Abstract
In this study, a chronic yet synchronized version of the K/BxN mouse, the KRN-cell transfer model (KRN-CTM), was developed and extensively characterized. The transfer of purified splenic KRN T cells into T cell-deficient B6.TCR.Cα−/−H-2b/g7 mice induced anti-glucose 6-phosphate isomerase antibody-dependent chronic arthritis in 100% of the mice with uniform onset of disease 7 days after T cell transfer. Cellular infiltrations were assessed by whole-ankle transcript microarray, cytokine and chemokine levels, and microscopic and immunohistochemical analyses 7 through 42 days after T cell transfer. Transcripts identified an influx of monocytes/macrophages and neutrophils into the ankles and identified temporal progression of cartilage damage and bone resorption. In both serum and ankle tissue there was a significant elevation in interleukin-6, whereas macrophage inflammatory protein-1 α and monocyte chemotactic protein-1 were only elevated in tissue. Microscopic and immunohistochemical analyses revealed a time course for edema, synovial hypertrophy and hyperplasia, infiltration of F4/80-positive monocytes/macrophages and myeloperoxidase-positive neutrophils, destruction of articular cartilage, pannus invasion, bone resorption, extra-articular fibroplasia, and joint ankylosis. The KRN cell transfer model replicates many features of chronic rheumatoid arthritis in humans in a synchronized manner and lends itself to manipulation of adoptively transferred T cells and characterizing specific genes and T cell subsets responsible for rheumatoid arthritis pathogenesis and progression.
Rheumatoid arthritis (RA) is characterized by chronic inflammation of the distal joints with hypertrophy and hyperplasia of the synovial epithelium, increased synovial fluid volume, and radiographic evidence of bone erosion. Infiltration of inflammatory cell types such as monocytes/macrophages, neutrophils, fibroblasts, T cells, and dendritic cells into the synovial, subsynovial, and peri-articular tissues leads to complement activation, production of pro-inflammatory chemokines and cytokines, and eventual destruction/remodeling of cartilage and bone. Indeed, it is the influx of monocytes/macrophages that mediates and amplifies this tissue damage, thereby maintaining the clinical signs of inflammation.1,2 This process begins when chemokines drive monocyte emigration out of the blood,3,4,5,6,7,8 and is amplified when activated macrophages produce additional cytokines once they enter the joint.9,10,11,12,13 The extent of macrophage infiltration and their activation state is known to correlate with joint pain and the general inflammatory status of the patient, and most therapies currently available for RA decrease the number of macrophages in synovial tissue.14,15,16 Despite this, a complete picture of how inflammatory cells and their mediators contribute to the stepwise pathogenesis of RA has not been established, and the reliance on rodent models for obtaining this information is substantial.
Murine models of arthritis are essential components of basic research and preclinical testing of disease-modifying antirheumatic drugs. One of these models, the K/BxN murine model, exists in two different forms. In the first form, KRN TCR transgenic mice are bred to nonobese diabetic mice and the resultant F1 (K/BxN) mice develop a severe, destructive arthritis by 4 to 5 weeks of age.17 In the second form, serum transferred from K/BxN mice into normal recipient mice induces arthritis due to the presence of arthritogenic antibodies specific for the ubiquitously expressed glycolytic enzyme glucose 6-phosphate isomerase (GPI).18,19 Arthritis in this second, K/BxN serum transfer model initiates within 1 to 2 days, peaks within 7 to 14 days, and wanes substantially by day 21. In either case, the K/BxN approach offers advantages over more traditional models of RA such as the rat adjuvant-induced arthritis and the mouse collagen-induced arthritis (CIA) models. For instance, anti-GPI antibodies, neutrophils, macrophages, TNF, IL-1, and the alternative complement pathway have all been demonstrated to contribute to K/BxN disease induction.20,21,22,23 Also, the K/BxN serum transfer model is much less strain dependent than the mouse CIA model. Both of these features greatly facilitate potential elucidation of key effector molecules and cells; however, there are some limitations to the K/BxN approach. In K/BxN mice, because arthritis initiates by 4 to 5 weeks of age, it is difficult to assess the relationship between the temporal progression of disease and inflammatory cell infiltrates. Second, although the K/BxN serum transfer model is more synchronized in its initiation, the disease is not sustainable beyond 12 days post serum transfer and, notably, bypasses the participation of CD4+ T cells.
To overcome the limitations of both K/BxN mice and the K/BxN serum transfer model we developed a novel modification of the K/BxN mouse, the KRN cell transfer model (KRN-CTM). The KRN-CTM is based on the established approach of transferring autoreactive T cells into lymphopenic hosts,24,25 and was designed to retain the utility of the K/BxN serum transfer model (namely, the ability to separate the inductive and effector phases) yet, importantly, allow the investigator to model the chronic stages of RA when joint effusion and pain have subsided and subclinical synovial inflammation and structural deterioration remain.26 In this study, we found that the transfer of naive KRN T cells into T cell-deficient B6.TCR.Cα−/− H-2b/g7 mice induced a chronic, destructive arthritis. Disease was monitored by measuring ankle swelling, anti-GPI antibody production, and histopathology. The role of inflammatory cells in the early pathogenesis and sustained pathophysiology of the KRN-CTM was assessed by whole ankle gene transcriptional profiling, protein cytokine and chemokine analysis, and immunohistochemistry.
Materials and Methods
Animals
KRN TCR Tg mice on a C57BL/6 background were originally obtained from Mathis and Benoist as described.17 K/BxN mice were generated by crossing KRN males with nonobese diabetic females. The B6.G7 mouse strain was purchased from The Jackson Laboratory (JAX, Bar Harbor, ME). B6.TCR.Cα−/− (H-2b) mice used in these studies have been previously described27,28 and were purchased from JAX. B6.TCR.Cα−/− were crossed with B6.G7 mice to create B6.TCR.Cα−/−.H-2bxg7 mice. These mice were T cell deficient and expressed H-2bxg7 allowing the transferred KRN T cells to 1) recognize GPI peptide presented on the I-Ag7 molecule, and 2) be histocompatible with the recipients. All mice were bred and housed under specific pathogen-free conditions in the animal facility at the Washington University Medical Center (St. Louis, MO). Studies were performed in accordance with National Institutes of Health and Association for Assessment and Accreditation of Laboratory Animal Care guidelines.
Reagents
Anti-F4/80 antibody was obtained from AbD Serotec, Oxford UK (cat # MCA 497G, 1:400 dilution), anti-myeloperoxidase (MPO) antibody was obtained from Ventana Medical, Tucson AZ (cat # 760-2659 RTU).
Adoptive T Cell Transfer and Measurement of Joint Swelling
Naive KRN T cells (H-2b) were isolated from the spleens of male mice age 8 to 10 weeks old by using CD4 (L3T4) MicroBeads (Miltenyi Biotec, Auburn, CA) according to the manufacturer’s instructions. By passing the T cells through a second column, they were purified to >98% CD4+ T cells. The percentage of these T cells that expressed the KRN TCR was determined by fluorescence-activated cell sorting (FACS) analysis using anti-Vβ6 and ranged from 85% to 95%. The T cells were washed extensively with 1× HBSS then resuspended in 1× HBSS at a concentration of 3.5 × 106 KRN T cells/ml. Male B6.TCR.Cα−/−H-2b/g7 (recipient) mice age 10 to 14 weeks old were anesthetized with a mixture of ketamine/xylazine, and 100 μl of the cell suspension was injected into the retro-orbital sinus. The two rear ankles of the recipient B6.TCR.Cα−/−H-2b/g7 mice were measured starting at day 0 post T cell transfer. Measurement of ankle thickness was made axially across the tibiotarsal (ankle) joint using a Kafer Dial Thickness Gauge J15 (Long Island Indicator Service Inc., Hauppauge, NY). Ankle thickness was rounded off to the nearest 0.1 mm.
Anti-GPI Antibody Enzyme-Linked Immunosorbent Assay
Mice were bled once a week starting on day 0 post T cell transfer. Sera were stored at −20°C before analysis. Sera were plated at an initial dilution of 1:100 followed by serial half log dilutions in Immulon II plates (Fisher Scientific, Pittsburgh, PA) coated with 5 μg/ml recombinant murine GPI.29 Donkey anti-rabbit total Ig-horseradish peroxidase (HRP) (Jackson ImmunoResearch, West Grove, PA), goat anti-mouse IgM-HRP, IgG1-HRP, IgG2b-HRP, IgG2c-HRP, or IgG3-HRP (Southern Biotechnology Associates, Birmingham, AL) were used as secondary antibodies. Serum antibody was detected using ABTS substrate (Roche Molecular Biochemicals, Indianapolis, IN). Absorbance was measured at 414 nm. The serum titer was defined as the reciprocal of the last dilution, which gave an OD >3 times higher than that of the background.
Flow Cytometry
Popliteal lymph nodes were harvested from n = 5 recipient mice 35 days post T cell transfer and evaluated for Vβ6 cellular expression. Cells were surface-stained according to standard protocols using the biotin-conjugated rat anti-mouse Vβ6 antibody (BD Pharminogen, San Diego, CA), which represents >60% of the TCR in the K/BxN model,30 and streptavidin-phycoerythrin-Cy7 (BD Pharmingen, San Diego, CA). All samples were analyzed on a FACSCanto/FACSDiva platform (BD Biosciences, San Diego, CA), data were analyzed using FlowJo software (Tree Star, Inc., Ashland, OR).
RNA Isolation and Amplification
Right-sided hind paws, sectioned just proximal to the tibiotarsal joint, were individually flash-frozen in liquid nitrogen and stored at −80°C. Frozen tissue was later powdered using a SPEX 6750 freezer mill and 6753 microvials (Wolf Laboratories, York UK). Total RNA was prepared from each paw using a modified TRIzol (Invitrogen, Carlsbad CA) isolation protocol incorporating additional purification steps. Total RNA integrity (size and quality) was assessed with an Agilent HP2100 Electrophoresis Bioanalyzer (Agilent Technologies, Santa Clara CA) and quantified using a NanoDrop 1000 UV spectrophotometer by measuring the absorbance at A260 and A280 nm (Thermo Fisher Scientific, Waltham MA). Typical yields ranged from ∼ 200 to >800 ng/ul. Ten ng of total RNA from each paw was amplified for two rounds using a modified amplification protocol (Message Amp II aRNA Amplification Kit, Ambion, Austin TX).
Preparation and Analysis of Agilent Whole Mouse Genome Arrays
Amplified RNA was labeled using a modified protocol with the Kreatech ULS aRNA Fluorescent Cy3/Cy5 labeling kit (Kreatech Diagnostics, Amsterdam, The Netherlands); specifically, 2 μl of labeling buffer and 4 μl of ULS were added to 5 μg of aRNA in a total volume of 20 μl. Once combined, samples were incubated for 15 minutes at 85°C and after incubation samples were returned to room temperature and spun down to collect condensate. All samples were labeled with each dye to allow for generation of fluoro-reverse duplicates. Labeled samples were purified using Zymo Research RNA Clean and Concentrate spin columns according to manufacturer’s protocol (Zymo Research Corporation, Orange CA). Samples were eluted in 100 μl of RNase/DNase-free water. Labeled samples were quantified on the Nanodrop ND-1000 spectrophotometer, and 750 ng of each labeled aRNA from a comparison was added to a total volume of 230 μl water and 9 μl of fragmentation buffer (Agilent Technologies, Santa Clara, CA). Hybridization mixtures were then incubated for 30 minutes at 60°C. Hybridization mixtures were cooled to room temperature and spun down to collect condensate. 240 μl of 2× Hi-RPM hybridization buffer (Agilent Technologies) was added to each mixture. These probes were then hybridized to 4x44K Mouse Whole Genome Microarrays for 18 hours at 65°C in Sure-Hyb Hybridization chambers (Agilent Technologies). Chambers were disassembled in a 0.6× SSC 0.005% Triton X-102 wash bath at room temperature and washed for 10 minutes in wash bath 1. Microarrays were transferred to a 0.01× SSC 0.005% Triton X-102 wash for 5 minutes at 40°C. Microarrays were then dried with HEPA-filtered compressed nitrogen and scanned on the Agilent Technologies DNA Microarray Scanner at 5 μm resolution. Data were extracted from the scanned image using Agilent Technologies Feature Extraction Software version 9.5.
Background subtraction was performed and normalization was completed using the LOWESS method.31 After quality control assessments at the spot, chip, and experiment levels, the fold increase or decrease of mRNA expression compared with normal tissue was calculated. The fold value reported for each oligonucleotide was the average of two arrays with swapped fluorophores for experimental and control RNA. Transcriptional array data have been deposited to Array Express (http://www.ebi.ac.uk/microarray-as/ae/browse.html; accession number E-TABM-966). The oligonucleotides where the values did not flip in the fluoro-reverse were excluded from the analysis. A 1.7-fold cutoff was applied, and genes that did not show differential regulation in at least one condition were removed from the analysis. TaqMan confirmation of reported changes in transcript was completed (data not shown).
Whole Joint Tissue Protein Analyses
Protein cytokine analysis was performed using a portion of the same right ankle powdered tissue used for whole genome microarray analysis. Protein was extracted using Pierce T-Per tissue extraction buffer in the presence of HALT protease inhibitors (Thermo Fisher Scientific, Rockford IL). The soluble fraction of paw extracts was analyzed using murine 22-plex cytokine Luminex immunoassays (Linco/Millipore, Billerica MA). Cytokine concentrations were normalized to the total protein concentration in the same sample using the bicinchoninic acid total protein determination assay (Thermo Fisher Scientific, Rockford IL).
Microscopic and Immunohistochemical Analyses
The left hind paw from each animal was fixed in 10% neutral buffered formalin for 24 hours at 20°C, followed by decalcification in Immunocal (Decal Chemical Corporation, Tallman NY) for 7 days at 20°C. Decalcified joints were then paraffin-embedded with medial aspect down for longitudinal view, sectioned twice (4 μm each), and stained with H&E for general evaluation or toluidine blue for specific assessment of cartilage changes. For F4/80 immunohistochemistry staining, tissue slides were treated with proteinase K (Dako, Carpinteria CA) for 3 minutes followed by blocking solution (Rodent Block M, Biocare Medical, Concord MA) for 15 minutes. This was followed by 30 minutes incubation with F4/80 working dilution. Rat on mouse Polymer kit was used for detection (Biocare), followed by DAB+ (Dako) and a light hematoxylin counterstain. Matched IgG was used for negative control. For MPO immunohistochemistry staining, a CC1 mild cell conditioner was used for pretreatment, followed by a 60-minute incubation with the anti-MPO antibody (Ventana Medical Systems Inc., Tucson AZ). The OmniMap anti-rabbit kit (Ventana) was used for detection followed by hematoxylin counterstain (Discovery XT automated stainer, Ventana).
Joints were evaluated microscopically by a board-certified veterinary pathologist (TPL) and scored for inflammation, cartilage damage, and bone resorption according to previously published criteria.32 For inflammation, scores were as follows: 0 = normal; 1 = minimal infiltration of inflammatory cells in the synovial and/or peri-articular tissues; 2 = mild infiltration with mild edema; 3 = moderate infiltration (including joint space) with moderate edema; 4 = marked infiltration with marked edema; and 5 = severe infiltration with severe edema. For cartilage damage, scores were as follows: 0 = normal; 1 = loss of toluidine blue staining with no chondrocyte degeneration/loss and/or matrix disruption; 2 = loss of toluidine blue staining with minimal chondrocyte degeneration/loss and/or mild matrix disruption in some affected joints; 3 = loss of toluidine blue staining with moderate chondrocyte loss and obvious (depth to deep zone) matrix loss in affected joints; 4 = loss of toluidine blue staining with marked (depth to tide mark) chondrocyte and matrix loss; 5 = loss of toluidine blue staining with severe (depth to subchondral bone) chondrocyte loss and matrix loss in affected joints. For bone resorption, scores were as follows: 0 = normal; 1 = minimal (small areas of resorption in the medullary trabecular or cortical bone, not readily apparent on low magnification, and rare osteoclasts); 2 = mild (increasing areas of resorption in medullary trabecular or cortical bone, not readily apparent on low magnification, with osteoclasts more numerous); 3 = moderate (obvious resorption of the medullary trabecular and cortical bone, without full-thickness defects, lesion apparent on low magnification, and osteoclasts more numerous); 4 = marked (full-thickness defects in the cortical bone, marked loss of medullary trabecular bone, numerous osteoclasts); and 5 = severe (full-thickness defects in the cortical bone, severe loss of medullary trabecular bone). For F4/80 and MPO immunohistochemistry, scores were as follows: 0 = normal; 1 = minimal (small numbers of cells within the synovial epithelium and in proximity to blood vessels within the subsynovium); 2 = mild (increasing numbers of cells evenly distributed within synovial tissues); 3 = moderate (moderate numbers of cells within the synovial tissues and joint space, low numbers of cells within the local peri-articular soft tissues); 4 = marked (obvious cell population within the synovial tissues, joint space, and peri-articular soft tissues with extension to the subcutis and skin); and 5 = severe (high numbers of cells within the synovial tissues, joint space, peri-articular soft tissues, and skin).
Statistical Analyses
Prism 5.01 (GraphPad Software, Inc. La Jolla, CA) was used for all graphical representation of data, including calculation of mean and SEM.
Results
Naive KRN T Cells Induce a Chronic, Destructive Arthritis in T Cell-Deficient Mice
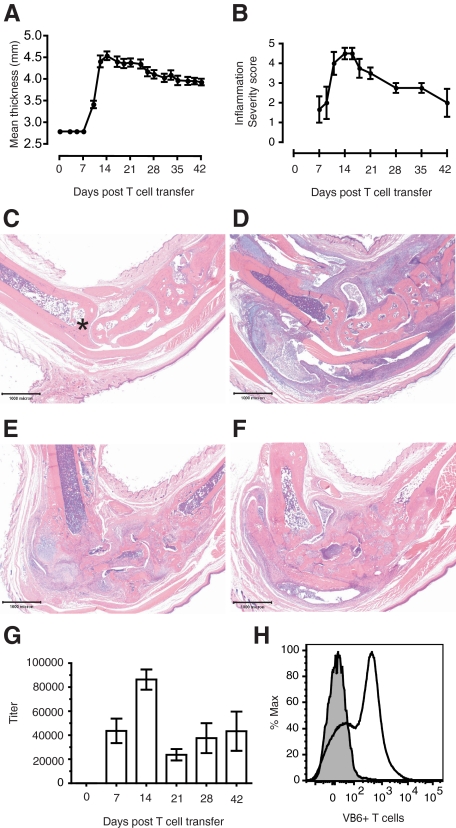
To study the role of T cells in the development of arthritis we transferred naive KRN T cells into T cell deficient B6.TCR.Cα−/−H-2b/g7 mice. These mice expressed the autoantigen GPI on I-Ag7 and were histocompatible with the transferred KRN T cells (H-2b). On intravenous transfer of purified naive KRN T cells, 100% of the recipient male mice developed arthritis; female mice were also susceptible to disease induction (data not shown). There was a visible erythema and increase in the thickness of the ankles, and the mean thickness of both rear ankles as measured via caliper (mean ± SEM, n = 9) peaked 14 to 21 days post T cell transfer (Figure 1A). A decrease in ankle thickness was present 24 days post T cell transfer and beyond, indicating that the inflammatory response to the cartilage damage shifted to a chronic (decreasing edema) stage. This was consistent with the microscopic presentation (Figures 1, B–F). For instance, Figure 1B (a semiquantitative histopathology summary of general inflammation in the ankle over time) demonstrates a decrease in edema and cell influx after 14 days post transfer. Figures 1, C–F provide representative photomicrographs (H&E, 2× magnification) that highlight the progression from normal (Figure 1C), to acute severe inflammatory cell influx 14 days post T cell transfer (Figure 1D), to severe articular cartilage damage with cartilaginous metaplasia and periosteal new bone formation 29 days post T cell transfer (Figure 1E), to joint ankylosis with severe bone remodeling, ulcerated articular cartilage, extra-articular fibroplasia and few remaining inflammatory cells 42 days post T cell transfer (Figure 1F).
Figure 1.
The KRN-CTM model of RA presents a chronic, destructive arthritis that is dependent on anti-GPI IgG antibodies. On day 0, 3.5 × 105 CD4+ KRN T cells were injected intravenously into B6.TCR.Cα−/−H-2b/g7 mice. A: Ankle thickness (mean ± SEM, n = 9) correlates with B, semiquantitative microscopic scoring of general inflammation (mean ± SEM, n = 4). There is progression from Control (C; ×2 magnification; asterisk, distal tibia) to acute severe inflammatory cell influx 14 days post T cell transfer (D; ×2 magnification) to severe articular cartilage damage with cartilaginous metaplasia and periosteal new bone formation 29 days post T cell transfer (E; ×2 magnification), to joint ankylosis with severe bone remodeling, loss of articular cartilage, and extra-articular fibroplasia with few inflammatory cells 42 days post T cell transfer (F; ×2 magnification). G: Total IgG-GPI specific titers as measured by enzyme-linked immunosorbent assay (mean ± SEM, n = 9). H: Identification of popliteal lymph node cellular staining for Vβ6+ T cells in T cell-recipient mice via FACS analysis. Representative histogram of n = 5 mice, 35 days post T cell transfer.
The development of arthritis in the K/BxN mouse model is dependent on antibodies directed against the ubiquitously expressed protein GPI. Therefore, we assayed for total anti-GPI IgG by enzyme-linked immunosorbent assay (mean ± SEM, n = 9). Anti-GPI antibody levels preceded the increase in ankle thickness, yet peaked at the same time (14 days post T cell transfer) (Figure 1G). Antibody levels in KRN-CTM mice persisted for at least 7 weeks, and when their serum was injected into BALB/c mice disease was transferred (data not shown), thereby demonstrating the pathogenic relevance of autoantibodies in this new model. The role of popliteal lymph node lymphocytes in facilitating the production of arthritogenic autoantibodies in K/BxN mice has previously been described.33 In this study, popliteal lymph node cellular expression of Vβ6 was assessed in n = 5 KRN-CTM mice 35 days post T cell transfer via FACS analysis. The results suggest T cell recruitment to lymph nodes once transferred (Figure 1H).
T Cells Induce Monocyte/Macrophage Infiltration in the Ankles of KRN-CTM Mice
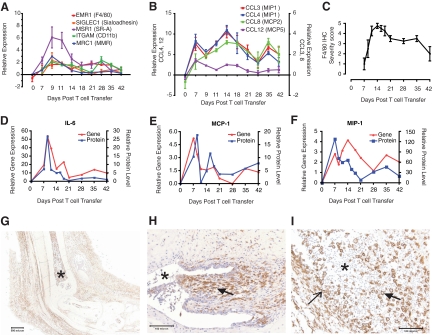
Myeloid cells such as monocytes, macrophages and neutrophils are found in the joints of RA patients and have been previously shown to be important in animal models of arthritis, including the K/BxN model. To determine the role of monocytes/macrophages in KRN-CTM, microarray analysis of macrophage and monocyte transcripts was performed on ankle tissues. Gene transcripts selective for macrophages illustrated a peak 9 days post T cell transfer (Figure 2A), while in Figure 2B, transcripts for monocyte/macrophage CCL cytokines peaked 14 days post T cell transfer with levels maintained through 35 days post T cell transfer. In Figure 2C, semiquantitative scoring of immunohistochemical staining for F4/80 (marker for murine monocytes, macrophages, and dendritic cells) correlated with transcriptional profiling in the illustration of an early influx of cells (peak 14 days post T cell transfer) followed by sustained levels of cells through 42 days post T cell transfer; data points are mean ± SEM (n = 4). In Figure 2, D–F, ankle tissue levels of several protein cytokines and chemokines (interleukin-6, monocyte chemotactic protein (MCP)-1, and macrophage inflammatory protein [MIP]-1α) peaked 7 to 9 days post T cell transfer, with strong correlation between the protein and gene transcript. In Figure 2, G–H (×2 and ×20 magnification, respectively), photomicrographs revealed a uniform distribution of F4/80-positive cells within the joint space, the synovial tissues, the subcutis and skin, while in Figure 2I many large, activated macrophages are present in the tibiotarsal joint space against a backdrop of (unstained) neutrophils (×20 magnification).
Figure 2.
Monocytes/macrophages infiltrate into the ankles of KRN-CTM mice. On the day of transfer 3.5 × 105 CD4+ KRN T cells were injected intravenously into B6.TCR.Cα−/−H-2b/g7 mice. mRNA expression of macrophage markers (A) and monocyte/macrophage CCL cytokines (B) were determined by microarray analysis. C: Semiquantitative microscopic scoring of immunohistochemical staining for F4/80 (mean ± SEM, n = 4). Levels of interleukin-6 (D), MCP-1 (E), and MIP-1α (F) transcript and protein in ankle tissue (mean, n = 2 to 4). G–I: Ankle sections stained for F4/80 from representative mice sampled between 7 and 14 days post T cell transfer. In G, large numbers of F4/80-positive cells are evident in the joint space, the extra-articular soft tissues, and skin (×2 magnification; asterisk, distal tibia). In H, the hyperplastic synovium is raised by a subsynovium, which is expanded by F4/80-positive cells (×20 magnification; asterisk, joint space; closed arrow, subsynovium). In I, large, polygonal macrophages with ample cytoplasm are seen within the tibiotarsal joint space (×20 magnification; open arrow, denuded synovium; asterisk, joint space; closed arrow, macrophage).
Well-described protein chemoattractants of neutrophils granulocyte-colony stimulating factor, IL-1β, and KC peaked in ankle tissue 7 to 9 days post T cell transfer and were strongly correlated with transcript (Figure 3, A–C). Semiquantitative microscopic scoring of immunohistochemical staining for the neutrophil-specific azurophilic granule enzyme MPO correlated with cytokine levels in identifying an early influx of cells (peak 14 days post T cell transfer), followed by a rapid loss of cell numbers through 42 days post T cell transfer (Figure 3D). During the early influx period large numbers of MPO-positive cells were present within the tibiotarsal joint space with fewer numbers within the subsynovial tissues (Figure 3E, ×10 magnification), while higher power examination of the same image revealed particularly concentrated MPO staining due to phagocytosis of apoptotic neutrophils by activated macrophages (Figure 3F, ×100 magnification).
Figure 3.
Neutrophils infiltrate into the ankles of KRN-CTM mice. On day 0, 3.5 × 105 CD4+ KRN T cells were injected intravenously into B6.TCR.Cα−/− H-2b/g7 mice. Levels of granulocyte-colony stimulating factor (A), IL-1 (B), and KC (C) protein and transcript in ankle tissue (mean, n = 2–4). D: Semiquantitative microscopic scoring of immunohistochemical staining for MPO (mean ± SEM, n = 4). E–F: Ankle sections stained for MPO from a representative mouse sampled 14 days post T cell transfer. In E, large numbers of MPO-positive neutrophils are present within the dorsal aspect of the tibiotarsal joint (×10 magnification; asterisk, joint space; open arrow, denuded synovium). A higher magnification view of the same affected joint space, (F), shows apoptotic MPO-positive neutrophils inside large, polygonal macrophages with ample cytoplasm (×100 magnification; closed arrows, macrophages with phagocytized MPO-positive neutrophils).
There was no microscopic evidence of appreciable numbers of lymphocytes in ankle tissues. Also, evidence of an increased lymphoid (T cell and B cell) signature in ankle tissues relative to normal animals was not identified via mRNA expression (Supplemental Figures S1 and S2 at http://ajp.amjpathol.org).
Cartilage Damage and Osteoclast-Mediated Bone Resorption in KRN-CTM Mice Is Comparable to Other Established Rodent Models of RA
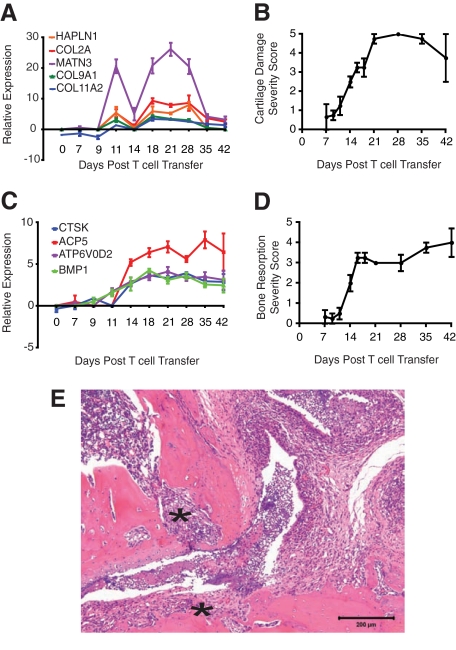
Transcriptional evidence of de novo production of cartilage matrix in response to insult (enzymatic degradation by neutrophils and macrophages), peaked 21 days post T cell transfer then decreased significantly, presumably as the loss of chondrocytes outpaced the repair response (Figure 4A). Ultimately, this resulted in ulceration of articular cartilage 28 to 42 days post T cell transfer (Figure 4B). In Figure 4C, transcripts in the ankle selective for osteoclasts increased after 14 days and were sustained through 42 days post T cell transfer, correlating with the microscopic observations of active bone resorption by multinucleated osteoclasts (Figure 4D) and invasion by fibrovascular “pannus” tissue (Figure 4E, ×10 magnification).
Figure 4.
Cartilage damage and osteoclast-mediated bone resorption is evident in KRN-CTM mice. On day 0, 3.5 × 105 CD4+ KRN T cells were injected intravenously into B6.TCR.Cα−/−H-2b/g7 mice. A: mRNA expression of transcripts related to cartilage matrix production was determined by microarray analysis. B: Semiquantitative microscopic scoring of cartilage damage (mean ± SEM, n = 4). C: mRNA expression of transcripts for osteoclasts was determined by microarray analysis. D: Semiquantitative microscopic scoring of osteoclast-mediated bone resorption (mean ± SEM, n = 4). E: Histological section of ankle from a representative mouse sampled 29 days post T cell transfer; note invasion of the cartilage, subchondral bone and bone marrow space by fibrovascular “pannus” tissue (asterisk, ×10 magnification).
Discussion
Using clinical, gene transcriptional profiling, protein cytokine and chemokine, and microscopic and immunohistochemical endpoints this study provides a temporal account of how transferring 3.5 × 105 purified KRN splenic CD4+ T cells (H-2b) into B6.TCR.Cα−/−H-2b/g7 recipient male mice resulted in 100% arthritis incidence, with a high degree of disease synchronization through 42 days post T cell transfer. Korganow et al,18 in a limited study, observed similar arthritis induction with the transfer of KRN spleen cells into a Cα−/− host, but did not characterize the disease pathology or progression. In this study, arthritis was more severe in the ankle and wrist than in either the knee, elbow, or shoulder joints (data not shown), and progressed from an acute inflammatory cell influx with edema (initiated 7 to 9 days post T cell transfer, peaked 12 to 14 days) to articular cartilage damage with periosteal new bone formation (14 to 21 days post T cell transfer). Bone resorption, with loss of articular cartilage and sustained low numbers of inflammatory cells were present 21 to 42 days post T cell transfer.
Demonstrating CD4+ T cell-induced myeloid cell recruitment into the joints, immunohistochemistry revealed uniform distribution of F4/80-positive macrophages and MPO-positive neutrophils within the joint space, monocytes/macrophages and neutrophils within the vascular-laden subsynovial tissue, and F4/80-positive dendritic cells within both the superficial and deep layers of the skin. However, appreciable numbers of T cells were not observed in the joint or surrounding tissues via microscopy, a finding supported by transcriptional array analysis performed on ankle tissues (no increased expression of CD4, CD3d, CD3e, CD3g, or CD28 relative to normal mice; Supplemental Figure S1 at http://ajp.amjpathol.org). Instead, the T cells orchestrating this response likely resided in the popliteal lymph nodes, where Vβ6 expression was strong. Demonstrating that transferred T cells provided B cell help in autoantibody production, KRN-CTM mice produced high titer, arthritogenic anti-GPI antibodies that peaked at the same time as tissue inflammation.
There was a significant increase in macrophage-specific gene transcription markers 9 to 14 days post T cell transfer, which is 3 to 5 days before the observed increase in macrophage numbers via microscopy. This burst of transcription is likely due to the differentiation of recruited monocytes into macrophages, and the expression continued as newly recruited monocytes presumably replaced exiting macrophages. Also, several protein cytokines tracked well with both the microscopic and transcriptional evidence of monocyte/macrophage infiltration into the joint including, but not limited to, CCL3 (MIP-1α), CCL4 (MIP-1β), CCL8 (MCP-2), and CCCL12 (MCP-5). Overall, these findings indicate that the KRN-CTM recapitulated many features of the K/BxN model.18
The KRN-CTM has several key advantages over other murine models of RA, particularly when studying the role of T cells in disease pathology. For instance, one can manipulate the adoptively transferred T cells (use of knockouts, transgenics, or transfer of either polarized T cell subsets or naive T cells subjected to selective gene knockdown) to characterize specific genes and/or Th1 vs. Th2 vs. Th17 cells in the recruitment and activation of inflammatory cells. Second, given the mouse CIA model is more strain specific (DBA) in terms of disease penetrance and severity, and given that a multitude of knockouts and transgenics are available with the (recipient mice) B6 background the KRN-CTM may be a more flexible platform than the mouse CIA. Third, KRN-CTM arthritis is synchronized, completely penetrant, involves all four paws, and persists to at least 42 days post T cell transfer, thereby requiring substantially fewer animals to run a disease characterization or efficacy study than mouse CIA. The transfer of autoreactive T cells into lymphopenic hosts can generate an exuberant autoimmune response, which may not accurately phenocopy the spontaneous diseases, such as in diabetes models34; however, in this study the KRN-CTM’s presentation of disease was very similar to the spontaneous disease in K/BxN mice and limited to distal joints.
Based on our previous studies33,35 and the results presented here, we propose the following events in KRN-CTM arthritis pathogenesis: 1) transferred KRN T cells expand and are activated in peripheral lymphoid organs; 2) activated KRN T cells provide B cell help inducing autoantibody production; and, 3) recognition of GPI on the articular cartilage surface induces activation of the alternative complement pathway as well as the migration of monocytes/macrophages and neutrophils to the joints. Although the utility of the KRN-CTM model resides in its development of a chronic disease phenotype, it is important to further validate this new model in the context of both prophylactic and therapeutic efficacy studies and compare the results against the models that are currently used most frequently (mouse CIA and rat adjuvant-induced arthritis). The effort described here is the union of complementary scientific disciplines, including transcriptional profiling, chemokine and cytokine protein level analysis and histopathology, and provides a more complete characterization and understanding of the development and progression of disease in the KRN-CTM than any one approach could produce. Based on these results, we propose that the KRN-CTM not only retains the utility of the K/BxN serum transfer and mouse CIA models, but also adds significant logistical and platform advantages. The KRN-CTM presents an opportunity for investigators to study pathways and mechanisms involved in human RA pathogenesis and progression, enabling the validation of druggable targets and biopharmaceuticals.
Acknowledgments
We thank Fei Shih, M.D., Ph.D. for her thoughtful recommendations and critical review of this manuscript.
Footnotes
Address reprint requests to Timothy P. LaBranche, DVM., Ph.D., Dipl. ACVP, Global Research & Development, Pfizer Inc., 35 Cambridgepark Drive, Cambridge MA 02140. E-mail: timothy.labranche@pfizer.com.
Supported by Pfizer, Inc.
T.P.L. is employed by and has stock options in Pfizer, Inc. None of the other authors declared any relevant financial relationships.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Burmester GR, Stuhlmuller B, Keyszer G, Kinne RW. Mononuclear phagocytes and rheumatoid synovitis. Mastermind or workhorse in arthritis? Arthritis Rheum. 1997;40:5–18. doi: 10.1002/art.1780400104. [DOI] [PubMed] [Google Scholar]
- Tak PP, Smeets TJ, Daha MR, Kluin PM, Meijers KA, Brand R, Meinders AE, Breedveld FC. Analysis of the synovial cell infiltrate in early rheumatoid synovial tissue in relation to local disease activity. Arthritis Rheum. 1997;40:217–225. doi: 10.1002/art.1780400206. [DOI] [PubMed] [Google Scholar]
- Koch AE, Kunkel SL, Harlow LA, Johnson B, Evanoff HL, Haines GK, Burdick MD, Pope RM, Strieter RM. Enhanced production of monocyte chemoattractant protein-1 in rheumatoid arthritis. J Clin Invest. 1992;90:772–779. doi: 10.1172/JCI115950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galligan CL, Baig E, Bykerk V, Keystone EC, Fish EN. Distinctive gene expression signatures in rheumatoid arthritis synovial tissue fibroblast cells: correlates with disease activity. Genes Immun. 2007;8:480–491. doi: 10.1038/sj.gene.6364400. [DOI] [PubMed] [Google Scholar]
- Haringman JJ, Smeets TJ, Reinders-Blankert P, Tak PP. Chemokine and chemokine receptor expression in paired peripheral blood mononuclear cells and synovial tissue of patients with rheumatoid arthritis, osteoarthritis, and reactive arthritis. Ann Rheum Dis. 2006;65:294–300. doi: 10.1136/ard.2005.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haringman JJ, Ludikhuize J, Tak PP. Chemokines in joint disease: the key to inflammation? Ann Rheum Dis. 2004;63:1186–1194. doi: 10.1136/ard.2004.020529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph GJ, Furie MB. A soluble gradient of endogenous monocyte chemoattractant protein-1 promotes the transendothelial migration of monocytes in vitro. J Immunol. 1995;155:3610–3618. [PubMed] [Google Scholar]
- Vergunst CE, Gerlag DM, Lopatinskaya L, Klareskog L, Smith MD, van den Bosch F, Dinant HJ, Lee Y, Wyant T, Jacobson EW, Baeten D, Tak PP. Modulation of CCR2 in rheumatoid arthritis: a double-blind, randomized, placebo-controlled clinical trial. Arthritis Rheum. 2008;58:1931–1939. doi: 10.1002/art.23591. [DOI] [PubMed] [Google Scholar]
- Rico MC, Manns JM, Driban JB, Uknis AB, Kunapuli SP, Dela Cadena RA. Thrombospondin-1 and transforming growth factor beta are pro-inflammatory molecules in rheumatoid arthritis. Transl Res. 2008;152:95–98. doi: 10.1016/j.trsl.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torikai E, Kageyama Y, Suzuki M, Ichikawa T, Nagano A. The effect of infliximab on chemokines in patients with rheumatoid arthritis. Clin Rheumatol. 2007;26:1088–1093. doi: 10.1007/s10067-006-0453-5. [DOI] [PubMed] [Google Scholar]
- Cutolo M, Sulli A, Barone A, Seriolo B, Accardo S. Macrophages, synovial tissue and rheumatoid arthritis. Clin Exp Rheumatol. 1993;11:331–339. [PubMed] [Google Scholar]
- Moore BA, Aznavoorian S, Engler JA, Windsor LJ. Induction of collagenase-3 (MMP-13) in rheumatoid arthritis synovial fibroblasts. Biochim Biophys Acta. 2000;1502:307–318. doi: 10.1016/s0925-4439(00)00056-9. [DOI] [PubMed] [Google Scholar]
- Thornton S, Duwel LE, Boivin GP, Ma Y, Hirsch R. Association of the course of collagen-induced arthritis with distinct patterns of cytokine and chemokine messenger RNA expression. Arthritis Rheum. 1999;42:1109–1118. doi: 10.1002/1529-0131(199906)42:6<1109::AID-ANR7>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Gerlag DM, Haringman JJ, Smeets TJ, Zwinderman AH, Kraan MC, Laud PJ, Morgan S, Nash AF, Tak PP. Effects of oral prednisolone on biomarkers in synovial tissue and clinical improvement in rheumatoid arthritis. Arthritis Rheum. 2004;50:3783–3791. doi: 10.1002/art.20664. [DOI] [PubMed] [Google Scholar]
- Haringman JJ, Gerlag DM, Zwinderman AH, Smeets TJ, Kraan MC, Baeten D, McInnes IB, Bresnihan B, Tak PP. Synovial tissue macrophages: a sensitive biomarker for response to treatment in patients with rheumatoid arthritis. Ann Rheum Dis. 2005;64:834–838. doi: 10.1136/ard.2004.029751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijbrandts CA, Vergunst CE, Haringman JJ, Gerlag DM, Smeets TJ, Tak PP. Absence of changes in the number of synovial sublining macrophages after ineffective treatment for rheumatoid arthritis: implications for use of synovial sublining macrophages as a biomarker. Arthritis Rheum. 2007;56:3869–3871. doi: 10.1002/art.22964. [DOI] [PubMed] [Google Scholar]
- Kouskoff V, Korganow AS, Duchatelle V, Degott C, Benoist C, Mathis D. Organ-specific disease provoked by systemic autoimmunity. Cell. 1996;87:811–822. doi: 10.1016/s0092-8674(00)81989-3. [DOI] [PubMed] [Google Scholar]
- Korganow AS, Ji H, Mangialaio S, Duchatelle V, Pelanda R, Martin T, Degott C, Kikutani H, Rajewsky K, Pasquali JL, Benoist C, Mathis D. From systemic T cell self-reactivity to organ-specific autoimmune disease via immunoglobulins. Immunity. 1999;10:451–461. doi: 10.1016/s1074-7613(00)80045-x. [DOI] [PubMed] [Google Scholar]
- Maccioni M, Zeder-Lutz G, Huang H, Ebel C, Gerber P, Hergueux J, Marchal P, Duchatelle V, Degott C, van Regenmortel M, Benoist C, Mathis D. Arthritogenic monoclonal antibodies from K/BxN mice. J Exp Med. 2002;195:1071–1077. doi: 10.1084/jem.20011941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipke BT, Allen PM. Essential role of neutrophils in the initiation and progression of a murine model of rheumatoid arthritis. J Immunol. 2001;167:1601–1608. doi: 10.4049/jimmunol.167.3.1601. [DOI] [PubMed] [Google Scholar]
- Lee DM, Friend DS, Gurish MF, Benoist C, Mathis D, Brenner MB. Mast cells: a cellular link between autoantibodies and inflammatory arthritis. Science. 2002;297:1689–1692. doi: 10.1126/science.1073176. [DOI] [PubMed] [Google Scholar]
- Ji H, Ohmura K, Mahmood U, Lee DM, Hofhuis FM, Boackle SA, Takahashi K, Holers VM, Walport M, Gerard C, Ezekowitz A, Carroll MC, Brenner M, Weissleder R, Verbeek JS, Duchatelle V, Degott C, Benoist C, Mathis D. Arthritis critically dependent on innate immune system players. Immunity. 2002;16:157–168. doi: 10.1016/s1074-7613(02)00275-3. [DOI] [PubMed] [Google Scholar]
- Ji H, Pettit A, Ohmura K, Ortiz-Lopez A, Duchatelle V, Degott C, Gravallese E, Mathis D, Benoist C. Critical roles for interleukin 1 and tumor necrosis factor alpha in antibody-induced arthritis. J Exp Med. 2002;196:77–85. doi: 10.1084/jem.20020439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafaille JJ, Keere FV, Hsu AL, Baron JL, Haas W, Raine CS, Tonegawa S. Myelin basic protein-specific T helper 2 (Th2) cells cause experimental autoimmune encephalomyelitis in immunodeficient hosts rather than protect them from the disease. J Exp Med. 1997;186:307–312. doi: 10.1084/jem.186.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakala SV, Kurrer MO, Katz JD. T helper 2 (Th2) T cells induce acute pancreatitis and diabetes in immune-compromised nonobese diabetic (NOD) mice. J Exp Med. 1997;186:299–306. doi: 10.1084/jem.186.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AK, Conaghan PG, Karim Z, Quinn MA, Ikeda K, Peterfy CG, Hensor E, Wakefield RJ, O'Connor PJ, Emery P. An explanation for the apparent dissociation between clinical remission and continued structural deterioration in rheumatoid arthritis. Arthritis Rheum. 2008;58:2958–2967. doi: 10.1002/art.23945. [DOI] [PubMed] [Google Scholar]
- Philpott KL, Viney JL, Kay G, Rastan S, Gardiner EM, Chae S, Hayday AC, Owen MJ. Lymphoid development in mice congenitally lacking T cell receptor alpha beta-expressing cells. Science. 1992;256:1448–1452. doi: 10.1126/science.1604321. [DOI] [PubMed] [Google Scholar]
- Wen L, Roberts SJ, Viney JL, Wong FS, Mallick C, Findly RC, Peng Q, Craft JE, Owen MJ, Hayday AC. Immunoglobulin synthesis and generalized autoimmunity in mice congenitally deficient in alpha beta(+) T cells. Nature. 1994;369:654–658. doi: 10.1038/369654a0. [DOI] [PubMed] [Google Scholar]
- Mandik-Nayak L, Racz J, Sleckman BP, Allen PM. Autoreactive marginal zone B cells are spontaneously activated but lymph node B cells require T cell help. J Exp Med. 2006;203:1985–1998. doi: 10.1084/jem.20060701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih FF, Mandik-Nayak L, Wipke BT, Allen PM. Massive thymic deletion results in systemic autoimmunity through elimination of CD4+ CD25+ T regulatory cells. J Exp Med. 2004;199:323–335. doi: 10.1084/jem.20031137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni TT, Lemon WJ, Shyr Y, Zhong TP. Use of normalization methods for analysis of microarrays containing a high degree of gene effects. BMC Bioinformatics. 2008;9:505. doi: 10.1186/1471-2105-9-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendele A, McComb J, Gould T, McAbee T, Sennello G, Chlipala E, Guy M. Animal models of arthritis: relevance to human disease. Toxicol Pathol. 1999;27:134–142. doi: 10.1177/019262339902700125. [DOI] [PubMed] [Google Scholar]
- Mandik-Nayak L, Wipke BT, Shih FF, Unanue ER, Allen PM. Despite ubiquitous autoantigen expression, arthritogenic autoantibody response initiates in the local lymph node. Proc Natl Acad Sci USA. 2002;99:14368–14373. doi: 10.1073/pnas.182549099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri A, Katz JD. Dissecting the role of CD4+ T cells in autoimmune diabetes through the use of TCR transgenic mice. Immunological Reviews. 1999;169:55–65. doi: 10.1111/j.1600-065x.1999.tb01306.x. [DOI] [PubMed] [Google Scholar]
- Wipke BT, Wang Z, Nagengast W, Reichert DE, Allen PM. Staging the initiation of autoantibody-induced arthritis: a critical role for immune complexes. J Immunol. 2004;172:7694–7702. doi: 10.4049/jimmunol.172.12.7694. [DOI] [PubMed] [Google Scholar]