Abstract
Up to one third of human melanomas are characterized by an oncogenic mutation in the gene encoding the small GTPase NRAS. Ras proteins activate three primary classes of effectors: Rafs, PI3Ks, and RalGEFs. In melanomas lacking NRAS mutations, the first two effectors can still be activated vis-à-vis oncogenic BRAF mutations coupled with a loss of the PI3K negative regulator PTEN. This suggests that Ras effectors promote melanoma, regardless of whether they are activated by oncogenic NRas. The only major Ras effector pathway not explored for its role in melanoma is the RalGEF-Ral pathway, in which the Ras activation of RalGEFs converts the small GTPases RalA and RalB to an active GTP-bound state. We report that RalA is activated in a number of human melanoma cancer cell lines harboring an oncogenic NRAS allele, an oncogenic BRAF allele, or wild type NRAS and BRAF alleles. Furthermore, shRNA-mediated knock down of RalA, and to a lesser extent RalB, inhibited the tumorigenic growth of melanoma cell lines having these three genotypes. Thus, as is the case for Raf and PI3K signaling, Rals also contribute to melanoma tumorigenesis.
Keywords: melanoma, RalA, RalB, NRas
Up to a third of human melanomas harbor an activating mutation, typically at Q61, in the gene NRAS (Gray-Schopfer et al., 2007), which traps Ras proteins in the active GTP-bound state, resulting in constitutive activation of the effectors Rafs, PI3Ks and RalGEFs to promote many tumorigenic phenotypes (Downward, 2003). In melanomas lacking an NRAS mutation, the first two pathways can be activated through a combination of a V600E activating mutation in the gene BRAF coupled with a loss of or decreased expression of the PI3K negative regulator PTEN (Gray-Schopfer et al., 2007). These results suggest a general requirement to activate Ras effector pathways in melanoma, either through an activating mutation in NRAS itself, or by individually activating the Ras effector pathways through mutations to BRAF and PTEN. Following this logic, we explored whether the third major effector of Ras signaling in cancer, the RalGEF pathway (Bodemann & White, 2008; Feig, 2003), was similarly activated in melanoma, and whether this activation occurred in the absence of an oncogenic NRAS mutation.
Activated Ras binds directly to Ral guanine exchange factors (RalGEFs), recruiting these proteins principally to the plasma membrane where they convert the small GTPases RalA and RalB to the active GTP-bound state (Bodemann & White, 2008; Feig, 2003). Activated versions of RalGEFs are transforming in human (Hamad et al., 2002), and to a lesser extent rodent cells (D’Adamo et al., 1997; Khosravi-Far et al., 1996; Miller et al., 1998; Urano et al., 1996; White et al., 1996), and have been shown to promote the metastatic growth of prostate DU145 cancer cells (Yin et al., 2007). Consistent with these observations, RalGDS−/− mice exhibit more than a twofold reduction in the number of skin papillomas induced by chemical carcinogens, which were also significantly smaller and less aggressive, compared to wild type mice (Gonzalez-Garcia et al., 2005). Both RalA and RalB have also been implicated in oncogenic Ras-driven cancers. shRNA-mediated knockdown of RalA uniformly reduced the anchorage independent and tumorigenic growth in oncogenic KRAS mutation-positive pancreatic cancer cell lines (Lim et al., 2006), although this can be context-dependent, as a similar knockdown of RalA enhanced the tumorigenic properties of the immortalized human keratinocyte cell line HaCaT when engineered to over-express oncogenic HRas (Sowalsky et al., 2009). Conversely, activated versions of RalA weakly transform human (Lim et al., 2005), canine (Wu et al., 2005) and in the presence of other Ras effectors, rodent cells (Urano et al., 1996). In the case of RalB, shRNA targeting this gene transcript can decrease survival of transformed cells, particularly when in suspension (Chien et al., 2006; Chien & White, 2003), cell migration (de Gorter et al., 2008; Oxford et al., 2005; Rosse et al., 2006), cell invasion (Lim et al., 2006) and, consistent with these in vitro phenotypes, metastasis of KRAS mutation-positive pancreatic cancer cell lines (Lim et al., 2006). Both RalA and RalB are activated in a number of oncogenic KRas- and HRas-driven human cancer cell lines and tumor specimens (Lim et al., 2005; Lim et al., 2006; Smith et al., 2007). Rals have also been shown to be both activated and promote tumorigenesis in tumors lacking Ras mutations (Bodempudi et al., 2009). Given that the two other major Ras effector pathways, Rafs and PI3K, are commonly activated in melanoma, regardless of the mutational status of NRAS(Gray-Schopfer et al., 2007), that the RalGEF-Ral pathway typically promotes oncogenic Ras-driven transformation and tumorigenesis (Bodemann & White, 2008), and that Rals can be activated in tumors lacking an oncogenic Ras mutation (Bodempudi et al., 2009), we assayed whether Rals were activated and required for tumor growth of human melanoma cancer cell lines with or without an oncogenic NRAS allele.
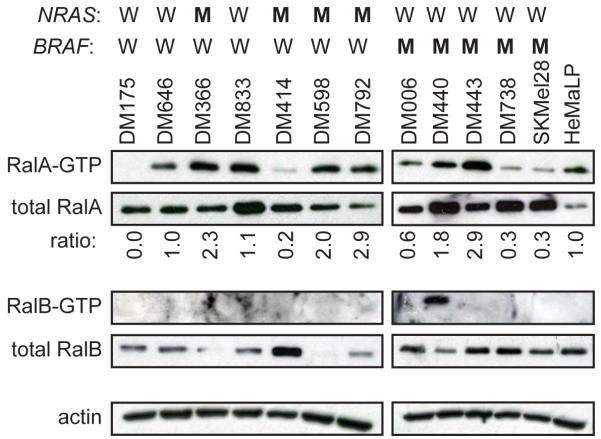
To explore the role of Rals in melanoma we first assessed the activation status of RalA and RalB in a panel of independently derived human melanoma cancer cell lines having wild type BRAF and NRAS (DM175, DM646 and DM833), wild type BRAF and oncogenic NRASQ61L or NRASQ61R, hereafter referred jointly as NRASQ61L/R for ease of description (DM366, DM414, DM598 and DM792), or oncogenic BRAFV600E and wild type NRAS (DM006, DM440, DM443, DM738 and SKMel28) alleles, as assessed by PCR (not shown), compared to control normal human primary melanocytes (HeMaLP). Recombinant GST-RBD encoding the Ral binding domain of the Ral effector protein RalBP1 was used to capture endogenous GTP-bound active RalA and RalB from cellular lysates, which were detected by immunoblot with α-RalA and α-RalB antibodies (Wolthuis et al., 1998). Total levels of RalA, but not RalB, were elevated relative to control melanocytes in all the melanoma cell lines, regardless of the mutation status of NRAS and BRAF. While RalB-GTP levels were generally difficult to detect, with only one cell line exhibiting detectable RalB-GTP, either the amount of RalA-GTP (normalized to actin) or the ratio of RalA-GTP to total RalA (normalized to control cells) was higher in one of three NRAS/BRAF, three of four NRASQ61L/R/BRAF and two of five NRAS/BRAFV600E human melanoma cell lines compared to the control melanocytes (Figure 1). Thus, there was elevated GTP-bound RalA, which was a product of either or both more RalA protein and/or elevated activation in melanoma cell lines compared to the normal melanocyte cells.
Figure 1.
Activation state and expression of Rals in human melanoma cell lines. Equal levels of total protein from whole cell extracts were assayed for GTP-bound and total RalA and RalB in the indicated melanoma cell lines characterized by wild type (W) or oncogenic mutant (M) NRAS and BRAF alleles and in control HEMaLP normal human melanocytes (Invitrogen) by either pulldown with GST-RBD followed by immunoblot (GTP-Ral) or immunoblot (total Ral) with an α-RalA (BD Biosciences) or α-RalB (Millipore) antibody as previously described (Lim et al., 2005; Wolthuis et al., 1998). Actin, detected with an α-actin antibody (Sigma) serves as a loading control. Ratio: RalA-GTP/total RalA of each cancer cell line relative to the control HeMaLP cells.
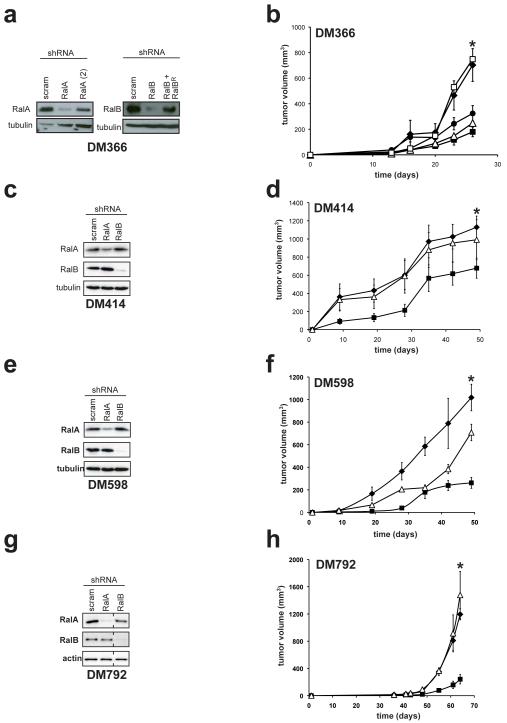
We next assessed the requirement of RalA and RalB in oncogenic NRas-driven melanomagenesis. To begin, the human melanoma cell line DM366, characterized by mutant NRASQ61L/R and wild type BRAF alleles, was stably infected with a retrovirus encoding RalA shRNA, RalB shRNA, or as a control a scramble sequence (Lim et al., 2005). Retroviral introduction of these shRNAs was used to generate polyclonal cell lines to limit the effects of clonal variability. Appropriate reduction of the desired endogenous RalA or RalB protein was confirmed in the resultant cell lines by immunoblot (Figure 2A). We note that while immunoblot exposures were dictated by the cell line with the highest RalA or RalB levels when comparing different melanoma cell lines (Figure 1A), in experiments in which RalA or RalB were knocked down, exposure times were often extended to optimize detection of endogenous RalA or RalB in the control scramble control lines in order to visualize knock down of the Ral proteins (Figure 2A). Lastly, neither RalA shRNA nor RalB shRNA altered the level of NRas or BRaf, as assessed by immunoblot (not shown). The three derived DM366 cell lines were injected into the flank of immunocompromised mice, and tumor growth was monitored over time. The scramble control cell line readily formed tumors, reaching maximum tumor size in less than one month. Knockdown of RalA or RalB similarly reduced tumor growth, as evidenced by a 75% and 55% reduction in the average tumor volume, respectively, by the termination of the experiment compared to scramble control cells (Figure 2B).
Figure 2.
Rals are required for tumor growth of NRASQ61L/R-positive melanoma cells. (a,c,e,g) Protein expression of endogenous RalA, RalB, or as a control actin or tubulin, as assessed by immunoblot with the aforementioned α-RalA, α-RalB, α-actin and α-tubulin (Sigma) antibodies and (b,d,f,h) mean tumor volume (mm3) ± SEM versus time (days) of the indicated melanoma cell lines stably infected with retroviruses encoding RalA shRNA (■), a second RalA (2) shRNA (●), RalB shRNA in the absence (△) or presence of RalBR, RalB cDNA engineered with the silent mutations T129→C, A132→A, T135→A and C138→G to make it resistant to RalB shRNA(□), or a scramble (scram) sequence (◆) using constructs and approaches previously described (Lim et al., 2005). All cell lines were injected subcutaneously into the flanks of four immunocompromised mice each. * p≤0.05 difference in tumor size at the termination of the experiment between scramble control and RalA shRNA expressing cells.
To validate that the reduced tumor growth was not an off-target effect of RalA shRNA, DM366 cells were also stably infected with retrovirus encoding a second shRNA targeting RalA (Lim et al., 2006), which was validated by immunoblot to reduce the level of endogenous RalA, although admittedly not as well as the first RalA shRNA (Figure 2A). Tumors arising from these cells displayed reduced tumor growth, as evidenced by a 65% decrease in tumor size compared to scramble control tumors at the termination of the experiment, although consistent with the higher residual RalA expression in these cells, the reduction in tumor size was not as dramatic as that observed with the first RalA shRNA (Figure 2B). Thus, two independent RalA shRNAs cause a reduction in tumor size relative to the degree of RalA knockdown, indicating that the effect of knocking RalA down is not an off-target effect. Because we were unable to generate a second RalB shRNA that robustly reduced RalB expression (not shown), off-target effects of RalB shRNA were investigated by re-infecting the DM366 RalB knockdown cells with a retrovirus we generated to encode an shRNA-resistant version of RalB, termed RalBR, which was confirmed by immunoblot to restore RalB expression (Figure 2A). When the resultant cells were injected into immunocompromised mice, the derived tumors grew with identical kinetics to scramble control cells (Figure 2B), indicating that restoring RalB expression completely rescued the reduction in tumor growth induced by RalB shRNA. The ability of RalB shRNA to retard tumor growth is thus not an off-target effect. We conclude that reduction in RalA or RalB expression inhibits tumorigenesis of DM366 cells.
To ascertain if the reduction in tumor growth was unique to this cell line, or could be observed in other NRAS-mutation positive melanoma cell lines, the aforementioned DM414, DM598, and DM792 human melanoma cell lines characterized by mutant NRASQ61L/R and wild type BRAF alleles were stably infected with a retrovirus encoding a scramble control sequence, RalA shRNA or RalB shRNA. DM414, DM598, and DM792 cell lines stably infected with a retrovirus encoding RalA shRNA or RalB shRNA were confirmed by immunoblot to have reduced expression of RalA or RalB, respectively, compared to the same cell lines stably infected with a retrovirus encoding a scramble control shRNA sequence (Figures 2C,E,G). The resultant nine cell lines were then injected into the flank of immunocompromised mice, and tumor growth monitored over time. Scramble control DM414, DM598 and DM792 cell lines readily formed tumors, with variable latency and growth kinetics (Figures 2D,F,H). Knockdown of RalB reduced tumor growth in only DM598 cells (Figure 2F), where as knock down of RalA inhibited tumor growth more uniformly, as evidenced by up to an 80% reduction in the average tumor volume by the termination of the experiment of mice injected with RalA shRNA-treated cells compared to scramble control counterparts in all three cell lines (Figure 2D,F,H). We thus conclude that knockdown of RalA, but less so RalB, inhibits the tumorigenic growth of NRASQ61L/R-driven human melanoma cell lines.
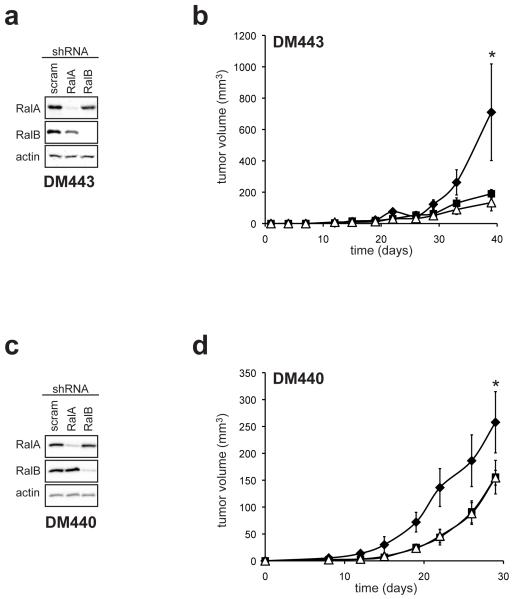
Given that the other two major Ras effector pathways, the Raf and PI3K pathways, are activated in melanomas lacking an NRAS mutation (Gray-Schopfer et al., 2007), and RalA-GTP levels are elevated in NRAS mutation-negative melanoma cell lines (Figure 1), we tested whether RalA or RalB expression was required for the tumorigenic growth of melanoma cell lines lacking an NRAS mutation. We first focused on BRAF V600E mutation-positive human melanoma cell lines, due to the prevalence of BRAFV600E mutations in this cancer (Gray-Schopfer et al., 2007). DM443 and DM440 melanoma cell lines characterized by mutant BRAFV600E and wild type NRAS alleles were stably infected with a retrovirus encoding RalA shRNA, RalB shRNA, or as a control a scramble sequence, and appropriate reduction of the targeted endogenous RalA or RalB protein confirmed by immunoblot (Figure 3A,C) and the cells lines assayed for tumor growth when injected into immunocompromised mice. Knock down of either RalA or RalB reduced tumor growth in both cell lines (Figure 3B,D), which was the most obvious in the DM443 cells (Figure 3B). Thus, RalA and RalB contribute to the tumorigenic growth of BRAFV600E-driven human melanoma cell lines.
Figure 3.
Rals are required for tumor growth of BRAFV600E-positive melanoma cells. (a,c) Protein expression of RalA, RalB, or as a control actin or tubulin, as assessed by immunoblot with the aforementioned α-RalA, α-RalB and α-actin antibodies and (b,d) mean tumor volume (mm3) ± SEM versus time of the indicated melanoma cell lines stably infected with retrovirus derived encoding RalA shRNA (■), RalB shRNA (△) or a scramble (scram) sequence (◆) using constructs and approaches previously described (Lim et al., 2005). All cell lines were injected subcutaneously into the flanks of four immunocompromised mice each. * p≤0.05, difference in tumor size at the termination of the experiment between scramble control and RalA shRNA or RalB shRNA expressing cells.
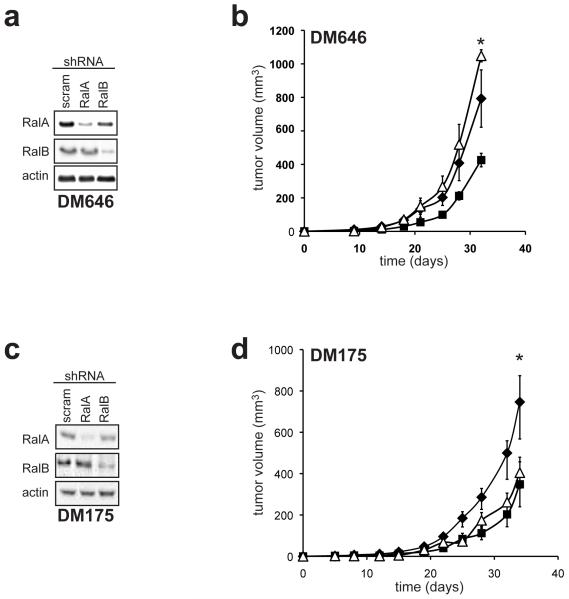
We last tested whether Rals would be required in the absence of both oncogenic NRAS and BRAF alleles. Human melanoma cell lines DM646 and DM175 stably infected with a retrovirus encoding RalA shRNA or RalB shRNA to reduce RalA or RalB protein levels compared to the same cells infected with the control scramble expressing retrovirus, as assessed by immunoblot (Figure 4A,C) were assayed for tumor growth in immunocompromised mice. Knockdown of RalA negatively impacted the tumor growth of both cell lines (Figure 4B,D), whereas knockdown of RalB only reduced tumor growth in DM175 cells (Figure 4D) compared to the scramble control counterparts, although the effect of reducing Rals in these cell lines appeared to be less severe than that observed in the other melanoma cell lines.
Figure 4.
Rals are required for tumor growth of BRAF and NRAS wild type melanoma cells. (a) Protein expression of RalA, RalB, or as a control actin or tubulin, as assessed by immunoblot with the aforementioned α-RalA, α-RalB, α-actin and α-tubulin (Sigma) antibodies and (b) mean tumor volume (mm3) ± SEM versus time of the indicated melanoma cell lines stably infected with retrovirus encoding RalA shRNA (■), RalB shRNA (△) or a scramble (scram) sequence (◆) using constructs and approaches previously described (Lim et al., 2005). All cell lines were injected subcutaneously into the flanks of four immunocompromised mice each. * p≤0.05 difference in tumor size at the termination of the experiment between scramble control and RalA shRNA expressing cells.
In summary, we show that RalA-GTP levels are high in human melanoma cancer cell lines, regardless of the mutation status of NRAS and BRAF, whereas RalB-GTP was difficult to detect. Why RalA is preferentially activated is unclear, although this was the isoform most commonly required for tumor growth in melanoma cell lines. It also remains to be determined how RalA-GTP levels are elevated in melanomas lacking NRAS oncogenic mutations, since activating mutations of Rals in tumors have yet to be reported. The change leading to RalA activation presumably lies upstream of Rals, perhaps by inappropriate activation of RalGEFs. In this regard, mutations have been detected in RalGEFs in cancers, one of which has oncogenic activity (Bodemann & White, 2008). Mounting evidence also suggests that RalGEFs can be activated by the PI3K pathway (Bodemann & White, 2008), and proteins of this pathway are commonly mutated and activated in melanoma (Gray-Schopfer et al., 2007).
Knockdown of RalA or RalB reduced tumor growth of all or half, respectively, of the tested human melanoma cell lines. Knockdown of RalA or RalB did not cause any significant change in the proliferative potential over the course of four days in DM366, DM414, DM443 and DM598 cell lines, as measured by the MTT assay (Supplemental Figure 1), suggesting Ral activation contributes to oncogenic signaling. While RalB shRNA did not reduce the tumorigenic growth in a panel of ten human pancreatic cancer cell lines (Lim et al., 2006), knockdown of RalB has been found to reduce viability of transformed, but not normal cell in suspension (Chien & White, 2003). Taken together, these results suggest that the requirement for RalB may vary in different settings. On the other hand, knockdown of RalA inhibited the tumor growth, at least to some degree, of all the melanoma cancer cell lines. An activated version of RalA, but not RalB, promotes anchorage-independent growth of virally transformed human cell lines (Lim et al., 2005), and particularly relevant to melanoma, Arf-deficient immortalized murine melanocytes (Mishra et al., 2010). As knockdown of RalA has now been shown to reduce tumor growth of oncogenic NRas- (Figures 2-4), KRas- and HRas-driven (Lim et al., 2006) human cell lines, RalA may be broadly required, irrespective of Ras isoform, for oncogenic Ras-mediated tumorigenesis. Nevertheless, like RalB, the requirement for RalA may still be context dependant, as it was recently reported that knockdown of RalA enhanced the tumorigenic growth of the HaCaT keratinocyte cell line transformed by ectopic oncogenic HRas (Sowalsky et al., 2009).
The finding that RalA-GTP levels are high and that RalA shRNA reduced tumorigenic growth in both wild type NRAS/mutant BRAF and wild type NRAS/wild type BRAF cell lines suggests that, like Raf and PI3K signaling, Rals may also be required for melanomagenesis, even in the absence of an oncogenic Ras mutations. Targeting this signaling pathway may have therapeutic value for the treatment of melanoma of different genotypes. In this regard geranyl geranyl transferase inhibitors reduced the tumorigenic growth of pancreatic cancer cell lines in a manner dependent, at least in part, on Rals (Falsetti et al., 2007). Alternatively, RalA was recently shown to be phosphorylated by Aurora A, and moreover, this phosphorylation was required for the oncogenic activity of the protein (Lim et al., 2010; Sablina et al., 2007; Wu et al., 2005), raising the possibility of targeting RalA by inhibiting a druggable kinase.
Supplementary Material
Acknowledgements
This work is supported in part by an American Cancer Society Grant to P.A.Z, a Leukemia & Lymphoma Fellowship to D.K., a Veterans Affairs Merit Review Grant to D.S.T. and a National Institutes of Health grant (CA94184) to C.M.C. We thank Dr. Hillard Siegler (Duke University Medical Center) for all melanoma cell lines except SKMel28 cells.
Footnotes
Conflict of interest statement
The authors declare that there are not any competing financial interests in relation to the work described.
References
- Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer. 2008;8:133–140. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- Bodempudi V, Yamoutpoor F, Pan W, Dudek AZ, Esfandyari T, Piedra M, et al. Ral overactivation in malignant peripheral nerve sheath tumors. Mol Cell Biol. 2009;29:3964–3974. doi: 10.1128/MCB.01153-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127:157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- Chien Y, White MA. RAL GTPases are linchpin modulators of human tumour-cell proliferation and survival. EMBO Rep. 2003;4:800–806. doi: 10.1038/sj.embor.embor899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Adamo DR, Novick S, Kahn JM, Leonardi P, Pellicer A. rsc: a novel oncogene with structural and functional homology with the gene family of exchange factors for Ral. Oncogene. 1997;14:1295–1305. doi: 10.1038/sj.onc.1200950. [DOI] [PubMed] [Google Scholar]
- de Gorter DJ, Reijmers RM, Beuling EA, Naber HP, Kuil A, Kersten MJ, et al. The small GTPase Ral mediates SDF-1-induced migration of B cells and multiple myeloma cells. Blood. 2008;111:3364–3372. doi: 10.1182/blood-2007-08-106583. [DOI] [PubMed] [Google Scholar]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- Falsetti SC, Wang DA, Peng H, Carrico D, Cox AD, Der CJ, et al. Geranylgeranyltransferase I inhibitors target RalB to inhibit anchorage-dependent growth and induce apoptosis and RalA to inhibit anchorage-independent growth. Mol Cell Biol. 2007;27:8003–8014. doi: 10.1128/MCB.00057-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig LA. Ral-GTPases: approaching their 15 minutes of fame. Trends Cell Biol. 2003;13:419–425. doi: 10.1016/s0962-8924(03)00152-1. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Garcia A, Pritchard CA, Paterson HF, Mavria G, Stamp G, Marshall CJ. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell. 2005;7:219–226. doi: 10.1016/j.ccr.2005.01.029. [DOI] [PubMed] [Google Scholar]
- Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- Hamad NM, Elconin JH, Karnoub AE, Bai W, Rich JN, Abraham RT, et al. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 2002;16:2045–2057. doi: 10.1101/gad.993902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi-Far R, White MA, Westwick JK, Solski PA, Chrzanowska-Wodnicka M, Van Aelst L, et al. Oncogenic Ras activation of Raf/mitogen-activated protein kinase-independent pathways is sufficient to cause tumorigenic transformation. Mol Cell Biol. 1996;16:3923–3933. doi: 10.1128/mcb.16.7.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KH, Baines AT, Fiordalisi JJ, Shipitsin M, Feig LA, Cox AD, et al. Activation of RalA is critical for Ras-induced tumorigenesis of human cells. Cancer Cell. 2005;7:533–545. doi: 10.1016/j.ccr.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Lim KH, Brady DC, Kashatus DF, Ancrile BB, Der CJ, Cox AD, et al. Aurora-A phosphorylates, activates, and relocalizes the small GTPase RalA. Mol Cell Biol. 2010;30:508–523. doi: 10.1128/MCB.00916-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KH, O’Hayer K, Adam SJ, Kendall SD, Campbell PM, Der CJ, et al. Divergent roles for RalA and RalB in malignant growth of human pancreatic carcinoma cells. Curr Biol. 2006;16:2385–2394. doi: 10.1016/j.cub.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Miller MJ, Rioux L, Prendergast GV, Cannon S, White MA, Meinkoth JL. Differential effects of protein kinase A on Ras effector pathways. Mol Cell Biol. 1998;18:3718–3726. doi: 10.1128/mcb.18.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PJ, Ha L, Rieker J, Sviderskaya EV, Bennett DC, Oberst MD, et al. Dissection of RAS downstream pathways in melanomagenesis: A role for Ral in transformation. Oncogene. 2010 doi: 10.1038/onc.2009.521. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxford G, Owens CR, Titus BJ, Foreman TL, Herlevsen MC, Smith SC, et al. RalA and RalB: antagonistic relatives in cancer cell migration. Cancer Res. 2005;65:7111–7120. doi: 10.1158/0008-5472.CAN-04-1957. [DOI] [PubMed] [Google Scholar]
- Rosse C, Hatzoglou A, Parrini MC, White MA, Chavrier P, Camonis J. RalB mobilizes the exocyst to drive cell migration. Mol Cell Biol. 2006;26:727–734. doi: 10.1128/MCB.26.2.727-734.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablina AA, Chen W, Arroyo JD, Corral L, Hector M, Bulmer SE, et al. The tumor suppressor PP2A Abeta regulates the RalA GTPase. Cell. 2007;129:969–982. doi: 10.1016/j.cell.2007.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SC, Oxford G, Baras AS, Owens C, Havaleshko D, Brautigan DL, et al. Expression of ral GTPases, their effectors, and activators in human bladder cancer. Clin Cancer Res. 2007;13:3803–3813. doi: 10.1158/1078-0432.CCR-06-2419. [DOI] [PubMed] [Google Scholar]
- Sowalsky AG, Alt-Holland A, Shamis Y, Garlick JA, Feig LA. RalA suppresses early stages of Ras-induced squamous cell carcinoma progression. Oncogene. 2009;29:45–55. doi: 10.1038/onc.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano T, Emkey R, Feig LA. Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 1996;15:810–816. [PMC free article] [PubMed] [Google Scholar]
- White MA, Vale T, Camonis JH, Schaefer E, Wigler MH. A role for the Ral guanine nucleotide dissociation stimulator in mediating Ras-induced transformation. J Biol Chem. 1996;271:16439–16442. doi: 10.1074/jbc.271.28.16439. [DOI] [PubMed] [Google Scholar]
- Wolthuis RM, Franke B, van Triest M, Bauer B, Cool RH, Camonis JH, et al. Activation of the small GTPase Ral in platelets. Mol Cell Biol. 1998;18:2486–2491. doi: 10.1128/mcb.18.5.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JC, Chen TY, Yu CT, Tsai SJ, Hsu JM, Tang MJ, et al. Identification of V23RalA-Ser194 as a critical mediator for Aurora-A-induced cellular motility and transformation by small pool expression screening. J Biol Chem. 2005;280:9013–9022. doi: 10.1074/jbc.M411068200. [DOI] [PubMed] [Google Scholar]
- Yin J, Pollock C, Tracy K, Chock M, Martin P, Oberst M, et al. Activation of the RalGEF/Ral pathway promotes prostate cancer metastasis to bone. Mol Cell Biol. 2007;27:7538–7550. doi: 10.1128/MCB.00955-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.