mammalian stress-activated protein kinase (SAPK)-interacting protein 1) [32, 33, 127]. In addition, protor (protein observed with rictor), DEPTOR, PRR5 (proline-rich protein 5) and Hsp70 are other novel components of mTORC2 [Fig. (2)] [128-130].
mLST8 is a stable component of both mTOR complexes. mLST8-knockout embryos are developmentally delayed and die around E10.5 [50]. Knockout of mLST8 only disrupts mTORC2 assembly and results in the loss of Akt phosphorylation on S473, indicating that mLST8 is functionally essential only for mTORC2 in mammalian development, but may not be important for mTORC1.
Rictor is a novel mTOR-associated protein that is unique to mTORC2 [33]. The rictor-containing mTOR complex is not bound by FKBP12-rapamycin and is insensitive to acute rapamycin treatment. It was found that the rictor-mTOR complex does not regulate the mTOR effectors S6K1 and 4E-BP1, but molulates phosphorylation of Akt, PKCα and the focal adhesion proteins, as well as activities of the small GTPases, which links to cell survival and migration [33-35, 131, 132]. The data suggest that mTORC2 has different physiological functions from mTORC1.
mSin1 is an essential subunit of mTORC2 because it is important for mTORC2 integrity and mTOR activity toward Akt-S473 phosphorylation [127]. In the absence of mSin1, the interaction between rictor and mTOR was impaired and the phosphorylation of Akt on S473 was diminished [127, 133].
Protor-1 (protein observed with rictor-1) and protor-2 interact with rictor, but they are not essential for the assembly of other mTORC2 subunits into the complex [128]. To date, the function of protor is unclear. Similar with protor, PRR5 interacts with rictor, but it is not important for the interaction of mTOR-rictor and mTOR activity toward Akt phosphorylation at S473. However, PRR5 plays a crucial role in the modulation of PDGF signaling by regulating PDGFRβ expression [129]. Hsp70, a heat shock protein, is required for proper formation and the kinase activity of mTORC2 under basal conditions and following heat shock [130]. In addition, a recent study identified DEPTOR as an mTOR-interacting protein [41]. DEPTOR interact with both mTORC1 and mTORC2, negatively regulating their activities.
As mTORC2 complex was discovered only recently, its functions and regulatory mechanisms are less well understood than mTORC1 [32]. The first study on the mTOR-rictor complex revealed that one of the functions of mTORC2 is to regulate the actin cytoskeleton by mediating the PKCα phosphorylation state [33]. Susequently, the important finding that mTORC2 directly phosphorylates Akt on S473 adds a new insight into the role of mTOR in cancer [36]. mTORC2 was originally thought to be rapamycin-insensitive. However, recent studies showed that in some cell lines, prolonged rapamycin treatment inhibits the assembly and function of mTORC2 to inhibit Akt [34]. Most recently, one striking observation revealed that in adipose tissue, mTORC2 negatively controls whole-body growth by controlling an adipose to pancreas/liver signaling axis [134]. In adipose-specific rictor knockout (rictorad-/-) mice, the body size was increased due to an increased size of non-adipose tissue, including heart, kidney, spleen and bone [134]. However, it remains to be determined how mTORC2 in adipose regulates the pancreas and liver.
3.1. Upstream regulators of mTORC2
It was suggested that mTORC2 lies downstream of PI3K signaling, since growth factors stimulate mTORC2 activity and low concentrations of wortmannin, a specific PI3K inhibitor, inhibits Akt S473 phosphorylation [36]. However, the mechanism by which mTORC2 is activated is not entirely clear. Rheb has been demonstrated as a key upstream activator of mTORC1. However, Rheb showed negative and indirect effect on the regulation of mTORC2. In Drosophila S2 cells, knockdown of Drosophila Rheb (dRheb) decreased dS6K phosphorylation and elevated dAkt phosphorylation, suggesting that dRheb has opposite effects on dTORC1 and dTORC2 activity [135]. Moreover, it was proposed that the negative effect of dRheb on dTORC2 may probably through a negative feedback loop involving dS6K [135]. In many cell types, activation of mTORC1 by Rheb activates S6K1, which in turn suppresses the PI3K-Akt signaling pathway by phosphorylation and inhibition of IRS [136]. Taken together, these data indicate that Rheb activates TORC1 in a more direct manner, whereas the inhibition of Rheb toward TORC2 may be indirect.
3.2. Downstream targets of mTORC2
The best-characterized substrate of mTORC2 is the Akt kinase, since growing evidence showed that mTORC2 phosphorylates Akt on S473. Akt belongs to the AGC kinase family, which also includes S6Ks, glucocorticoid-induced protein kinase (SGKs), p90 ribosomal protein S6 kinase (RSKs), and PKCs. The members in AGC kinase family have similar structures, which contain the hydrophobic motif phosphorylation site and an activation loop site near or within the kinase domain [137, 138]. Compare to Akt or other family members, S6K1 possesses an additional C-terminal inhibitory domain, which may be preferentially be recognized by mTORC1 complex [139]. Because of the difference between the structures, S6K1 and Akt are phosphorylated by mTORC1 and mTORC2, respectively. Of interest, it has been reported that S6K1 can be phosphorylated by mTORC2 by deleting the C-terminal domain [140]. Therefore, it would be interesting to unveil whether other AGC kinases, which are structurally similar to Akt, are also regulated by mTORC2.
Akt is one of the most important survival kinases. At the membrane, active PI3K recruits Akt and PDK1 to the plasma membrane. Akt is subsequently phosphorylated at the activation loop site T308 and the C-terminal hydrophobic motif site S473 by PDK1 and PDK2, respectively [141]. Active Akt in turn regulates different cellular processes including cell growth, proliferation, apoptosis and glucose metabolism [142]. Numerous studies attempted to identify the crucial kinase(s) (often referred to as PDK2) responsible for the phosphorylation of S473 in Akt. Several enzymes are in the candidate list, including PDK1, integrin-linked kinase (ILK), Akt itself, DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and mTORC2 [143-146]. Since mTORC2 complex fulfills the role of the Akt S473 kinase, mTORC2 has been identified as the long-sought PDK2 [36]. In human cancer cells, knockdown of mTOR or rictor expression using lentiviral short hairpin RNA (shRNA) decreased phosphorylation of S473 of Akt [36]. In rictor- and mLST8-deficient mice, the mLST8 and rictor were demonstrated to be necessary for the basal, serum-induced, and insulin-induced phosphorylation of Akt on S473 [50]. The results from subsequent studies in Drosophila cultured cells further confirm these observations, substantiating the notion that mTORC2 is the principal S473 kinase of Akt [127, 133]. Considering the importance of Akt signaling in disease and the critical role of mTORC2 in Akt activation, mTORC2 complex has received great attention as a novel drug target, especially for treating cancers characterized by hyperactive Akt.
In addition to Akt/PKB, PKCα, paxillin and Rho GTPases are also involved in the regulation of the actin cytoskeleton by mTORC2 [Fig. (4)]. Several knockdown and knockout studies demonstrated that mTORC2 regulates PKCα phosphorylation [32, 33, 50]. The phosphorylation of PKCα on S657 is dramatically reduced in rictor null MEFs [50]. In Drosophila, reduction in rictor by using dsRNAs also decreases the phosphorylation of dPKCα [33]. Paxillin, which functions as a docking protein, localizes to the focal adhesions of adherent cells [147]. Jacinto et al. showed that paxillin was highly phosphorylated at Tyr 118 in control cells, whereas knockdown of mTORC2 inhibited the phosphorylation of paxillin [32]. Rho, Rac and Cdc42, three best-characterized members of the Rho family of small GTPases, were demonstrated to be involved in actin cytoskeleton assembly and disassembly [148]. It was reported that mTORC2 may function as upstream of Rho GTPases to regulate the actin cytoskeleton [32]. In mTOR, mLST8 or rictor siRNA-transfected cells, expression of constitutively active form of Rac (Rac1-L61) or Rho (RhoA-L63) restored organization of the actin cytoskeleton, indicating that mTORC2 may regulate the actin cytoskeleton through Rho and Rac [32].
Most recently, the serum glucocorticoid-induced protein kinase 1 (SGK1), a member of the AGC family, was identified as a novel substrate of mTORC2 [149-151]. In MEFs lacking rictor, mSin1 or mLST8, which are the critical components of mTORC2, both the activity and hydrophobic motif phosphorylation of SGK1 are abolished [149]. Moreover, S422 can also be phosphorylated by immunoprecipitated mTORC2 in vitro, further confirming that mTORC2 regulates SGK1 [149]. The SGK family has three members in mammals, SGK1, SGK2 and SGK3. The phosphorylation and activity of the SGK isoforms can be stimulated by oxidation, insulin and IGF-1 through the PI3K-PDK1/2 signaling pathway [152, 153]. It was presumed that SGK isoforms may probably share substrates in common with Akt, but they may also have distinct targets and specific functions [154]. It was reported that the ubiquitin ligase Nedd4-2 (neuronal precursor cells expressed developmentally downregulated 4-2) is a target of SGK1 [155]. SGK1 phosphorylates Nedd4-2 mainly on S444 via a PY motif-dependent mechanism and this phosphorylation reduces the binding of Nedd4-2 to the epithelial sodium channel (ENaC), thus leading to the enhanced cell surface expression of ENaC channel protein [155]. Most recent work showed that SGK1 is an important mediator of TORC2 signaling. Meanwhile, this study identified new functions of TORC2, which are mediated by SGK, in the regulation of fat accumulation, size and growth in Caenorhabditis elegans (C. elegans) [150]. Viable loss-of-function mutants in the C. elegans homolog of the TORC2-specific component, rictor (Cerictor) showed a developmental delay and reduced body size, but increased fat accumulation [150]. Thus, the view that Akt is the primary effector of TORC2 function is challenged by these findings. However, in contrast to Akt, most of the substrates and functions of SGKs are not well characterized. Particularly the physiological functions of SGK2 and SGK3 are presently poorly understood. Therefore, in order to better understand the roles of these kinases, more studies are needed to identify novel SGK substrates.
4. mTOR and human diseases
Hyperactivation of PI3K/mTORC1 signaling is proposed to be a common cause of human cancers and several hamartoma syndromes, including tuberous sclerosis complex (TSC) syndrome, the PTEN-related hamartoma syndromes and Peutz-Jeghers syndrome (PJS) (Table 1) [156-179]. These hamartoma syndromes are characterized by benign, focal tumors in which the tumor cells differentiate normally but grow in a disorganized mass [156, 180]. Hamartomas may occur in many different parts of the body, including lung, heart, skin, kidneys, hypothalamus and other vascular organs. Although hamartomas are benign tumors, they may progress to certain malignant cancers, or even cause death. During the last 10 years, the identification of genes mutated in these diseases and the findings of the functions of these genes revealed that mutational inactivation of the tumor suppressors, TSC, PTEN, and LKB1, three negative regulators of mTORC1 signaling, stimulates mTORC1 activity and ultimately results in the development of TSC syndrome, PTEN associated hamartoma syndromes and PJS, respectively [181-183]. Therefore, inhibition of mTORC1 signaling has been considered to be a promising therapeutic strategy for treatment of some hamartoma syndromes. Thus, rapamycin, a specific inhibitor of mTORC1, attracts great interest and is most well studied. By forming a complex with its intracellular receptor FKBP12, rapamycin binds to mTORC1 and inhibits mTORC1 signaling. So far, four mTOR inhibitors are currently being evaluated in clinical trials for cancer treatments: the prototype rapamycin and three rapamycin derivatives, CCI-779 (temsirolimus), RAD001 (everolimus) and AP23573. Preliminary data indicate that mTORC1 inhibitors have potent inhibitory effects against a broad range of malignant tumors with relatively minor toxicities [184, 185]. However, the results from ongoing clinical trials of these compounds are less than satisfactory. Normally the benign nature of hamartomas is partially explained by the existence of an auto-regulatory pathway which is defined as S6K1-dependent negative feedback inhibition [136]. In many cell types, hyperactive mTORC1 signaling caused by genetic mutations activates S6K1. Active S6K1 strongly represses Akt signaling by directly phosphorylating and inhibiting IRS, which is important for tumor growth [136]. These findings cast a certain doubt as to the feasibility of long-term treatment with rapamycin in hamartoma patients, since such treatment may sensitize the PI3K-Akt signaling through the feedback loop and put the patients under higher risk of malignant development. Thus, the combinational use of mTORC1 inhibitors with antagonists of other upstream signaling, such as IGF-1 receptor inhibitors and PI3K inhibitors, would be an alternative strategy to overcome this feedback effect and for the treatment of hamartoma syndromes [186].
Table 1.
Diseases commonly linked to mTOR signaling
| Disease name | Mutated gene which might affect mTOR signaling | Functional link to mTOR signaling | References |
|---|---|---|---|
| tuberous sclerosis complex (TSC) | TSC1 or TSC2 | TSC1 and TSC2 function as a heterodimer to negatively regulate mTOR | [158,162] |
| Lymphangioleiomyomatosis (LAM) | TSC2 | TSC1 and TSC2 function as a heterodimer to negatively regulate mTOR | [159] |
| Cowden’s disease | PTEN | PTEN is a negative regulator of PI3K; PTEN loss activates AKT signaling, leading to hyperactive mTOR signaling through inhibition of TSC2 | [156] |
| Lhermitte-Duclos disease | PTEN | PTEN is a negative regulator of PI3K | [160] |
| Bannayan-Riley-Ruvalcaba syndrome (BRRS) | PTEN | PTEN is a negative regulator of PI3K | [161] |
| Proteus syndrome | PTEN | PTEN is a negative regulator of PI3K | [163] |
| Peutz-Jeghers syndrome (PJS) | LKB1 (STK11) | LKB1 (STK11) phosphorylate and activate AMPK in response to ATP depletion | [164,165] |
| familial hypertrophic cardiomyopathy (HCM) | AMPKγ2 (encoded by PRKAG2) | AMPK can activate TSC2 on ATP depletion; mutations in PRKAG2 reduce AMPK catalytic activity | [166] |
| Wolff-Parkinson-White syndrome | AMPKγ2 (encoded by PRKAG2) | AMPK can activate TSC2 on ATP depletion; Mutations in PRKAG2 reduce AMPK catalytic activity | [167] |
| Breast cancer | PTEN; PIK3CA mutation or amplification; Her2 amplification | PTEN is a negative regulator of PI3K; PIK3CA mutation or amplification results in abnormal activation of mTOR | [168,169] |
| Prostate cancer | PTEN; PIK3CA mutation or amplification | PTEN is a negative regulator of PI3K; PIK3CA mutation or amplification results in abnormal activation of mTOR | [168] |
| Ovarian cancer | PTEN; PIK3CA mutation or amplification; Her2 amplification | PTEN is a negative regulator of PI3K; PIK3CA mutation or amplification results in abnormal activation of mTOR | [169] |
| Lung cancer | PTEN; PIK3CA mutation or amplification; Her2 amplification | PTEN is a negative regulator of PI3K; PIK3CA mutation or amplification results in abnormal activation of mTOR | [170] |
| Bladder cancer | PTEN | PTEN is a negative regulator of PI3K | [171,172] |
| Melanoma | PTEN | PTEN is a negative regulator of PI3K | [173] |
| Endometrial cancer | PTEN | PTEN is a negative regulator of PI3K | [174] |
| Thyroid | PTEN; PIK3CA mutation or amplification; Her2 amplification | PTEN is a negative regulator of PI3K; PIK3CA mutation or amplification results in abnormal activation of mTOR | [168,175] |
| Glioblastoma | PTEN; PIK3CA mutation or amplification | PTEN is a negative regulator of PI3K; PIK3CA mutation or amplification results in abnormal activation of mTOR | [176,177] |
| Renal carcinomas | PTEN | PTEN is a negative regulator of PI3K | [178,179] |
Besides tumorigenesis, deregulation of mTORC1 is also involved in many other human diseases and metabolic disorders, including cardiac hypertrophy, type II diabetes and obesity [156]. Cardiac hypertrophy, a consequence of increased cell size and number of cardiomyocytes, is one of the major risk factors for heart failure [156]. Strong evidence showed that hyperactivation of PI3K/Akt/mTOR signaling caused enlargement of the heart [187]. In load-induced cardiac hypertrophy, mTOR is also demonstrated to be involved in [188]. In mouse models, rapamycin effectively suppresses both PI3K/Akt/mTOR- and load-induced cardiac hypertrophy, indicating that mTOR plays a key role in the development of cardiac hypertrophy [187, 188].
It was suggested that both type II diabetes and obesity are associated with insulin resistance, and deregulation of mTORC1 may contribute to both of the disorders. Nutrients and hormonal factors mediate tissue or organ cellular metabolism through the mTOR signaling pathway. Hyperactive mTORC1 which is induced by high level of insulin or amino acid down-regulates IRS-1 activity through the S6K1-dependent negative feedback inhibition and results in the development of insulin resistance [189]. Long-term insulin resistance is one of the common causes of type II diabetes [189]. In S6K-/- mice, the weight accumulation was significantly reduced compared with wild-type mice [190]. The metabolic rate of in the 4E-BP1-/- male mice is significantly increased [191]. In addition, it was also shown that both S6K and 4E-BP mutant mice are resistant to diet-induced obesity. These data strongly suggest a potential involvement of deregulated mTOR signaling pathway in a variety of important human diseases. mTOR inhibitors may become effective tools for the treatment of heart disease and/or metabolic disorders, such as type II diabetes and obesity.
5. mTOR inhibitors
So far, rapamycin and its analogs (termed rapalogs) are the most well studied mTOR inhibitors (Table 2) [192]. As described early, rapamycin was first isolated from the soil bacterium Streptomyces hygroscopicus as a fungicide, and subsequently discovered to have equally potent immunosuppressive and anti-tumor properties [1-5]. As an immunosuppresive drug, rapamycin (rapamune, sirolimus) was approved by the Food and Drug Administration (FDA) in USA in 1999 for prevention of renal allograft rejection [193]. Subsequent studies from many laboratories found that rapamycin can also act as a cytostatic agent, slowing or arresting growth of cell lines derived from different tumor types such as rhabdomyosarcoma [194, 195], glioblastoma [196], small cell lung cancer [197], osteoscarcoma [198], pancreatic cancer [199], breast cancer, prostate cancer [200, 201] and B-cell lymphoma [202, 203]. However, the poor aqueous solubility and chemical stability of rapamycin restricts its development as an anticancer agent. Therefore, several derivatives of rapamycin with more favorable pharmaceutical characteristics have been synthesized, such as CCI-779 (Temsirolimus, Wyeth, Madison, NJ, USA), RAD001 (Everolimus, Novartis, Novartis, Basel, Switzerland), AP23573 (Deforolimus, ARIAD, Cambridge, MA, USA), 32-deoxorapamycin (SAR943) or zotarolimus (ABT-578, Abbott Laboratories, Abbott Park, IL, USA). Like rapamycin, these rapamycin analogs forms a complex with the intracellular receptor FKBP12, resulting in potent inhibition on mTORC1 signaling. CCI-779, RAD001 and AP23573, which are currently being tested in clinical development as anticancer agents, have shown antiproliferative activity against a diverse range of cancer types in preclinical studies, and clinical evaluations also have demonstrated promising results in a subset of cancers [204-206]. However, increasing studies also suggested that the antiproliferative effects of the rapalogs are variable in cancer cells due to failure to inhibit mTORC2 in some tumor types. O’ Reilly et al. [207] showed that the specific inhibition of mTORC1 by RAD001 might induce upstream receptor tyrosine kinase signaling and Akt upregulation, leading to the attenuation of its therapeutic effects. Thus, the combination therapy or a dual-specificity agent that targets both of mTOR function and Akt activation may have improved antitumor activity.
Table 2.
mTOR inhibitors
| mTOR inhibitors | Structure | Mechanism of action | References |
|---|---|---|---|
| Rapamycins | |||
| Rapamycin and rapamycin analogs | Macrolide ester | Functions by binding to the immunophilin FKBP12 Partial mTORC1 inhibitor Cell-type specific mTORC2 inhibitor |
[192] |
| Small molecule inhibitors of kinases | |||
| Torin1 | Pyridinonequinoline | mTOR kinase inhibitor | [208] |
| PP242 | Pyrazolopyrimidines | mTOR kinase inhibitor | [209] |
| PP30 | Pyrazolopyrimidines | mTOR kinase inhibitor | [209] |
| WYE-354 | Pyrazolopyrimidine | ATP competitive inhibitor of mTOR | [210] |
| Ku-0063794 | pyridopyrimidin | specific mTORC1 and mTORC2 inhibitor | [211] |
| mTOR and PI3K dual-specificity inhibitors | |||
| NVP-BEZ235 | Imidazoquinazoline | ATP-competitive inhibitor of PI3K and mTOR | [212] |
| PI-103 | Tricyclic pyridofuropyrimidine | ATP competitive inhibitor of DNA-PK, PI3K and mTOR | [213-215] |
| XL765 | Not available | ATP-competitive inhibitor of DNA-PK, PI3K and mTOR | [216] |
| Diet-derived chemopreventive agents | |||
| Curcumin | Diferuloylmethane | Disrupts the mTOR-Raptor Complex | [217] |
| Resveratrol epigallocatechin gallate (EGCG) | Trans-3,4’, 5-trihydroxystilbene | Inhibits PI3K/Akt/mTOR signaling pathway | [218,219] |
| Polyphenol | Inhibits PI3K/Akt/mTOR signaling pathway | [220] | |
| Genistein | Isoflavone | Inhibits PI3K/Akt/mTOR signaling pathway | [221,212] |
| 3,3-Diindolylmethane (DIM) | Indole-3-carbinol | Inhibits both mTOR and Akt activity | [223] |
Recently, a new generation of mTOR inhibitors, which binds to the ATP-binding site of mTOR and inhibits the catalytic activity of mTORC1 and mTORC2, is being developed (Table 2) [208-223]. Since mTORC2 was demonstrated to be a positive regulator of Akt, inhibition of mTORC2 would effectively minimize the feedback activation of PI3K and suppress the activation of Akt, finally preventing malignant progression of tumors. Two active-site inhibitors of mTOR, PP242 and PP30, which inhibited insulin-stimulated phosphorylation of Akt at S473, showed potent inhibitory effects on protein synthesis and cell proliferation [209]. Torin1, another selective ATP-competitive mTOR inhibitor, which directly inhibited both mTORC1 and mTORC2, also inhibited cell growth and proliferation more effectively than rapamycin [208]. However, more studies are required to further evaluate the efficacy of these agents in the treatment of cancers caused by hyperactive PI3K/mTOR pathway as well as the distinct effects of such broad-spectrum mTOR inhibitors on cellular and organismal physiology.
Recent preliminary studies showed that some diet-derived chemopreventive agents (e.g. curcumin, resveratrol, 3, 3-Diindolylmethane, epigallocatechin gallate, or genistein) may inhibit mTOR signaling through direct or indirect mTOR inhibition (Table 2). For example, curcumin, a polyphenol natural product isolated from the rhizome of the plant Curcuma longa, showed effectiveness as a chemopreventive agent in animal models of carcinogenesis and is undergoing preclinical trial evaluation for anti-cancer drug development [224]. In numerous cancer cell lines, curcumin inhibited phosphorylation of mTOR and its downstream effector molecules, S6K1 and 4E-BP1, suggesting that curcumin may execute its anticancer activity primarily by blocking mTOR mediated signaling pathways [225, 226]. Recently, the mechanism study found that curcumin was able to dissociate raptor from mTOR, leading to inhibition of mTORC1 activity [217].
6. Conclusions and perspectives
Despite the discovery of mTOR for over 15 years, the complexity of the mTOR network is just beginning to be understood. Currently it is known that mTOR functions as two distinct multiprotein complexes: mTORC1 and mTORC2. mTOR is the core component of the two complexes, each having distinct interacting proteins. With great possibility, more interacting proteins will be identified in the future. mTORC1 regulates physiological functions, including cell growth, proliferation and survival by integrating hormonal factors, nutrients, stressors and energy signals. In contrast to mTORC1, mTORC2 is less well studied and is largely insensitive to nutrients and energy conditions. mTORC2 has been shown to be involved in the regulation of actin cytoskeleton and plays an important role in Akt phosphorylation at S473, indicating a possible role of mTORC2 in cell migration and survival. In recent years, significant progress has been achieved in understanding the TOR field, but several important questions remain to be elucidated. How do cells sense amino acid sufficiency and transmit the signal to mTOR in order to regulate cell growth? How is mTORC2 activated and regulated? What are the undiscovered functions of mTORC1 and mTORC2? Are there additional substrates of mTORC2 besides Akt and SGK1? Given the increasing evidence that deregulation of mTOR occurs in many human diseases, the answers to these questions will not only bring novel insights into mTOR biology but also help us develop more effective therapeutic strategies for treating mTOR-related diseases, including cancer, cardiac hypertrophy and metabolic disorders. Importantly, new evidence that mTORC1 might have rapamycin-resistant functions may explain the modest clinical efficacy of rapamycin. The growing appreciation of mTOR’s central role in cancer opens a new door in the search for promising anti-cancer drugs.
Acknowledgments
The authors’ work cited in this review was supported in part by NIH (CA115414 to S.H.) and American Cancer Society (RSG-08-135-01-CNE to S.H.).
Footnotes
Conflict of interest statement None declared.
References
- 1.Vezina C, Kudelski A, Sehgal SN. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 1975;28:721–726. doi: 10.7164/antibiotics.28.721. [DOI] [PubMed] [Google Scholar]
- 2.Eng CP, Sehgal SN, Vezina C. Activity of rapamycin (AY-22,989) against transplanted tumors. J Antibiot (Tokyo) 1984;37:1231–1237. doi: 10.7164/antibiotics.37.1231. [DOI] [PubMed] [Google Scholar]
- 3.Douros J, Suffness M. New antitumor substances of natural origin. Cancer Treat Rev. 1981;8:63–87. doi: 10.1016/s0305-7372(81)80006-0. [DOI] [PubMed] [Google Scholar]
- 4.Linhares MM, Gonzalez AM, Trivino T, Melaragno C, Moura RM, Garcez MH, Sa JR, Aguiar WF, Succi T, Barbosa CS, Pestana JO. Simultaneous pancreas-kidney transplantation initial experience. Transplant Proc. 2003;35:1109. doi: 10.1016/s0041-1345(03)00328-2. [DOI] [PubMed] [Google Scholar]
- 5.Sehgal SN, Baker H, Vezina C. Rapamycin (AY-22,989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. J Antibiot (Tokyo) 1975;28:727–732. doi: 10.7164/antibiotics.28.727. [DOI] [PubMed] [Google Scholar]
- 6.Cafferkey R, Young PR, McLaughlin MM, Bergsma DJ, Koltin Y, Sathe GM, Faucette L, Eng WK, Johnson RK, Livi GP. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol Cell Biol. 1993;13:6012–6023. doi: 10.1128/mcb.13.10.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Zheng XF, Brown EJ, Schreiber SL. Identification of an 11-kDa FKBP12-rapamycin-binding domain within the 289-kDa FKBP12-rapamycin-associated protein and characterization of a critical serine residue. Proc Natl Acad Sci USA. 1995;92:4947–4951. doi: 10.1073/pnas.92.11.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 10.Sabers CJ, Martin MM, Brunn GJ, Williams JM, Dumont FJ, Wiederrecht G, Abraham RT. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J Biol Chem. 1995;270:815–822. doi: 10.1074/jbc.270.2.815. [DOI] [PubMed] [Google Scholar]
- 11.Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 12.Chiu MI, Katz H, Berlin V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc Natl Acad Sci USA. 1994;91:12574–12578. doi: 10.1073/pnas.91.26.12574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Chen H, Rhoad AE, Warner L, Caggiano TJ, Failli A, Zhang H, Hsiao CL, Nakanishi K, Molnar-Kimber KL. A putative sirolimus (rapamycin) effector protein. Biochem Biophys Res Commun. 1994;203:1–7. doi: 10.1006/bbrc.1994.2140. [DOI] [PubMed] [Google Scholar]
- 15.Keith CT, Schreiber SL. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 16.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 17.Thomas G, Hall MN. TOR signalling and control of cell growth. Curr Opin Cell Biol. 1997;9:782–787. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- 18.Brown EJ, Schreiber SL. A signaling pathway to translational control. Cell. 1996;86:517–520. doi: 10.1016/s0092-8674(00)80125-7. [DOI] [PubMed] [Google Scholar]
- 19.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 20.Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–1105. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- 21.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 22.Chakraborty S, Mohiyuddin SM, Gopinath KS, Kumar A. Involvement of TSC genes and differential expression of other members of the mTOR signaling pathway in oral squamous cell carcinoma. BMC Cancer. 2008;8:163. doi: 10.1186/1471-2407-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darb-Esfahani S, Faggad A, Noske A, Weichert W, Buckendahl AC, Muller B, Budczies J, Roske A, Dietel M, Denkert C. Phospho-mTOR and phospho-4EBP1 in endometrial adenocarcinoma: association with stage and grade in vivo and link with response to rapamycin treatment in vitro. J Cancer Res Clin Oncol. 2009;135:933–941. doi: 10.1007/s00432-008-0529-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenwald IB. The role of translation in neoplastic transformation from a pathologist’s point of view. Oncogene. 2004;23:3230–3247. doi: 10.1038/sj.onc.1207552. [DOI] [PubMed] [Google Scholar]
- 25.Ekstrand AI, Jonsson M, Lindblom A, Borg A, Nilbert M. Frequent alterations of the PI3K/AKT/mTOR pathways in hereditary nonpolyposis colorectal cancer. Fam Cancer. 2009 doi: 10.1007/s10689-009-9293-1. [DOI] [PubMed] [Google Scholar]
- 26.Chan JA, Zhang H, Roberts PS, Jozwiak S, Wieslawa G, Lewin-Kowalik J, Kotulska K, Kwiatkowski DJ. Pathogenesis of tuberous sclerosis subependymal giant cell astrocytomas: biallelic inactivation of TSC1 or TSC2 leads to mTOR activation. J Neuropathol Exp Neurol. 2004;63:1236–1242. doi: 10.1093/jnen/63.12.1236. [DOI] [PubMed] [Google Scholar]
- 27.Riemenschneider MJ, Betensky RA, Pasedag SM, Louis DN. AKT activation in human glioblastomas enhances proliferation via TSC2 and S6 kinase signaling. Cancer Res. 2006;66:5618–5623. doi: 10.1158/0008-5472.CAN-06-0364. [DOI] [PubMed] [Google Scholar]
- 28.Xu G, Zhang W, Bertram P, Zheng XF, McLeod H. Pharmacogenomic profiling of the PI3K/PTEN-AKT-mTOR pathway in common human tumors. Int J Oncol. 2004;24:893–900. [PubMed] [Google Scholar]
- 29.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 30.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 31.Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 32.Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 33.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 34.Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 35.Hresko RC, Mueckler M. mTOR.RICTOR is the Ser473 kinase for Akt/protein kinase B in 3T3-L1 adipocytes. J Biol Chem. 2005;280:40406–40416. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- 36.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 37.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 38.Polak P, Hall MN. mTOR and the control of whole body metabolism. Curr Opin Cell Biol. 2009;21:209–218. doi: 10.1016/j.ceb.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 39.Kim DH, Sarbassov DD, Ali SM, Latek RR, Guntur KV, Erdjument-Bromage H, Tempst P, Sabatini DM. GbetaL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- 40.Sancak Y, Thoreen CC, Peterson TR, Lindquist RA, Kang SA, Spooner E, Carr SA, Sabatini DM. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol Cell. 2007;25:903–915. doi: 10.1016/j.molcel.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abraham RT. Mammalian target of rapamycin: immunosuppressive drugs uncover a novel pathway of cytokine receptor signaling. Curr Opin Immunol. 1998;10:330–336. doi: 10.1016/s0952-7915(98)80172-6. [DOI] [PubMed] [Google Scholar]
- 43.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 44.Janus A, Robak T, Smolewski P. The mammalian target of the rapamycin (mTOR) kinase pathway: its role in tumourigenesis and targeted antitumour therapy. Cell Mol Biol Lett. 2005;10:479–498. [PubMed] [Google Scholar]
- 45.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 46.Perry J, Kleckner N. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell. 2003;112:151–155. doi: 10.1016/s0092-8674(03)00033-3. [DOI] [PubMed] [Google Scholar]
- 47.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carriere A, Cargnello M, Julien LA, Gao H, Bonneil E, Thibault P, Roux PP. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr Biol. 2008;18:1269–1277. doi: 10.1016/j.cub.2008.07.078. [DOI] [PubMed] [Google Scholar]
- 49.Wang L, Lawrence JC, Jr, Sturgill TW, Harris TE. Mammalian target of rapamycin complex 1 (mTORC1) activity is associated with phosphorylation of raptor by mTOR. J Biol Chem. 2009;284:14693–14697. doi: 10.1074/jbc.C109.002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 51.Kovacina KS, Park GY, Bae SS, Guzzetta AW, Schaefer E, Birnbaum MJ, Roth RA. Identification of a proline-rich Akt substrate as a 14-3-3 binding partner. J Biol Chem. 2003;278:10189–10194. doi: 10.1074/jbc.M210837200. [DOI] [PubMed] [Google Scholar]
- 52.Oshiro N, Takahashi R, Yoshino K, Tanimura K, Nakashima A, Eguchi S, Miyamoto T, Hara K, Takehana K, Avruch J, Kikkawa U, Yonezawa K. The proline-rich Akt substrate of 40 kDa (PRAS40) is a physiological substrate of mammalian target of rapamycin complex 1. J Biol Chem. 2007;282:20329–20339. doi: 10.1074/jbc.M702636200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Harris TE, Roth RA, Lawrence JC., Jr PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J Biol Chem. 2007;282:20036–20044. doi: 10.1074/jbc.M702376200. [DOI] [PubMed] [Google Scholar]
- 54.Wang L, Harris TE, Lawrence JC., Jr Regulation of proline-rich Akt substrate of 40 kDa (PRAS40) function by mammalian target of rapamycin complex 1 (mTORC1)-mediated phosphorylation. J Biol Chem. 2008;283:15619–15627. doi: 10.1074/jbc.M800723200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valentinis B, Baserga R. IGF-I receptor signalling in transformation and differentiation. Mol Pathol. 2001;54:133–137. doi: 10.1136/mp.54.3.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang S, Houghton PJ. Targeting mTOR signaling for cancer therapy. Curr Opin Pharmacol. 2003;3:371–377. doi: 10.1016/s1471-4892(03)00071-7. [DOI] [PubMed] [Google Scholar]
- 57.Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, Pan D. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat Cell Biol. 2002;4:699–704. doi: 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- 58.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 59.Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci USA. 2002;99:13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- 61.Manning BD, Cantley LC. Rheb fills a GAP between TSC and TOR. Trends Biochem Sci. 2003;28:573–576. doi: 10.1016/j.tibs.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
- 63.Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 64.Bai X, Ma D, Liu A, Shen X, Wang QJ, Liu Y, Jiang Y. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science. 2007;318:977–980. doi: 10.1126/science.1147379. [DOI] [PubMed] [Google Scholar]
- 65.Wang Y, Huang BP, Luciani DS, Wang X, Johnson JD, Proud CG. Rheb activates protein synthesis and growth in adult rat ventricular cardiomyocytes. J Mol Cell Cardiol. 2008;45:812–820. doi: 10.1016/j.yjmcc.2008.07.016. [DOI] [PubMed] [Google Scholar]
- 66.Reiter AK, Bolster DR, Crozier SJ, Kimball SR, Jefferson LS. Repression of protein synthesis and mTOR signaling in rat liver mediated by the AMPK activator aminoimidazole carboxamide ribonucleoside. Am J Physiol Endocrinol Metab. 2005;288:E980–988. doi: 10.1152/ajpendo.00333.2004. [DOI] [PubMed] [Google Scholar]
- 67.Shaw RJ, Bardeesy N, Manning BD, Lopez L, Kosmatka M, DePinho RA, Cantley LC. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;6:91–99. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 68.Towler MC, Hardie DG. AMP-activated protein kinase in metabolic control and insulin signaling. Circ Res. 2007;100:328–341. doi: 10.1161/01.RES.0000256090.42690.05. [DOI] [PubMed] [Google Scholar]
- 69.Hong SP, Momcilovic M, Carlson M. Function of mammalian LKB1 and Ca2+/calmodulin-dependent protein kinase kinase alpha as Snf1-activating kinases in yeast. J Biol Chem. 2005;280:21804–21809. doi: 10.1074/jbc.M501887200. [DOI] [PubMed] [Google Scholar]
- 70.Hahn-Windgassen A, Nogueira V, Chen CC, Skeen JE, Sonenberg N, Hay N. Akt activates the mammalian target of rapamycin by regulating cellular ATP level and AMPK activity. J Biol Chem. 2005;280:32081–32089. doi: 10.1074/jbc.M502876200. [DOI] [PubMed] [Google Scholar]
- 71.Arsham AM, Howell JJ, Simon MC. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J Biol Chem. 2003;278:29655–29660. doi: 10.1074/jbc.M212770200. [DOI] [PubMed] [Google Scholar]
- 72.Schneider A, Younis RH, Gutkind JS. Hypoxia-induced energy stress inhibits the mTOR pathway by activating an AMPK/REDD1 signaling axis in head and neck squamous cell carcinoma. Neoplasia. 2008;10:1295–1302. doi: 10.1593/neo.08586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ellisen LW, Ramsayer KD, Johannessen CM, Yang A, Beppu H, Minda K, Oliner JD, McKeon F, Haber DA. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol Cell. 2002;10:995–1005. doi: 10.1016/s1097-2765(02)00706-2. [DOI] [PubMed] [Google Scholar]
- 74.Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 2005;25:5834–5845. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, Witters LA, Ellisen LW, Kaelin WG., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shoshani T, Faerman A, Mett I, Zelin E, Tenne T, Gorodin S, Moshel Y, Elbaz S, Budanov A, Chajut A, Kalinski H, Kamer I, Rozen A, Mor O, Keshet E, Leshkowitz D, Einat P, Skaliter R, Feinstein E. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol Cell Biol. 2002;22:2283–2293. doi: 10.1128/MCB.22.7.2283-2293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Long X, Ortiz-Vega S, Lin Y, Avruch J. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem. 2005;280:23433–23436. doi: 10.1074/jbc.C500169200. [DOI] [PubMed] [Google Scholar]
- 78.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 79.Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi JH, Bertram PG, Drenan R, Carvalho J, Zhou HH, Zheng XF. The FKBP12-rapamycin-associated protein (FRAP) is a CLIP-170 kinase. EMBO Rep. 2002;3:988–994. doi: 10.1093/embo-reports/kvf197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Redpath NT, Foulstone EJ, Proud CG. Regulation of translation elongation factor-2 by insulin via a rapamycin-sensitive signalling pathway. Embo J. 1996;15:2291–2297. [PMC free article] [PubMed] [Google Scholar]
- 84.Seidel ER, Ragan VL. Inhibition by rapamycin of ornithine decarboxylase and epithelial cell proliferation in intestinal IEC-6 cells in culture. Br J Pharmacol. 1997;120:571–574. doi: 10.1038/sj.bjp.0700936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Azpiazu I, Saltiel AR, DePaoli-Roach AA, Lawrence JC. Regulation of both glycogen synthase and PHAS-I by insulin in rat skeletal muscle involves mitogen-activated protein kinase-independent and rapamycin-sensitive pathways. J Biol Chem. 1996;271:5033–5039. doi: 10.1074/jbc.271.9.5033. [DOI] [PubMed] [Google Scholar]
- 86.Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, Giaccia AJ, Abraham RT. Regulation of hypoxia-inducible factor 1alpha expression and function by the mammalian target of rapamycin. Mol Cell Biol. 2002;22:7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Treins C, Giorgetti-Peraldi S, Murdaca J, Semenza GL, Van Obberghen E. Insulin stimulates hypoxia-inducible factor 1 through a phosphatidylinositol 3-kinase/target of rapamycin-dependent signaling pathway. J Biol Chem. 2002;277:27975–27981. doi: 10.1074/jbc.M204152200. [DOI] [PubMed] [Google Scholar]
- 88.Huffman TA, Mothe-Satney I, Lawrence JC., Jr Insulin-stimulated phosphorylation of lipin mediated by the mammalian target of rapamycin. Proc Natl Acad Sci USA. 2002;99:1047–1052. doi: 10.1073/pnas.022634399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Parekh D, Ziegler W, Yonezawa K, Hara K, Parker PJ. Mammalian TOR controls one of two kinase pathways acting upon nPKCdelta and nPKCepsilon. J Biol Chem. 1999;274:34758–34764. doi: 10.1074/jbc.274.49.34758. [DOI] [PubMed] [Google Scholar]
- 90.Peterson RT, Desai BN, Hardwick JS, Schreiber SL. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycinassociated protein. Proc Natl Acad Sci USA. 1999;96:4438–4442. doi: 10.1073/pnas.96.8.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang S, Liu LN, Hosoi H, Dilling MB, Shikata T, Houghton PJ. p53/p21(CIP1) cooperate in enforcing rapamycin-induced G(1) arrest and determine the cellular response to rapamycin. Cancer Res. 2001;61:3373–3381. [PubMed] [Google Scholar]
- 92.Nourse J, Firpo E, Flanagan WM, Coats S, Polyak K, Lee MH, Massague J, Crabtree GR, Roberts JM. Interleukin-2-mediated elimination of the p27Kip1 cyclin-dependent kinase inhibitor prevented by rapamycin. Nature. 1994;372:570–573. doi: 10.1038/372570a0. [DOI] [PubMed] [Google Scholar]
- 93.Usui I, Haruta T, Iwata M, Takano A, Uno T, Kawahara J, Ueno E, Sasaoka T, Kobayashi M. Retinoblastoma protein phosphorylation via PI 3-kinase and mTOR pathway regulates adipocyte differentiation. Biochem Biophys Res Commun. 2000;275:115–120. doi: 10.1006/bbrc.2000.3201. [DOI] [PubMed] [Google Scholar]
- 94.Yokogami K, Wakisaka S, Avruch J, Reeves SA. Serine phosphorylation and maximal activation of STAT3 during CNTF signaling is mediated by the rapamycin target mTOR. Curr Biol. 2000;10:47–50. doi: 10.1016/s0960-9822(99)00268-7. [DOI] [PubMed] [Google Scholar]
- 95.Reinhard C, Thomas G, Kozma SC. A single gene encodes two isoforms of the p70 S6 kinase: activation upon mitogenic stimulation. Proc Natl Acad Sci USA. 1992;89:4052–4056. doi: 10.1073/pnas.89.9.4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martin KA, Blenis J. Coordinate regulation of translation by the PI 3-kinase and mTOR pathways. Adv Cancer Res. 2002;86:1–39. doi: 10.1016/s0065-230x(02)86001-8. [DOI] [PubMed] [Google Scholar]
- 97.Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. Embo J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Park IH, Bachmann R, Shirazi H, Chen J. Regulation of ribosomal S6 kinase 2 by mammalian target of rapamycin. J Biol Chem. 2002;277:31423–31429. doi: 10.1074/jbc.M204080200. [DOI] [PubMed] [Google Scholar]
- 99.Price DJ, Grove JR, Calvo V, Avruch J, Bierer BE. Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science. 1992;257:973–977. doi: 10.1126/science.1380182. [DOI] [PubMed] [Google Scholar]
- 100.Pearson RB, Dennis PB, Han JW, Williamson NA, Kozma SC, Wettenhall RE, Thomas G. The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. Embo J. 1995;14:5279–5287. doi: 10.1002/j.1460-2075.1995.tb00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moser BA, Dennis PB, Pullen N, Pearson RB, Williamson NA, Wettenhall RE, Kozma SC, Thomas G. Dual requirement for a newly identified phosphorylation site in p70s6k. Mol Cell Biol. 1997;17:5648–5655. doi: 10.1128/mcb.17.9.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saitoh M, Pullen N, Brennan P, Cantrell D, Dennis PB, Thomas G. Regulation of an activated S6 kinase 1 variant reveals a novel mammalian target of rapamycin phosphorylation site. J Biol Chem. 2002;277:20104–20112. doi: 10.1074/jbc.M201745200. [DOI] [PubMed] [Google Scholar]
- 103.Alessi DR, Kozlowski MT, Weng QP, Morrice N, Avruch J. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- 104.Pullen N, Dennis PB, Andjelkovic M, Dufner A, Kozma SC, Hemmings BA, Thomas G. Phosphorylation and activation of p70s6k by PDK1. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 105.Isotani S, Hara K, Tokunaga C, Inoue H, Avruch J, Yonezawa K. Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase alpha in vitro. J Biol Chem. 1999;274:34493–34498. doi: 10.1074/jbc.274.48.34493. [DOI] [PubMed] [Google Scholar]
- 106.Cheatham L, Monfar M, Chou MM, Blenis J. Structural and functional analysis of pp70S6k. Proc Natl Acad Sci USA. 1995;92:11696–11700. doi: 10.1073/pnas.92.25.11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jeno P, Ballou LM, Novak-Hofer I, Thomas G. Identification and characterization of a mitogen-activated S6 kinase. Proc Natl Acad Sci USA. 1988;85:406–410. doi: 10.1073/pnas.85.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dufner A, Thomas G. Ribosomal S6 kinase signaling and the control of translation. Exp Cell Res. 1999;253:100–109. doi: 10.1006/excr.1999.4683. [DOI] [PubMed] [Google Scholar]
- 109.Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: a regulator of cell size. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- 110.Radimerski T, Montagne J, Rintelen F, Stocker H, van der Kaay J, Downes CP, Hafen E, Thomas G. dS6K-regulated cell growth is dPKB/dPI(3)K-independent, but requires dPDK1. Nat Cell Biol. 2002;4:251–255. doi: 10.1038/ncb763. [DOI] [PubMed] [Google Scholar]
- 111.Avruch J, Belham C, Weng Q, Hara K, Yonezawa K. The p70 S6 kinase integrates nutrient and growth signals to control translational capacity. Prog Mol Subcell Biol. 2001;26:115–154. doi: 10.1007/978-3-642-56688-2_5. [DOI] [PubMed] [Google Scholar]
- 112.Jefferies HB, Reinhard C, Kozma SC, Thomas G. Rapamycin selectively represses translation of the “polypyrimidine tract” mRNA family. Proc Natl Acad Sci USA. 1994;91:4441–4445. doi: 10.1073/pnas.91.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Terada N, Patel HR, Takase K, Kohno K, Nairn AC, Gelfand EW. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc Natl Acad Sci USA. 1994;91:11477–11481. doi: 10.1073/pnas.91.24.11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stolovich M, Tang H, Hornstein E, Levy G, Cohen R, Bae SS, Birnbaum MJ, Meyuhas O. Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol Cell Biol. 2002;22:8101–8113. doi: 10.1128/MCB.22.23.8101-8113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5’-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Holz MK, Ballif BA, Gygi SP, Blenis J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 117.Satoh MS, Poirier GG, Lindahl T. Dual function for poly(ADP-ribose) synthesis in response to DNA strand breakage. Biochemistry. 1994;33:7099–7106. doi: 10.1021/bi00189a012. [DOI] [PubMed] [Google Scholar]
- 118.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5’-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 119.Poulin F, Gingras AC, Olsen H, Chevalier S, Sonenberg N. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J Biol Chem. 1998;273:14002–14007. doi: 10.1074/jbc.273.22.14002. [DOI] [PubMed] [Google Scholar]
- 120.Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr, Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 121.Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem. 1997;272:26457–26463. doi: 10.1074/jbc.272.42.26457. [DOI] [PubMed] [Google Scholar]
- 122.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang DQ, Kastan MB. Participation of ATM in insulin signalling through phosphorylation of eIF-4E-binding protein 1. Nat Cell Biol. 2000;2:893–898. doi: 10.1038/35046542. [DOI] [PubMed] [Google Scholar]
- 124.Shah OJ, Anthony JC, Kimball SR, Jefferson LS. 4E-BP1 and S6K1: translational integration sites for nutritional and hormonal information in muscle. Am J Physiol Endocrinol Metab. 2000;279:E715–729. doi: 10.1152/ajpendo.2000.279.4.E715. [DOI] [PubMed] [Google Scholar]
- 125.Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell. 1999;3:707–716. doi: 10.1016/s1097-2765(01)80003-4. [DOI] [PubMed] [Google Scholar]
- 126.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 127.Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, Sabatini DM. mSin1 is necessary for Akt/PKB phosphorylation, and its isoforms define three distinct mTORC2s. Curr Biol. 2006;16:1865–1870. doi: 10.1016/j.cub.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 128.Pearce LR, Huang X, Boudeau J, Pawlowski R, Wullschleger S, Deak M, Ibrahim AF, Gourlay R, Magnuson MA, Alessi DR. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 2007;405:513–522. doi: 10.1042/BJ20070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Woo SY, Kim DH, Jun CB, Kim YM, Haar EV, Lee SI, Hegg JW, Bandhakavi S, Griffin TJ, Kim DH. PRR5, a novel component of mTOR complex 2, regulates platelet-derived growth factor receptor beta expression and signaling. J Biol Chem. 2007;282:25604–25612. doi: 10.1074/jbc.M704343200. [DOI] [PubMed] [Google Scholar]
- 130.Martin J, Masri J, Bernath A, Nishimura RN, Gera J. Hsp70 associates with Rictor and is required for mTORC2 formation and activity. Biochem Biophys Res Commun. 2008;372:578–583. doi: 10.1016/j.bbrc.2008.05.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu L, Li F, Cardelli JA, Martin KA, Blenis J, Huang S. Rapamycin inhibits cell motility by suppression of mTOR-mediated S6K1 and 4E-BP1 pathways. Oncogene. 2006;25:7029–7040. doi: 10.1038/sj.onc.1209691. [DOI] [PubMed] [Google Scholar]
- 132.Liu L, Chen L, Chung J, Huang S. Rapamycin inhibits F-actin reorganization and phosphorylation of focal adhesion proteins. Oncogene. 2008;27:4998–5010. doi: 10.1038/onc.2008.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 134.Cybulski N, Polak P, Auwerx J, Ruegg MA, Hall MN. mTOR complex 2 in adipose tissue negatively controls whole-body growth. Proc Natl Acad Sci USA. 2009;106:9902–9907. doi: 10.1073/pnas.0811321106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang Q, Inoki K, Kim E, Guan KL. TSC1/TSC2 and Rheb have different effects on TORC1 and TORC2 activity. Proc Natl Acad Sci USA. 2006;103:6811–6816. doi: 10.1073/pnas.0602282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Manning BD. Balancing Akt with S6K: implications for both metabolic diseases and tumorigenesis. J Cell Biol. 2004;167:399–403. doi: 10.1083/jcb.200408161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Scheid MP, Woodgett JR. Unravelling the activation mechanisms of protein kinase B/Akt. FEBS Lett. 2003;546:108–112. doi: 10.1016/s0014-5793(03)00562-3. [DOI] [PubMed] [Google Scholar]
- 138.Newton AC. Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem J. 2003;370:361–371. doi: 10.1042/BJ20021626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schalm SS, Tee AR, Blenis J. Characterization of a conserved C-terminal motif (RSPRR) in ribosomal protein S6 kinase 1 required for its mammalian target of rapamycin-dependent regulation. J Biol Chem. 2005;280:11101–11106. doi: 10.1074/jbc.M413995200. [DOI] [PubMed] [Google Scholar]
- 140.Ali SM, Sabatini DM. Structure of S6 kinase 1 determines whether raptor-mTOR or rictor-mTOR phosphorylates its hydrophobic motif site. J Biol Chem. 2005;280:19445–19448. doi: 10.1074/jbc.C500125200. [DOI] [PubMed] [Google Scholar]
- 141.Andjelkovic M, Alessi DR, Meier R, Fernandez A, Lamb NJ, Frech M, Cron P, Cohen P, Lucocq JM, Hemmings BA. Role of translocation in the activation and function of protein kinase B. J Biol Chem. 1997;272:31515–31524. doi: 10.1074/jbc.272.50.31515. [DOI] [PubMed] [Google Scholar]
- 142.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Balendran A, Casamayor A, Deak M, Paterson A, Gaffney P, Currie R, Downes CP, Alessi DR. PDK1 acquires PDK2 activity in the presence of a synthetic peptide derived from the carboxyl terminus of PRK2. Curr Biol. 1999;9:393–404. doi: 10.1016/s0960-9822(99)80186-9. [DOI] [PubMed] [Google Scholar]
- 144.Persad S, Attwell S, Gray V, Mawji N, Deng JT, Leung D, Yan J, Sanghera J, Walsh MP, Dedhar S. Regulation of protein kinase B/Akt-serine 473 phosphorylation by integrin-linked kinase: critical roles for kinase activity and amino acids arginine 211 and serine 343. J Biol Chem. 2001;276:27462–27469. doi: 10.1074/jbc.M102940200. [DOI] [PubMed] [Google Scholar]
- 145.Toker A, Newton AC. Akt/protein kinase B is regulated by autophosphorylation at the hypothetical PDK-2 site. J Biol Chem. 2000;275:8271–8274. doi: 10.1074/jbc.275.12.8271. [DOI] [PubMed] [Google Scholar]
- 146.Feng J, Park J, Cron P, Hess D, Hemmings BA. Identification of a PKB/Akt hydrophobic motif Ser-473 kinase as DNA-dependent protein kinase. J Biol Chem. 2004;279:41189–41196. doi: 10.1074/jbc.M406731200. [DOI] [PubMed] [Google Scholar]
- 147.Schaller MD. Paxillin: a focal adhesion-associated adaptor protein. Oncogene. 2001;20:6459–6472. doi: 10.1038/sj.onc.1204786. [DOI] [PubMed] [Google Scholar]
- 148.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 149.Garcia-Martinez JM, Alessi DR. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1) Biochem J. 2008;416:375–385. doi: 10.1042/BJ20081668. [DOI] [PubMed] [Google Scholar]
- 150.Jones KT, Greer ER, Pearce D, Ashrafi K. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 2009;7:e60. doi: 10.1371/journal.pbio.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kobayashi T, Cohen P. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem J. 1999;339(Pt 2):319–328. [PMC free article] [PubMed] [Google Scholar]
- 153.Park J, Leong ML, Buse P, Maiyar AC, Firestone GL, Hemmings BA. Serum and glucocorticoid-inducible kinase (SGK) is a target of the PI 3-kinase-stimulated signaling pathway. Embo J. 1999;18:3024–3033. doi: 10.1093/emboj/18.11.3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tessier M, Woodgett JR. Serum and glucocorticoid-regulated protein kinases: variations on a theme. J Cell Biochem. 2006;98:1391–1407. doi: 10.1002/jcb.20894. [DOI] [PubMed] [Google Scholar]
- 155.Debonneville C, Flores SY, Kamynina E, Plant PJ, Tauxe C, Thomas MA, Munster C, Chraibi A, Pratt JH, Horisberger JD, Pearce D, Loffing J, Staub O. Phosphorylation of Nedd4-2 by Sgk1 regulates epithelial Na(+) channel cell surface expression. Embo J. 2001;20:7052–7059. doi: 10.1093/emboj/20.24.7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- 157.Tee AR, Blenis J. mTOR, translational control and human disease. Semin Cell Dev Biol. 2005;16:29–37. doi: 10.1016/j.semcdb.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 158.Kenerson HL, Aicher LD, True LD, Yeung RS. Activated mammalian target of rapamycin pathway in the pathogenesis of tuberous sclerosis complex renal tumors. Cancer Res. 2002;62:5645–5650. [PubMed] [Google Scholar]
- 159.Johnson SR, Tattersfield AE. Lymphangioleiomyomatosis. Semin Respir Crit Care Med. 2002;23:85–92. doi: 10.1055/s-2002-25298. [DOI] [PubMed] [Google Scholar]
- 160.Abel TW, Baker SJ, Fraser MM, Tihan T, Nelson JS, Yachnis AT, Bouffard JP, Mena H, Burger PC, Eberhart CG. Lhermitte-Duclos disease: a report of 31 cases with immunohistochemical analysis of the PTEN/AKT/mTOR pathway. J Neuropathol Exp Neurol. 2005;64:341–349. doi: 10.1093/jnen/64.4.341. [DOI] [PubMed] [Google Scholar]
- 161.Lachlan KL, Lucassen AM, Bunyan D, Temple IK. Cowden syndrome and Bannayan Riley Ruvalcaba syndrome represent one condition with variable expression and age-related penetrance: results of a clinical study of PTEN mutation carriers. J Med Genet. 2007;44:579–585. doi: 10.1136/jmg.2007.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kwiatkowski DJ. Tuberous sclerosis: from tubers to mTOR. Ann Hum Genet. 2003;67:87–96. doi: 10.1046/j.1469-1809.2003.00012.x. [DOI] [PubMed] [Google Scholar]
- 163.Biesecker LG, Rosenberg MJ, Vacha S, Turner JT, Cohen MM. PTEN mutations and proteus syndrome. Lancet. 2001;358:2079–2080. doi: 10.1016/S0140-6736(01)07109-4. [DOI] [PubMed] [Google Scholar]
- 164.Foley TR, McGarrity TJ, Abt AB. Peutz-Jeghers syndrome: a clinicopathologic survey of the “Harrisburg family” with a 49-year follow-up. Gastroenterology. 1988;95:1535–1540. doi: 10.1016/s0016-5085(88)80074-x. [DOI] [PubMed] [Google Scholar]
- 165.Choi HS, Park YJ, Park JG. Peutz-Jeghers syndrome: a new understanding. J Korean Med Sci. 1999;14:2–7. doi: 10.3346/jkms.1999.14.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Blair E, Redwood C, Ashrafian H, Oliveira M, Broxholme J, Kerr B, Salmon A, Ostman-Smith I, Watkins H. Mutations in the gamma(2) subunit of AMP-activated protein kinase cause familial hypertrophic cardiomyopathy: evidence for the central role of energy compromise in disease pathogenesis. Hum Mol Genet. 2001;10:1215–1220. doi: 10.1093/hmg/10.11.1215. [DOI] [PubMed] [Google Scholar]
- 167.Gollob MH, Green MS, Tang AS, Gollob T, Karibe A, Ali Hassan AS, Ahmad F, Lozado R, Shah G, Fananapazir L, Bachinski LL, Roberts R. Identification of a gene responsible for familial Wolff-Parkinson-White syndrome. N Engl J Med. 2001;344:1823–1831. doi: 10.1056/NEJM200106143442403. [DOI] [PubMed] [Google Scholar]
- 168.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 169.Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB, Phillips WA. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–7681. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 170.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 171.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 172.Rotterud R, Fossa SD, Nesland JM. Protein networking in bladder cancer: immunoreactivity for FGFR3, EGFR, ERBB2, KAI1, PTEN, and RAS in normal and malignant urothelium. Histol Histopathol. 2007;22:349–363. doi: 10.14670/HH-22.349. [DOI] [PubMed] [Google Scholar]
- 173.Madhunapantula SV, Robertson GP. The PTEN-AKT3 signaling cascade as a therapeutic target in melanoma. Pigment Cell Melanoma Res. 2009;22:400–419. doi: 10.1111/j.1755-148X.2009.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lacey JV, Jr, Mutter GL, Ronnett BM, Ioffe OB, Duggan MA, Rush BB, Glass AG, Richesson DA, Chatterjee N, Langholz B, Sherman ME. PTEN expression in endometrial biopsies as a marker of progression to endometrial carcinoma. Cancer Res. 2008;68:6014–6020. doi: 10.1158/0008-5472.CAN-08-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Rasheed BK, Wiltshire RN, Bigner SH, Bigner DD. Molecular pathogenesis of malignant gliomas. Curr Opin Oncol. 1999;11:162–167. doi: 10.1097/00001622-199905000-00004. [DOI] [PubMed] [Google Scholar]
- 176.Haas-Kogan D, Shalev N, Wong M, Mills G, Yount G, Stokoe D. Protein kinase B (PKB/Akt) activity is elevated in glioblastoma cells due to mutation of the tumor suppressor PTEN/MMAC. Curr Biol. 1998;8:1195–1198. doi: 10.1016/s0960-9822(07)00493-9. [DOI] [PubMed] [Google Scholar]
- 177.Gallia GL, Rand V, Siu IM, Eberhart CG, James CD, Marie SK, Oba-Shinjo SM, Carlotti CG, Caballero OL, Simpson AJ, Brock MV, Massion PP, Carson BS, Sr, Riggins GJ. PIK3CA gene mutations in pediatric and adult glioblastoma multiforme. Mol Cancer Res. 2006;4:709–714. doi: 10.1158/1541-7786.MCR-06-0172. [DOI] [PubMed] [Google Scholar]
- 178.Kondo K, Yao M, Kobayashi K, Ota S, Yoshida M, Kaneko S, Baba M, Sakai N, Kishida T, Kawakami S, Uemura H, Nagashima Y, Nakatani Y, Hosaka M. PTEN/MMAC1/TEP1 mutations in human primary renal-cell carcinomas and renal carcinoma cell lines. Int J Cancer. 2001;91:219–224. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1034>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 179.Hager M, Haufe H, Kemmerling R, Mikuz G, Kolbitsch C, Moser PL. PTEN expression in renal cell carcinoma and oncocytoma and prognosis. Pathology. 2007;39:482–485. doi: 10.1080/00313020701570012. [DOI] [PubMed] [Google Scholar]
- 180.Krymskaya VP, Goncharova EA. PI3K/mTORC1 activation in hamartoma syndromes: therapeutic prospects. Cell Cycle. 2009;8:403–413. doi: 10.4161/cc.8.3.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Goncharova EA, Goncharov DA, Eszterhas A, Hunter DS, Glassberg MK, Yeung RS, Walker CL, Noonan D, Kwiatkowski DJ, Chou MM, Panettieri RA, Jr, Krymskaya VP. Tuberin regulates p70 S6 kinase activation and ribosomal protein S6 phosphorylation. A role for the TSC2 tumor suppressor gene in pulmonary lymphangioleiomyomatosis (LAM) J Biol Chem. 2002;277:30958–30967. doi: 10.1074/jbc.M202678200. [DOI] [PubMed] [Google Scholar]
- 182.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Hoglund P, Jarvinen H, Kristo P, Pelin K, Ridanpaa M, Salovaara R, Toro T, Bodmer W, Olschwang S, Olsen AS, Stratton MR, de la Chapelle A, Aaltonen LA. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184–187. doi: 10.1038/34432. [DOI] [PubMed] [Google Scholar]
- 183.Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, Eng C, Parsons R. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 184.Bjornsti MA, Houghton PJ. The TOR pathway: a target for cancer therapy. Nat Rev Cancer. 2004;4:335–348. doi: 10.1038/nrc1362. [DOI] [PubMed] [Google Scholar]
- 185.Vignot S, Faivre S, Aguirre D, Raymond E. mTOR-targeted therapy of cancer with rapamycin derivatives. Ann Oncol. 2005;16:525–537. doi: 10.1093/annonc/mdi113. [DOI] [PubMed] [Google Scholar]
- 186.Meric-Bernstam F, Gonzalez-Angulo AM. Targeting the mTOR signaling network for cancer therapy. J Clin Oncol. 2009;27:2278–2287. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shioi T, McMullen JR, Kang PM, Douglas PS, Obata T, Franke TF, Cantley LC, Izumo S. Akt/protein kinase B promotes organ growth in transgenic mice. Mol Cell Biol. 2002;22:2799–2809. doi: 10.1128/MCB.22.8.2799-2809.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Shioi T, McMullen JR, Tarnavski O, Converso K, Sherwood MC, Manning WJ, Izumo S. Rapamycin attenuates load-induced cardiac hypertrophy in mice. Circulation. 2003;107:1664–1670. doi: 10.1161/01.CIR.0000057979.36322.88. [DOI] [PubMed] [Google Scholar]
- 189.White MF. IRS proteins and the common path to diabetes. Am J Physiol Endocrinol Metab. 2002;283:E413–422. doi: 10.1152/ajpendo.00514.2001. [DOI] [PubMed] [Google Scholar]
- 190.Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, Thomas G. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431:200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 191.Tsukiyama-Kohara K, Poulin F, Kohara M, DeMaria CT, Cheng A, Wu Z, Gingras AC, Katsume A, Elchebly M, Spiegelman BM, Harper ME, Tremblay ML, Sonenberg N. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat Med. 2001;7:1128–1132. doi: 10.1038/nm1001-1128. [DOI] [PubMed] [Google Scholar]
- 192.Ballou LM, Lin RZ. Rapamycin and mTOR kinase inhibitors. J Chem Biol. 2008;1:27–36. doi: 10.1007/s12154-008-0003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Huang S, Houghton PJ. Resistance to rapamycin: a novel anticancer drug. Cancer Metastasis Rev. 2001;20:69–78. doi: 10.1023/a:1013167315885. [DOI] [PubMed] [Google Scholar]
- 194.Dilling MB, Dias P, Shapiro DN, Germain GS, Johnson RK, Houghton PJ. Rapamycin selectively inhibits the growth of childhood rhabdomyosarcoma cells through inhibition of signaling via the type I insulin-like growth factor receptor. Cancer Res. 1994;54:903–907. [PubMed] [Google Scholar]
- 195.Hosoi H, Dilling MB, Shikata T, Liu LN, Shu L, Ashmun RA, Germain GS, Abraham RT, Houghton PJ. Rapamycin causes poorly reversible inhibition of mTOR and induces p53-independent apoptosis in human rhabdomyosarcoma cells. Cancer Res. 1999;59:886–894. [PubMed] [Google Scholar]
- 196.Geoerger B, Kerr K, Tang CB, Fung KM, Powell B, Sutton LN, Phillips PC, Janss AJ. Antitumor activity of the rapamycin analog CCI-779 in human primitive neuroectodermal tumor/medulloblastoma models as single agent and in combination chemotherapy. Cancer Res. 2001;61:1527–1532. [PubMed] [Google Scholar]
- 197.Seufferlein T, Rozengurt E. Rapamycin inhibits constitutive p70s6k phosphorylation, cell proliferation, and colony formation in small cell lung cancer cells. Cancer Res. 1996;56:3895–3897. [PubMed] [Google Scholar]
- 198.Ogawa T, Tokuda M, Tomizawa K, Matsui H, Itano T, Konishi R, Nagahata S, Hatase O. Osteoblastic differentiation is enhanced by rapamycin in rat osteoblast-like osteosarcoma (ROS 17/2.8) cells. Biochem Biophys Res Commun. 1998;249:226–230. doi: 10.1006/bbrc.1998.9118. [DOI] [PubMed] [Google Scholar]
- 199.Grewe M, Gansauge F, Schmid RM, Adler G, Seufferlein T. Regulation of cell growth and cyclin D1 expression by the constitutively active FRAP-p70s6K pathway in human pancreatic cancer cells. Cancer Res. 1999;59:3581–3587. [PubMed] [Google Scholar]
- 200.van der Poel HG, Hanrahan C, Zhong H, Simons JW. Rapamycin induces Smad activity in prostate cancer cell lines. Urol Res. 2003;30:380–386. doi: 10.1007/s00240-002-0282-1. [DOI] [PubMed] [Google Scholar]
- 201.Pang H, Faber LE. Estrogen and rapamycin effects on cell cycle progression in T47D breast cancer cells. Breast Cancer Res Treat. 2001;70:21–26. doi: 10.1023/a:1012570204923. [DOI] [PubMed] [Google Scholar]
- 202.Busca R, Bertolotto C, Ortonne JP, Ballotti R. Inhibition of the phosphatidylinositol 3-kinase/p70(S6)-kinase pathway induces B16 melanoma cell differentiation. J Biol Chem. 1996;271:31824–31830. doi: 10.1074/jbc.271.50.31824. [DOI] [PubMed] [Google Scholar]
- 203.Muthukkumar S, Ramesh TM, Bondada S. Rapamycin, a potent immunosuppressive drug, causes programmed cell death in B lymphoma cells. Transplantation. 1995;60:264–270. doi: 10.1097/00007890-199508000-00010. [DOI] [PubMed] [Google Scholar]
- 204.Rizzieri DA, Feldman E, Dipersio JF, Gabrail N, Stock W, Strair R, Rivera VM, Albitar M, Bedrosian CL, Giles FJ. A phase 2 clinical trial of deforolimus (AP23573, MK-8669), a novel mammalian target of rapamycin inhibitor, in patients with relapsed or refractory hematologic malignancies. Clin Cancer Res. 2008;14:2756–2762. doi: 10.1158/1078-0432.CCR-07-1372. [DOI] [PubMed] [Google Scholar]
- 205.Rini BI. Temsirolimus, an inhibitor of mammalian target of rapamycin. Clin Cancer Res. 2008;14:1286–1290. doi: 10.1158/1078-0432.CCR-07-4719. [DOI] [PubMed] [Google Scholar]
- 206.Wolpin BM, Hezel AF, Abrams T, Blaszkowsky LS, Meyerhardt JA, Chan JA, Enzinger PC, Allen B, Clark JW, Ryan DP, Fuchs CS. Oral mTOR inhibitor everolimus in patients with gemcitabine-refractory metastatic pancreatic cancer. J Clin Oncol. 2009;27:193–198. doi: 10.1200/JCO.2008.18.9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.O’Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, Lane H, Hofmann F, Hicklin DJ, Ludwig DL, Baselga J, Rosen N. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer Res. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Yu K, Toral-Barza L, Shi C, Zhang WG, Lucas J, Shor B, Kim J, Verheijen J, Curran K, Malwitz DJ, Cole DC, Ellingboe J, Ayral-Kaloustian S, Mansour TS, Gibbons JJ, Abraham RT, Nowak P, Zask A. Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Res. 2009;69:6232–6240. doi: 10.1158/0008-5472.CAN-09-0299. [DOI] [PubMed] [Google Scholar]
- 211.Garcia-Martinez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, Alessi DR. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009 doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Liu TJ, Koul D, LaFortune T, Tiao N, Shen RJ, Maira SM, Garcia-Echevrria C, Yung WK. NVP-BEZ235, a novel dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor, elicits multifaceted antitumor activities in human gliomas. Mol Cancer Ther. 2009;8:2204–2210. doi: 10.1158/1535-7163.MCT-09-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Raynaud FI, Eccles S, Clarke PA, Hayes A, Nutley B, Alix S, Henley A, Di-Stefano F, Ahmad Z, Guillard S, Bjerke LM, Kelland L, Valenti M, Patterson L, Gowan S, de Haven Brandon A, Hayakawa M, Kaizawa H, Koizumi T, Ohishi T, Patel S, Saghir N, Parker P, Waterfield M, Workman P. Pharmacologic characterization of a potent inhibitor of class I phosphatidylinositide 3-kinases. Cancer Res. 2007;67:5840–5850. doi: 10.1158/0008-5472.CAN-06-4615. [DOI] [PubMed] [Google Scholar]
- 214.Fan QW, Knight ZA, Goldenberg DD, Yu W, Mostov KE, Stokoe D, Shokat KM, Weiss WA. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell. 2006;9:341–349. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Westhoff MA, Kandenwein JA, Karl S, Vellanki SH, Braun V, Eramo A, Antoniadis G, Debatin KM, Fulda S. The pyridinylfuranopyrimidine inhibitor, PI-103, chemosensitizes glioblastoma cells for apoptosis by inhibiting DNA repair. Oncogene. 2009;28:3586–3596. doi: 10.1038/onc.2009.215. [DOI] [PubMed] [Google Scholar]
- 216.Molckovsky A, Siu LL. First-in-class, first-in-human phase I results of targeted agents: Highlights of the 2008 American Society of Clinical Oncology meeting. J Hematol Oncol. 2008;1:20. doi: 10.1186/1756-8722-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Beevers CS, Chen L, Liu L, Luo Y, Webster NJ, Huang S. Curcumin disrupts the Mammalian target of rapamycin-raptor complex. Cancer Res. 2009;69:1000–1008. doi: 10.1158/0008-5472.CAN-08-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Jiang H, Shang X, Wu H, Gautam SC, Al-Holou S, Li C, Kuo J, Zhang L, Chopp M. Resveratrol downregulates PI3K/Akt/mTOR signaling pathways in human U251 glioma cells. J Exp Ther Oncol. 2009;8:25–33. [PMC free article] [PubMed] [Google Scholar]
- 219.Brito PM, Devillard R, Negre-Salvayre A, Almeida LM, Dinis TC, Salvayre R, Auge N. Resveratrol inhibits the mTOR mitogenic signaling evoked by oxidized LDL in smooth muscle cells. Atherosclerosis. 2009;205:126–134. doi: 10.1016/j.atherosclerosis.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 220.Zhang Q, Kelly AP, Wang L, French SW, Tang X, Duong HS, Messadi DV, Le AD. Green tea extract and (-)-epigallocatechin-3-gallate inhibit mast cell-stimulated type I collagen expression in keloid fibroblasts via blocking PI-3K/AkT signaling pathways. J Invest Dermatol. 2006;126:2607–2613. doi: 10.1038/sj.jid.5700472. [DOI] [PubMed] [Google Scholar]
- 221.Anastasius N, Boston S, Lacey M, Storing N, Whitehead SA. Evidence that low-dose, long-term genistein treatment inhibits oestradiol-stimulated growth in MCF-7 cells by down-regulation of the PI3-kinase/Akt signalling pathway. J Steroid Biochem Mol Biol. 2009;116:50–55. doi: 10.1016/j.jsbmb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 222.Nakamura Y, Yogosawa S, Izutani Y, Watanabe H, Otsuji E, Sakai T. A combination of indol-3-carbinol and genistein synergistically induces apoptosis in human colon cancer HT-29 cells by inhibiting Akt phosphorylation and progression of autophagy. Mol Cancer. 2009;8:100. doi: 10.1186/1476-4598-8-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kong D, Banerjee S, Huang W, Li Y, Wang Z, Kim HR, Sarkar FH. Mammalian target of rapamycin repression by 3,3’-diindolylmethane inhibits invasion and angiogenesis in platelet-derived growth factor-D-overexpressing PC3 cells. Cancer Res. 2008;68:1927–1934. doi: 10.1158/0008-5472.CAN-07-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Chan MM, Huang HI, Fenton MR, Fong D. In vivo inhibition of nitric oxide synthase gene expression by curcumin, a cancer preventive natural product with anti-inflammatory properties. Biochem Pharmacol. 1998;55:1955–1962. doi: 10.1016/s0006-2952(98)00114-2. [DOI] [PubMed] [Google Scholar]
- 225.Beevers CS, Li F, Liu L, Huang S. Curcumin inhibits the mammalian target of rapamycin-mediated signaling pathways in cancer cells. Int J Cancer. 2006;119:757–764. doi: 10.1002/ijc.21932. [DOI] [PubMed] [Google Scholar]
- 226.Johnson SM, Gulhati P, Arrieta I, Wang X, Uchida T, Gao T, Evers BM. Curcumin inhibits proliferation of colorectal carcinoma by modulating Akt/mTOR signaling. Anticancer Res. 2009;29:3185–3190. [PMC free article] [PubMed] [Google Scholar]