Abstract
Breast cancer preferentially metastasizes to bone. We therefore addressed the role of Notch signaling in osteoblast-cancer cell interactions and in bone metastasis. Human bone marrow osteoblasts selectively enhanced the expression of Notch3 and its ligand Jagged1 in human breast cancer cell lines. Osteoblasts also stimulated cancer cell colony formation in soft agar, which was reduced by a chemical inhibitor of Notch signaling and anti-transforming growth factor β1 (TGFβ1) antibody. TGFβ1, a major prometastatic product of osteoblasts, also stimulated cancer cell Notch3 expression. Notch3 knockdown in the cancer cells by stable short hairpin RNA interference decreased the osteoblast- and TGFβ1-stimulated colony formation as well as TGFβ1-mediated Smad3/Smad2 phosphorylation; Jagged1 level was coordinately reduced. In addition, expression of snail, a regulator of epithelial-mesenchymal transition, and the mesenchymal markers fibronectin and vimentin was attenuated by reducing Notch3 levels. To study the role of Notch3 signaling in bone metastasis, cancer cells were inoculated into athymic mice, either into femoral bone marrow cavities or into the systemic circulation via the left ventricle. Compared with robust osteolysis in mice receiving control cells, osteolytic lesions were significantly reduced following inoculation of cells with constitutively reduced Notch3 expression. Taken together, our results suggest that enhanced Notch3 expression in breast cancer cells, triggered by osteoblasts and their secretion of TGFβ1 in the bone marrow niche, may stand as a novel mechanism for promoting bone metastasis.
Notch signaling has been highlighted as a pathway involved in the development of breast cancer and is frequently dysregulated in invasive breast cancer.1 Activation of Notch signaling is initiated by the interaction of a Notch ligand such as Jagged1 with the extracellular domain of a Notch receptor. Sequential proteolytic cleavages produce a fragment, Notch Intracellular Domain, which then enters the nucleus and regulates expression of specific genes including Hes and Hey family transcription factors.2
Transgenic mice that overexpress activated forms of Notch receptors 1, 3, and 4 in mammary glands develop mammary tumors.3,4 Elevated expression of Jagged1 and Notch receptors 1, 3, and 4 in breast cancer is correlated with poor prognosis,1,5,6,7 whereas elevated Notch2 is correlated with a higher chance of survival.8 Epithelial-mesenchymal transition (EMT) is characterized by loss of cell adhesion, and is associated with tumor invasion and metastasis, particularly in breast cancer.9 Notch activation mediated by Jagged1 promotes EMT.10 EMT is accompanied by specific changes in gene expression, such as loss of E-cadherin and gain of mesenchymal markers vimentin and fibronectin,11,12,13 and controlled by elevated transcriptional activity involving snail and slug.10,13,14,15,16
Bone is the first site of metastasis in about 50% of breast cancer patients at the time of their first relapse. Bone metastasis is a leading cause of pathological fracture, hypercalcemia of malignancy, nerve compression, pain, morbidity and death. Tumor cells, osteoblasts, osteoclasts and bone extracellular matrix are the four components of a vicious cycle necessary for the initiation and development of metastatic lesions in the skeleton.17 A well documented mechanism in this cycle involves osteoblast-secreted transforming growth factor (TGF)β, which enhances the local tumor cell expression of Parathyroid Hormone related Protein (PTHrP), leading to elevated osteoblastic expression of Receptor Activator of Nuclear Factor kappa B Ligand (RANKL), more osteoclasts, and increased osteolytic bone metastasis of breast cancer.17,18,19,20,21
Osteoblasts lining the bone marrow endosteal surface support long-term hematopoietic stem cells, possibly through direct cell-cell contact, or osteoblast-secreted factors.22,23 Notch signaling is implicated in osteoblast-regulated long-term hematopoietic stem cell homeostasis.22,24 Increasing evidence suggests that the osteoblast niche also controls malignant cell growth and survival.25 In the present work, we demonstrate that bone marrow osteoblast co-culture enhances the soft agar colony formation by human breast cancer cells, and this effect is mediated by the osteoblast product TGFβ1. Both osteoblasts and TGFβ1 independently increase breast cancer gene expression of Notch3. Inhibition of Notch3 expression in breast cancer cells significantly decreases the enhancement of colony formation in soft agar by osteoblasts and TGFβ1 and reduces osteolytic bone metastasis in xenograft animal models.
Materials and Methods
Reagents and Antibodies
Suppliers of reagents include human TGFβ1, recombinant human fibroblast growth factor-2, anti-TGFβ1 antibody, human TGFβ1 ELISA kit (R&D Systems, Minneapolis, MN); anti-Notch3 antibody (Santa Cruz Biotechnology, Santa Cruz, CA); anti-Snail, anti-phospho-Smad2, and anti-phospho-Smad3 antibodies (Cell Signaling Technology, Beverly, MA); anti-vimentin antibody (Millipore, Billerica, MA); anti-fibronectin antibody (BD Biosciences, San Jose, CA); anti-V5 antibody (Invitrogen, Carlsbad, CA); and γ-secretase inhibitor L685,458 (EMD Chemicals, Gibbstown, NJ); all other reagents were from Sigma-Aldrich (St. Louis, MO).
Cells
Human Bone Marrow Osteoblasts
Human bone marrow osteoblasts (hBMOB) were generated as described previously with minor modifications.26 The mononuclear cells were isolated from fresh bone marrow aspirates (Lonza, Allendale, NJ) and cultured in Iscove’s modified Dulbecco’s medium supplemented with 10% fetal bovine serum and nonessential amino acids. Adherent monolayers were then cultured in media containing 5 ng/ml fibroblast growth factor-2 to generate bone marrow stromal cells. The hBMOB were differentiated by culturing bone marrow stromal cells with osteogenic media (0.1 mmol/L ascorbic acid-2-phosphate, 10 mmol/L β-glycerophosphate, and 100 nmol/L dexamethasone). Osteoblast differentiation was confirmed by staining for alkaline phosphatase and positive von Kossa staining of the phosphate-rich mineralized extracellular matrix.27
Human Breast Cancer Cell Lines
MDA-MET28 was derived from MDA-MB-231 by in vivo selection for murine bone metastasis and was generously provided by M. Bendre and L. Suva (University of Arkansas, Little Rock, AR). MDA-MB-231 was purchased from the American Type Culture Collection. MDA-MET and MDA-MB-231 were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% FBS and penicillin/streptomycin. MDA-MB-43529 was kindly provided by D. R. Welch (University of Alabama, Birmingham, AL), and cultured in Dulbecco’s modified Eagle’s medium/F12 (Invitrogen number 11330) supplemented with 5% FBS, 1 mmol/L sodium pyruvate, and 0.02 mmol/L nonessential amino acids. To facilitate cell detection and FACS purification following osteoblast co-culture experiments, the MDA-MET cells were tagged with enhanced Green Fluorescent Protein (eGFP), and the eGFP-labeled cells were used for the experiments described below. Similarly, the eGFP-tagged MDA-MB-435 cells were used for in vitro studies.
Co-Culture of Breast Cancer Cells and Bone Marrow Osteoblasts
Co-culture with Cell-Cell Contact: hBMOB cells (passages 2–4) were seeded at a density of 4 × 105 cells/10-cm culture dish and cultured until confluent and mineralized. The eGFP-labeled MDA-MET or MDA-MB-435 cells were then seeded in the dish with or without an underlying hBMOB layer at a density of 1.5 × 106 cells/dish and cultured for 48 hours. The eGFP-labeled cancer cells were purified for further analysis by flow cytometry (MoFlo Legacy; Beckman Coulter, Brea, CA). Transwell Co-Culture without Cell-Cell Contact: 4 × 103 hBMOB were seeded in 24-well cell culture inserts (0.4-μm pore, Falcon; BD Labware, Franklin Lakes, NJ) and cultured until confluent. Then 5 × 104 breast cancer cells were seeded in 24-well plates and cultured with the inserts with or without hBMOB for 48 hours.
RNA Extraction, Reverse Transcription, and PCR
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA). cDNA was synthesized with Ready-To-Go You-Prime First-Strand Beads (GE Healthcare, Piscataway, NJ) and oligo dT. Quantitative RT-PCR (qRT-PCR) was conducted using SYBR GREEN PCR Master Mix (Applied Biosystems, Foster City, CA) and the ABI Prism 7700 Sequence Detection System. Semiquantitative RT-PCR was performed using Platinum PCR SuperMix (Invitrogen).
The following pairs of primers were usedL hJagged-1 primers for qRT-PCR (5′-CCCACATGCTCTACAAAC-3′ and 5′-CAGCGATAACCATTAACC-3′); hJagged-1 primers for semiquantitative RT-PCR (5′-TCGCTGTATCTGTCCACCTG-3′ and 5′-AGTCACTGGCACGGTTGTAG-3′); hHey-1 (5′-GCTGGTACCCAGTGCTTTTGA-3′G and 5′-TGCAGGATCTCGGCTTTTTCT-3′); hHes-1 (5′-ACACCGGATAAACCAAAGAC-3′ and 5′-TGATCTGGGTCATGCAGTTG-3′); hSnail (5′-CATCCTTCTCACTGCCATGGA-3′ and 5′-AGGCAGAGGACACAGAACCAGA-3′); and hSlug (5′-AGATGCATATTCGGACCCAC-3′ and 5′-CCTCATGTTTGTGCAGGAGA-3′). Primer sequences for other Notch receptors and ligands were described previously.30
Notch3-Specific Short Hairpin RNA Stable Interference
The human Notch3-specific short hairpin RNA (shRNA) and the control shRNA in pSuper-Puro retroviral vectors were provided by M. Bonafe and C. Giovannini (University of Bologna, Bologna, Italy). The control shRNA does not match to any known human transcript (5′-gatccccAATATCCTTGGACACAAGTTGttcaagagaCAACTTGTGTCCAAGGATATTtttttggaac-3′). Retroviral gene transfer was performed as previously described31 with minor modifications. Briefly, the human Phoenix Ampho packaging cell line (Orbigen, San Diego, CA) was transfected with the retroviral DNAs. Stable retroviral infection of breast cancer cells was performed using standard procedures, and stable clones were selected with 5 μg/ml puromycin. Alternately, Notch3 was knocked down using Notch3 shRNA lentiviral particles according to the manufacturer’s instruction (Santa Cruz Biotechnology, Santa Cruz, CA). The bulk population was selected with 5 μg/ml puromycin. For controls, cultures were transduced with lentiviral particles encoding a scrambled shRNA sequence.
Anchorage-Independent Colony Assay in Soft Agar
The colony formation was performed in 24-well plates. A single-cell suspension of MDA-MET cells (3 × 102 suspended in 0.5 ml 0.4% bactoagar) was added over a base layer of 0.8% bactoagar; both gel media contained complete culture medium. After 7 to 8 days of incubation, colonies composed of equal to or more than four cells were counted by microscopy. To study the role of osteoblasts in colony formation, hBMOB were seeded on 24-well plates and cultured to an endpoint of confluent, mineralized layers. The plates were then layered with soft agar containing the suspension of MDA-MET cells and incubated for 7 to 8 days.
Cell Proliferation, Apoptosis, and Invasion Assays
Cells were seeded in 24-well plates at a density of 2 × 104 cells/well. On the days indicated, cells were trypsinized, washed in PBS, and counted with a Cellometer Automated Cell Counter (Nexcelom Bioscience, Lawrence, MA). Apoptosis was assessed using Annexin V-Allophycocyanin (APC) staining (BD Biosciences, San Diego, CA), according to the protocol recommended by the manufacturer. Briefly, cells were trypsinized, washed twice with PBS, resuspended in 1× binding buffer and stained with Annexin V-APC for 15 minutes in the dark. Fluorescence was analyzed by flow cytometry (MoFlo Legacy; Beckman Coulter). Invasion assays were performed with BD BioCoat Matrigel Invasion Chambers (BD Biosciences, Bedford, MA). Cells were seeded in the 24-well chambers at a density of 1 × 105 cells per chamber. After 48 hours of incubation in the presence of TGFβ1 or vehicle, the invaded cells were fixed, stained with 1% toluidine blue, and counted under the light microscope.
Animals and Xenograft Models
Six- to 7-week-old female athymic mice (Hsd: Athymic Nude-Foxn1nu) were purchased from Harlan (Indianapolis, IN). All surgical procedures were conducted in accordance with the protocol approved by the Institutional Animal Care and Use Committee of Children’s Hospital Boston. The animal studies were monitored by the staff veterinarians.
Animal model of intraosseous tumorigenesis by intrafemoral implantation of cancer cells: for intraosseous tumor implant, a 30-gauge needle was inserted into the bone cortex of the distal femur. A single-cell suspension (2 × 104 cells in 20 μl of HBSS) was slowly injected in the bone marrow cavity.
Animal model of bone metastasis by ultrasound-guided intracardiac injection of cancer cells: A single-cell suspension (3 × 105 cells in 200 μl of HBSS) was injected into the left cardiac ventricle with a 25-gauge needle. The injection was aided by ultrasound guidance using the Vevo 770 imaging system. To confirm injection of the entire volume into the arterial system, partial withdrawal of the syringe piston showed needle uptake of bright red blood before and following each inoculation.
MicroCT scanning (SIEMENS micro-CAT II scanner, ImTek, Knoxville, TN) and radiographical imaging were performed on anesthetized mice. After euthanasia, hind limbs were fixed in 10% buffered neutral formalin, decalcified in EDTA, and paraffin embedded; 5-μm sections of long bones were then cut with a microtome and stained with Goldner’s Trichrome.
Statistical Analysis
Statistical analyses were performed by unpaired Student’s t-test; P values <0.05 were considered statistically significant.
Results
Osteoblasts Enhance Notch3 Expression by Breast Cancer Cells
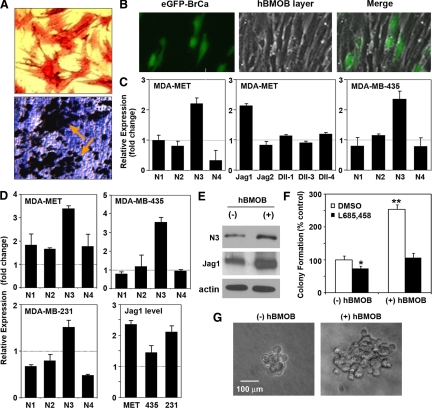
The interaction between breast cancer cells and the bone microenvironment contributes to the initiation and progression of bone metastases. To test the hypothesis that bone marrow osteoblasts support bone colonization of disseminated breast cancer, human breast cancer cell lines were co-cultured with hBMOB. The differentiated osteoblasts were alkaline phosphatase positive (Figure 1A, upper panel) and produced a well-mineralized von Kossa-positive extracellular matrix (Figure 1A, lower panel).
Figure 1.
Osteoblasts enhance Notch signaling in breast cancer cells. A: Generation of primary human osteoblasts from fresh human bone marrow aspirates. The differentiated osteoblasts (hBMOB) were stained for alkaline phosphatase activity (red, upper panel) and by von Kossa staining for mineralization (black, lower panel). B: Images showing co-culture of eGFP-labeled breast cancer cells (green) and hBMOB (phase contrast) with direct cell-cell contact. In these co-cultures, the hBMOB were confluent and positive for von Kossa staining. C: Regulation of Notch and Notch ligands in breast cancer cells by hBMOB with direct cell-cell contact. Following 48 hours of co-culture, the eGFP-labeled MDA-MET and MDA-MB-435 cells were isolated from hBMOB by FACS sorting. The cells were then processed for qRT-PCR to determine the mRNA levels of Notch signaling molecules as indicated. The results are expressed as fold changes relative to the control breast cancer cells that were cultured without hBMOB; mean ± SEM (n = 3), a representative of two experiments. D: Notch pathway gene regulation by hBMOB with noncontact (Transwell) co-culture. Breast cancer cells (MDA-MET, MDA-MB-435, and MDA-MB-231) were co-cultured with cell culture inserts containing hBMOB for 48 hours. The cancer cells were then processed for qRT-PCR as described in C. The cancer cells cultured with empty inserts without hBMOB served as the normalization control; mean ± SEM (n = 3), a representative of two experiments. E: Western blot analysis showing enhanced protein expression of Notch3 and Jagged1 in MDA-MET cells by the Transwell co-culture with hBMOB. F: Colony formation assay showing that an underlying, noncontacting layer of hBMOB enhances the number of colonies formed in soft agar in MDA-MET cells, and this effect is inhibited by 1 μmol/L of the γ-secretase inhibitor L685,458. Colony formation was also slightly inhibited by L685,458 in the absence of hBMOB. Data are normalized to vehicle DMSO-treated culture in the absence of hBMOB; *P < 0.05 and **P < 0.01. G: Phase contrast images of representative single colonies in the absence or presence of hBMOB.
The co-culture was initially performed with direct cell-cell contact between MDA-MET cells and hBMOB (Figure 1B). Compared with cancer cells that were cultured alone, cell-cell contact with hBMOB caused a twofold enhancement in gene expression of Notch3, as well as the Notch ligand Jagged1 in MDA-MET cells (Figure 1C). However, there were no significant changes in expression of other Notch isoforms (Notch1, 2, and 4), or Notch ligands Jagged2 and δ-like ligands 1, 3, and 4.
Since it is well documented that soluble cytokines and growth factors present in bone play a critical role in cancer metastasis,32,33 we then evaluated whether the direct cell-cell contact with hBMOB is necessary for the enhancement of Notch3 expression in MDA-MET cells. hBMOB were cultured in hanging inserts that allowed free diffusion of osteoblast-derived factors into the underlying media layer containing the breast cancer cells. Similar to what was observed in cell-cell contact co-culture, hBMOB selectively enhanced the mRNA expresson of Notch3 but not other Notch receptors in MDA-MET cells. The mRNA level of the Notch ligand Jagged1 was also increased (Figure 1D). Furthermore, we demonstrated enhanced protein expression of Notch3 and Jagged1 caused by Transwell hBMOB (Figure 1E). These results suggest the existence of a soluble factor released from hBMOB that enhances the expression of Notch3, as well as its ligand Jagged1 by breast cancer cells.
MDA-MB-231 is the parental line from which MDA-MET was selected for its in vivo bone-metastatic properties.28 MDA-MB-231 in co-culture with hBMOB also showed selective elevation in the expression of Notch3 and Jagged1 (Figure 1D). MDA-MB-435 is another human breast cancer line that showed selective increase in Notch3 both in direct contact with hBMOB (Figure 1C) and Transwell co-culture (Figure 1D), but Jagged1 was only weakly elevated.
To test the specificity of osteoblast enhancement of Notch3/Jagged1 expression, we performed co-culture of breast cancer cells with nonosteoblastic human bone marrow stromal cells (Supplemental Figure 1, see http://ajp.amjpathol.org). Co-culturing with human bone marrow stromal cells also increased the Notch3 expression in both MDA-MET and MDA-435 cells. However, this was accompanied by an increase in Notch1 in MDA-MET and Notch1 and Notch4 in MDA-MB-435. No enhanced Jagged1 expression was detected by co-culturing with human bone marrow stromal cells. Therefore, compared with undifferentiated bone marrow stromal cells (human bone marrow stromal cells), the bone marrow-derived osteoblasts (hBMOB) behave differently in regards to the stimulation of Notch signaling in breast cancer cells. A recent report shows that by production and secrection of interleukin-6, fibroblasts isolated from lung, bone, and breast enhance the growth and invasion of ER+HER2+ (estrogen receptor-positive and human epidermal growth factor receptor 2-positive) MCF7 breast cancer cells, through STAT3 and downstream effectors such as Notch3, Jagged1, and Carbonic Anhydrase IX. In contrast, ER−HER2− MDA-MB-231 cells are less responsive to paracrine interleukin-6 signaling.34 A unique role for Notch3 has been implicated in the proliferation of HER2− breast cancer cell lines,35 and our study focuses on three cell lines that are ER−HER2−: MDA-MET, MDA-MB-435, and MDA-MB-231. The enhancement of Notch3 and Jagged1 that we observed for these cell lines in the presence of osteoblasts may play a unique role in selective colonization of the bone marrow microenvironment by ER−HER2− breast cancer.
Notch3 Regulates Osteoblast-Stimulated Anchorage-Independent Growth
Suspensions of single cancer cells were cultured in soft agar in the presence or absence of hBMOB to evaluate anchorage-independent colony formation. The colony formation of MDA-MET breast cancer cells is significantly increased in both colony number and colony size by factors from hBMOB, compared with cancer cells cultured in the absence of hBMOB (Figures 1, F and G). Treatment with L685,458, a γ-secretase inhibitor that inhibits the proteolytic cleavage involved in Notch activation, completely blocked the osteoblast-mediated increase in colony number (Figure 1F), whereas the colony size was unaffected (data not shown).
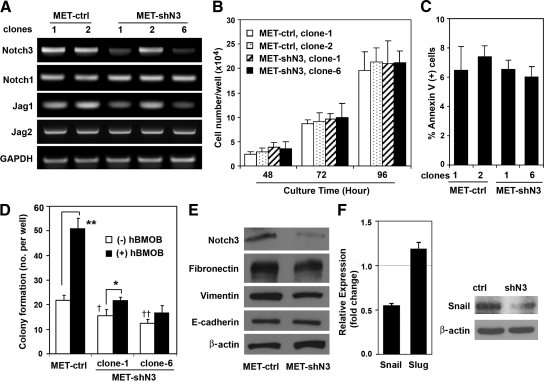
Notch3 was recently reported to control self-renewal and hypoxic survival of primary breast carcinoma cells and the human MCF7 breast cancer cell line.31 To specifically address the function of Notch3 in breast cancer colonization of bone, we generated MDA-MET cells stably expressing Notch3-specific shRNAs. Two independent clones featuring a strong decrease of Notch3 and Jagged1 expression (clone 1 and clone 6) were tested (Figure 2A). We assessed the effect of decreased Notch3 expression on cell growth and survival. As shown in Figure 2B, Notch3 knockdown did not affect the growth of MDA-MET cells. The effect of reduced Notch3 signaling on cell apoptosis was determined through Annexin V staining (Figure 2C). No significant changes in percentage of APC Annexin V-positive cells were observed in the two MET-shN3 clones (6.54 and 6.02%) compared with the MET-ctrl clones (6.48 and 7.15%). Staging of apoptosis by staining with APC Annexin V and 7-aminoactinomycin confirmed the similar viability of the MET-shN3 and MET-ctrl clones. Since Notch3 was reported to play a role in proliferation of other breast cancer cell lines such as HCC1143 and BT-549,31 we also studied Notch3 knockdown in the MDA-MET parental cell line MDA-MB-231, but found no effect on proliferation. Notch3 was knocked down by shRNA interference and the bulk population was selected with puromycin. Cells were plated at 2 × 104 cells/well in 24-well plates (n = 4 wells). After 72 hours of culture, the total cell number of MDA-MB-231 with reduced Notch3 expression was not significantly different from the scrambled shRNA control cells (1.22 × 105 ± 1.95 × 103 versus l.23 × 105 ± 1.10 × 103 cells/well, respectively; mean ± SEM, P = 0.48).
Figure 2.
Notch3 regulates osteoblast-stimulated colony formation and expression of mesenchymal markers. A: Stable retroviral infection of MDA-MET cells with the retroviral vector expressing Notch3-specific shRNA (MET-shN3) or control shRNA (MET-ctrl). Total RNA was extracted from the cells of indicated -shN3 and -ctrl clones, and processed for semiquantitative RT-PCR to determine the mRNA levels of Notch signaling molecules as indicated. B: Effect of Notch3 knockdown on cell growth. Clones of MET-shN3 or MET-ctrl were plated at 2 × 104 cells/well and cultured for indicated times. Cells were then trypsinized and counted with Cellometer Automated Cell Counter; mean ± SEM (n = 2), a representative of two experiments. C: Effect of Notch3 knockdown on cell apoptosis assessed by Annexin V-APC staining; fluorescence of 1 × 104 cells/group was analyzed by flow cytometry. D: Inhibition of the hBMOB-stimulated colony formation by decreased Notch3 expression. The hBMOB-stimulated colony formation was evaluated for two clones of MET-shN3 and compared with MET-ctrl; mean ± SEM (n = 4), a representative of two experiments. Tests for significance included the effect of hBMOB stimulation (*P < 0.05 and **P < 0.01) and the effect of Notch3 knockdown on basal levels of colony formation (without hBMOB) in MET-shN3 clone-1 and clone-6 compared with MET-ctrl (†P < 0.05 and ††P < 0.01). E: Whole-cell lysates of MET-shN3 and MET-ctrl were immunoblotted with antibodies against indicated proteins. The membranes were also blotted with anti-actin antibody to verify equal protein loading. F: qRT-PCR analysis showing decreased mRNA level of snail (left panel) in MET-shN3 compared with MET-ctrl, whereas slug is relatively unaffected; data are expressed as the mean ± SEM (n = 3) and represent two independent experiments. Western blot showing reduced protein expression of Snail in MET-shN3 compared with MET-ctrl (right panel).
We then asked whether the decrease in Notch3 expression affected anchorage-independent colony formation. Knockdown of Notch3 significantly inhibited the osteoblast-stimulated colony formation in soft agar of MET-shN3 compared with MET-ctrl (Figure 2D). These data indicate a requirement for Notch3 in the osteoblast enhancement of tumor cell colony formation, as would occur in the bone microenvironment. The role of Notch3 was also assessed by overexpressing full-length Notch3 with pcDNA3.1/V5-His plasmid (provided by T. P. Dang, Vanderbilt University Medical Center, Nashville, TN).31 The construct was confirmed by DNA sequencing, and the overexpression of Notch3 was demonstrated by Western blot analysis using anti-V5 and anti-Notch3 antibodies. Compared with empty vector control cells, overexpression of Notch3 enhanced the anchorage-independent colony formation in both MDA-MET and MDA-MB-435 cells (Supplemental Figure 2, see http://ajp.amjpathol.org).
Possible Role of Notch3 in EMT
Decreasing Notch3 expression in MDA-MET cells reduced the expression of vimentin and fibronectin, although E-cadherin expression remained unchanged (Figure 2E). The level of snail, a transcription factor that controls EMT,36 was also decreased by Notch3 knockdown (Figure 2F). To control for the possibility that the selected clones were not representative, we also performed stable transduction of MDA-MET cells with Notch3 shRNA lentiviral particles. Parallel studies confirmed that reduced Notch3 expression in MDA-MET cells inhibited the expression of Snail but had no effect on E-cadherin expression (data not shown). These results suggest that blocking Notch3 expression could play a role in inhibiting the invasive mesenchymal traits and decreasing the invasion of cancer cells. The function of Notch3 could be mediated by the expression level of Notch ligands. Jagged1 expression was reported to be regulated by Notch3,31,37 and TGF-β-induced EMT is blocked by RNA silencing of Jagged1.38 Indeed, we detected reduced Jagged1 expression in clones with lower Notch3 expression, whereas the levels of Jagged2 and Notch1 were unchanged (Figure 2A).
Notch3 and TGFβ Signaling
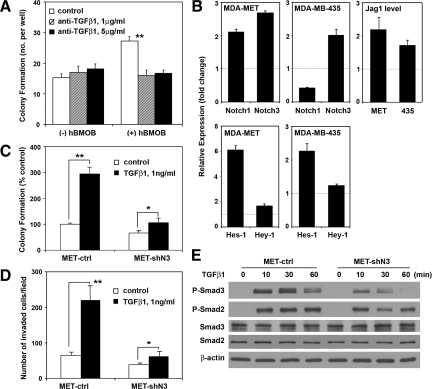
TGFβ is a prometastatic growth factor released from osteoblasts and resorbing bone matrix.39 We therefore explored the possible role of TGFβ as the active paracrine regulator from osteoblasts. Anti-TGFβ1 antibody at 1 and 5 μg/ml blocked the osteoblast-stimulated colony formation of MDA-MET cells (Figure 3A). We next examined whether Notch3 signaling plays a role in the effect of TGFβ1 on breast cancer cells. Compared with vehicle-treated cells, TGFβ1 at 1 ng/ml enhanced the mRNA level of Notch3, as well as Notch ligand Jagged1, in both MDA-MET and MDA-MB-435 cells (Figure 3B, upper panel). In MDA-MET cells, TGFβ1 also caused a sixfold elevation in expression of Hes-1, a direct target of Notch signaling, whereas Hey-1 expression remained unchanged. The increased Hes-1 level was also observed in MDA-MB-435 cells (Figure 3B, lower panel). Knockdown of Notch3 expression significantly inhibited the TGFβ1-stimulated anchorage-independent colony formation (Figure 3C) and cell invasion in vitro (Figure 3D). Smad2/Smad3 are key intracellular mediators of TGFβ signals. TGFβ1 at 1 ng/ml stimulated Smad3 phosphorylation at 10 minutes, reaching a maximum at 30 minutes. Decreasing the Notch3 expression in MET-shN3 cells significantly reduced TGFβ1-stimulated Smad3 phosphorylation at 10 and 30 minutes, and Smad3 phosphorylation was undetectable at 60 minutes. Also, Smad2 phosphorylation was slightly attenuated in breast cancer cells with decreased Notch3 expression, although expression levels of Smad3 and Smad2 protein remained unchanged (Figure 3E). To control for the possibility that the selected MET-shN3 and MET-ctrl clones were not representative, we studied the effect of TGFβ1 on MDA-MET bulk cell populations with decreased Notch3 level caused by shRNA lentiviral transduction. The results confirmed the marked inhibition of TGFβ1-stimulated Smad3 phosphorylation by reduction of Notch3 expression (data not shown).
Figure 3.
Positive feedback between TGFβ and Notch3 signaling. A: The hBMOB-stimulated colony formation by MDA-MET cells is inhibited by anti-TGFβ1 blocking antibody at the indicated concentrations; **P < 0.01 (n = 4). B: qRT-PCR analysis of Notch signaling molecules in the presence or absence of TGFβ1. MDA-MET and MDA-MB-435 cells were treated with TGFβ1 at 1 ng/ml for two and five hours, respectively. The cells were lysed, and relative mRNA levels were determined for Notch1, Notch3, and Jagged1 and for the transcription factors Hes-1 and Hey-1. Cells treated with vehicle alone served as normalization controls. C: Decreased Notch3 expression (MET-shN3) inhibits the TGFβ1-stimulated colony formation. Cultures treated with vehicle alone served as the control; *P < 0.05 and **P < 0.01 (n = 4). D: Effect of Notch3 knockdown on the invasiveness of MDA-MET cells. After 48 hours of incubation in the presence of TGFβ1 or vehicle, the invaded cells were fixed, stained, and counted. Mean values for three randomly selected fields were obtained for each well. Data are expressed as mean ± SEM (n = 3 wells) and represent two independent experiments. E: Western blot analysis showing the decreased phosphorylation of Smad3 and Smad2 in MET-shN3 cells versus MET-ctrl following treatment with 1 ng/ml TGFβ1 for 0 to 60 minutes.
TGFβ1 stimulation of colony formation was detected over a dose range of 0.01 to 10 ng/ml, and exhibited a maximum at 1 ng/ml, which established the dose used in these experiments. To determine the level of TGFβ1 released from hBMOB, ELISA was performed on conditioned media and the TGFβ1 background in control media was subtracted. Cultured confluent hBMOB osteoblasts were measured to release ∼50 to 250 pg/ml TGFβ1 in 24 hours (Supplemental Figure 3, see http://ajp.amjpathol.org). Therefore, our in vitro results indicate that TGFβ1 released from osteoblasts enhances the breast cancer cell expression of Notch3, which facilitates TGFβ signaling and TGFβ-stimulated colony formation and cancer cell invasion.
Notch3 Expression and Osteolytic Bone Metastasis
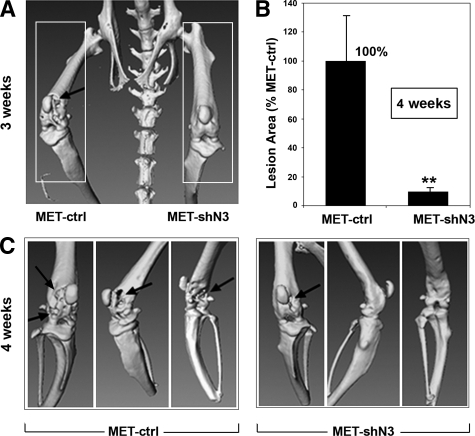
To further investigate the pathophysiological function of Notch3 in vivo, we inoculated MDA-MET cells expressing Notch3-specific shRNA (MET-shN3) or control plasmid (MET-ctrl) directly into the distal bone marrow cavities of the left and right femurs, respectively, of athymic mice. This procedure allows comparison of tumor-mediated osteolytic potentials within one animal, enabling testing of the role of Notch3 in the interaction between breast cancer cells and the bone microenvironment. Three weeks after tumor inoculation, femurs bearing MET-ctrl cells exhibited significant osteolytic lesions as detected by three-dimensional microCT scanning, whereas bones bearing MET-shN3 cells with Notch3 knockdown appeared normal (Figure 4A). Four weeks after tumor inoculation, bones bearing MET-ctrl cells manifested severe osteolysis, whereas only mild osteolysis was observed in bones bearing MET-shN3 cells (Figure 4, B and C).
Figure 4.
Notch3 and intraosseous tumor growth. MDA-MET cells expressing Notch3-specific shRNA (MET-shN3) or control shRNA (MET-ctrl) were injected into bone marrow cavities of left and right femurs, respectively (n = 6 mice/group). A: Representative microCT scanning (anterior view) was performed three weeks after tumor cell inoculation. Note the extensive osteolytic lesions in the cortical bone of the distal femurs inoculated with MET-ctrl, whereas no significant osteolysis followed MET-shN3 inoculation. B and C: Osteolysis determined four weeks after tumor cell inoculation. B: Quantification of the radiolucent lesion area. Bones bearing MET-ctrl served as 100% control; **P < 0.01. C: Representative microCT images showing dorsal view and two lateral views. Note the mild osteolytic lesions by MET-shN3 inoculation compared with MET-ctrl (arrows).
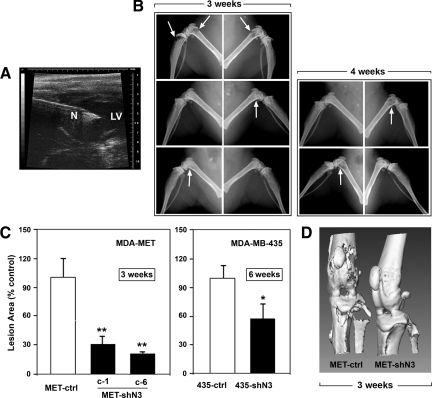
To model the process of bone metastasis by circulating tumor cells,40 we inoculated tumor cells into the left ventricles of athymic mice with ultrasound guidance (Figure 5A). As shown in Figure 5B (left panel), mice that received MET-ctrl cells (top) exhibited severe osteolysis in both femurs and tibias 3 weeks after the injection. In contrast, decreased Notch3 expression in both clone 1 and clone 6 of MET-shN3 cells significantly inhibited the bone destruction, and only mild osteolysis was detected 3 weeks after the injection. Osteolytic lesions from MET-shN3 cells required 4 weeks to develop (Figure 5B, right panel). Quantitation of radiolucent osteolytic area in hindlimbs 3 weeks after tumor inoculation showed significantly decreased osteolysis in mice bearing either clone-1 or clone-6 of MET-shN3 cells, compared with mice receiving MET-ctrl cells (Figure 5C, left panel). Furthermore, three-dimensional micro-CT images of hindlimbs showed numerous perforations of cortical bone around the knee in the MET-ctrl group. Decreased Notch3 expression significantly reduced the bone destruction in the MET-shN3 group and the cortical bone surface remained intact (Figure 5D).
Figure 5.
Notch3 and osteolytic bone metastasis. A: Bone metastasis was modeled by intracardiac tumor cell injection. Two-dimensional ultrasound guidance image showing the left ventricle (LV), and the tip of the needle (N) positioned in the LV for tumor cell injection. B: Representative radiographs of hindlimbs three weeks (left panel) and four weeks (right panel) after tumor inoculation. Mice were inoculated with MET-ctrl (top), MET-shN3 (clone-1, middle), and MET-shN3 (clone-6, bottom). Sites with osteolytic lesions are indicated by arrows. C: Quantitation of the osteolytic lesion areas in both femur and tibia, three weeks after inoculation of MDA-MET (left panel) and six weeks after inoculation of MDA-MB-435 (right panel) expressing Notch3-specific shRNA versus control shRNA; n = 4–7 mice/group; *P < 0.05 and **P < 0.01. D: Representative microCT images of the knee joint region of hind limbs 3 weeks after intracardiac injection of tumor cells. Note the strong attenuation of metastatic osteolysis by knockdown of Notch3.
Similar experiments were performed with another human breast cancer cell line. MDA-MB-435 cells expressing Notch3-specific shRNA (435-shN3) or control shRNA (435-ctrl) were inoculated into left ventricles of athymic mice. Six weeks after the inoculation, decreased osteolysis was detected in mice bearing 435-shN3 compared with mice bearing 435-ctrl cells (Figure 5C, right panel).
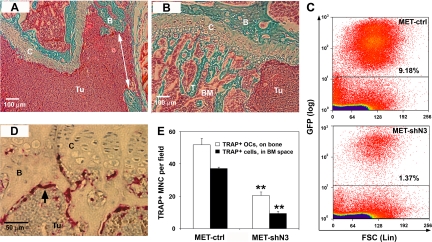
Histological analysis was performed on the distal femurs 3 weeks after the intracardiac injection. In mice injected with MET-ctrl cells, the bone marrow cavity was filled with breast cancer cells with few bone trabeculae remaining. The normal structure of cartilage was also destroyed, and perforated cortical bone was observed in the metaphyseal area (Figure 6A). Mice bearing MET-shN3 cells exhibited markedly reduced tumor burden, and the normal structure of the epiphyseal growth plate cartilage and bone marrow cavity was partially preserved (Figure 6B). This reduced tumor cell burden in MET-shN3 mice was confirmed by FACS analysis of eGFP+ tumor cells. Although equal numbers of cells had been inoculated, by 2 weeks the total number of MET-shN3 cells in the femoral bone marrow was reduced by about sixfold compared with MET-ctrl mice (Figure 6C).
Figure 6.
Histological analysis of bone metastasis. Three weeks after intracardiac tumor cell inoculation, hind limbs were fixed, decalcified, and embedded in paraffin. A and B: Goldner’s Trichrome staining of distal femurs from MET-ctrl (A) and MET-shN3 (B) recipient mice. Note that in bones bearing MET-ctrl cells (A), the bone marrow cavity is filled with tumor cells (Tu, solid red), and the cortical bone (green) has been perforated (arrow), whereas in bones with MET-shN3 (B), the tumor burden is significantly decreased with preservation of growth plate cartilage and trabecular bone. Tissue labels: B, bone; BM, bone marrow; C, cartilage; T, bone trabecular bone; Tu, tumor. C: Two weeks after the injection, single-cell suspensions of femoral bone marrow were analyzed for eGFP-positive MET cells by flow cytometry. Note that Notch3 knockdown reduces the tumor cell population by over sixfold. D: TRAP-eosin staining of bone sections bearing MET-ctrl cells. Numerous TRAP-positive osteoclasts (red, arrow) are observed at the trabecular bone surface adjacent to the tumor cells of the metastatic site. E: Quantitation of osteoclasts in the distal femurs of mice (n = 2/group) 3 weeks after intracardiac injection of MET-ctrl versus MET-shN3 tumor cells. Cells were counted by light microscopy using a ×15 objective and a standard field of the distal femur; serial sections (n = 3) were examined. TRAP+ multinucleated (MNC) osteoclasts on mineralized surface are decreased ∼2.5-fold by Notch3 knockdown. Other TRAP+ cells in the bone marrow space, possibly osteoclast precursors, are decreased ∼3.7-fold; **P < 0.01.
Osteoclasts are the major bone-resorbing cells activated in tumor osteolysis. In mice injected with MET-ctrl, large numbers of TRAP+ osteoclasts were observed on bone surfaces at the margin of the tumor mass (Figure 6D). Comparative histomorphometry of distal femurs from MET-ctrl and MET-shN3 groups showed that Notch3 knockdown caused significantly fewer TRAP+ osteoclasts to be present on the mineralized surfaces, as well as in the bone marrow space (Figure 6E).
Discussion
Although tumor-specific gene mutations and tumor cell phenotype are major determinants of metatastic efficiency, understanding the unique role the bone microenvironment plays in metastasis is essential for treating and preventing bone-metastatic breast cancer. The present work reveals a novel osteoblast activation pathway for breast cancer, involving Notch3 signaling in breast cancer cells driven by TGFβ1 from osteoblasts. This paracrine pathway contributes to aggressive osteolytic metastasis and bone destruction in vivo.
Growing Role for Notch3 in Breast Cancer
There is increasing evidence suggesting a role for Notch3 in breast cancer development. Notch3 has been identified as an oncogene that is amplified in primary breast tumors.41 Furthermore, the 66-kDa isoform of the Shc (Src homology 2 domain containing) gene, which is induced in hypoxic breast cancer mammospheres controls Notch3 and Jagged1 expression, thereby modulating self-renewal of human breast cancer cells.31 Also, interleukin 6 has been shown to regulate a Notch3-dependent signaling pathway that promotes self-renewal, hypoxic survival, and the invasive potential of mammosphere cells derived from aggressive ductal breast carcinomas.37 In addition, enhanced Notch3 expression, rather than other Notch receptors, is detected in an inflammatory breast cancer model.42
Unlike the reported activation of Notch signaling in hematopoietic stem cells that may be regulated by endosteal osteoblasts expressing Jagged1 on the cell surface,22 the osteoblast-mediated enhancement of Notch3 signaling in breast cancer cells does not appear to require direct cell-cell contact (Figure 1, C and D). Reduced expression of Notch3 also inhibited the Jagged1 expression in MDA-MET breast cancer cells, whereas expression of other Notch signaling molecules was unaffected (Figure 2A). This finding agrees with recent reports that Notch3 up-regulates its own ligand Jagged1 in human primary mammospheres from normal and ductal breast carcinoma and in MCF7 breast cancer cells.31,37 Administration of an antibody that inhibits the Notch3/Jagged1 interaction was shown to reduce the self-renewal of mammospheres.31 Thus, the activation of Notch3 signaling may involve an autocrine or paracrine Jagged1/Notch3 canonical ligand-receptor interaction within clusters of breast cancer cells, thereby promoting their tumorigenic properties and metastasis.
Our results suggest that Notch3 signaling regulates bone metastasis of breast cancer. Decreased Notch3 expression in breast cancer cells significantly reduced the osteolytic lesions in athymic mice using two independent routes of inoculation: intrafemoral bone marrow space, or systemic circulation via the left ventricle. Importantly, our Notch3 knockdown strategy did not affect cancer cell proliferation rate or viability in vitro yet caused a dramatic sixfold decrease in MET-shN3 cell recovery from the femoral compartment compared with MET-ctrl cells (Figure 6C).
Role of the Osteoblast/TGFβ Axis in Tumor Progression
TGFβ secreted by osteoblasts in the bone microenvironment plays major roles in osteolytic bone metastasis of breast cancer.18,19,20,21,39,43 A functional interaction between TGFβ family growth factors and Notch pathways has recently been described in other cell systems.38,44 We showed here that TGFβ1 enhances the expression of Notch3 and the Notch target gene Hes-1. Knockdown of Notch3 expression inhibits the TGFβ1-stimulated colony formation and phosphorylation of Smad3/Smad2, suggesting that the enhancement of bone metastasis by TGFβ1 is mediated through activation of the Notch3 signaling pathway.
Notch3 and Its Special Role in HER2− Breast Cancer
Notch3 signaling appears to have a unique cell proliferative and antiapoptotic role in HER2− but not in HER2+ breast cancer.35,45 HER2− incidence is 70 to 80% in primary and metastatic disease. A highly malignant subset of HER2− breast cancer is “triple negative” (ER−, PR−(Progesterone Receptor-negative), and HER2−), and these patients have few therapeutic options and a very poor prognosis.46 The human breast carcinoma cell lines in the present study are all HER2−: MDA-MB-231, MDA-MET,28 and MDA-MB-435.29,47 We observed that Notch3 expression level positively regulates the anchorage-independent colony formation in soft agar by these HER2− breast cancer cells in response to osteoblasts and their secreted prometastatic factor TGFβ1. Decreased Notch3 expression also inhibits tumor growth in bone and bone metastasis modeled in athymic mice by intraosseous and intracardiac tumor cell inoculations. Thus, characterization of Notch3 function in breast cancer bone metastasis could lead to better therapies for the majority of patients who suffer from HER2− disease.
In summary, our findings suggest that an increase in Notch3 expression, induced by osteoblasts and osteoblast-derived TGFβ1, contributes to the bone metastasis of breast cancer. Interference with this paracrine pathway in bone may represent a new therapeutic approach.
Acknowledgments
We thank Massimiliano Bonafe and Catia Giovannini (University of Bologna), Jiang Yang (Children’s Hospital Boston), Thao P. Dang (Vanderbilt University), and Josef Zustin (Institute of Pathology, Universitätsklinikum Hamburg-Eppendorf (UKE), Hamburg, Germany) for helpful discussions and reagents. We thank Patricia Dunning, Erxi Wu, Kenneth M. Kozloff, Ye Sun, and Chaoshe Guo for technical advice and support.
Footnotes
Address reprint requests to Peter V. Hauschka, Ph.D., Department Orthopaedic Surgery, Children’s Hospital Boston, 300 Longwood Ave., Enders- 226, Boston, MA 02115. E-mail: peter.hauschka@childrens.harvard.edu.
Supported by grant BCTR0600935 from Susan G. Komen for the Cure, and the Department of Defense/Congressionally Directed Medical Research Program (CDMRP) Breast Cancer Research Program under grants managed by the U.S. Army Research and Materiel Command (W81XWH-06-1-0626, W81XWH-06-1-0422, and W81XWH-09-1-0292).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Farnie G, Clarke RB. Mammary stem cells and breast cancer—role of Notch signalling. Stem Cell Rev. 2007;3:169–175. doi: 10.1007/s12015-007-0023-5. [DOI] [PubMed] [Google Scholar]
- Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107:2223–2233. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- Hu C, Dievart A, Lupien M, Calvo E, Tremblay G, Jolicoeur P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am J Pathol. 2006;168:973–990. doi: 10.2353/ajpath.2006.050416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi K, Feirt N, Kitajewski J. Notch in mammary gland development and breast cancer. Semin Cancer Biol. 2004;14:341–347. doi: 10.1016/j.semcancer.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004;6:R605–R615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, Lockwood G, Egan SE. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- Dickson BC, Mulligan AM, Zhang H, Lockwood G, O'Malley FP, Egan SE, Reedijk M. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod Pathol. 2007;20:685–693. doi: 10.1038/modpathol.3800785. [DOI] [PubMed] [Google Scholar]
- Parr C, Watkins G, Jiang WG. The possible correlation of Notch-1 and Notch-2 with clinical outcome and tumour clinicopathological parameters in human breast cancer. Int J Mol Med. 2004;14:779–786. doi: 10.3892/ijmm.14.5.779. [DOI] [PubMed] [Google Scholar]
- Vincent-Salomon A, Thiery JP. Host microenvironment in breast cancer development: epithelial-mesenchymal transition in breast cancer development. Breast Cancer Res. 2003;5:101–106. doi: 10.1186/bcr578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong KG, Niessen K, Kulic I, Raouf A, Eaves C, Pollet I, Karsan A. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J Exp Med. 2007;204:2935–2948. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Yang J, Bielenberg DR, Rodig SJ, Doiron R, Clifton MC, Kung AL, Strong RK, Zurakowski D, Moses MA. Lipocalin 2 promotes breast cancer progression. Proc Natl Acad Sci USA. 2009;106:3913–3918. doi: 10.1073/pnas.0810617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci USA. 2008;105:6392–6397. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlow W, Guise TA. Breast cancer metastasis to bone: mechanisms of osteolysis and implications for therapy. J Mammary Gland Biol Neoplasia. 2005;10:169–180. doi: 10.1007/s10911-005-5399-8. [DOI] [PubMed] [Google Scholar]
- Yin JJ, Selander K, Chirgwin JM, Dallas M, Grubbs BG, Wieser R, Massague J, Mundy GR, Guise TA. TGF-β signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J Clin Invest. 1999;103:197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakonen SM, Selander KS, Chirgwin JM, Yin JJ, Burns S, Rankin WA, Grubbs BG, Dallas M, Cui Y, Guise TA. Transforming growth factor-β stimulates parathyroid hormone-related protein and osteolytic metastases via Smad and mitogen-activated protein kinase signaling pathways. J Biol Chem. 2002;277:24571–24578. doi: 10.1074/jbc.M202561200. [DOI] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massague J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Ehata S, Hanyu A, Fujime M, Katsuno Y, Fukunaga E, Goto K, Ishikawa Y, Nomura K, Yokoo H, Shimizu T, Ogata E, Miyazono K, Shimizu K, Imamura T. Ki26894, a novel transforming growth factor-β type I receptor kinase inhibitor, inhibits in vitro invasion and in vivo bone metastasis of a human breast cancer cell line. Cancer Sci. 2007;98:127–133. doi: 10.1111/j.1349-7006.2006.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, Milner LA, Kronenberg HM, Scadden DT. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- Arai F, Hirao A, Ohmura M, Sato H, Matsuoka S, Takubo K, Ito K, Koh GY, Suda T. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118:149–161. doi: 10.1016/j.cell.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Mancini SJ, Mantei N, Dumortier A, Suter U, MacDonald HR, Radtke F. Jagged1-dependent Notch signaling is dispensable for hematopoietic stem cell self-renewal and differentiation. Blood. 2005;105:2340–2342. doi: 10.1182/blood-2004-08-3207. [DOI] [PubMed] [Google Scholar]
- Nefedova Y, Cheng P, Alsina M, Dalton WS, Gabrilovich DI. Involvement of Notch-1 signaling in bone marrow stroma-mediated de novo drug resistance of myeloma and other malignant lymphoid cell lines. Blood. 2004;103:3503–3510. doi: 10.1182/blood-2003-07-2340. [DOI] [PubMed] [Google Scholar]
- Meury T, Verrier S, Alini M. Human endothelial cells inhibit BMSC differentiation into mature osteoblasts in vitro by interfering with osterix expression. J Cell Biochem. 2006;98:992–1006. doi: 10.1002/jcb.20818. [DOI] [PubMed] [Google Scholar]
- Duarte WR, Shibata T, Takenaga K, Takahashi E, Kubota K, Ohya K, Ishikawa I, Yamauchi M, Kasugai S. S100A4: a novel negative regulator of mineralization and osteoblast differentiation. J Bone Miner Res. 2003;18:493–501. doi: 10.1359/jbmr.2003.18.3.493. [DOI] [PubMed] [Google Scholar]
- Bendre MS, Gaddy-Kurten D, Mon-Foote T, Akel NS, Skinner RA, Nicholas RW, Suva LJ. Expression of interleukin 8 and not parathyroid hormone-related protein by human breast cancer cells correlates with bone metastasis in vivo. Cancer Res. 2002;62:5571–5579. [PubMed] [Google Scholar]
- Phadke PA, Mercer RR, Harms JF, Jia Y, Frost AR, Jewell JL, Bussard KM, Nelson S, Moore C, Kappes JC, Gay CV, Mastro AM, Welch DR. Kinetics of metastatic breast cancer cell trafficking in bone. Clin Cancer Res. 2006;12:1431–1440. doi: 10.1158/1078-0432.CCR-05-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- Sansone P, Storci G, Giovannini C, Pandolfi S, Pianetti S, Taffurelli M, Santini D, Ceccarelli C, Chieco P, Bonafe M. p66Shc/Notch-3 interplay controls self-renewal and hypoxia survival in human stem/progenitor cells of the mammary gland expanded in vitro as mammospheres. Stem Cells. 2007;25:807–815. doi: 10.1634/stemcells.2006-0442. [DOI] [PubMed] [Google Scholar]
- Bartholin L, Wessner LL, Chirgwin JM, Guise TA. The human Cyr61 gene is a transcriptional target of transforming growth factor β in cancer cells. Cancer Lett. 2007;246:230–236. doi: 10.1016/j.canlet.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Kingsley LA, Fournier PG, Chirgwin JM, Guise TA. Molecular biology of bone metastasis. Mol Cancer Ther. 2007;6:2609–2617. doi: 10.1158/1535-7163.MCT-07-0234. [DOI] [PubMed] [Google Scholar]
- Studebaker AW, Storci G, Werbeck JL, Sansone P, Sasser AK, Tavolari S, Huang T, Chan MW, Marini FC, Rosol TJ, Bonafe M, Hall BM. Fibroblasts isolated from common sites of breast cancer metastasis enhance cancer cell growth rates and invasiveness in an interleukin-6-dependent manner. Cancer Res. 2008;68:9087–9095. doi: 10.1158/0008-5472.CAN-08-0400. [DOI] [PubMed] [Google Scholar]
- Yamaguchi N, Oyama T, Ito E, Satoh H, Azuma S, Hayashi M, Shimizu K, Honma R, Yanagisawa Y, Nishikawa A, Kawamura M, Imai J, Ohwada S, Tatsuta K, Inoue J, Semba K, Watanabe S. NOTCH3 signaling pathway plays crucial roles in the proliferation of ErbB2-negative human breast cancer cells. Cancer Res. 2008;68:1881–1888. doi: 10.1158/0008-5472.CAN-07-1597. [DOI] [PubMed] [Google Scholar]
- Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P, Marcu KB, Chieco P, Bonafe M. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavadil J, Cermak L, Soto-Nieves N, Bottinger EP. Integration of TGF-beta/Smad and Jagged1/Notch signalling in epithelial-to-mesenchymal transition. EMBO J. 2004;23:1155–1165. doi: 10.1038/sj.emboj.7600069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guise TA, Chirgwin JM. Transforming growth factor-β in osteolytic breast cancer bone metastases. Clin Orthop Relat Res. 2003:S32–S38. doi: 10.1097/01.blo.0000093055.96273.69. [DOI] [PubMed] [Google Scholar]
- Arguello F, Baggs RB, Frantz CN. A murine model of experimental metastasis to bone and bone marrow. Cancer Res. 1988;48:6876–6881. [PubMed] [Google Scholar]
- Kadota M, Sato M, Duncan B, Ooshima A, Yang HH, Diaz-Meyer N, Gere S, Kageyama S, Fukuoka J, Nagata T, Tsukada K, Dunn BK, Wakefield LM, Lee MP. Identification of novel gene amplifications in breast cancer and coexistence of gene amplification with an activating mutation of PIK3CA. Cancer Res. 2009;69:7357–7365. doi: 10.1158/0008-5472.CAN-09-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Ye Y, Yearsley K, Jones S, Barsky SH. The lymphovascular embolus of inflammatory breast cancer expresses a stem cell-like phenotype. Am J Pathol. 2008;173:561–574. doi: 10.2353/ajpath.2008.071214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Hiraga T. Crosstalk between cancer cells and bone microenvironment in bone metastasis. Biochem Biophys Res Commun. 2005;328:679–687. doi: 10.1016/j.bbrc.2004.11.070. [DOI] [PubMed] [Google Scholar]
- Blokzijl A, Dahlqvist C, Reissmann E, Falk A, Moliner A, Lendahl U, Ibanez CF. Cross-talk between the Notch and TGF-β signaling pathways mediated by interaction of the Notch intracellular domain with Smad3. J Cell Biol. 2003;163:723–728. doi: 10.1083/jcb.200305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose H, Ishii H, Mimori K, Ohta D, Ohkuma M, Tsujii H, Saito T, Sekimoto M, Doki Y, Mori M. Notch pathway as candidate therapeutic target in Her2/Neu/ErbB2 receptor-negative breast tumors. Oncol Rep. 2010;23:35–43. [PubMed] [Google Scholar]
- Nishimura R, Arima N. Is triple negative a prognostic factor in breast cancer? Breast Cancer. 2008;15:303–308. doi: 10.1007/s12282-008-0042-3. [DOI] [PubMed] [Google Scholar]
- Harms JF, Welch DR. MDA-MB-435 human breast carcinoma metastasis to bone. Clin Exp Metastasis. 2003;20:327–334. doi: 10.1023/a:1024062911144. [DOI] [PubMed] [Google Scholar]