Abstract
The hallmark pathologies of sporadic inclusion-body myositis (s-IBM) muscle fibers are autophagic vacuoles and accumulation of ubiquitin-positive multiprotein aggregates that contain amyloid-β or phosphorylated tau in a β-pleated sheet amyloid configuration. Endoplasmic reticulum stress (ERS) and 26S proteasome inhibition, also associated with s-IBM, putatively aggrandize the accumulation of misfolded proteins. However, autophagosomal-lysosomal pathway formation and function, indicated by autophagosome maturation, have not been previously analyzed in this system. Here we studied the autophagosomal-lysosomal pathway using 14 s-IBM and 30 disease control and normal control muscle biopsy samples and our cultured human muscle fibers in a microenvironment modified to resemble aspects of s-IBM pathology. We report for the first time that in s-IBM, lysosomal enzyme activities of cathepsin D and B were decreased 60% (P < 0.01) and 40% (P < 0.05), respectively. We also detected two indicators of increased autophagosome maturation, the presence of LC3-II and decreased mammalian target of rapamycin-mediated phosphorylation of p70S6 kinase. Moreover, in cultured human muscle fibers, ERS induction significantly decreased activities of cathepsins D and B, increased levels of LC3-II, decreased phosphorylation of p70S6 kinase, and decreased expression of VMA21, a chaperone for assembly of lysosomal V-ATPase. We conclude that in s-IBM muscle, decreased lysosomal proteolytic activity might enhance accumulation of misfolded proteins, despite increased maturation of autophagosomes, and that ERS is a possible cause of s-IBM-impaired lysosomal function. Thus, unblocking protein degradation in s-IBM muscle fibers may be a desirable therapeutic strategy.
Sporadic inclusion-body myositis (s-IBM) is the most common progressive muscle disease of older persons (reviewed in 1, 2). It leads to severe disability, and there is no enduring treatment currently available.1,2
The characteristic pathologies of s-IBM muscle fibers are autophagic vacuoles, accumulation of ubiquitin-positive multiprotein aggregates containing misfolded proteins in the β-pleated sheet conformation of amyloid and mononuclear cell inflammation (recently reviewed in 3, 4). In addition to their vacuolization, s-IBM muscle fibers have other intriguing features, such as their phenotypic similarities to Alzheimer’s disease and Parkinson’s disease brains.3 Several proteins accumulated in Alzheimer’s and Parkinson’s disease brains are accumulated within s-IBM muscle fibers, such as aggregated amyloid-β (Aβ), phosphorylated tau occurring in paired helical filaments, α-synuclein, and parkin.3,4 In addition, similarly to Alzheimer’s and Parkinson’s disease brains,5,6,7 endoplasmic reticulum stress, and oxidative stress, inhibition of the 26S proteasome and mitochondrial abnormalities are important aspects of the s-IBM pathology.8,9,10,11,12
Vacuoles in apparently living s-IBM muscle fibers are considered autophagic, because they often contain i) lysosomal membranous debris by light and electron microscopy, which is considered a result of indigestibility of normally turned-over or pathologically damaged cellular proteins and organelles, ii) acid phosphatase positivity, and iii) increased immunoreactivity of some lysosomal enzymes (13, 14 also reviewed in 1, 4). Although the pathogenesis of s-IBM seems complex and multifactorial,1,2,3,4 accumulation of the nondegraded, misfolded proteins and putatively toxic oligomers has been proposed to be an important aspect.4
Recently, increased synthesis and accumulation of p62/SQSTM1, a ubiquitin-binding protein allegedly shuttling polyubiquitinated proteins for their degradation by both proteasome and lysosomes,15,16 have been demonstrated in i) s-IBM muscle fibers17 and ii) cultured human muscle fibers in which activity of either the lysosomes or proteasome was experimentally inhibited.17 Increased accumulation of p62/SQSTM1 in s-IBM muscle fibers also indicated an inadequate protein disposal system.
The two major pathways of cellular protein degradation relate to the 26S proteasome and the autophagic/lysosomal systems.18 The 26S proteasome, also called the ubiquitin-proteasome system, is a major degradation mechanism for i) normal regulatory and other short-lived proteins and ii) misfolded proteins exported from the endoplasmic reticulum (ER) through a ubiquitin-mediated ATP-independent process.6,18,19 However, long-lived, structural proteins and/or damaged or misfolded proteins and obsolescent cellular organelles are degraded through “autophagy.”20,21,22,23 “Macroautophagy” refers to formation and maturation of autophagosomes, which are structures carrying proteins and organelles destined for lysosomal degradation. After an autophagosome fuses with the lysosomal membrane, it disposes its cargo into the lysosome, where it is then degraded by the lysosomal enzymes.20,21,22,23 In normal cells, the correctly functioning ubiquitin-proteasome system and the autophagosomal-lysosomal pathways (ALP) assure proper control of protein quality and quantity. Abnormal accumulation within a cell of misfolded or damaged proteins, which might occur due either to their excessive production, proteasome inhibition, oxidative stress, and/or other stressors, increases the need for autophagic degradation, which is accompanied by increased formation and maturation of autophagosomes.18,21 However, when lysosomal degradation is impaired or the amount of material to be degraded exceeds lysosomal capability, formation of autophagosomes dramatically increases, leading to formation of autophagic vacuoles.23 In mammalian cells, the presence on immunoblots of LC3-II protein, a lipidated form of the autophagosomal protein LC3 (microtubule-associated protein light chain 3B) is considered the most important indicator of increased formation and maturation of autophagosomes.21,23,24
In contrast to several neurodegenerative diseases in which ALP functions have been extensively studied,20,21,22,23,25 ALP function in s-IBM muscle fibers is virtually unexplored. In the present article, we demonstrate for the first time that i) in s-IBM muscle, increased proliferation and maturation of autophagosomes, as indicated by the presence of LC3-II protein and a decreased ratio of phosphorylated p70S6 kinase to total p70S6 kinase, is associated with impaired activities of the two major lysosomal enzymes, cathepsin D and B and ii) in cultured human muscle fibers, experimentally induced ER stress leads to impaired lysosomal enzyme activities, suggesting that ER stress, perhaps among other factors, might contribute to malfunction of the ALP in patients with s-IBM.
Materials and Methods
Muscle Biopsy Samples
Studies were performed on fresh-frozen diagnostic muscle biopsy samples obtained, with informed consent, from 14 patients with s-IBM, 6 with polymyositis (PM), 1 with dermatomyositis, 1 with morphologically nonspecific myopathy, 2 with amyotrophic lateral sclerosis, and 2 with peripheral neuropathy, and on 18 control muscle biopsy samples from patients who, after all tests were performed, were considered free of muscle disease. Patients with s-IBM were aged 46 to 79 years (median 68 years); control patients for s-IBM were aged 49 to 84 years (median 63 years). (Not all studies were performed on all biopsy samples [details below].)
Patient diagnoses were based on clinical and laboratory investigations, including our routinely performed 16-reaction diagnostic histochemical analysis of the muscle biopsy samples. All s-IBM biopsy samples met s-IBM diagnostic criteria.1,3 Clinically all of our patients with s-IBM had very weak and atrophic quadriceps muscle (making them unlike patients with GNE mutation hereditary inclusion-body myopathy), and none had a positive family history for any neuromuscular disorder. In addition, none had any family members with Paget’s disease or frontotemporal dementia, which often occur in patients with valosin-containing protein (VCP) mutations. Pathologically, all of our patients with s-IBM had various-sized foci of mononuclear cell inflammation in their muscle biopsy samples, and they did not have foci of myofibrillar disorganization (as occur in myofibrillar myopathy). Accordingly, we are confident that none of our patients had myofibrillar myopathy or hereditary inclusion-body myopathy.
Light Microscopic Immunocytochemistry
Immunofluorescence detection was performed, as described previously8,9,10,17 on 10-μm-thick transverse sections of four s-IBM, four age-matched-control, two polymyositis, and six other disease control muscle biopsy samples, as specified above. The sources and dilutions of antibodies used are listed in Table 1. Incubations in each antibody were performed overnight at 4°C. For double immunofluorescence an antibody against LC3 (which recognizes both LC3-I and LC3-II) combined with an antibody against p62 was used (Table 1), as described previously for immuno-colocalizing other proteins.8,9,10,17 To block nonspecific binding of an antibody to Fc receptors, sections were preincubated with normal goat serum diluted 1:10.8,9,10,17 Controls for staining specificity were i) omission of the primary antibody or ii) its replacement with nonimmune sera or irrelevant antibody. Results for these were always negative.
Table 1.
Antibodies: Their Dilution and Sources
| Name | Specificity | Source | Host | Application | Dilution |
|---|---|---|---|---|---|
| LC3 | N terminus of LC3-B of human origin | NanoTools/Axxora, LLC (San Diego, CA) | Mouse | IHC, EM | 1:100 |
| LC3 | N-terminal portion of human LC3 protein | Novus Biologicals (Littleton, CO) | Rabbit | IB | 1:500 |
| Phosphorylated-p70S6 kinase | aa surrounding Thr389 of human p70S6K | Cell Signaling (Danvers, MA) | Rabbit | IB | 1:500 |
| p70S6 kinase | C terminus of p70S6K of human origin | Cell Signaling | Rabbit | IB | 1:100 |
| p62/SQSTM1 | aa 151-440 of SQSTM1 of human origin | Santa Cruz Biotechnology (Santa Cruz, CA) | Mouse | IB, IP | 1:100 |
| p62/SQSTM1 | aa 151-440 of SQSTM1 of human origin | Santa Cruz Biotechnology | Rabbit | IHC, EM | 1:100 |
| Cathepsin D (C-20) | C terminus of cathepsin D of human origin | Santa Cruz Biotechnology | Goat | IB | 1:5000 |
| Cathepsin B (FL-339) | aa 1-339 representing full-length cathepsin B of human origin | Santa Cruz Biotechnology | Rabbit | IB | 1:100 |
| VMA21 (M-22) | LOC203547 of human origin | Santa Cruz Biotechnology | Rabbit | IB | 1:100 |
| Actin (C-2) | C terminus of actin of human origin | Santa Cruz Biotechnology | Mouse | IB | 1:500 |
IHC, immunohistochemistry; EM, electron microscopy; IB, immunoblotting; aa, amino acids; IP, immunoprecipitation.
Gold Immunoelectron Microscopy
This was performed on adhered to the bottom of 35-mm Petri dishes 10 μm fresh-frozen biopsy sections from three different patients with s-IBM, as described.8,9,10,11,17 In brief, sections were prefixed in 2% paraformaldehyde for 2 minutes and preincubated with 10% normal goat serum, followed by incubation with mouse monoclonal antibody against LC3 used in combination with rabbit polyclonal antibody against p62. After incubation with appropriate secondary antibodies conjugated to 6- and 12-nm gold particles (Jackson ImmunoResearch Laboratories Inc., West Grove, PA), sections were processed for electron microscopy.8,9,10,11,17
Immunoblots
Immunoblots were performed on muscle biopsy samples from 7 patients with s-IBM, 6 patients with polymyositis, and 10 control patients as recently detailed.8,9,10,11,17 Twenty to 50 μg of protein was loaded into the gel and electrophoretically separated. After electrophoresis, samples were transferred to a nitrocellulose membrane. All reagents were obtained from Invitrogen (Carlsbad, CA). To prevent nonspecific binding of antibodies, the nitrocellulose membranes were blocked in blocking reagent (Invitrogen); they were then incubated overnight at 4°C with an appropriate primary antibody (Table 1). Blots were developed using Western Breeze chemiluminescent kits (Invitrogen). Protein loading was evaluated by the actin band (Santa Cruz Biotechnology, Santa Cruz, CA). Quantification of immunoreactivity was performed by densitometric analysis using NIH ImageJ 1.310 software.
Combined Immunoprecipitation/Immunoblot Procedure
To evaluate whether LC3-I/LC3-II and p62 physically associate in s-IBM muscle fibers, a combined immunoprecipitation/immunoblot technique was used, as detailed previously.8,9,26 In brief, 150 μg of total muscle protein were immunoprecipitated in precipitation buffer containing 10 μg of anti-p62 antibody (Table 1). The immunoprecipitated complexes, containing IgG antibody along with its bound target antigen and all proteins bound to that antigen, were pulled down using Protein G Sepharose 4 Fast Flow (GE Healthcare, Piscataway, NJ) during 4 hours of incubation at 4°C. The solution was then centrifuged for 5 minutes (16,000 × g at 4°C), and the supernatant was removed. The precipitated immunocomplexes were washed five times with the precipitation buffer by centrifuging for 5 minutes each (16,000 × g at 4°C). Immunoprecipitates were then electrophoresed and immunoprobed with the anti-LC3 antibody (which recognizes both LC3-I and LC3-II), followed by an appropriate secondary antibody, and developed using the Western Breeze anti-rabbit chemiluminescence kit (Invitrogen). To confirm specificity of the physical association identified by the immunoprecipitation-immunoblot reaction, the primary antibody was omitted from the immunoprecipitation solution. To confirm the identity of the immunoprecipitate, the immunoprecipitate was also probed with the anti-p62 antibody.
Measurement of Cathepsin D and B Activities
Cathepsin D (EC 3.4.23.5) activity was measured using a Cathepsin D Assay Kit (Sigma-Aldrich, St. Louis, MO) according to the manufacturer’s instructions. In brief, 10 10-μm-thick fresh-frozen sections of 5 s-IBM, 4 PM, and 13 control muscle biopsy samples were individually homogenized in PBS with addition of 0.05% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid (CHAPS) (Sigma-Aldrich). The reaction mixture contained a cathepsin D assay buffer, muscle extracts (30 μg of protein from each biopsy), and an internally quenched fluorescent substrate (included in the kit). Because other enzymatic activities might possibly cleave the substrate, the samples were also measured in the presence of the specific inhibitor pepstatin A to inhibit cathepsin D activity. The activity in the presence of pepstatin A (equivalent to the non-cathepsin D activity in the samples) was subtracted from the total activity measured to obtain cathepsin D activity. The positive control was purified cathepsin D from bovine spleen (Sigma-Aldrich kit) used instead of our muscle homogenates; and the negative control was that reaction with pepstatin A. All samples were measured in duplicate. They were incubated at 37°C, and the fluorescence emission excited at 355 nm was recorded at 444 nm every 2 minutes. Activity of cathepsin D measured in the linear phase of the reaction was expressed as fluorescence intensity per minute and was normalized to the amount of cathepsin D protein in the same homogenate as measured by immunoblotting and expressed per actin (actin was used as the protein loading reference).
Cathepsin B (EC 3.4.22.1) activity was measured as described.27 Ten 10-μm-thick fresh-frozen sections of 3 s-IBM, 6 PM, and 11 control muscle biopsy samples were homogenized in the PBS with addition of 0.05% CHAPS. The reaction mixture contained cathepsin B assay buffer (0.1 mol/L 4-morpholineethanesulfonic acid, 1 mmol/L EDTA, and 2 mmol/L dithiothreitol, pH 6.0), muscle extracts (50 μg of protein from each biopsy), and 1 mmol/L cysteine. The reaction was initiated by addition of the specific synthetic substrate Z-ARG-ARG-AMC (1 μmol/L). Because other enzymatic activities might cleave the substrate, the specific inhibitor CA-074 (10 μmol/L) (Enzo Life Sciences, Plymouth Meeting, PA) was added to the samples to inhibit cathepsin B activity. The activity in the presence of CA-074 (equivalent to the non-cathepsin B activity present in the samples) was subtracted from the total activity measured. Samples were measured in duplicate. All samples were incubated for 30 minutes at 37°C, and the fluorescence emission excited at 355 nm was recorded at 444 nm. The positive control was a purified cathepsin B from bovine spleen used instead of the muscle homogenates, and the negative control was the reaction with CA-074. Activity of cathepsin B was expressed as arbitrary fluorescence units. Because cathepsin B protein was not studied in the same biopsy homogenate as activity, we do not express cathepsin B activity per its protein.
All reagents used for this assay were purchased from Sigma-Aldrich, unless indicated otherwise.
Cultured Human Muscle Fibers
Primary cultures of normal human muscle were established as routinely performed in our laboratory,28 from archived satellite cells of portions of diagnostic muscle biopsy samples from patients who, after all tests had been performed, were considered free of muscle disease. Each experiment was performed on at least five different culture sets, each established from satellite cells derived from a different muscle biopsy sample. Twelve to 18 days after fusion of myoblasts, well differentiated myotubes were exposed for 24 hours to either i) an established ER stress inducer tunicamycin, which is an N-glycosylation inhibitor29 (4 μg/ml; Sigma-Aldrich) that in our previous studies was shown to induce ER stress and its consequences,10,30,31 ii) an irreversible proteasome inhibitor epoxomicin8,26,32 (1 μmol/L; BIOMOL Research Laboratories, Plymouth Meeting, PA), iii) bafilomycin A1, an inhibitor of lysosomal V-ATPase, causing an increase of lysosomal pH33 and inhibiting activities of cathepsins (25 nmol/L; Sigma-Aldrich), or iv) chloroquine (20 μmol/L; Sigma-Aldrich) for 24 and 48 hours, a lysosomotropic agent raising lysosomal pH33 and inhibiting activities of cathepsins. After these various treatments, cultures were processed for immunoblots as described previously9,10,26,30 or measurement of enzymatic activities as above for muscle biopsy samples. In each experiment, treated cultures were compared with their untreated sister control cultures.
Statistical Analysis
Statistical significance was determined by Student’s t-test. The level of significance was set at P < 0.05. Data are presented as means ± SEM.
Results
Macroautophagy is Activated in s-IBM Muscle Fibers
Morphology
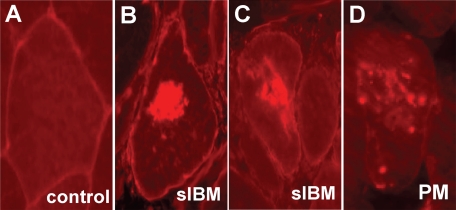
Light microscopic immunohistochemistry was used to evaluate the distribution and intracellular localization of LC3-I/LC3-II protein in s-IBM and control muscle fibers. In normal human muscle biopsy samples there was no detectable LC3-I/LC3-II immunoreactivity, except for occasional slight sarcolemmal staining on a rare muscle fiber (Figure 1A). In contrast, in s-IBM muscle fibers, LC3-I/LC3-II was accumulated in the cytoplasm in the form of aggregates that were often very large (Figure 1, B and C) and were commonly adjacent to or surrounding vacuoles; these were not evident in PM. In PM there were very rare muscle fibers that contained one or several small areas of increased LC3-I/LC3-II immunoreactivity (Figure 1D). In PM, dermatomyositis, and morphologically nonspecific myopathy, there were occasional small muscle fibers that contained slight diffuse LC3-I/LC3-II immunoreactivity (not shown). Endomysial and perivascular mononucleated cells in s-IBM, polymyositis, and dermatomyositis muscle biopsy samples were not immunoreactive for LC3-I/LC3-II. None of the normal or disease control biopsy samples had muscle fibers containing large LC3-I/LC3-II immunoreactive aggregates like those present in s-IBM.
Figure 1.
LC3 in control, s-IBM and PM muscle fibers. Although a representative fiber in a control biopsy (A) does not contain any inclusions, within s-IBM muscle fibers (B and C) there are large LC3-immunoreactive cytoplasmic inclusions. D: In PM there are small various-sized LC3-immunoreactive inclusions. In A, B, and C there is some degree of muscle-fiber sarcolemmal staining. Original magnification, ×900.
Immunoblots
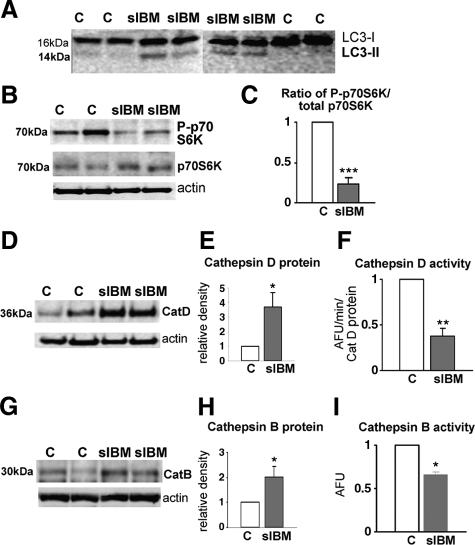
In s-IBM muscle biopsy samples, we studied by immunoblot two important markers of activated macroautophagy, the process that involves proliferation and maturation of autophagosomes. Those markers were i) LC3-II, a lipidated form of LC324,25 that is distinguished from LC3-I by its electrophoretic mobility, and ii) the ratio of phospho-p70S6 kinase (phosphorylated on threonine 389) to total p70S6 kinase.24,25 A decrease in the ratio of mammalian target of rapamycin-mediated phosphorylation of p70S6 kinase to total p70S6 kinase is considered an important marker of macroautophagy induction.24,25 On immunoblots, s-IBM muscle showed a distinct LC3-II band, whereas normal age-matched controls did not manifest an LC3-II band (Figure 2A). In addition, in s-IBM the ratio of phospho-p70S6 kinase to total p70S6 kinase was decreased by 60% (P < 0.001) (Figure 2, B and C). Both the presence of the LC3-II and the decreased ratio of the phosphorylated p70S6 to total p70S6 kinase indicate that macroautophagy and mechanisms leading to autophagosome formation are activated in s-IBM.
Figure 2.
LC3-II, p70S6 kinase, and cathepsins D and B in control and s-IBM muscle biopsies. A: On representative immunoblots of four s-IBM and four control muscle biopsies, there is a LC3-II band in s-IBM but not in control samples. B: Representative immunoblot of phosphorylated p70S6 kinase (P-p70S6K) shows a clearly decreased band of a P-p70S6K in s-IBM. C: Densitometric analysis of P-p70S6K per total p70S6K, based on six s-IBM and four control muscle biopsy samples, indicates a 60% decrease of P-p70S6K in s-IBM. Representative immunoblots (D) and a densitometric analysis (E) of cathepsin D (CatD) in five s-IBM and six control muscle biopsy samples show a fourfold increase in cathepsin D protein in s-IBM, whereas its activity (F) is decreased by 60% compared with that of control biopsy samples. Representative immunoblots of cathepsin B (CatB) protein (G) and densitometric analysis (H) based on seven s-IBM and five control muscle biopsy samples show a twofold increase in cathepsin B compared with that in control samples, but its activity (I) is decreased by 40% compared with controls. Actin bands in D and E are used as protein loading controls. AFU, arbitrary fluorescence units. *P < 0.05, **P < 0.01, ***P < 0.001.
Decreased Activity of Lysosomal Cathepsin D and Cathepsin B in s-IBM
To evaluate whether the lysosomal enzymatic system functions normally in s-IBM muscle, we studied two major lysosomal proteases, cathepsin D and B. By immunoblots, in s-IBM muscle cathepsin D protein was increased approximately fourfold (P < 0.05) (Figure 2, D and E), and cathepsin B protein was increased twofold (P < 0.05) (Figure 2, G and H).
However, in s-IBM muscle biopsy samples, despite increased cathepsin D and B protein, cathepsin D enzymatic activity, compared with age-matched controls, was surprisingly decreased by 60% (P < 0.01) (Figure 2F), and cathepsin B activity was decreased by 40% (P < 0.05) (Figure 2I). This decreased enzymatic activity could impair lysosomal protein degradation in s-IBM.
Increased Macroautophagy but Unimpaired Lysosomal Enzymatic Activity in Polymyositis
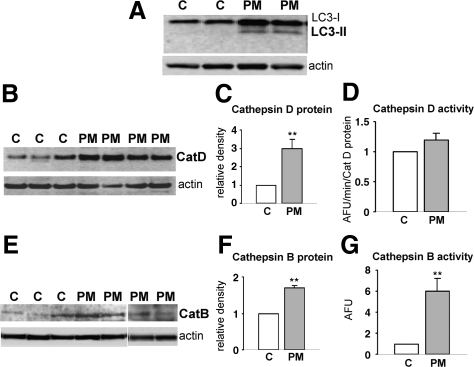
To assess whether the abnormalities we observed in the ALP are specific to s-IBM, we studied LC3-II and cathepsin D and B activities in PM. In PM, but not in control muscle biopsy samples, an occasional muscle fiber contained very small LC3-I/LC3-II immunoreactive inclusions (Figure 1D), and LC3-II was evident in immunoblots (Figure 3A). In PM the protein levels of cathepsin D and B were increased compared with those in PM age-matched controls (Figure 3, B, C, E, and F), similarly to s-IBM. But, in contrast to s-IBM, in PM muscle biopsy samples the enzymatic activities of cathepsin D and B were not decreased; in fact, the activity of cathepsin D was slightly increased (Figure 3D) and that of cathepsin B was significantly increased (Figure 3G). Hypothetically, the increased macroautophagy in PM may result from an increased demand for lysosomal degradation of damaged or excessively produced proteins resulting from the muscle fiber degeneration and regeneration characteristic of PM muscle biopsy samples. In PM, probably because the lysosomal degradation itself seems to be functioning properly, autophagic vacuoles do not form.
Figure 3.
LC3-II and cathepsins D and B in PM muscle biopsy samples. A: By immunoblots, there is a LC3-II band present in PM samples, whereas the control samples do not manifest a LC3-II band. Representative immunoblots (B) and (C) a densitometric analysis of cathepsin D (CatD), based on four PM and eight control muscle biopsy samples show a threefold increase of cathepsin D protein in PM, and its activity is slightly increased (D). In PM muscle biopsy samples, representative immunoblots of cathepsin B (CatB) protein (E) and their densitometric analysis (F) based on six PM and seven control muscle biopsy samples show a 1.7-fold increase of cathepsin B compared with control samples; its enzymatic activity is increased sixfold (G). AFU, arbitrary fluorescence units. **P < 0.01.
Lack of Colocalization and Physical Association between LC3 and p62/SQSTM1 in s-IBM
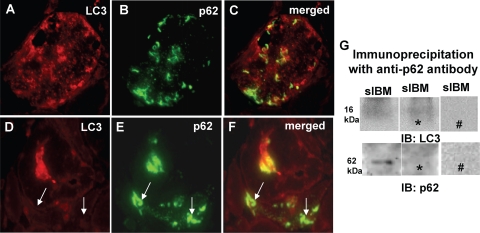
p62/SQSTM1 is a shuttle protein shown to deliver ubiquitinated proteins to both the proteasomal and autophagic degradation systems. It was shown to bind to LC3 in various LC3-overexpressing cultured cells13,34 and mouse liver.35 In the present study, we investigated immunohistochemical colocalization and physical association between LC3 and p62 in s-IBM muscle fibers. By light microscopy, LC3 and p62 were in close proximity but did not colocalize with each other (Figure 4, A–C). In addition, there were fibers that contained strongly immunoreactive p62, but were LC3-negative (Figure 4, D–F, arrows).
Figure 4.
Light microscopic immunohistochemical colocalization and immunoprecipitation of p62/SQSTM1 and LC3/LC3-II in s-IBM muscle biopsy samples. A–F: p62- and LC3-immunoreactive inclusions seem to be near each other, but they do not absolutely colocalize; there are a number of inclusions that are positive only for one or the other protein (A–C). In E there is a muscle fiber that contains p62-immunoreactive inclusions but is LC3-negative (D–F, arrows). Original magnification, ×1100 (A–F). G: Immunoprecipitation of s-IBM biopsy sample using an antibody against p62 followed by immunoblotting (IB) with an antibody recognizing LC3 or p62. There is no immunoprecipitation band visible using an antibody against LC3, but there is a clear band present using an antibody against p62. This results indicates a successful immunoprecipitation reaction and a lack of association between p62 and LC3. ∗, omitting the primary antibody in the immunoprecipitation reaction; #, omitting the secondary antibody in the immunoblot.
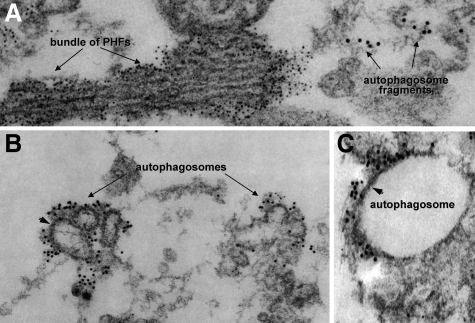
Our double immunolocalization of p62 and LC3 by gold-immunoelectron microscopy showed that they were in very close proximity to each other but were not immunodecorating the same structures (Figure 5, A–C). LC3 seemed to immunodecorate autophagosomal fragments, and, as we have shown previously,17 p62 was immunodecorating paired helical filaments.
Figure 5.
Double-label gold-immunoelectron microscopy of p62 and LC3 in s-IBM muscle biopsies. p62 and LC3 were in a very close proximity to each other but were not immunodecorating the same subcellular structures. A: p62 antibody (6-nm gold particles) is immunodecorating a bundle of paired helical filaments (PHFs). However, LC3 antibody (12-nm gold particles) seems to be associated with autophagosomal fragments (A–C), some of which seem to have an autophagosome-characteristic double membrane (B and C, arrowheads). Original magnification: ×79,000 (A); ×63,000 (B); ×95,000 (C).
In our immunoprecipitation of s-IBM biopsy samples using an antibody against p62 followed by immunoblotting with an antibody recognizing LC3-I/LC3-II, we were not able to detect an immunoprecipitation band in our material (Figure 4G); this result suggests that there is no physical association between p62 and LC3-I or LC3-II in s-IBM muscle fibers. Immunoblotting of p62 immunoprecipitates using an anti-p62 antibody, performed as a positive control of the immunoprecipitation reaction, revealed a 62-kDa p62 band, confirming the validity of the immunoprecipitation experiment (Figure 4G).
Experimentally Evoked ER Stress in Cultured Human Muscle Fibers Induced Macroautophagy but Impaired Cathepsin D and B Activities
To study possible molecular mechanisms involved in the s-IBM pathogenesis, we routinely modify the cellular microenvironment of cultured human muscle fibers (CHMFs) to mimic various aspects of the s-IBM pathogenesis, thereby providing experimental “IBM-cultured human muscle models.” Those models have proved useful in our previous studies.8,9,10,26,30,31,36
In the present study, to seek mechanisms that might lead to the lysosomal impairment in s-IBM, we used our IBM culture models with perturbed protein degradation, achieved experimentally by i) inhibiting proteasome activity and ii) inducing ER stress. For each study, the proteasome-inhibited and ER stress-provoked cultures were compared with their sister nonaltered cultures and with sister cultures treated with either chloroquine or bafilomycin, both being known inhibitors of lysosomal function.
Macroautophagy
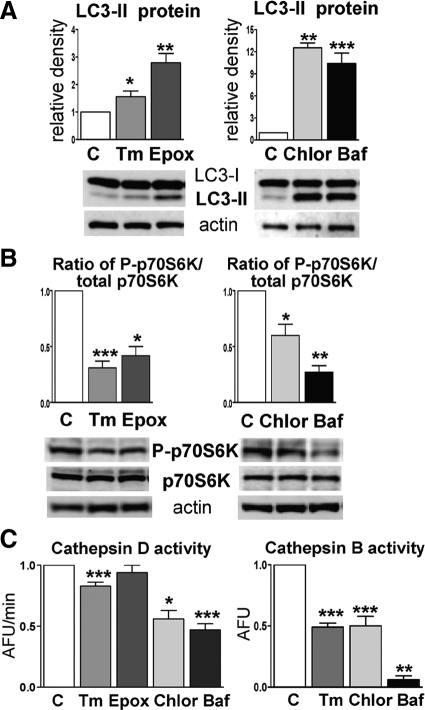
Compared with controls, all four experimental conditions led to increased LC3-II protein by immunoblots; the strongest increase was achieved after treatment with chloroquine or bafilomycin (Figure 6A). The ratio of phosphorylated p70S6 kinase to total p70S6 kinase was significantly decreased after all treatments: tunicamycin by 70% (P < 0.001), epoxomycin by 70% (P < 0.05), chloroquine by 40% (P < 0.05), and bafilomycin by 70% (P < 0.01) (Figure 6B). These data provide evidence of activated macroautophagy.
Figure 6.
Macroautophagy and activities of cathepsins D and B in CHMFs studied under different conditions. A: Treatments with tunicamycin (Tm), epoxomycin (Epox), chloroquine (Chlor), or bafilomycin (Baf) significantly increase LC3-II compared with their untreated sister control cultures (details in the text). Densitometric analysis based on 10 different experimental culture sets is shown above the representative blots. *P < 0.05; **P < 0.01; ***P < 0.001. B: Phosphorylated p70S6K (P-p70S6K), expressed per total p70S6K, is significantly decreased under all four experimental conditions. *P < 0.05; **P < 0.01; ***P < 0.001. A and B: Activated macroautophagy. C: Treatments with tunicamycin, chloroquine, or bafilomycin significantly decrease cathepsin D and B activities. *P < 0.05; **P < 0.01; ***P < 0.001 (details in the text). Because exposure to epoxomycin did not influence cathepsin D activity, cathepsin B was not measured under this experimental condition. AFU, arbitrary fluorescence units.
Cathepsins D and B
Both chloroquine and bafilomycin significantly decreased cathepsin D and B enzymatic activities (Figure 6C). In tunicamycin-treated CHMFs, cathepsin D and B activities were decreased by 17% (P < 0.001) and 50% (P < 0.001), respectively (Figure 6C). Proteasome inhibition did not influence cathepsin D activity (Figure 6C); thus, we did not measure cathepsin B activity under this condition because of the scarcity of human muscle material for the primary cultures.
Morphological Evaluation
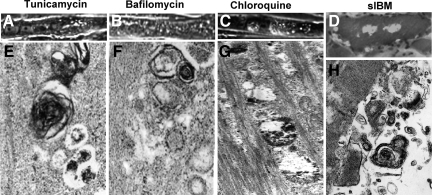
By phase-contrast light microscopy, tunicamycin-, chloroquine-, and bafilomycin-treated cultures were highly vacuolated (Figure 7, A–C). Transmission electron microscopy revealed vacuolar structures containing partially digested cytoplasmic contents, electron-dense material, and membranous whorls (Figure 7, E–G), abnormalities resembling those observed in s-IBM muscle fibers (Figure 7, D and H).38
Figure 7.
Morphology of CHMFs exposed to tunicamycin, bafilomycin, or chloroquine and of biopsied s-IBM muscle. A–C: Phase-contrast light microscopy illustrating vacuolization of CHMFs under all these challenges. E–G: Transmission electron microscopy of CHMFs shows that all these treatments induced pronounced abnormalities, as evidenced by various lysosomal debris, myelin-like whorls, and autophagosome-like structures. D and H: Biopsied s-IBM muscle. D: Bright-field light microscopy of an abnormal muscle fiber stained with Engel-Gomori trichrome37 illustrates various-sized vacuoles. H: Electron microscopy shows a vacuole containing inclusions consisting of numerous various-sized membranous whorls of autophagosomal/lysosomal debris. Original magnification: ×1800 (A–C); ×2100 (D); ×95,000 (E–G); ×50,000 (H).
ER Stress Decreases VMA21 Protein in CHMFs
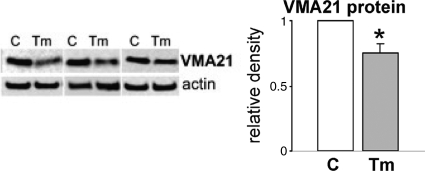
A mutation in VMA21, a chaperone essential for assembly of lysosomal V-ATPase, which is responsible for maintaining lysosomal pH, was recently described in a human X-linked myopathy with excessive autophagy.38,39 It was associated with muscle fiber vacuolization, increased macroautophagy, and impaired lysosomal degradation. In regard to s-IBM and the possible mechanisms causing impaired lysosomal function associated with ER stress, we studied VMA21 protein in our IBM culture model. Treatment with tunicamycin decreased VMA21 protein by 25% (P < 0.01) (Figure 8), whereas this effect was not induced by inhibition of proteasome or inhibition of lysosomal functions with either chloroquine or bafilomycin (not shown).
Figure 8.
VMA21 in tunicamycin-treated cultures. On the representative immunoblots of three different culture sets, VMA21 is clearly decreased in tunicamycin (Tm)-treated cultures compared with controls (C). Densitometric analysis based on 10 culture sets indicates a 25% decrease of VMA21 in Tm-treated cultures. *P < 0.01.
Discussion
In addition to s-IBM, impaired lysosomal degradation has been implicated in an X-linked myopathy with excessive autophagy,38,39 in hereditary inclusion-body myopathy with VCP mutation,40 and in other myopathies containing autophagic vacuoles, such as Danon disease and acid maltase deficiency (reviewed in 41). Even though the existence of autophagic vacuoles associated with accumulated lysosomal-membranous structures in s-IBM muscle biopsy samples is well known (13, 14, 42 and reviewed in 1, 4), the mechanism of their formation is not well understood. We now demonstrate, for the first time, increased formation and maturation of vacuolar autophagosomes in s-IBM muscle fibers, as indicated by i) the autophagosomal marker LC3-II24,25 and ii) mammalian target of rapamycin-mediated phosphorylation of p70S6K.23,24,25 These observations strongly suggest that activated macroautophagy might be an important factor leading to formation of the vacuoles.
We also demonstrate for the first time that enzymatic activities of the two major lysosomal proteases, cathepsin D and B, are decreased in s-IBM muscle fibers. Accordingly, our studies strongly suggest that total autophagy, composed of the autophagosomal-lysosomal pathway, is impaired in s-IBM muscle fibers.
Aβ-immunoreactive inclusions were previously found by immunohistochemistry to colocalize with LC3 in s-IBM muscle fibers; however, LC3-II, an indicator of autophagosome proliferation and maturation, was not examined.43 Our present study, in addition to immunohistochemically identifying large LC3-I/LC3-II-imunoreactive inclusions, also demonstrated, by immunoblots of s-IBM muscle, the presence of LC3-II protein, a reliable marker of increased macroautophagy. The LC3-II increase reflects increased synthesis and formation of autophagosomes, which might occur either when i) there is an increased burden of undigested, misfolded, or damaged proteins and organelles for the delivery to the lysosomes or ii) when there is an impairment (block) at a later step, for example, involving impaired lysosomal activity, a delayed/impaired trafficking to lysosomes, or reduced fusion between autophagosomes and lysosomes.24 Our studies strongly suggest that in s-IBM increased formation and maturation of autophagosomes are, at least partially, due to impaired lysosomal enzymatic activity. Thus, in s-IBM muscle fibers, there is i) impaired autophagy involving at least two key lysosomal enzymes, cathepsin D and B, and ii) attempted compensation by excessive proliferation of autophagosomes and lysosomes, as indicated by the increase in the lysosomal enzymes cathepsin D and cathepsin B protein per immunoblots (but not of their enzymatic activity) and increased acid phosphatase by histochemistry. The decreased lysosomal cathepsin D and B enzymatic activities seem to be specific to s-IBM, because in PM muscle fibers, cathepsin D and B activities were found to be increased in this study and by others.44,45 Our demonstration of a macroautophagy increase in PM suggests a need of the PM muscle for increased protein degradation but, perhaps because the lysosomal system may be functioning adequately, autophagic vacuoles and inclusions do not form. Our results also suggest that inflammation, which is present in both s-IBM and PM, does not contribute to impairment of autophagic/lysosomal degradation in s-IBM. Possible explanations for the decreased enzymatic activity of the increased cathepsin D and B proteins in s-IBM might include their inhibition due to i) changes in lysosomal pH or ii) a detrimental influence on cathepsins by the products of oxidative stress,46 which is known to occur in s-IBM11,47,48 or, in the case of cathepsin B, increased cystatin C, a recognized inhibitor of serine proteases49 that is increased in s-IBM muscle fibers.50
It has been reported that when the ALP functions properly, p62, a receptor for ubiquitinated proteins,51 is incorporated into autophagosomes and subsequently degraded by lysosomes.15,24,34 One possible explanation for the increased p62 is impaired autophagic degradation.15,24,34 We previously demonstrated that p62 is significantly increased and aggregated in s-IBM muscle fibers, but not in PM.17 In contrast to studies by others in various LC3-overexpressing cells and in mouse liver,15,34,35 we were not able to demonstrate a physical association between p62 and LC3 in the present study of s-IBM muscle fibers. Whether this lack of association, which conceivably could contribute to accumulation of ubiquitinated proteins i) reflects a characteristic of s-IBM muscle or ii) is specific for human muscle tissue is not known.
Another interesting and novel aspect of our study is the identification of ER stress as a possible cause of the impaired lysosomal degradation: ER stress is known to be an important aspect of the s-IBM pathogenesis.9,10 Previously, in various species including mammals, ER stress was reported to induce macroautophagy, as evidenced by increased LC3-II,51,52,53,54 putatively to eliminate misfolded/unfolded proteins accumulated in the ER lumen.54 In agreement with those reports, ER stress induced in our primary cultures of human muscle increased macroautophagy as reflected by the LC3-II increase and phosphorylated p70S6 kinase decrease; however, concurrently, ER stress impaired enzymatic activities of the lysosomal proteases cathepsin D and B. Even though the exact mechanism involving the ER stress reduction of cathepsin D and B enzymatic activities in CHMFs is not known, our demonstrated ER stress-dependent decrease of VMA21 in them might raise the lysosomal pH, thereby decreasing cathepsin D and B activities and perhaps activities of other unexamined lysosomal enzymes. Whether or not a similar mechanism occurs in s-IBM muscle fibers is not known. (Because of an inherently small amount of VMA21 in human muscle, we were not able, with the antibody used, to adequately evaluate it by immunoblots in either normal or s-IBM biopsy samples.)
Cathepsin D and B are two major proteases participating in normal lysosomal degradation. Detrimental consequences of their impaired activities might be several, including abnormalities known to occur in s-IBM muscle fibers. For example, autophagy-lysosomal perturbations have been shown to enhance tau aggregation,55 and autophagosomes have been demonstrated as an important locus for Aβ production.21,56,57 Accordingly, the impaired ALP system in s-IBM muscle fibers might contribute to the well known increase in Aβ and aggregation of tau within s-IBM muscle fibers (58, 59 and reviewed in 3, 4). Interestingly, the skeletal muscle of chloroquine-treated rats was reported to accumulate Aβ60,61 and to display some aspects of ER stress.61 However, our preliminary studies of CHMFs treated with either chloroquine or bafilomycin have not revealed evidence of ER stress.
Several additional proteins accumulated in s-IBM muscle fibers; for example, BACE1, α-synuclein and tau, have been reported to be degraded by both lysosomes and the 26S proteasome.19,20,55,62,63 Aβ42, a more toxic form of Aβ that is preferentially increased in s-IBM muscle fibers,58 was shown to be degraded in lysosomes by cathepsin B.21,64
In CHMFs, AβPP and its phosphorylated form are both degraded by proteasome.26 Accordingly, inhibition of both proteasomal and lysosomal systems probably has very detrimental consequences. Moreover, both AβPP and Aβ have been shown to inhibit proteasome.8,65,66
In summary, we propose that a double impairment of protein disposal, caused by inhibition of both lysosomal and proteasomal degradation, contributes importantly to s-IBM pathogenesis. Although the igniting mechanisms leading to the s-IBM muscle fiber degeneration are still not known, our studies suggest that attempts to unblock protein degradation might be a therapeutic strategy for patients with s-IBM.
Acknowledgments
We thank Dr. Kelvin J. Davies from the USC Andrus Gerontology Center for allowing us to use his fluorometer to measure enzyme activities. Maggie Baburyan provided technical assistance in electron microscopy. Margherita Simonetti participated in preliminary tissue culture experiments.
Footnotes
Address reprint requests to Valerie Askanas, M.D., Ph.D., USC Neuromuscular Center, Good Samaritan Hospital, 637 S. Lucas Ave., Los Angeles, CA 90017-1912. E-mail: askanas@usc.edu.
Supported by grants from the National Institutes of Health (AG 16768 Merit Award), the Muscular Dystrophy Association, and the Myositis Association and the Helen Lewis Research Fund (to V.A.).
None of the authors declare any relevant financial relationships.
A.N. is on leave from Department of Biochemistry, Medical University of Gdansk, Gdansk, Poland. C.T. was on leave from Department of Neuroscience, University of Tor Vergata and Fondazione S. Lucia, Rome, Italy.
References
- Engel WK, Askanas V. Inclusion-body myositis: clinical, diagnostic, and pathologic aspects. Neurology. 2006;66:S20–S29. doi: 10.1212/01.wnl.0000192260.33106.bb. [DOI] [PubMed] [Google Scholar]
- Dalakas MC. Sporadic inclusion body myositis—diagnosis, pathogenesis and therapeutic strategies. Nat Clin Pract Neurol. 2006;2:437–447. doi: 10.1038/ncpneuro0261. [DOI] [PubMed] [Google Scholar]
- Askanas V, Engel WK. Inclusion-body myositis: muscle-fiber molecular pathology and possible pathogenic significance of its similarity to Alzheimer’s and Parkinson’s disease brains. Acta Neuropathol. 2008;116:583–595. doi: 10.1007/s00401-008-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askanas V, Engel WK, Nogalska A. Inclusion-body myositis: a degenerative muscle disease associated with intra-muscle-fiber multiprotein aggregates, proteasome inhibition, endoplasmic reticulum stress, and decreased lysosomal degradation. Brain Pathol. 2009;19:493–506. doi: 10.1111/j.1750-3639.2009.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholm D, Wootz H, Korhonen L. ER stress and neurodegenerative diseases. Cell Death Differ. 2006;13:385–392. doi: 10.1038/sj.cdd.4401778. [DOI] [PubMed] [Google Scholar]
- Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer’s disease. J Neurochem. 2000;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- Bossy-Wetzel E, Schwarzenbacher R, Lipton SA. Molecular pathways to neurodegeneration. Nat Med. 2004;10:S2–S9. doi: 10.1038/nm1067. [DOI] [PubMed] [Google Scholar]
- Fratta P, Engel WK, McFerrin J, Davies KJ, Lin SW, Askanas V. Proteasome inhibition and aggresome formation in sporadic inclusion-body myositis and in amyloid-β precursor protein-overexpressing cultured human muscle fibers. Am J Pathol. 2005;167:517–526. doi: 10.1016/s0002-9440(10)62994-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattemi G, Engel WK, McFerrin J, Askanas V. Endoplasmic reticulum stress and unfolded protein response in inclusion body myositis muscle. Am J Pathol. 2004;164:1–7. doi: 10.1016/S0002-9440(10)63089-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogalska A, Engel WK, McFerrin J, Kokame K, Komano H, Askanas V. Homocysteine-induced endoplasmic reticulum protein (Herp) is up-regulated in sporadic inclusion-body myositis and in endoplasmic reticulum stress-induced cultured human muscle fibers. J Neurochem. 2006;96:1491–1499. doi: 10.1111/j.1471-4159.2006.03668.x. [DOI] [PubMed] [Google Scholar]
- Terracciano C, Nogalska A, Engel WK, Wojcik S, Askanas V. In inclusion-body myositis muscle fibers Parkinson-associated DJ-1 is increased and oxidized. Free Radic Biol Med. 2008;45:773–779. doi: 10.1016/j.freeradbiomed.2008.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santorelli FM, Sciacco M, Tanji K, Shanske S, Vu TH, Golzi V, Griggs RC, Mendell JR, Hays AP, Bertorini TE, Pestronk A, Bonilla E, DiMauro S. Multiple mitochondrial DNA deletions in sporadic inclusion body myositis: a study of 56 patients. Ann Neurol. 1996;39:789–795. doi: 10.1002/ana.410390615. [DOI] [PubMed] [Google Scholar]
- Tsuruta Y, Furuta A, Furuta K, Yamada T, Kira J, Iwaki T. Expression of the lysosome-associated membrane proteins in myopathies with rimmed vacuoles. Acta Neuropathol. 2001;101:579–584. doi: 10.1007/s004010000329. [DOI] [PubMed] [Google Scholar]
- Kumamoto T, Ueyama H, Tsumura H, Toyoshima I, Tsuda T. Expression of lysosome-related proteins and genes in the skeletal muscles of inclusion body myositis. Acta Neuropathol. 2004;107:59–65. doi: 10.1007/s00401-003-0774-2. [DOI] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogalska A, Terracciano C, D'Agostino C, King Engel W, Askanas V. p62/SQSTM1 is overexpressed and prominently accumulated in inclusions of sporadic inclusion-body myositis muscle fibers, and can help differentiating it from polymyositis and dermatomyositis. Acta Neuropathol. 2009;118:407–413. doi: 10.1007/s00401-009-0564-6. [DOI] [PubMed] [Google Scholar]
- Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- Oddo S. The ubiquitin-proteasome system in Alzheimer’s disease. J Cell Mol Med. 2008;12:363–373. doi: 10.1111/j.1582-4934.2008.00276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vicente M, Cuervo AM. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 2007;6:352–361. doi: 10.1016/S1474-4422(07)70076-5. [DOI] [PubMed] [Google Scholar]
- Nixon RA. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci. 2007;120:4081–4091. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- Shacka JJ, Roth KA, Zhang J. The autophagy-lysosomal degradation pathway: role in neurodegenerative disease and therapy. Front Biosci. 2008;13:718–736. doi: 10.2741/2714. [DOI] [PubMed] [Google Scholar]
- Nixon RA. Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends Neurosci. 2006;29:528–535. doi: 10.1016/j.tins.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, Bamber BA, Bassham DC, Bergamini E, Bi X, Biard-Piechaczyk M, Blum JS, Bredesen DE, Brodsky JL, Brumell JH, Brunk UT, Bursch W, Camougrand N, Cebollero E, Cecconi F, Chen Y, Chin LS, Choi A, Chu CT, Chung J, Clarke PG, Clark RS, Clarke SG, Clave C, Cleveland JL, Codogno P, Colombo MI, Coto-Montes A, Cregg JM, Cuervo AM, Debnath J, Demarchi F, Dennis PB, Dennis PA, Deretic V, Devenish RJ, Di Sano F, Dice JF, Difiglia M, Dinesh-Kumar S, Distelhorst CW, Djavaheri-Mergny M, Dorsey FC, Droge W, Dron M, Dunn WA, Jr, Duszenko M, Eissa NT, Elazar Z, Esclatine A, Eskelinen EL, Fesus L, Finley KD, Fuentes JM, Fueyo J, Fujisaki K, Galliot B, Gao FB, Gewirtz DA, Gibson SB, Gohla A, Goldberg AL, Gonzalez R, Gonzalez-Estevez C, Gorski S, Gottlieb RA, Haussinger D, He YW, Heidenreich K, Hill JA, Hoyer-Hansen M, Hu X, Huang WP, Iwasaki A, Jaattela M, Jackson WT, Jiang X, Jin S, Johansen T, Jung JU, Kadowaki M, Kang C, Kelekar A, Kessel DH, Kiel JA, Kim HP, Kimchi A, Kinsella TJ, Kiselyov K, Kitamoto K, Knecht E, Komatsu M, Kominami E, Kondo S, Kovacs AL, Kroemer G, Kuan CY, Kumar R, Kundu M, Landry J, Laporte M, Le W, Lei HY, Lenardo MJ, Levine B, Lieberman A, Lim KL, Lin FC, Liou W, Liu LF, Lopez-Berestein G, Lopez-Otin C, Lu B, Macleod KF, Malorni W, Martinet W, Matsuoka K, Mautner J, Meijer AJ, Melendez A, Michels P, Miotto G, Mistiaen WP, Mizushima N, Mograbi B, Monastyrska I, Moore MN, Moreira PI, Moriyasu Y, Motyl T, Munz C, Murphy LO, Naqvi NI, Neufeld TP, Nishino I, Nixon RA, Noda T, Nurnberg B, Ogawa M, Oleinick NL, Olsen LJ, Ozpolat B, Paglin S, Palmer GE, Papassideri I, Parkes M, Perlmutter DH, Perry G, Piacentini M, Pinkas-Kramarski R, Prescott M, Proikas-Cezanne T, Raben N, Rami A, Reggiori F, Rohrer B, Rubinsztein DC, Ryan KM, Sadoshima J, Sakagami H, Sakai Y, Sandri M, Sasakawa C, Sass M, Schneider C, Seglen PO, Seleverstov O, Settleman J, Shacka JJ, Shapiro IM, Sibirny A, Silva-Zacarin EC, Simon HU, Simone C, Simonsen A, Smith MA, Spanel-Borowski K, Srinivas V, Steeves M, Stenmark H, Stromhaug PE, Subauste CS, Sugimoto S, Sulzer D, Suzuki T, Swanson MS, Tabas I, Takeshita F, Talbot NJ, Talloczy Z, Tanaka K, Tanida I, Taylor GS, Taylor JP, Terman A, Tettamanti G, Thompson CB, Thumm M, Tolkovsky AM, Tooze SA, Truant R, Tumanovska LV, Uchiyama Y, Ueno T, Uzcategui NL, van der Klei I, Vaquero EC, Vellai T, Vogel MW, Wang HG, Webster P, Wiley JW, Xi Z, Xiao G, Yahalom J, Yang JM, Yap G, Yin XM, Yoshimori T, Yu L, Yue Z, Yuzaki M, Zabirnyk O, Zheng X, Zhu X, Deter RL. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terracciano C, Nogalska A, Engel WK, Askanas V. In AβPP-overexpressing cultured human muscle fibers proteasome inhibition enhances phosphorylation of AβPP751 and GSK3β activation: effects mitigated by lithium and apparently relevant to sporadic inclusion-body myositis. J Neurochem. 2010;112:389–396. doi: 10.1111/j.1471-4159.2009.06461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley WH, Lima PH, Rodgers L, Blomquist CH, Downs LS. Endometrial carcinoma expresses an increased cathepsin B/D ratio. Gynecol Oncol. 2008;108:84–89. doi: 10.1016/j.ygyno.2007.08.094. [DOI] [PubMed] [Google Scholar]
- Askanas V, Engel WK. Cultured normal and genetically abnormal human muscle. Rowland LP, Di Mauro S, editors. Amsterdam: North Holland,; The Handbook of Clinical Neurology, Myopathies. 1992:pp 85–116. [Google Scholar]
- Lee AS. The ER chaperone and signaling regulator GRP78/BiP as a monitor of endoplasmic reticulum stress. Methods. 2005;35:373–381. doi: 10.1016/j.ymeth.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Nogalska A, D'Agostino C, Engel WK, Davies KJ, Askanas V: Decreased SIRT1 deacetylase activity in sporadic inclusion-body myositis muscle fibers. Neurobiol Aging, 2008, doi: 10.1016/j.neurobiolaging.2008.08.021 [DOI] [PubMed] [Google Scholar]
- Nogalska A, Wojcik S, Engel WK, McFerrin J, Askanas V. Endoplasmic reticulum stress induces myostatin precursor protein and NF-κB in cultured human muscle fibers: relevance to inclusion body myositis. Exp Neurol. 2007;204:610–618. doi: 10.1016/j.expneurol.2006.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci USA. 1999;96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shacka JJ, Klocke BJ, Shibata M, Uchiyama Y, Datta G, Schmidt RE, Roth KA. Bafilomycin A1 inhibits chloroquine-induced death of cerebellar granule neurons. Mol Pharmacol. 2006;69:1125–1136. doi: 10.1124/mol.105.018408. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Wojcik S, Nogalska A, McFerrin J, Engel WK, Oledzka G, Askanas V. Myostatin precursor protein is increased and associates with amyloid-β precursor protein in inclusion-body myositis culture model. Neuropathol Appl Neurobiol. 2007;33:238–242. doi: 10.1111/j.1365-2990.2006.00821.x. [DOI] [PubMed] [Google Scholar]
- Engel WK, Cunningham GG. Rapid examination of muscle tissue. An improved trichrome method for fresh-frozen biopsy sections. Neurology. 1963;13:919–923. doi: 10.1212/wnl.13.11.919. [DOI] [PubMed] [Google Scholar]
- Ramachandran N, Munteanu I, Wang P, Aubourg P, Rilstone JJ, Israelian N, Naranian T, Paroutis P, Guo R, Ren ZP, Nishino I, Chabrol B, Pellissier JF, Minetti C, Udd B, Fardeau M, Tailor CS, Mahuran DJ, Kissel JT, Kalimo H, Levy N, Manolson MF, Ackerley CA, Minassian BA. VMA21 deficiency causes an autophagic myopathy by compromising V-ATPase activity and lysosomal acidification. Cell. 2009;137:235–246. doi: 10.1016/j.cell.2009.01.054. [DOI] [PubMed] [Google Scholar]
- Hirano M, DiMauro S. VMA21 deficiency: a case of myocyte indigestion. Cell. 2009;137:213–215. doi: 10.1016/j.cell.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. Cell Biol. 2009;187:875–888. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malicdan MC, Noguchi S, Nonaka I, Saftig P, Nishino I. Lysosomal myopathies: an excessive build-up in autophagosomes is too much to handle. Neuromuscul Disord. 2008;18:521–529. doi: 10.1016/j.nmd.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Carpenter S, Karpati G, Heller I, Eisen A. Inclusion body myositis: a distinct variety of idiopathic inflammatory myopathy. Neurology. 1978;28:8–17. doi: 10.1212/wnl.28.1.8. [DOI] [PubMed] [Google Scholar]
- Lünemann JD, Schmidt J, Schmid D, Barthel K, Wrede A, Dalakas MC, Munz C. β-Amyloid is a substrate of autophagy in sporadic inclusion body myositis. Ann Neurol. 2007;61:476–483. doi: 10.1002/ana.21115. [DOI] [PubMed] [Google Scholar]
- Kar NC, Pearson CM. Early elevation of cathepsin B1 in human muscle disease. Biochem Med. 1977;18:126–129. doi: 10.1016/0006-2944(77)90059-x. [DOI] [PubMed] [Google Scholar]
- Pearson CM, Kar NC. Muscle breakdown and lysosomal activation (biochemistry). Ann NY Acad Sci. 1979;317:465–477. doi: 10.1111/j.1749-6632.1979.tb56562.x. [DOI] [PubMed] [Google Scholar]
- Krohne TU, Kaemmerer E, Holz FG, Kopitz J. Lipid peroxidation products reduce lysosomal protease activities in human retinal pigment epithelial cells via two different mechanisms of action. Exp Eye Res. 2010;90:261–266. doi: 10.1016/j.exer.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Broccolini A, Engel WK, Alvarez RB, Thomas C, Yang CC, Askanas V. Possible pathogenic role of malondialdehyde, a toxic product of lipid peroxidation, in sporadic inclusion-body myositis. Neurology. 1998;50:A367–A368. [Google Scholar]
- Yang CC, Alvarez RB, Engel WK, Askanas V. Increase of nitric oxide synthases and nitrotyrosine in inclusion-body myositis. Neuroreport. 1996;8:153–158. doi: 10.1097/00001756-199612200-00031. [DOI] [PubMed] [Google Scholar]
- Grubb AO. Cystatin C—properties and use as diagnostic marker. Adv Clin Chem. 2000;35:63–99. doi: 10.1016/S0065-2423(01)35015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattemi G, Engel WK, McFerrin J, Askanas V. Cystatin C colocalizes with amyloid-beta and coimmunoprecipitates with amyloid-β precursor protein in sporadic inclusion-body myositis muscles. J Neurochem. 2003;85:1539–1546. doi: 10.1046/j.1471-4159.2003.01798.x. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Ni HM, Gao W, Hou YF, Melan MA, Chen X, Stolz DB, Shao ZM, Yin XM. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J Biol Chem. 2007;282:4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- Li J, Ni M, Lee B, Barron E, Hinton DR, Lee AS. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ. 2008;15:1460–1471. doi: 10.1038/cdd.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høyer-Hansen M, Jaattela M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- Hamano T, Gendron TF, Causevic E, Yen SH, Lin WL, Isidoro C, Deture M, Ko LW. Autophagic-lysosomal perturbation enhances tau aggregation in transfectants with induced wild-type tau expression. Eur J Neurosci. 2008;27:1119–1130. doi: 10.1111/j.1460-9568.2008.06084.x. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, Lee JH, Mohan PS, Mercken M, Farmery MR, Tjernberg LO, Jiang Y, Duff K, Uchiyama Y, Naslund J, Mathews PM, Cataldo AM, Nixon RA. Macroautophagy—a novel β-amyloid peptide-generating pathway activated in Alzheimer’s disease. J Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vattemi G, Nogalska A, Engel WK, D'Agostino C, Checler F, Askanas V. Amyloid-β42 is preferentially accumulated in muscle fibers of patients with sporadic inclusion-body myositis. Acta Neuropathol. 2009;117:569–574. doi: 10.1007/s00401-009-0511-6. [DOI] [PubMed] [Google Scholar]
- Mirabella M, Alvarez RB, Bilak M, Engel WK, Askanas V. Difference in expression of phosphorylated tau epitopes between sporadic inclusion-body myositis and hereditary inclusion-body myopathies. J Neuropathol Exp Neurol. 1996;55:774–86. doi: 10.1097/00005072-199607000-00003. [DOI] [PubMed] [Google Scholar]
- Tsuzuki K, Fukatsu R, Takamaru Y, Yoshida T, Hayashi Y, Yamaguchi H, Fujii N, Takahata N. Amyloid β protein in rat soleus muscle in chloroquine-induced myopathy using end-specific antibodies for Aβ40 and Aβ42: immunohistochemical evidence for amyloid beta protein. Neurosci Lett. 1995;202:77–80. doi: 10.1016/0304-3940(95)12205-2. [DOI] [PubMed] [Google Scholar]
- Ikezoe K, Furuya H, Arahata H, Nakagawa M, Tateishi T, Fujii N, Kira J. Amyloid-β accumulation caused by chloroquine injections precedes ER stress and autophagosome formation in rat skeletal muscle. Acta Neuropathol. 2009;117:575–582. doi: 10.1007/s00401-009-0488-1. [DOI] [PubMed] [Google Scholar]
- Qing H, Zhou W, Christensen MA, Sun X, Tong Y, Song W. Degradation of BACE by the ubiquitin-proteasome pathway. FASEB J. 2004;18:1571–1573. doi: 10.1096/fj.04-1994fje. [DOI] [PubMed] [Google Scholar]
- Koh YH, von Arnim CA, Hyman BT, Tanzi RE, Tesco G. BACE is degraded via the lysosomal pathway. J Biol Chem. 2005;280:32499–32504. doi: 10.1074/jbc.M506199200. [DOI] [PubMed] [Google Scholar]
- Mueller-Steiner S, Zhou Y, Arai H, Roberson ED, Sun B, Chen J, Wang X, Yu G, Esposito L, Mucke L, Gan L. Antiamyloidogenic and neuroprotective functions of cathepsin B: implications for Alzheimer’s disease. Neuron. 2006;51:703–714. doi: 10.1016/j.neuron.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Tseng BP, Green KN, Chan JL, Blurton-Jones M, LaFerla FM. Aβ inhibits the proteasome and enhances amyloid and tau accumulation. Neurobiol Aging. 2008;29:1607–1618. doi: 10.1016/j.neurobiolaging.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecarini V, Bonfili L, Amici M, Angeletti M, Keller JN, Eleuteri AM. Amyloid peptides in different assembly states and related effects on isolated and cellular proteasomes. Brain Res. 2008;1209:8–18. doi: 10.1016/j.brainres.2008.03.003. [DOI] [PubMed] [Google Scholar]