Abstract
Integrins are a family of receptors for extracellular matrix proteins that have critical roles in human tissue development. Previous studies identified down-regulation and/or mutations of integrin α7 (ITGA7) in prostate cancer, liver cancer, soft tissue leiomyosarcoma, and glioblastoma multiforme. Here we report that expression of ITGA7 induced apoptosis in the human prostate cancer cell lines PC3 and DU145. Yeast two-hybrid analysis revealed that the C-terminus of ITGA7 interacts with high temperature requirement A2 (HtrA2), a serine protease with a critical role in apoptosis. Expression of ITGA7 increases the protease activity of HtrA2 both in vitro and in vivo. Deletion of the HtrA2 interaction domain abrogates the cell death activity of ITGA7, whereas down-regulation of HtrA2 dramatically reduced cell death mediated by ITGA7. In addition, site-directed protease-null mutant HtrA2S306A expression blocked apoptosis induced by ITGA7. Interestingly, interaction between ITGA7 and its ligand laminin 2 appears to protect against cell death, since depleting laminin β2 with a small-interfering RNA significantly exacerbated apoptosis induced by ITGA7 expression. This report provides a novel insight into the mechanism by which ITGA7 acts as a tumor suppressor.
As a major class of cell adhesion molecules in mammalian cells, the integrins are involved in many cellular processes, including development, immune responses, leukocyte traffic, and hemostasis.1 Integrin knock-out mice have distinctive developmental defects, including kidney tubule defects, severe skin blistering, chylothorax, and muscular dystrophy.2,3,4,5,6,7 The integrin superfamily consists of 24 members, each of which mediates a unique function in mammals. Regulation of integrin expression is critical for certain aspects of tissue differentiation and regeneration (eg, keratinocyte differentiation, hair follicle formation, and skeletal muscle development),8,9,10 and abnormal integrin expression is associated with several human diseases (eg, muscular dystrophy, Glanzmann thrombasthenia, and congenital cardiac myopathy).10,11,12 Integrin α7 (ITGA7) forms a heterodimer with integrin β1 in the plasma membrane and is responsible for communication between the extracellular matrix and cells.13 The ITGA7/integrin β1 heterodimer binds laminin 1 and 2. Physiologically, ITGA7 mediates cell-extracellular matrix interaction and regulates cell differentiation and migration.
Recent mutational analysis of ITGA714 revealed ITGA7 mutations in prostate cancer (PC), hepatocellular carcinoma, soft tissue leiomyosarcoma, and glioblastoma multiforme with frequencies ranging from 25% to 83%. Many of these mutations cause truncation, microdeletion, or frameshift of the protein. Interestingly, patients with ITGA7 mutations had higher rate of clinical relapse in both PC and hepatocellular carcinoma. Meta-analysis of previously published microarray data indicated that ITGA7 was down-regulated in nonmetastatic prostate cancer and leiomyosarcoma, but the magnitude of the down-regulation was larger in metastatic cancers. PC and soft tissue leiomyosarcoma with focal or no ITGA7 expression were associated with less metastasis-free survival time. Forced expression of normal ITGA7 in PC and leiomyosarcoma cell lines suppressed tumor growth and cancer cell migration in vitro. A mouse model of PC3 and DU145 xenograft prostate tumors showed a dramatic reduction in tumor volume, metastatic rate, and mortality rate when ITGA7 expression was restored. However, the molecular mechanism of ITGA7-mediated tumor suppressor activity remains unclear. In this study, we report that high temperature requirement A2 (HtrA2), a serine protease,15,16 interacts with the C-terminus of ITGA7. We show that activation of ITGA7 leads to increased HtrA2 protease activity and apoptotic cell death.
Materials and Methods
Cells, Culture Conditions, and Antibodies
All cell lines, including PC-3 (prostate cancer), Du145 (prostate cancer), LNCaP (prostate cancer), and RWPE-1 (immortalized prostate epithelial cells), were purchased from American Type Cell Culture (Manassas, VA). PC-3 cells were cultured with F12K medium supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA). Du145 cells were cultured with modified Eagle medium supplemented with 10% fetal bovine serum (Invitrogen). RWPE-1 cells were cultured with K-SFM medium supplemented with 10% fetal bovine serum (Invitrogen).
Rabbit anti-integrin α7 serum (polyclonal) was raised through immunization of a rabbit with the synthetic peptide GTILRNNWGSPRREGPDAH. Mouse anti-integrin α7 antibody was purchased from Novus Biologicals Inc. (Littleton, CO). Mouse monoclonal anti-laminin β2 was previously described,17 and mouse anti–β-actin monoclonal antibody and goat anti-HtrA2 antibodies were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA).
Plasmid Construction
For construction of pGBKT7-ITGA7c fusion proteins, a mutagenic primer set (5′-TGTGGCCTATAATCATATGACCTTTCTGGAGGAGTACTCA-3′ and 5′-TTCATTTCAAGCAAAGTCGACGCCTGGATCTGCTCTGCGCCCCTC-3′) was designed to create two restriction sites, NdeI and SalI (New England Biolabs, Ipswich, MA), in the C-terminus (252 amino acids) of ITGA7, so that the PCR product could be ligated into a pGBKT7 vector (Clontech, Mountain View, CA). PCR was performed on the cDNA template from the donor prostate (Clontech) by using the following conditions: 94°C for 1 minute followed by 35 cycles of 94°C for 30 seconds, 68°C for 3 minutes, and a final 3-minute extension step at 68°C. The PCR product was cut with NdeI and SalI, gel purified, and ligated into a similarly cut pGBKT7 vector. The fusion protein contained 156 amino acids from the ITGA7 C-terminus. Similar strategy was used for pGBKT7-ITGA7full construction except the forward primer being replaced as 5′-TGTGGCCTATAATCATATGGCCGGGGCTCGGAGCCGCGAC-3′. The constructs were transformed into One Shot competent cells (Invitrogen). Plasmid DNA was extracted from selected transformed cells and digested with NdeI and SalI to detect the presence of the insert. The coding frame was confirmed by automated sequencing.
For construction of pGST-ITGA7, a glutathione S-transferase (GST) fusion protein, a mutagenic primer set (5′-AGG AATTCCCGGGTCGACGCCGGGGCTCGGAGCCGCGAC-3′/5′-AGTCACGATGCG GCCGCTGCCCCTGAGGAAGCCGATCCT-3′) was designed to create a SalI and a NotI restricted site within the ITGA7 coding region that encodes 1136 amino acids of ITGA7. PCR was performed by using these primers under the following conditions: 94°C for 1 minute followed by 35 cycles of 94°C for 30 seconds, 68°C for 3 minutes, and a final 10-minute extension step at 68°C. The PCR product was gel purified and ligated into a pCR2.1 TA cloning vector (Invitrogen). A similar strategy was used for constructing pGST-ITGA7c (the forward primer sequence is 5′-AGGAATTCCCGGGTCGACGGGCA-3′ 5′-GGGGCCTGGGCAGAAA-3′) where 252 amino acids of ITGA7 C terminus were encoded. The plasmid DNA was transformed into Escherichia coli. The plasmid DNA from the selected transformants was restricted with SalI and NotI, and ligated into a similarly restricted pGEX-5x-3 vector in frame. A series of deletions, including 5′ or 3′ deletions of pGST-ITGA7c, were performed by using the primer sets listed in Table 1 (also see Figure 1D for amino acid sequences of the constructs). The procedures for generating these mutants were similar to those described for pGST-ITGA7c. The pGST-ITGA7c and its mutants were transformed into E. coli BL21 cells for recombinant protein production.
Table 1.
Primers for ITGA7 and HtrA2 Mutant Constructs
| Name | Sequence |
|---|---|
| GST-ITGA7cf | 5′-AGGAATTCCCGGGTCGACGGGCAGGGGCCTGGGCAGAAA-3′ |
| GST-ITGA7cr | 5′-AGTCACGATGCGGCCGCTGAATTAGCTTGGCTGCAAGCG-3′ |
| GST-ITGA7c1 | 5′-CACGATGCGGCCGCTCGAGTTGTTCCTCAGGA-3′ |
| GST-ITGA7c2 | 5′-CACGATGCGGCCGCTCGAGGCTTCGCCCGTTTGAA-3′ |
| GST-ITGA7c3 | 5′-CACGATGCGGCCGCTCGAGGATGACCCACCAGG-3′ |
| GST-ITGA7c4 | 5′-CACGATGCGGCCGCTCGAGCATCAAGTTCTTTATG-3′ |
| GST-ITGA7c5 | 5′-CACGATGCGGCCGCTCGAGACGGCCCCAGACA-3′ |
| GST-ITGA7n1 | 5′-TGGGATCCCCAGGAATTCCAGCATGTCCTGGTGGCCA-3′ |
| GST-ITGA7n2 | 5′-TGGGATCCCCAGGAATTCCTGGGGCCGTCTCTGGAAC-3′ |
| GST-ITGA7n3 | 5′-TGGGATCCCCAGGAATTCAGTGCCCTGGTGGGTCATC-3′ |
| GST-ITGA7n4 | 5′-TGGGATCCCCAGGAATTCGGTGAAGATTCCTCGGGAA-3′ |
| ITGAmf | 5′-GAGCCTGGTGAGCGGCAGGAGATCACCCTGGACTGCGCCCGG-3′ |
| ITGAmr | 5′-CCGGGCGCAGTCCAGGGTGATCTCCTGCCGCTCACCAGGCTC-3′ |
| pcDNA4-ITGAmf | 5′-AACTTAAGCTTGGTACCAACAAAAGCTGGAGCTCCAC-3′ |
| pcDNA4-ITGAmr | 5′-CCTCTAGACTCGAGGAGGCGGTGCCTGGCCCTGGATG-3′ |
| pCMV-HtrA2f | 5′-TGGAGCTCCACCGCGGCTACAATTAATACATAACCT-3′ |
| pCMV-HtrA2r | 5′-TGGTACCGGCTCGAGGTAATACGACTCACTATAGG-3′ |
| HtrA2(S306A)f | 5′-GCTATTGATTTTGGAAACGCTGGAGGTCCCCTGG-3′ |
| HtrA2(S306A)r | 5′-CCAGGGGACCTCCAGCGTTTCCAAAATCAATAGC-3′ |
Figure 1.
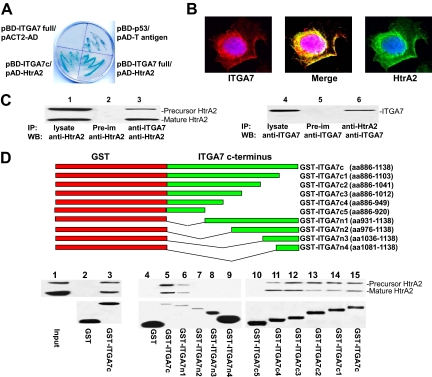
The C-terminus of integrin α7 interacts with HtrA2. A: β-galactosidase activity of yeast harboring pBD-ITGA7 full and pACT2-AD (negative control), pBD-ITGA7c and pAD-HtrA2, pBD-p53 and pAD-T antigen (positive control), or pBD-ITGA7 full and pAD-HtrA2. B: ITGA7 and HtrA2 co-localized in PITT1 cells. PITT1 cells were induced to express ITGA7 with 5 μg/ml tetracycline for 24 hours. Immunostaining was performed by using antibodies specific for ITGA7 or HtrA2 as described in Materials and Methods. C: Co-immunoprecipitation of ITGA7 and HtrA2. PITT1 cells were induced with 5 μg/ml tetracycline for 24 hours. The lysates were immunoprecipitated with the indicated antibodies and detected by Western blotting with either ITGA7 or HtrA2 antibodies. D: ITGA7 binds with HtrA2 in vitro. Upper panel: Diagrams of GST-ITGA7c mutants. Middle panel: Binding of GST-ITGA7c fusion proteins and its mutants with HtrA2 from PITT1 cells. After extensive washes, the bound proteins were eluted and immunoblotted with anti-HtrA2 antibodies. Lower panel: Coomassie staining of fusion proteins from the middle panel.
Yeast Transformation and Library Screening
The yeast AH109 competent cell preparation was described previously.18 Freshly prepared AH109 competent cells (100 μl) were mixed with plasmid pGBKT7-ITGA7c (0.25 to 0.50 μg), plus 0.5 μg plasmid DNA from a prostate cDNA library constructed in pACT2, in 0.6 ml of PEG/LiAc, and then incubated at 30°C for 30 minutes. Following this initial incubation with plasmid DNA, the cell solution was combined with 70 μl of dimethyl sulfoxide (Sigma, St. Louis, MO) and subjected to 15 minutes incubation at 42°C. The cells were pelleted, resuspended in 0.5 ml yeast extract peptone dextrose (YPDA) medium, and plated on different stringency SD agar plates. The transformants were plated either directly to the low, medium, and high stringent medium plates, or the colonies grown on the low and medium stringency plates of SD-Leu/-Trp and SD-Leu/-Trp/-His were replicated on high stringency plate (SD-Ade/-His/-Leu/-Trp and X-α-Gal). The grown colonies were subjected to the colony-lift filter β-galactosidase assay as described previously.19 pGBKT7–53 and pGADT7-T antigen were co-transformed into AH109 to be used as a positive control, and pGBKT7-Lamin C with pGADT7-T as a negative control. PCL1 transformed into AH109 was used as a positive control of galactosidase assay.
Validation of Protein Interactions in AH109
Plasmid DNA from positive clones (blue colonies in high stringency plate) were isolated from yeast, transformed into E. coli, and selected with ampicillin (100 μg/ml) to obtain gene-ACT2 fusion protein interacting with the bait-domain fusion protein. The individual purified pACT2/library plasmid DNA was then co-transformed with pGBKT7-ITGA7c, pGBKT7-ITGA7, or pGBKT7 into AH109 yeast cells and grown in a SD-Ade/-His/-Leu/-Trp high stringency medium. Colony-lift filter β-galactosidase activity was assayed on those cells grown in this medium. The positive clones were then sequenced.
GST Fusion Protein Pull-Down
The cells were grown in 5 ml of LB (ampicillin 100 μg/ml) overnight at 37°C and diluted in 20X Luria Broth, and then incubated with shaking until the solution reached an optical density of 0.6 to 1.0, then induced by isopropyl-β-D-thiogalactopyranoside (final concentration: 1 mmol/L) for 3 hours. The cells were then pelleted, resuspended in 1× PBS, and sonicated for 2 minutes. The proteins were solubilized in 1% triton X-100, then centrifuged at 15,000 × g for 5 minutes, and then the supernatant was collected. The GST and GST-ITGA7c fusion proteins were purified on a Glutathione Sepharose 4B column (Amersham Pharmacia Biotech, NJ). The PC3 cell protein extract was pre-incubated with the column for 15 minutes at 4°C. The effluent was collected after spinning at 3000 × g for 1 minute. This effluent was then incubated with GST fusion protein-packed Glutathione Sepharose 4B at 4°C for 2 hours. The column was spun at 3000 × g for 1 minute, and washed twice with PBS. The protein was eluted from the column with 40 μl of SDS-polyacrylamide gel electrophoresis gel sample loading dye. SDS-polyacrylamide gel electrophoresis and Western blot analysis were subsequently conducted.
Immunofluorescent Staining
PITT1 cells were cultured on chamber slides and induced with 5 μg/ml tetracycline for 24 hours. The slides were washed 3 times with PBS. The cells were fixed with 4% paraformaldehyde for 1 hour at room temperature. After washing the slides with PBS twice, the cells were blocked with 10% donkey serum containing 0.4% Triton X-100. The cells were then incubated with rabbit antisera against ITGA7 and goat antisera against HtrA2 (Santa Cruz Biotechnology) at room temperature for 1 hour. The slides were washed twice with PBS. Secondary antibodies from donkey directed against goat (fluorescein-conjugated) and against rabbit (rhodamine-conjugated) were added and incubated at room temperature for 1 hour. The slides were then washed twice with PBS before addition of 4′,6-diamidino-2-phenylindole dihydrochloride. After additional washes with PBS, slides were mounted with Prolong Gold Antifade Reagent (Invitrogen). Immunofluorescence staining was examined with a confocal microscope.
Detection of Apoptosis by Terminal Deoxynucleotidyl Transferase-Mediated dUTP Nick-End Labeling Assay
The ITGA7-inducible PITT1 cells were plated on 8-well chamber slides for 1 day, and were treated with tetracycline (5 μg/ml) for 48 hours. The slides were washed twice with PBS. The adherent cells were then fixed with 4% formaldehyde. The terminal addition of fluororescein labeled UTP was obtained by incubating the slides with 50 μl of terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling reaction buffer (Roche Applied Science, Nutley, NJ) for 60 minutes at 37°C in the dark. Then the slides were washed three times before the quantification of cell death by using an inverted fluorescence microscope.20 Experiments were performed in triplicate. Uninduced PITT1 cells were used as a negative control, and uninduced PITT1 cells exposed to recombinant DNase I before labeling procedure were used as a positive control.
Fluorescence Activated Cell Sorting Analysis of Apoptotic Cells
Cells with inducible ITGA7 clones were plated and treated with tetracycline (5 μg/ml) as described above. The cells were trypsinized and washed twice with cold PBS. The cells were then resuspended in 100 μl of annexin binding buffer (Invitrogen), and incubated with 5 μl of AlexaFluor 488-conjugated annexin V and 1 μl of 100 μg/ml propidium iodide for 15 minutes in the dark at room temperature. The binding assays were terminated by addition of 400 μl of cold annexin binding buffer. Fluorescence activated cell sorting (FACS) analysis was performed by using a BD-LSR-II flow cytometer (BD Science, San Jose, CA). The fluorescence stained cells were analyzed at the fluorescence emission at 530 nm (FL1) and >575 nm (FL3). The negative control, cells with neither Alexa Fluor 488 nor propidium iodide (PI) in the incubation medium, was used to set the background for the acquisition. Cells stained with either Alexa Fluor 488 or PI, but not both, were used for calibration and compensation before the acquisition. UV-treated cells were used as a positive control for apoptosis. For each acquisition, 10,000 to 20,000 cells were sorted based on the fluorescence color of the cells: only green (apoptotic cells), or only red, or both red and green stained (late apoptotic stage or dead cells), or without any staining (live cells). WinMDI 2.8 software (freeware from Joseph Trotter) was used to further analyze the data. For small-interfering RNA (siRNA) analysis, 125 pmol siRNA specific for HtrA2 (5′-rArArG rGrGrA rUrGrG rCrArA rArGrG rArGrA rUrUrC rCrArG rCrUrG/5Phos/rGrCrU rGrGrA rArUrC rUrCrC rUrUrU rGrCrC rArUrC rCrCT T-3′), siLam (5′-rGrArA rArUrG rArArA rCrUrC rArGrC rCrUrC rCrArG rGrUrC rCrArG/5Phos/rGrGrA rCrCrU rGrGrA rGrGrC rUrGrA rGrUrU rUrCrA rUrUT C-3′), or nonspecific control (uaauguauuggaacgcauauu/uaugcguuccaauacauua) were transfected into PIF1 or DIF1 cells by using the Lipofectamine 2000 transfection kit (Invitrogen). Immunoblots and FACS analyses were performed 24 hours after transfection.
Immunoprecipitation
Precleared protein extracts of PITT1 cells induced with tetracycline were incubated with ITGA7 or HtrA2 antibodies at 4°C for 16 hours, then with protein G sepharose for 3 hours. The immune complex was pelleted and washed five times with radioimmunoprecipitation assay buffer (50 mmol/L Tris-HCl, pH 7.4; 0.25% Na-deoxycholate; 150 mmol/L NaCl; 1 mmol/L EDTA; 1 mmol/L phenylmethylsulfonyl fluoride; 1 μg/ml aprotinin; 1 μg/ml leupeptin; 1 μg/ml pepstatin; 1 mmol/L Na3VO4; 1 mmol/L NaF), and the bound proteins were eluted with SDS-polyacrylamide gel electrophoresis sample buffer.
In Vitro Protease Assay
Cells were washed with PBS and harvested. The cell pellets were lysed by incubation for 15 minutes on ice with radioimmunoprecipitation assay buffer. After centrifugation at 14,000 × g for 20 minutes, the supernatants were transferred to a clean microcentrifuge tube, and 6 μl of anti-HtrA2 antibody was added for 1 hour at 4°C. Next, 20 μl of Protein A/G PLUS-Agarose at 4°C was added and the resulting mixture incubated on a rocker platform overnight. The immunoprecipitates were collected by centrifugation at 2500 × g for 5 minutes at 4°C. The agarose pellets were washed four times with PBS. A protease fluorescence detection kit (Sigma) was used to measure the protease activity of HtrA2. Briefly, each pellet was resuspended in 100 μl of incubation buffer from the kit. An aliquot of 10 μl immunoprecipitate was added, with 20 μl of fluorescein isothiocyanate-Casein Substrate, to a clean microfuge tube, and incubated at 37°C in the dark for 30 minutes, 60 minutes, and 120 minutes. Trichloric acid solution (0.6 N; 150 μl) was then added to each reaction for 30 minutes at 37°C. After centrifugation for 10 minutes at 10,000 × g, the supernatant containing the acid soluble fluorescein isothiocyanate-labeled fragments was aspirated. The fluorescence intensity was quantified by spectrofluorometry at 485 nm, monitoring the emission wavelength of 535 nm. A standard curve was generated by using known concentrations of the protease between 0.5 and 20 ng with fluorescein isothiocyanate-tagged casein as the substrate.
Morpholino siRNA Inhibition of ID2 Expression
A morpholino oligo (125 pmol) specific for HtrA2 (5′-CTCCTGAAGCAGCAGTTATGGAGCT-3′; mH2) or a scrambled control (5′-TAATGTATTGGAACGCATATT-3′; scr) was pipetted into electroporation cuvettes (2 mm), respectively. Opti-MEM (200 μl) were used to resuspend the PITT1 cell pellet and then transferred to electroporation cuvettes containing morpholino or control siRNA. Electroporation was performed in the GenePulser Xcell (Bio Rad, Hercules, CA) with one pulse of 175 V for 25 millisecond. After the electric pulse, the cells were transferred back to the culture disk with prewarmed minimum essential medium culture medium. Cells were harvested after 48 hours to examine HtrA2 expression.
Immunohistochemistry Staining
Formalin-fixed and paraffin-embedded human prostate tissues were used. For immunostaining, four-micron thick sections of tissue were cut and mounted on glass slides. The sections were heated at 60°C for 12 hours and deparaffinized in xylene and ethanol. Antigen retrieval was performed by using 25 mmol/L sodium citrate buffer (pH 9.0) at 90°C for 15 minutes, followed by treatment of 3% H2O2 to block endogenous peroxidase. The slides were incubated at room temperature for 2 hours with anti-laminin β2 at 1:500 dilution. The sections were then incubated with horseradish peroxidase conjugated anti-goat IgG for 30 minutes at room temperature. This was followed by incubating the section with 3,3′ diaminobenzidine solution (DAKO, Carpentaria, CA) to develop staining color. Hematoxylin was used for counterstaining. The specificity of immunostainings was verified by incubating the similar slides with pre-immune sera.
Results
Yeast two hybrid screening was used to detect intracellular proteins that interact with ITGA7. A full length ITGA7 cDNA was ligated into pGBKT7 vector to create a BD-ITGA7 fusion protein. Yeast two-hybrid screening of a prostate library using this vector identified 37 positive colonies after three rounds of metabolic screening. After several restriction enzyme digests, 15 unique clones were identified. One of these clones contains a cDNA encoding HtrA2, a critical protease in apoptosis.16,21 To validate the yeast two-hybrid screening results, pAD-HtrA2 and pBD-ITGA7 were co-transfected into Yeast AH109 cells, grown in high stringency medium, and tested for β-galactosidase activity. As shown in Figure 1A, yeast cells harboring pBD-ITGA7 and pAD-HtrA2 were positive for galactosidase activity, whereas co-transfection of pBD-ITGA7 and pACT2-AD (a negative control vector) resulted in yeast that failed to grow in the selective medium and were negative for β-galactosidase activity, confirming ITGA7/HtrA2 binding. Next, a fragment of cDNA encoding the 252 amino acids in the C-terminus of ITGA7 was ligated into pGBKT7 to create a BD-ITGA7c fusion protein. The co-transfection result indicated that the binding motif of ITGA7 was located in the C-terminus (Figure 1A).
Immunofluorescent staining using antibodies specific for ITGA7 and HtrA2 indicated that ITGA7 and HtrA2 were co-localized in the cytosol and membrane of prostate cancer cells (Figure 1B). To verify that the interaction occurred in prostate epithelial cells, an in vivo ITGA7 and HtrA2 binding analysis was performed in the protein extracts of PITT1 cells, a PC3 cell line transfected with pCDNA4-ITGA7/pCDNA6, which was induced with tetracycline (5 μg/ml). As shown in Figure 1C, co-immunoprecipitation of ITGA7 and HtrA2 was readily apparent, using antibodies specific for either ITGA7 or HtrA2. Similar results were also obtained for tetracycline-induced PITT2 cells (data not shown). Subsequently, we ligated the 252 amino acid sequence of the C-terminus of ITGA7 into pGEX-5X-3 to create a GST-ITGA7c fusion protein. A cell-free HtrA2/GST-ITGA7c binding analysis was performed. The binding assay indicated that HtrA2 and ITGA7 C-terminus bind each other in vitro (Figure 1D). Subsequently, we created a series of N-terminus and C-terminus deletion mutants of GST-ITGA7c fusion protein through mutational PCR. In vitro binding assays were performed with protein extracts from tetracycline-induced PITT1 cells. By overlapping the results of the binding assays, a 19-amino acid (931–949) motif was identified as critical for ITGA7/HtrA2 interaction (Figure 1D).
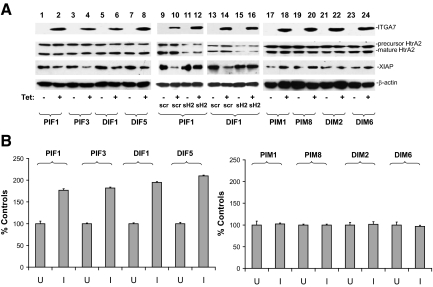
To investigate the functional significance of ITGA7/HtrA2 interaction, we created additional tetracycline inducible ITGA7 cell lines by transfecting pCDNA4-ITGA7 into DU145 cells, which have minimal ITGA7 expression due to a mutation. We then examined whether HtrA2 protease activity is activated in PC3 (PIF1 and PIF3) or DU145 (DIF1 and DIF5) cells by ITGA7. As demonstrated in Figure 2A, expression of ITGA7 significantly reduced the quantity of XIAP (X-linked inhibitor of apoptosis protein), a major target of HtrA2, in PIF1, PIF3, DIF1, and DIF5 cells. Knocking down HtrA2 in PIF1 and DIF1 cells, on the other hand, largely reversed the degradation of XIAP induced by ITGA7 expression (Figure 2A). To examine whether the increased HtrA2 protease activity due to ITGA7 expression occurs in vitro, cell-free protease activities were examined on HtrA2 immunoprecipitates by using fluorescein-conjugated casein as a substrate. As demonstrated in Figure 2B, induction of ITGA7 expression increased the protease activity of HtrA2 in PIF1 cells by 77%, PIF3 by 81%, DIF1 by 86%, and DIF5 by 101%, indicating that HtrA2 is a signaling target of ITGA7.
Figure 2.
ITGA7 activates HtrA2 protease activity in vivo and in vitro. A: Expression of ITGA7 reduces XIAP in vivo. pCDNA4-ITGA7/pCDNA6 transformed PC3 (PIF1 and PIF3), and DU145 cells (DIF1 and DIF5) were induced with (I) or without tetracycline (U) and treated with siRNA specific for HtrA2 (sH2) or a scrambled control (siScr). The lysates were immunoblotted with the indicated antibodies. An ITGA7 mutant (pcDNA4-ITGA7m), in which 19 amino acids representing the HtrA2 interaction motif were deleted, was transfected into PC3 and DU145 cells. Stably transformed PC3 (PIM1 and PIM8) and DU145 (DIM2 and DIM6) clones were induced with or without tetracycline. The protein extracts were similarly blotted with the indicated antibodies. B: In vitro HtrA2 protease activity. HtrA2 proteins were immunoprecipitated from protein extracts from PIF1, PIF3, DIF1, DIF5, PIM1, PIM8, DIM2, and DIM6 cell lines induced or not with tetracycline. In vitro protease activity was measured as described in Materials and Methods. HtrA2 protease activity from uninduced cell lines was used as the 100% control. Data represent an average of six individual assays. SDs are indicated.
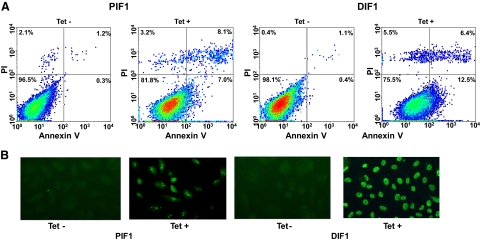
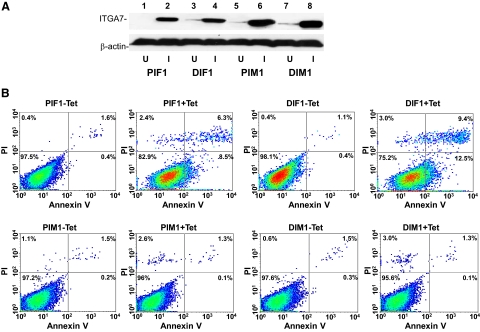
To investigate whether activation of ITGA7 expression induces cell death, PC3 (PIF1) or DU145 (DIF1) cell lines transfected with pCDNA4-ITGA7 cells were treated with 5 μg/ml tetracycline. Cell death was then quantified through Annexin V binding and propidium iodide staining. Since translocation of phosphatidylserine from the inner leaflet of membrane to outer membrane is an early event during apoptosis, detection of such translocation through binding of Annexin V to the intact cell membrane was used as indicator of cells undergoing apoptosis. On the other hand, propidium iodide readily detects cell necrosis by penetrating through leaky cell membrane and binds with genomic DNA. When cells are stained with both annexin V and propidium iodide, they are considered to be in late stages of death, making it impossible to distinguish between apoptosis and necrosis. As shown in Figure 3A and Table 2, treatment with tetracycline increased PIF1 and DIF1 cells death by more than 16-fold (P < 0.01) and 25-fold (P < 0.01), respectively. Most of the cell death is the result of apoptosis, either in early (8.5% to 12.9%) or late stages (6.7% to 6.9%). Similar to these findings, overexpression of ITGA7 in RWPE-1 cells, an immortalized prostate epithelial cell line, produced 3.2-fold increase of cell death (P < 0.01, Table 2). To corroborate these findings, terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling assays were performed on PIF1 and DIF1 cells. ITGA7 expression induced strong labeling of nuclear DNA by terminal transferase, consistent with apoptosis (Figure 3B). These experiments established that ITGA7 induces cell death in prostate cancer cell lines.
Figure 3.
Expression of ITGA7 induces apoptosis. A: FACS analysis of cell death induced by ITGA7 expression. PIF1 or DIF1 cells were treated with or without tetracycline for 48 hours, stained with fluorescein-conjugated Annexin V and propidium iodide, and analyzed in LSC-II flow cytometer. B: Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling assay of PIF1 and DIF1 cells treated with or without tetracycline.
Table 2.
Role of HtrA2 in ITGA7-Mediated Cell Deaths
| Clones | % Annexin V staining | % PI staining | % Annexin V + PI staining |
|---|---|---|---|
| PITT1 | |||
| Uninduced | 0.4% ± 0.7 | 2.0% ± 0.8 | 1.5% ± 0.5 |
| Induced | 7.1% ± 4.2 | 3.8% ± 2.3 | 8.3% ± 3.8 |
| Uninduced + pCMV-HtrA2 + mH2 | 0.3% ± 0.4 | 1.6% ± 0.6 | 2.0% ± 0.7 |
| Induced + pCMV-HtrA2+mH2 | 4.3% ± 1.7 | 9.1% ± 3.2 | 13.7% ± 2.4 |
| Uninduced + pCMV-HtrA2S306A +mH2 | 0.5% ± 0.4 | 2.4% ± 1.2 | 1.0% ± 0.8 |
| Induced + pCMV-HtrA2S306A +mH2 | 0.7% ± 0.3 | 0.8% ± 0.6 | 2.9% ± 1.0 |
| PIF1 | |||
| Uninduced | 0.5% ± 0.2 | 1.0% ± 0.9 | 1.2% ± 0.6 |
| Induced | 8.4% ± 3.6 | 3.8% ± 1.7 | 6.9% ± 1.5 |
| Uninduced + scr | 0.8% ± 0.7 | 3.0% ± 0.6 | 1.1% ± 0.7 |
| Induced + scr | 12.8% ± 3.6 | 2.3% ± 0.9 | 6.6% ± 2.5 |
| Uninduced + siRNA HtrA2 | 0.5% ± 0.3 | 1.1% ± 0.5 | 2.0% ± 0.9 |
| Induced + siRNA HtrA2 | 7.0% ± 1.8 | 0.7% ± 0.6 | 1.5% ± 0.5 |
| Uninduced + siRNA β2-laminin | 0.4% ± 0.2 | 1.2% ± 0.7 | 2.0% ± 1.1 |
| Induced + siRNA β2-laminin | 5.8% ± 3.3 | 11.4% ± 2.1 | 9.1% ± 1.9 |
| PIM1 | |||
| Uninduced | 0.5% ± 0.2 | 1.0% ± 0.9 | 1.2% ± 0.6 |
| Induced | 0.3% ± 0.6 | 2.8% ± 0.7 | 1.4% ± 0.3 |
| DIF1 | |||
| Uninduced | 0.5% ± 0.5 | 0.6% ± 0.3 | 1.3% ± 0.8 |
| Induced | 12.9% ± 1.3 | 5.3% ± 1.6 | 6.7% ± 2.4 |
| Uninduced + scr | 0.3% ± 0.1 | 3.3% ± 0.5 | 1.2% ± 0.4 |
| Induced + scr | 13.8% ± 2.4 | 3.3% ± 0.7 | 6.5% ± 2.1 |
| Uninduced + siH2 | 0.6% ± 0.4 | 1.5% ± 0.9 | 2.5% ± 1.7 |
| Induced + siH2 | 7.1% ± 2.1 | 0.7% ± 0.5 | 1.6% ± 0.6 |
| Uninduced + siRNA β2-laminin | 0.6% ± 0.5 | 1.0% ± 0.3 | 2.4% ± 1.5 |
| Induced + siRNA β2-laminin | 5.9% ± 3.1 | 18.4% ± 4.2 | 10.1% ± 2.3 |
| DIM1 | |||
| Uninduced | 0.3% ± 0.2 | 0.9% ± 0.3 | 1.5% ± 0.4 |
| Induced | 0.4% ± 0.4 | 2.9% ± 1.1 | 1.3% ± 0.3 |
| RWPE1+ pCMVscript | 1.4% ± 0.9 | 3.3% ± 1.8 | 2.2% ± 0.7 |
| RWPE1+ pCMV-ITGA7 | 8.0% ± 5.2 | 4.9% ± 3.1 | 9.2% ± 4.4 |
| RWPE1+ pCMVscript + scr | 2.1% ± 1.3 | 2.9% ± 0.7 | 2.5% ± 0.8 |
| RWPE1+ pCMV-ITGA7+ scr | 9.3%±4.5 | 5.5% ± 2.4 | 10.6% ± 3.7 |
| RWPE1+ pCMVscript + siH2 | 1.8%±0.7 | 2.5% ± 1.1 | 2.1% ± 0.5 |
| RWPE1+ pCMV-ITGA7+ siH2 | 2.3%±0.8 | 3.2% ± 0.9 | 2.3% ± 0.6 |
Average of three independent experiments with SD. Scr denotes scramble siRNA; siH2, siRNA for HtrA2; mH2, morpholino specific for HtrA2.
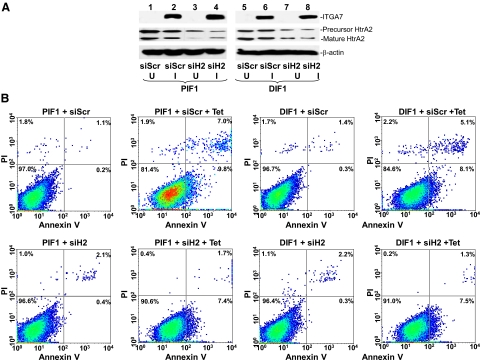
To investigate the role of HtrA2 in ITGA7-mediated cell deaths, we decreased HtrA2 expression by exposing PIF1 and DIF1 cells to HtrA2-specific small-interfering RNA (RNAi) These cells were subsequently induced to express ITGA7 with tetracycline (Figure 4A). As shown in Figure 4B and Table 2, down-regulation of HtrA2 reversed the cell death induced by ITGA7 × 55% (P = 0.003) in PIF1 cells, 51% (P = 0.001) in DIF1 cells and 91% (P < 0.001) in RWPE-1 cells, suggesting a significant role of HtrA2 in ITGA7-mediated cell death.
Figure 4.
ITGA7 mediated apoptosis is HtrA2 dependent. A: Suppression of HtrA2 expression by using siRNA (125 pmol/106 cells) specific for HtrA2 (siH2) or scrambled (siScr) on PIF1 (lanes 1 to 4) and DIF1 cells (lanes 5 to 8) induced with (I) or without (U) tetracycline. B: Representative FACS analysis of cell deaths of replicate samples from (A), stained with fluorescein-conjugated Annexin V and propidium iodide, and analyzed in a LSC-II flow cytometer.
Next, we examined whether interaction between ITGA7 and HtrA2 is required for ITGA7 induced apoptosis. We generated an ITGA7 mutant, in which the 19-amino acid HtrA2 interaction motif was deleted, in pCDNA4 (pCDNA4-ITGA7m). To examine the impact of the loss of the ITGA7/HtrA2 interaction on ITGA7-induced cell death, we transfected this mutant into PC3 and DU145 cells to generate the PIM1, PIM8 (for PC3), DIM2, and DIM6 (for DU145) cell lines. As shown in Figure 5, A and B, deletion of the HtrA2 interaction motif completely abrogated cell death induced by ITGA7 expression (> 95%, P < 0.001) for both PC3 and DU145 cell lines. These experiments indicate that HtrA2 is the major signaling target for ITGA7 cell death. Interestingly, transfection of pCMV-HtrA2, an expression vector that constitutively expresses HtrA2, into PC3 or DU145 cells induced apoptosis of these cells similar to ITGA7 expression activation (Figure 6, A and B). These results support a significant role of HtrA2 in ITGA7-mediated cell death.
Figure 5.
ITGA7/HtrA2 interaction is required for ITGA7 induced apoptosis. A: Immunoblotting of ITGA7 protein expression from the PIF1, DIF1, PIM1, and DIM1 cell lines treated with (I) or without (U) tetracycline. B: Representative FACS analysis of cell death of replicate samples from A, stained with fluorescein-conjugated Annexin V and propidium iodide, and analyzed in a LSC-II flow cytometer.
Figure 6.
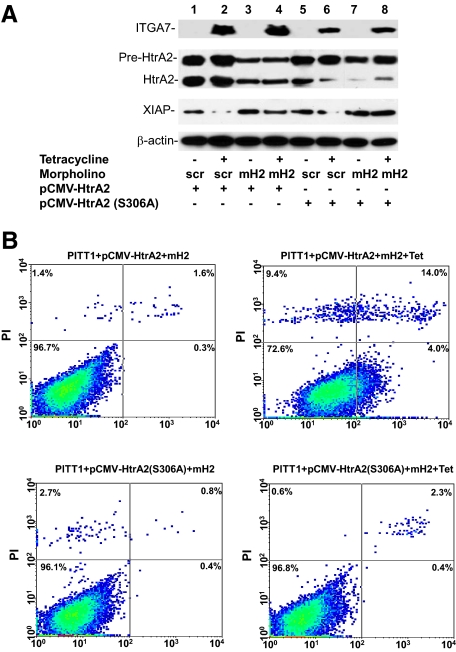
Protease activity of HtrA2 is required for ITGA7 mediated cell deaths. A: Immunoblotting analysis of PITT1 cells transfected with wild-type or protease-null mutant HtrA2 (S306A), treated with or without a morpholino oligo specific for the leader sequence of HtrA2 (mH2, 125 pmol) or its scramble control (scr), and induced with or without tetracycline. The protein extracts were immunoblotted with the indicated antibodies. B: Representative FACS analysis of cell death of replicate samples from A, stained with fluorescein-conjugated Annexin V and propidium iodide, and analyzed in a LSC-II flow cytometer.
To investigate whether HtrA2 protease activity is essential for ITGA7-induced cell death, we created a protease-null HtrA2 by mutating the protease motif (S306A) in the protein. This mutant was inserted into pCMV script to generate pCMV-HtrA2(S306A), which was transfected, along with a morpholino anti-sense specific for the untranslated sequence of endogenous HtrA2, into PITT1 cells. This dramatically reduced the background expression of wild-type HtrA2 while allowing the unimpeded expression of mutant HtrA2. These cells were stimulated with tetracycline to induce ITGA7expression. As shown in Figure 6, A and B, induction of mutant HtrA2 in transfected PITT1 cells resulted in minimal cell death (3.9% versus 4.4% cell death, P = 0.2), whereas a dramatic increase in cell death was observed in the wild-type HtrA2 control (3.9% versus 27.1%, P = 0.001). Therefore, HtrA2 protease activity appears to be critical in ITGA7 signaling.
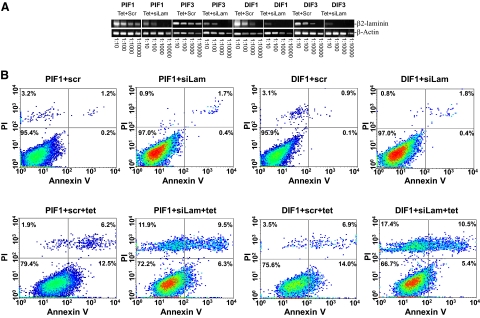
PC3 and DU145 cells generate no laminin 1 but abundant laminin 2 (α2β2Υ2), one of the ligands for the ITGA7/integrin β1 heterodimer. To determine whether ITGA7 signal transduction is ligand-dependent, we performed siRNA knock-down of laminin β2 mRNA to disrupt laminin 2 production in cells induced to express ITGA7. As shown in Figure 7, A and B, and Table 2, knocking down laminin β2 in PIF1 and DIF1 cells significantly (>34%, P < 0.01) enhanced the ITGA7 effect on cell death, albeit with a major necrotic component, suggesting an inhibitory role of laminin in ITGA7-mediated cell death.
Figure 7.
Knocking down β-laminin exacerbates ITGA7-mediated apoptosis. A: Semiquantitative RT-PCR analysis of β2-laminin in PIF1, PIF3, DIF1, and DIF5 cells. These cells were treated with tetracycline and siRNA (125 pmol/106 cells) specific for β2-laminin (siLam) or scrambled controls (scr). The cDNA templates from 1 μg total RNA of these cells were then diluted as indicated, and amplified in a PCR reaction by using primers specific for β2-laminin and β-actin. B: Representative FACS analysis of cell death of replicate samples from A, stained with fluorescein-conjugated Annexin V and propidium iodide, and analyzed in a LSC-II flow cytometer.
Discussion
To our knowledge, this is the first report demonstrating that induction of ITGA7 expression in prostate cancer cells causes cell death. There are several lines of evidences suggesting that the binding of HtrA2 and ITGA7 is critical for cell deaths. First, knocking down HtrA2 dramatically reduced cell death induced by ITGA7. Second, an ITGA7 mutant with the HtrA2 interaction site deleted does not induce cell death. Third, a protease-negative HtrA2 mutant blocks cell death induction by ITGA7. Fourth, activation of HtrA2 protease activity depends on interaction with ITGA7 both in vitro and in vivo.
Most of the studies on ITGA7 have focused on its role in myocyte development.10,22 The discovery of mutations and down-regulation of ITGA7 in human malignancies, as well as the tumor suppressor activity it confers to prostate and smooth muscle, suggests a much wider role. Because of a much higher level of ITGA7 expression in smooth and skeletal muscle, the physiological role of ITGA7 in these tissues might differ from its role in the prostate. Functional analysis suggests that ITGA7 suppresses growth and migration of multiple cancer cell lines. These analyses also indicate that ITGA7 is widely expressed among human organs, tissues, and cell lines, albeit at significantly lower levels. The expression of ITGA7 activates CDKN3 and RACGAP1 expression in both PC and soft tissue leiomyosarcoma cell lines, which in turn perform some of the tumor suppressor activity induced by ITGA7.14
Interestingly, the majority (84%, 27 of 32) of ITGA7 truncation mutations encompass the HtrA2 binding motif (supplemental Figure 1 at http://ajp.amjpathol.org), suggesting the functional significance and clinical relevance of HtrA2 binding motif in ITGA7 mediated tumor suppressor activity. Our findings show that HtrA2 and ITGA7 are bound and co-localized in the cytosol and cell membrane on induction of ITGA7 expression. HtrA2 is a serine protease found in the endoplasmic reticulum and mitochondria16,21,23 that is released into the cytoplasm on stimulation. HtrA2 promotes or induces cell death either by direct binding to and inhibition of inhibitor of apoptosis proteins, leading to an increase in caspase activity, or by inhibitor of apoptosis protein inhibition-independent, caspase-independent, and serine protease activity-dependent mechanism.
The current thinking that ITGA7 is involved primarily in smooth and skeletal muscle development probably represents only one aspect of ITGA7 function. ITGA7 is expressed widely among all tissues and organs. In a previous study, we found that ITGA7 is expressed abundantly in terminally differentiated prostate acinar cells, whereas its expression in basal cells is minimal.14 Herein, we demonstrate that ITGA7 expression induces cell death. Interestingly, interaction between laminin and ITGA7 inhibits its cell death signaling. Immunostaining of laminin β2 suggests more significant expression of laminin 2 in acinar cell layers than that of basal cells (supplemental Figure 2 at http://ajp.amjpathol.org). Based on these findings, it appears that one of the physiological functions of ITGA7 in prostate gland is to play a pivotal role in preventing hyperplasia and reducing the cell number of prostate epithelia. As a result, this may explain, in part, that the decrease in ITGA7 (either by mutation or protein expression) promotes the development of prostate cancer, hepatocellular carcinoma, glioblastoma multiformes, and soft tissue leiomyosarcoma.
Acknowledgments
We thank Chia-Yue Yen for technical support.
Footnotes
Address reprint requests to Jian-Hua Luo, M.D., Ph.D., Department of Pathology, University of Pittsburgh School of Medicine, 3550 Terrace St, Pittsburgh, PA 15261. E-mail: luoj@msx.upmc.edu.
Supported in part by grants from the American Cancer Society (RSG-08-137-01-CNE to Y.Y.P.), the National Cancer Institute (R56 CA098249 and RO1 CA098249 to J.H.L.), and the Urology Development Fund.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Pulkkinen L, Uitto J. Mutation analysis and molecular genetics of epidermolysis bullosa. Matrix Biol. 1999;18:29–42. doi: 10.1016/s0945-053x(98)00005-5. [DOI] [PubMed] [Google Scholar]
- Fassler R, Meyer M. Consequences of lack of beta 1 integrin gene expression in mice. Genes Dev. 1995;9:1896–1908. doi: 10.1101/gad.9.15.1896. [DOI] [PubMed] [Google Scholar]
- Georges-Labouesse E, Mark M, Messaddeq N, Gansmuller A. Essential role of alpha 6 integrins in cortical and retinal lamination. Curr Biol. 1998;8:983–986. doi: 10.1016/s0960-9822(98)70402-6. [DOI] [PubMed] [Google Scholar]
- Kreidberg JA, Donovan MJ, Goldstein SL, Rennke H, Shepherd K, Jones RC, Jaenisch R. Alpha 3 beta 1 integrin has a crucial role in kidney and lung organogenesis. Development. 1996;122:3537–3547. doi: 10.1242/dev.122.11.3537. [DOI] [PubMed] [Google Scholar]
- McHugh KP, Hodivala-Dilke K, Zheng MH, Namba N, Lam J, Novack D, Feng X, Ross FP, Hynes RO, Teitelbaum SL. Mice lacking beta3 integrins are osteosclerotic because of dysfunctional osteoclasts. J Clin Invest. 2000;105:433–440. doi: 10.1172/JCI8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna D, Disatnik MH, Rayburn H, Bronson RT, Yang J, Rando TA, Hynes RO. Dystrophic muscle in mice chimeric for expression of alpha5 integrin. J Cell Biol. 1998;143:849–859. doi: 10.1083/jcb.143.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakebusch C, Grose R, Quondamatteo F, Ramirez A, Jorcano JL, Pirro A, Svensson M, Herken R, Sasaki T, Timpl R, Werner S, Fassler R. Skin and hair follicle integrity is crucially dependent on beta 1 integrin expression on keratinocytes. EMBO J. 2000;19:3990–4003. doi: 10.1093/emboj/19.15.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner A, Willem M, Jones LL, Kreutzberg GW, Mayer U, Raivich G. Impaired axonal regeneration in alpha7 integrin-deficient mice. J Neurosci. 2000;20:1822–1830. doi: 10.1523/JNEUROSCI.20-05-01822.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer U, Saher G, Fassler R, Bornemann A, Echtermeyer F, von der Mark H, Miosge N, Poschl E, von der Mark K. Absence of integrin alpha 7 causes a novel form of muscular dystrophy. Nat Genet. 1997;17:318–323. doi: 10.1038/ng1197-318. [DOI] [PubMed] [Google Scholar]
- Basani RB, French DL, Vilaire G, Brown DL, Chen F, Coller BS, Derrick JM, Gartner TK, Bennett JS, Poncz M. A naturally occurring mutation near the amino terminus of alphaIIb defines a new region involved in ligand binding to alphaIIbbeta3. Blood. 2000;95:180–188. [PubMed] [Google Scholar]
- Hayashi YK, Chou FL, Engvall E, Ogawa M, Matsuda C, Hirabayashi S, Yokochi K, Ziober BL, Kramer RH, Kaufman SJ, Ozawa E, Goto Y, Nonaka I, Tsukahara T, Wang JZ, Hoffman EP, Arahata K. Mutations in the integrin alpha7 gene cause congenital myopathy. Nat Genet. 1998;19:94–97. doi: 10.1038/ng0598-94. [DOI] [PubMed] [Google Scholar]
- Echtermeyer F, Schober S, Poschl E, von der Mark H, von der Mark K. Specific induction of cell motility on laminin by alpha 7 integrin. J Biol Chem. 1996;271:2071–2075. doi: 10.1074/jbc.271.4.2071. [DOI] [PubMed] [Google Scholar]
- Ren B, Yu YP, Tseng GC, Wu C, Chen K, Rao UN, Nelson J, Michalopoulos GK, Luo JH. Analysis of integrin alpha7 mutations in prostate cancer, liver cancer, glioblastoma multiforme, and leiomyosarcoma. J Natl Cancer Inst. 2007;99:868–880. doi: 10.1093/jnci/djk199. [DOI] [PubMed] [Google Scholar]
- Verhagen AM, Silke J, Ekert PG, Pakusch M, Kaufmann H, Connolly LM, Day CL, Tikoo A, Burke R, Wrobel C, Moritz RL, Simpson RJ, Vaux DL. HtrA2 promotes cell death through its serine protease activity and its ability to antagonize inhibitor of apoptosis proteins. J Biol Chem. 2002;277:445–454. doi: 10.1074/jbc.M109891200. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell. 2001;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- Church HJ, Aplin JD. BeWo choriocarcinoma cells produce laminin 10. Biochem J. 1998;332(Pt 2):491–498. doi: 10.1042/bj3320491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YP, Luo JH. Myopodin-mediated suppression of prostate cancer cell migration involves interaction with zyxin. Cancer Res. 2006;66:7414–7419. doi: 10.1158/0008-5472.CAN-06-0227. [DOI] [PubMed] [Google Scholar]
- Yu YP, Tseng GC, Luo JH. Inactivation of myopodin expression associated with prostate cancer relapse. Urology. 2006;68:578–582. doi: 10.1016/j.urology.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Wijsman JH, Jonker RR, Keijzer R, van de Velde CJ, Cornelisse CJ, van Dierendonck JH. A new method to detect apoptosis in paraffin sections: in situ end-labeling of fragmented DNA. J Histochem Cytochem. 1993;41:7–12. doi: 10.1177/41.1.7678025. [DOI] [PubMed] [Google Scholar]
- Savopoulos JW, Carter PS, Turconi S, Pettman GR, Karran EH, Gray CW, Ward RV, Jenkins O, Creasy CL. Expression, purification, and functional analysis of the human serine protease HtrA2. Protein Expr Purif. 2000;19:227–234. doi: 10.1006/prep.2000.1240. [DOI] [PubMed] [Google Scholar]
- Flintoff-Dye NL, Welser J, Rooney J, Scowen P, Tamowski S, Hatton W, Burkin DJ. Role for the alpha7beta1 integrin in vascular development and integrity. Dev Dyn. 2005;234:11–21. doi: 10.1002/dvdy.20462. [DOI] [PubMed] [Google Scholar]
- Cilenti L, Lee Y, Hess S, Srinivasula S, Park KM, Junqueira D, Davis H, Bonventre JV, Alnemri ES, Zervos AS. Characterization of a novel and specific inhibitor for the pro-apoptotic protease Omi/HtrA2. J Biol Chem. 2003;278:11489–11494. doi: 10.1074/jbc.M212819200. [DOI] [PubMed] [Google Scholar]