Abstract
PRDM1/Blimp-1, a master regulator for B cell terminal differentiation, is a putative tumor suppressor in diffuse large B cell lymphomas (DLBCL). Inactivating mutations of PRDM1 have been previously identified in a subset of nongerminal center B cell–like (GCB) DLBCL. We investigated the presence of alternative mechanisms of down-regulating PRDM1 in a cohort of 25 primary DLBCL and six DLBCL cell lines. While some DLBCL, predominantly the GCB-type, showed low levels of both PRDM1α mRNA and protein, presumably as a result of direct transcription repression, discordant expressions between the two were identified in a subset of DLBCL without PRDM1 mutations, the primarily non-GCB type, consistent with translational down-regulation. This subset of DLBCL exhibits relatively high PRDM1α mRNA levels but low levels of PRDM1. Data obtained from expression analysis, luciferase reporter assays, and transfection experiments support a role of targeting of PRDM1 by microRNA let-7 family in mediating this down-regulation. Let-7, in particular let-7b, is overexpressed in DLBCL relative to normal GCB cells, suggesting that it is deregulated. Thus, abnormal epigenetic down-regulation of PRDM1 by let-7 and other microRNAs may represent an alternative mechanism of reducing normal PRDM1 function in a subset of DLBCL with relatively high PRDM1α mRNA expression and unmutated PRDM1. These findings provide further evidence for an important role of impairment of terminal B cell differentiation in DLBCL pathogenesis.
PRDM1/Blimp-1 is a master transcriptional regulator for terminal differentiation of B cells into plasma cells.1,2,3,4 It also plays a critical role in effector and memory T-cell differentiation,1,3,5 myeloid cell development,6 dendritic cell development,7 and epithelial cell differentiation.8 Inactivating mutations that result in the generation of a severely truncated nonfunctional PRDM1 are found in about 20% of the diffuse large B cell lymphomas (DLBCLs) of the nongerminal center B cell (GCB) subtype9,10 and also in about 19% of the primary DLBCL of the central nervous system.11 These mutations are associated with deletion of the other paired PRDM1 allele at chromosome 6q21. These findings are consistent with PRDM1 being a DLBCL tumor suppressor gene and imply an important role of impairment of terminal B cell differentiation in DLBCL pathogenesis. However, many DLBCLs lack genetic alterations in PRDM1. Posttranscriptional down-regulation of PRDM1 in DLBCL has been suggested, but no systematic evaluation was performed.10 In this study, we investigated this possibility in detail by comparative and quantitative analysis of PRDM1 mRNA and protein expression in different subgroups of DLBCL. We concluded that low or absent PRDM1 expression is a common phenomenon in DLBCL and documented the presence of translational down-regulation of PRDM1 in a subset of DLBCL by comparing PRDM1 mRNA and protein levels in these tumors. Recently it was demonstrated that the low levels of PRDM1 in Reed-Sternberg/Hodgkin cells in Hodgkin lymphoma can be at least partially attributed to translation inhibition by microRNAs (miRNAs) inside the cells,12 which are ∼22 nt RNAs that regulate gene expression by inhibiting protein translation and/or promoting mRNA degradation through binding to specific target sites in the 3′ untranslated region (UTR).13 In this study, we demonstrated that overexpression of let-7 may contribute to the down-regulation of PRDM1 expression in DLBCL by translation inhibition.
Materials and Methods
Patient Tissue Samples, Cell Lines, and Normal B Cells
Frozen archival tissue of 25 de novo DLBCL cases were obtained according to the protocols approved by the Institutional Review Board of Weill Medical College of Cornell University. All cases were reviewed and classified according to World Health Organization classification. All cases show >80% of tumor content. Classification into GCB and non-GCB types was performed according to Hans et al14 using immunohistochemical staining of paraffin-embedded formalin-fixed tissues sections against BCL6, CD10, and IRF4/MUM1. DLBCL cell lines (OCI-Ly1, OCI-LY7, SUDHL2, SUDHL6, and U2932) and myeloma cell line U266 were cultured in RPMI medium 1640 with 10% heat-inactivated fetal calf serum (Invitrogen, Carlsbad, CA). OCI-Ly3 was grown in Iscove modified Dulbecco medium supplemented with 10% human serum (NABI Biopharmaceuticals, Boca Raton, FL). Normal B cell subsets were sorted from fresh tonsillar CD19+ B cells by flow cytometry into naïve B cells (IgD+CD38−), GBC (IgD−CD38+), and memory B cells (IgD−CD38−). Three independent isolates were obtained from each B cell subset. Plasma cells were purchased from Stem Cell Technologies (Vancouver, Canada). Myeloma cells were isolated from frozen bone marrow aspirate samples of five different cases by CD138 immunomagnetic positive selection (Stem Cell Technologies, Vancouver, Canada).
Nucleic Acid Extraction
Total RNA was extracted from frozen tissue sections or tissue culture cells using the mirVana miRNA Isolation Kit (ABI/Ambion, Texas, USA) following the manufacturer’s protocol. High-molecular-weight genomic RNA was isolated using the salting-out method as described.
Quantitative Real-Time Reverse-Transcriptase PCR
Quantitative detection of PRDM1α mRNA was performed as previously described.12 Conditions of qRT-PCR for measurement of CD3ε expression are identical to those used for PRDM1, except that the primers and probes for real-time detection were purchased from Applied Biosystems as predeveloped assays.
MicroRNA Cloning and Quantitation
MicroRNA cloning was performed as previously described.15,16 Let-7a, let-7b, and let-7f expression were measured by Taqman MicroRNA Assay (Applied Biosystems) following manufacturer’s recommended protocols, using U6 RNA for normalization as determined by modified Invader assays.17 For each sample, three independent measurements were made with triplicates for each measurement. Relative levels were calculated with respect to let-7 content in normal plasma cells.
Antibodies
Monoclonal PRDM1 antibody clone ROS (ref) was kindly provided by Dr. Jose Francisco Garcia at Dako Diagnostics S.A., Barcelona, Spain. Other antibodies used were: PRDM1 (clone 3H2EB, Santa Cruz Biotechnology, Santa Cruz, CA), BCL-6 (clone PG-B6P, DAKO, USA, dilution 1:10), CD10 (clone 56C6, Vector Laboratories, Burlingame, CA, dilution 1: 30), MUM-1/IRF4 (polyclonal antibody, DAKO, USA, dilution 1:100).
Western Blotting and Immunohistochemistry
Immunoblotting was performed as previously described12 on total or nuclear extracts using clone ROS (1:1) and clone 3H2EB (1:200), respectively. Quantification of PRDM1α expression in the Western blot was done by densitometry and normalized with β-actin or lamin B loading control.
Immunohistochemistry for PRDM1 on paraffin-embedded formalin-fixed tissues was performed using the PRDM1 monoclonal antibody (clone ROS, 1:8) as described,18 except that primary and postprimary incubations were performed using the Bond Polymer Refine Detection System (Vision Biosystems, Bannockburn, IL).
Luciferase Reporter Assays
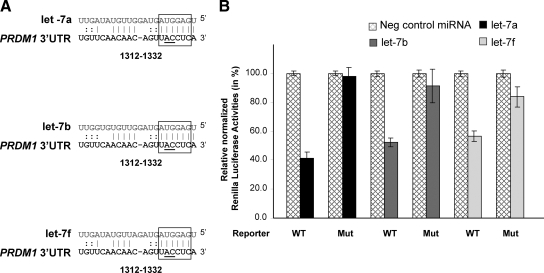
293T cells were transfected with 20 nmol/L of let-7a, let-7b, let-7f, or miRNA negative control (purchased from Ambion, Austin, TX) in 24-well plates using Lipofectin (Invitrogen). These mmiRNA analogs were cotransfected with 25 ng of pSIC.PRDM1.3′ UTR.538–2419 (wild-type or mutant). Construction of these plasmids has been described in detail.12 Briefly, the wild-type plasmid carries nucleotides 538 to 2419 of the PRDM1 3′ UTR downstream of the reporter gene. The mutant plasmid carries the same 3′ UTR genomic fragment, except that the fifth and sixth positions (from the 3′ end) of the putative let-7 binding site are mutated. Luciferase activities were measured 24 hours after transfection as described.
Bisulfite Sequencing
Promoter methylation in PRDM1 was assessed by bisulfite sequencing. Bisulfite-modified DNA from GCB-type primary DLBCL and DLBCL cell lines was amplified using a primer pair that contains no CpG sites. These primers were designed using the MethPrimer program19 and have the following sequences: 5′-GTGTTAGTAATTTGGGGGAAAGTTT-3′, forward primer; 5′-CAAACAAATATCCAACATCTAAAAAAAA-3′, reverse primer. This amplicon derived from these primers spans a sequence of 361 bp that contains 32 CpG sites within a CG island predicted at the PRDM1 promoter. The amplified sequence was cloned into pGEM-T cloning vector (Promega, Madison, WI), and at least 10 clones were sequenced per sample. The methylation status of each CpG site for each clone is indicated.
Results
Discordance of PRDM1 mRNA and Protein Expressions in Primary DLBCL
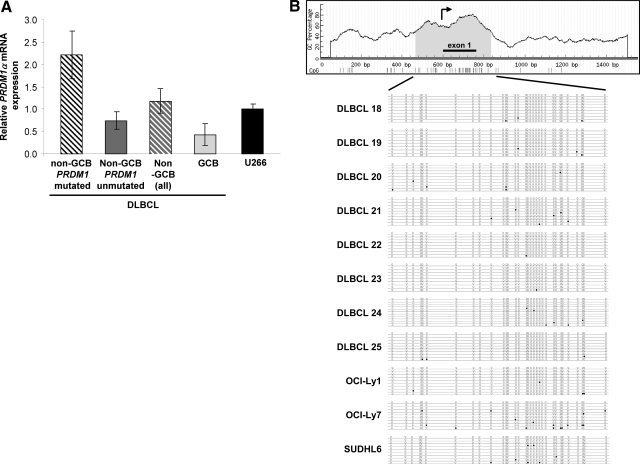
We analyzed PRDM1α mRNA expression by quantitative real-time PCR in five DLBCL with PRDM1 inactivating mutations, 12 non–GCB-type DLBCL without PRDM1 mutations, and eight GCB-type DLBCL (see Supplemental Table S1 at http://ajp.amjpathol.org). Overall, the non-GCB DLBCL cases tend to have a higher PRDM1α mRNA compared with GCB DLBCL (P = 0.08). In addition, the PRDM1-mutated subset of the non-GCB DLBCL cases have significantly higher levels of PRDM1α mRNA compared with the other two groups (P = 0.005) (Figure 1A). To ensure that the observed differences in PRDM1a mRNA are intrinsic to B cells and not due to variation in the quantity of reactive T cells in the biopsy specimen, the T cell contents of these representative cases were estimated by quantitative RT-PCR for CD3, which showed that they were low and similar among cases (see Supplemental Figure S1 at http://ajp.amjpathol.org). The low level of PRDM1α expression in GCB-type DLBCL is not a result of promoter hypermethylation (Figure 1B).
Figure 1.
A: Higher levels of PRDM1α mRNA in non-GCB-type DLBCL. Mean levels of PRDM1α transcripts in different groups of DLBCL (U266 = 1). Non-GCB DLBCL have higher PRDM1α mRNA expression compared with GCB DLBCL. B: Absence of PRDM1 promoter hypermethylation in GCB-type primary DLBCL and DLBCL cell lines. The CpG island identified in the PRDM1 promoter is depicted on the top of the figure (shaded). The approximate transcription start site is indicated by an arrow. A 361-bp sequence containing a total of 32 CpG sites within the CG island was amplified by a pair of primers that do not contain CpG sites from DNA extracted from GCB-type primary DLBCL (#18 to #25) and DLBCL cell lines (OCI-LY1, OCI-Ly7 and SUDHL-6). At least 10 clones were sequenced per sample, and the methylation status of each CpG site for each clone was indicated in the lollipop diagram. The open circle denotes an unmethylated CpG; the filled circle denotes a methylated CpG. None of the samples demonstrated promoter hypermethlyation in the PRDM1 gene.
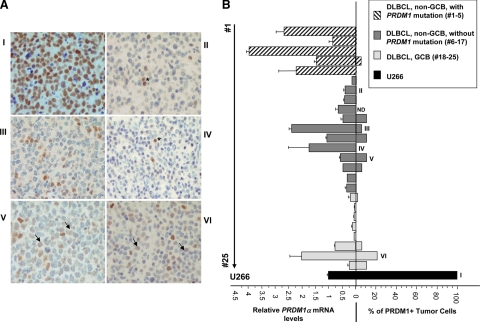
PRDM1α protein expression in these DLBCL cases was assessed by immunohistochemistry using the monoclonal anti-PRDM1 antibody. Though this antibody recognizes both PRDM1 isoforms, this immunohistochemical analysis was essentially a measure of the quantity of the PRDM1α isoform because PRDM1β is a minor isoform in DLBCL.20 Under our immunohistochemical conditions, all but one DLBCL showed PRDM1 positivity in less than 20% of tumor cells, with staining intensities often distinctly weaker compared with the myeloma U266 cells as well as the reactive T cells (see examples in Figure 2A). A previous study also demonstrated negative or weak PRDM1 staining in most DLBCL.18 Comparison of these immunohistochemistry results with the RNA expression data revealed a discordance between PRDM1α mRNA and protein expressions in virtually all of the non–GCB-type DLBCL. The lack of detectable full length PRDM1α expression in those DLBCL with PRDM1 mutations, which have high levels of PRDM1α mRNA, is expected as these mutations are inactivating and generate severe protein truncations.9,10 In the other cases without PRDM1 mutations, the degree of PRDM1α mRNA expression does not translate to the same extent of PRDM1 protein expression (Figure 2B), suggesting a translational or posttranslational down-regulation. This is well illustrated by cases #7, 11, 13, and 14 (II to IV, Figure 2B), in which the levels of PRDM1α mRNA is at least 40% that of U266 but the PRDM1 levels are markedly lower relative to U266. Overall, in the PRDM1-unmutated non-GCB DLBCL group, which has a mean PRDM1α mRNA level of 74% (±19%) that of U266, the mean percentage of PRDM1(+) tumor cells is only less than 5%.
Figure 2.
Discordance between PRDM1 mRNA and protein expressions in DLBCL. A: Examples of immunohistochemistry for PRDM1 in U266 myeloma cells (I) and selected DLBCL cases (II–VI). Under these immunohistochemical conditions, most of the DLBCL cells are negative for PRDM1. In cases II and IV, the tumor cells are essentially negative for PRDM1. Scattered small PRDM1(+) T cells serve as internal controls (asterisks). In case III, a small number of PRDM1(+) tumor cells are identified. Cases IV and V show higher percentage of PRDM1(+) lymphoma cells, but most of them are only weakly positive (arrows). In contrast, U266 cells show uniform strong PRDM1 staining. B: PRDM1α mRNA and the corresponding protein expression in 25 DLBCL cases. The PRDM1α mRNA levels were expressed relative to the myeloma cell line U266 (arbitrarily set as 1), which highly expressed the PRDM1 protein. The PRDM1 protein levels were determined by immunohistochemistry on formalin-fixed paraffin-embedded sections and expressed as percentage positivity of tumor cells. Cases that were illustrated in A were marked. ND, not determined.
Contrary to the non-GCB DLBCL, at least half of the GCB DLBCL (for example cases #19 to 22) demonstrated synchrony between levels of PRDM1α mRNA and PRDM1 protein. The mean PRDM1α mRNA expression among these four cases is about 7% that of U266, and the PRDM1 protein expression is barely detectable. These results imply that transcription repression is the major mode of PRDM1 regulation in many of the GCB DLBCL. However, in some of the GCB DLBCL, for example, case #24 (VI, Figure 2B), discordance between PRDM1 mRNA and protein expression is also observed.
Overexpression of Let-7 in Primary DLBCL
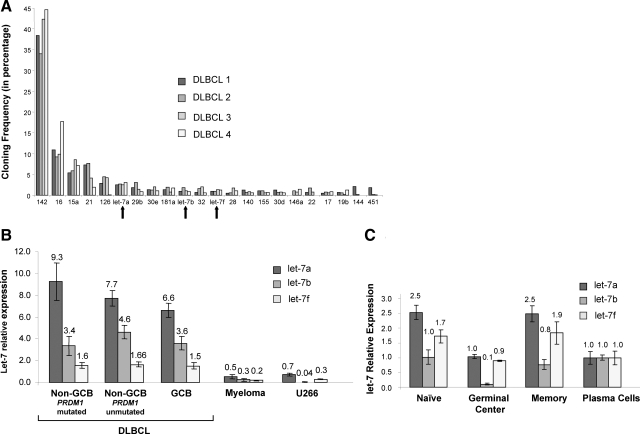
MiRNAs are small RNA regulators that are capable of modulating gene expression posttranscriptionally. Their functions depend on perfectly complementary interactions between the seed sequence at the 5′ end of the miRNAs and the 3′ region of the target sites.13 We examined the possibility that the miRNA milieu in DLBCL contributes to the posttranscriptional down-regulation of PRDM1 expression by searching for miRNAs that has the potential to target PRDM1 in DLBCL, as evidenced by a high expression and the presence of predicted miRNA binding sites in the PRDM1 3′ UTR. The miRNA expression profiles for four primary DLBCL cases have been established by direct miRNA cloning (Figure 3A).15 We identified from this analysis let-7 as a potential miRNA candidate to mediate PRDM1 down-regulation in DLBCL. Three members of the let-7 family, let-7a, let-7b, and let-7f, are among the most highly expressed miRNAs in these four DLBCL samples. An evolutionarily conserved let-7 binding site is present in the PRDM1 3′ UTR. Moreover, let-7a has been previously shown to down-regulate PRDM1 in Hodgkin lymphoma cell lines by translational inhibition.12
Figure 3.
Let-7 is overexpressed in DLBCL. A: MicroRNA profiles of four DLBCL samples based on direct miRNA cloning. The cloning frequency of each miRNA is shown as a percentage of total miRNA clones isolated for each DLBCL sample. MiRNA that represent >0.5% of the total miRNA population on average was indicated. Let-7a, let-7b, and let-7f (marked by arrows) were among the most highly expressed miRNA in DLBCL. B: Let-7a, let-7b, and let-7f expressions in DLBCL (25 cases), plasma cell myeloma (5 cases), and U266 myeloma cell line (plasma cells = 1 for each let-7 species). Overall, DLBCLs demonstrated significantly higher expressions of let-7 compared with U266 and plasma cells myeloma. Let-7 expressions, however, were similar among the DLBCL subgroups. Mean values are indicated on top of each bar. C: Let-7a, let-7b, and let-7f expressions in normal B cell subsets. Germinal center B cells have significantly lower levels of let-7 (P = 0.004, 0.03, 0.03 for let-7a, let-7b, and let-7f, respectively) relative to naïve and memory B cells. DLBCLs (see B) showed overexpression of let-7 relative to germinal center B cells. Mean values are indicated on top of each bar.
We confirmed the high levels of let-7 expression in DLBCL by measuring let-7a, let-7b, and let-7f levels in the current series of DLBCLs, as well as U266, plasma cell myeloma, and normal B cell subsets for comparison, using quantitative real-time PCR. In line with the miRNA profiling results, let-7a, let-7b, and let-7f are highly expressed in DLBCL, with levels 11, 115, and fivefold that of U266 (P = 0.001, 0.007 and 0.01) and 14, 16, and eightfold that of plasma cell myeloma (P < 0.0001, = 0.02, <0.0001, respectively; Figure 3B). There is no significant difference in let-7 expressions among the three DLBCL subgroups.
Let-7 expressions vary in B cells at different stages of differentiation (Figure 3C). The expression levels of let-7 are similar between naïve and memory B cells. All three members of let-7 are down-regulated in GC B cells relative to naïve B cells, with more marked decrease (∼10-fold) in let-7b (P = 0.004 for let-7a; P = 0.03 for let-7b and let-7f). There is a significant increase in let-7b (P = 0.001) as GC B cells differentiate into plasma cells, while differentiation from GC B cells into memory cells is associated with increased expression in all three let-7 members (P = 0.007 for let-7a; P = 0.02 for let-7b; P = 0.09 for let-7b). Comparison of let-7 expression between DLBCLs and normal GC B cells showed significant overexpression in the former. Overall, the levels of let-7a, let-7b, and let-7f in DLBCL are 7.7-, 41-, and 1.8-fold that of normal GC B cells, respectively (P = 0.0003, 0.0015, and 0.09).
Expression of PRDM1 and Let-7 in DLBCL Cell Lines
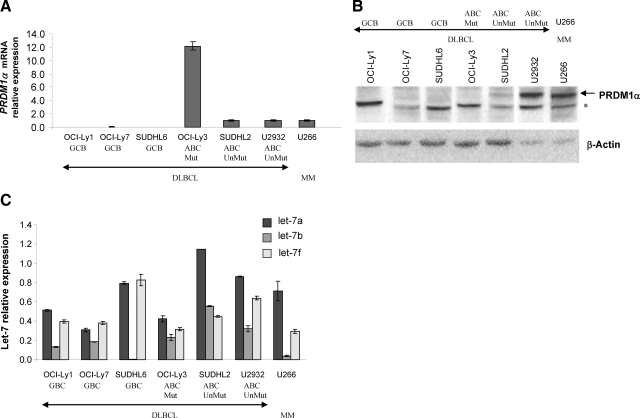
PRDM1 mRNA and protein expressions were also examined in DLBCL cell lines (Figure 4, A and B). In line with the observations for primary DLBCL, PRDM1α mRNA was expressed at barely detectable levels in GCB-type DLBCL cell lines (OCI-Ly1, OCI-Ly7, and SUDHL-6) but at much higher levels in DLBCL cell lines of the activated B cell (ABC) type (OCI-Ly3, SUDHL-2 and U2932). The low levels of PRDM1α mRNA in the GCB-type DLBCL cell lines is not due to promoter hypermethylation (see Figure 1B). In concordance with their low levels of PRDM1α mRNA transcription, PRDM1α protein was undetectable in the GCB-type DLBCL cell lines. However, we noted discordance in PRDM1α mRNA and protein expressions in two of three ABC-type DLBCL cell lines (OCI-Ly3 and SUDHL-2). The absence of PRDM1α in OCI-Ly3 can be attributed to the presence of inactivating mutations that resulted in premature translation termination and production of severely truncated nonfunctional protein.9,10 The existence of discordance in PRDM1 mRNA and protein expression in SUDHL-2 is evident when compared with U2932 and U266 cell lines. Though SUDHL-2 has a similar PRDM1α mRNA level as U2932 and U266, SUDHL2 has a much lower expression of PRDM1α protein compared with the other two cell lines. Sequencing of the PRDM1 coding region revealed no mutations in SUDHL-2 or U2932. These results support the presence of translational or posttranslational down-regulation of PRDM1 in the SUDHL-2 cell line and corroborate our findings in primary DLBCL cases.
Figure 4.
Expression of PRDM1α and let-7 in DLBCL cell lines. A: PRDM1α mRNA expression by real-time PCR analysis in six DLBCL cell lines and the myeloma cell line U266. The levels were expressed relative to U266 (arbitrarily set as 1). The PRDM1α transcript detected in OCI-Ly3 was truncated because of a rearrangement in the PRDM1 gene. B: Western blot analysis of total cell extracts prepared from the above cell lines using the monoclonal antibodies against PRDM1. The nonspecific band is indicated by an asterisk. C: Expression of let-7a, let-7b, and let-7f in DLBCL cell lines and U266 (plasma cells = 1 for each let-7 species). For A, B, and C, the DLBCL subtypes and PRDM1 mutation status for the DLBCL cell lines are indicated.
Examination of let-7 expression in DLBCL cell lines (Figure 4C) showed that let-7 is present at overall much lower levels in these cell lines compared with primary DLBCL cases (Figure 3B). Highest overall let-7 expressions are found in SUDHL-2, which shows discordance between PRDM1 mRNA and protein expressions. Compared with normal GCB (Figure 2B), we observed a two- to sixfold overexpression of let-7b in four of the six DLBCL cell lines. Let-7b expression in SUDHL-2 is the highest among all of the cell lines and is almost twice that of U2932 (P = 0.02), which has similar levels of PRDM1α mRNA as SUDHL-2 but lower levels of PRDM1 protein. Let-7a level in SUDHL-2 is also higher compared with U2932. These findings support a contribution of let-7, in particular let-7b, in down-regulating PRDM1 by translation inhibition in DLBCL cell lines.
Targeting of PRDM1 by Let-7
Let-7a has been shown to repress luciferase reporter activities by translation repression through specific binding to its predicted evolutionarily conserved target site in PRDM1 3′ UTR.12 Let-7b and let-7f share the same target site as let-7a, with identical seed pairing but slightly different pairing at the 3′ portion of the miRNAs (Figure 5A). To confirm that let-7b and let-7f can also functionally target PRDM1 3′ UTR, luciferase assays were performed in 293T cells using PRDM1 3′ UTR with wild-type or mutated let-7 binding site. Transfection of let-7a, let-7b or let-7f all repress luciferase activities by about 50 to 60%. This repression can be relieved by mutating the seed sequence of the putative let-7 binding site (Figure 5B). Thus, these results indicate that let-7a, let-7b and let-7f can all specifically interact with the PRDM1 3′UTR and down-regulate gene expression.
Figure 5.
Let-7 targets PRDM1 3′ UTR. A: Pairing between let-7 family (let-7a, let-7b, and let-7f) and their putative target site in PRDM1 3′ UTR. The highly conserved 6-bp “seed” pairing is highlighted by open boxes. The numbers indicate the locations of the putative binding sites in the PRDM1 3′ UTRs downstream from the PRDM1 stop codon. The nucleotides of the let-7 target site that were mutated (from AC to GT) to disrupt the “seed” pairing are underlined. B: Reporter plasmids harboring PRDM1 3′UTR with wild-type sequence (WT) or point mutations in the let-7 target site (MUT) were cotransfected into 293T cells with let-7a, let-7b, or let-7f (20 nmol/L, Ambion), or with miRNA Negative Control oligonucleotides (20 nmol/L, Ambion). Luciferase activities (in triplicates) were measured 24 hours after transfection. Renilla luciferase activities were normalized against firefly luciferase activities, and mean normalized Renilla luciferase activities (±SE) from three independent experiments were determined and expressed relative to control values. Transfection of let-7 members resulted in about 40% to 60% of inhibition of reporter gene expression, which was relieved by mutations in let-7 binding site.
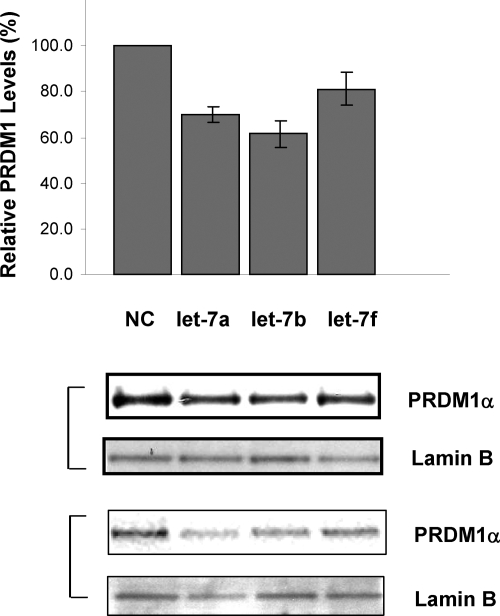
Previously, we have shown that transfection of let-7a into U266 myeloma cells can repress endogenous PRDM1 protein expression.12 To determine whether transfection of let-7 can also regulate PRDM1 in DLBCL cells, control miRNA, let-7a, or let-7b were transfected into U2932 cells and PRDM1α expression was measured by Western blotting 24 hours after transfection (Figure 6). Both let-7a and let-7b repress endogenous PRDM1 expression in U2932 cells by about 40% relative to negative controls. Let-7f also has repressive effects, but to a lesser extent. These results indicate that let-7 can also target endogenous PRDM1 in DLBCL cells.
Figure 6.
Let-7 down-regulated PRDM1 expression in U2932 DLBCL cell line. Control miRNA (NC), let-7a, and let-7b were transfected into U2932 cells by nucleofection. Nuclear extracts were prepared from each transfectant after 24 to 48 hours, and PRDM1α levels were measured by Western blot analysis using the anit-PRDM1 monoclonal antibody. Lamin B was used as a loading control. Normalized PRDM1α levels were expressed as a percentage relative to the negative control transfectants. The histogram was derived from two (let-7f) or four independent transfection experiments (let-7a and let-7b). Two representative Western blots are also shown.
Discussion
The expression of PRDM1α mRNA in DLBCL is heterogeneous, which is probably a result of the difference in signals and activities of transcription factors that regulate PRDM1 expression.1,21,22,23,24,25 Signals that can activate PRDM1 expression include those from cytokines (IL5, IL6, IL-10, and IL-21), toll-like receptors, and antigen receptors. AP-1, NF-κB, IRF4/MUM1, STAT3, PU1, and C/EBPβ induce PRDM1 transcription, while BCL6, PAX5, and BACH2 repress PRDM1 transcription. In the absence of promoter hypermethylation, the low level of PRDM1 mRNA present in some primary GCB-type DLBCL cases and GCB-type DLBCL cell lines is most likely due to transcription repression, for example, by BCL-6.26 The higher PRDM1α mRNA induction in the primary non-GCB DLBCL cases and in the ABC-type DLBLC cell lines may be a result of higher NF-κB activities.27
Despite the heterogeneity in PRDM1α mRNA levels, we observed in general a relatively low PRDM1α protein expression in primary DLBCL cases. Previously, Garcia et al18 has analyzed PRDM1 expression in a series of more than 200 DLBCL cases and observed negative or weak PRDM1 expression in most DLBCL. However, they also identified a group of DLBCL that demonstrated stronger PRDM1 expression. We were not able to detect these “high” PRDM1 expressors in our DLBCL series despite our ability to detect high PRDM1 expression in the normal GCB subset in reactive germinal centers and in the myeloma cell line U266 using identical immunohistochemical conditions throughout our study. The difference in results between the two studies may be due to case sampling and/or differences in immunohistochemical conditions. Because the study by Garcia et al did not explicitly compare PRDM1 expression in these “high” expressors with reference standards such as multiple myeloma it would be difficult to assess accurately the actual PRDM1 content in those DLBCL. We did identify a DLBCL cell line (U2932) that has similarly high PRDM1 level as U266 based on Western blot analysis. Interestingly, U2932 is CD138(+),28 indicative of plasmacytic differentiation. This finding suggests that those DLBCL with plasmacytic differentiation may tend to have higher levels of PRDM1 and provides additional supporting evidence for the established role of PRDM1 in plasma cell differentiation. In our series of primary DLBCL cases, none of the cases appear to demonstrate plasmacytic differentiation based on morphological evaluation and CD138 expression.
Comparative analysis of PRDM1α mRNA and protein in primary DLBCL cases and DLBCL cell lines demonstrated two distinct patterns. The first is present in a subset of GCB DLBCL, in which low PRDM1α mRNA expression is associated accordingly with low levels of PRDM1 protein. There are exceptions though (for example, in case #24), which may be ascribed to the possibility of misclassification of these cases to GCB-type by immunohistochemistry. The second one is present in the vast majority of non-GCB DLBCL and also in some GCB-DLBCL, which demonstrated discordance between PRDM1 mRNA and protein expressions. This discordance can be due to inactivating deletions and point mutations or translational/posttranslational down-regulation. The great majority of these DLBCL express IRF4/MUM1. Hence the low levels of PRDM1 seen in this group of DLBCL imply a pathogenetic dissociation between PRDM1 and IRF4 expressions, because these two transcription factors are normally co-expressed in GC centrocytes that are committed to plasma cell differentiation, and both factors are required for the generation of competent plasma cells.29,30 Similar to primary DLBCL, discordance between PRDM1 mRNA and protein is also evident in the DLBCL cell lines OCI-Ly3 and SUDHL-2. The former has mutations that inactivate PRDM1,9,10 while the latter is likely due to translational/posttranslational regulation.
Our current and previous studies support PRDM1 as a direct target of let-7 miRNAs. In line with our findings, Blimp-1/prdm1 was demonstrated by elegant genetic experiments to be an in vivo target for let-7 in primordial germ-cell development in mice.31 Down-regulation of PRDM1 by let-7 appears to be mediated by translation inhibition.12 Although let-7 has also been shown to be able to promote mRNA degradation,32 this effect may be dependent on target genes and cell types. For example, let-7 inhibits translation of RAS with no effects on Ras mRNA levels,33,34 while it primarily promotes mRNA degradation of HMGA2.35 Its regulatory effects on c-myc vary depending on cell types. In colon cancer cells, it primarily inhibits translation of c-MYC,34 while in Burkitt lymphoma cells, let-7 can down-regulate both c-myc mRNA and protein expressions.36
Let-7 members have been demonstrated by microarray analysis to exhibit temporal changes during B cell differentiation,37,38 implying their role in this process. Consistent with these results, our study using qRT-PCR also shows down-regulation of let-7 members in germinal center B cells. Let-7, in particular let-7b, appears overexpressed in DLBCL relative to normal GCB cells, implying that it may function as an oncogene in DLBCL. A tumor suppressor role of let-7 has been well documented in many human cancers by its ability to target several oncogenes, inhibit mitogenic pathways, and affect cell differentiation and apoptosis.39 However, the tumor suppressor functions of let-7 described to date are limited to solid tumors and may not be applicable in lymphoid malignancies. There are also reports that suggest an oncogenic function for let-7.40,41 Thus, it is conceivable that let-7 may have pleiotropic cell-type specific biological effects depending on the milieu of expressed genes inside the cells. In DLBCL, abnormal down-regulation of the tumor suppressor PRDM1 due to an overexpressed let-7 may represent one of the mechanisms by which the latter promotes lymphomagenesis. Moreover, let-7 is shown to regulate components of the translation machinery.42 Global changes in translation may contribute to lymphoma development by altering levels of critical oncogenes and tumor suppressors. Moreover, let-7 has the capacity to down-regulate DICER, which mediates processing of pre-miRNA to mature miRNA in the cytoplasm.43 Thus, let-7 overexpression may affect the steady state levels of many other miRNAs with tumorigenic potentials. Interestingly, while let-7a, let-7b, and let-7f are all overexpressed in primary DLBCL cases, only let-7b is overexpressed in some DLBCL cell lines relative to GCB. The difference in let-7 expression between in vivo lymphomas and cultured lymphoma cells are in line with the previous observations that DLBCL cell lines are not faithful replica for primary DLBCL with respect to miRNA expression profiles.
By measuring endogenous PRDM1 changes on let-7 transfections in U2932 cells, we demonstrated directly a role of let-7 in modulating PRDM1 expression in DLBCL. Though the modulatory effects of let-7 appear to be modest, it might have been underestimated owing to the limitations on the transfection efficiency (about 50%) and the artificial conditions of the experiments. The degree of suppression in PRMD1 expression observed in this experiment may also be influenced by the half-life of the protein. Furthermore, the levels of PRDM1 in DLBCL are possibly modulated by additional inputs besides let-7. This is suggested by the observation that despite markedly higher levels of let-7b in the SUDHL-2 and U2932 cell lines compared with U266 (see Figure 3C), only SUDHL2 demonstrates a reduction in PRDM1 expression. For example, miRNAs other than let-7 may play a role in down-regulating PRDM1. Several other miRNAs are predicted to have potential binding sites in the 3′ UTR of the PRDM1 gene, though only two of them, let-7 and miR-30, are among the most highly expressed miRNAs in DLBCL based on direct cloning. It is conceivable that besides let-7, miR-30 may also target PRDM1 in DLBCL. MiR-30 has been shown to target PRDM1 3′ UTR by luciferase assays and has been postulated to regulate PRDM1 expression during germinal center reaction.36,38,44 Mechanisms other than miRNA, for example, those related to protein degradation, might also contribute to PRDM1 posttranscriptional down-regulation. The steady state of PRDM1 in the cells may depend on an integrated summation of these inputs as well as the steady state levels of PRDM1 transcripts. It is interesting that higher levels of PRDM1α mRNA are found in DLBCL harboring PRDM1 inactivating mutations compared with those without such mutations. Although this observation may simply be attributed to a loss of the negative feedback signals due to total absence of PRDM1,45 the possibility that stronger PRDM1 induction signals are received by these cells before inactivating gene mutation events cannot be excluded. If the later scenario is indeed true, mutational inactivation rather than miRNA alone may be necessary to down-regulate PRDM1 to levels that promote outgrowth of lymphoma cells owing to the higher levels of PRDM1 transcripts.
In summary, we show that protein levels of the tumor suppressor gene PRDM1 may be down-regulated in DLBCL by miRNA-mediated down-regulation, in addition to mutational inactivation and transcription repression. MiRNA-mediated down-regulation of PRDM1 as a potential pathogenetic event in lymphomagenesis has also been implicated in Hodgkin lymphoma12 and EBV-positive Burkitt lymphoma.46 Because B cell terminal differentiation is characterized by quantitative changes in PRDM1 levels,47 its down-regulation may prevent it from reaching a threshold for terminal differentiation to proceed. Our studies provide supporting data on the importance of abnormal down-regulation of PRDM1 and the consequent impairment of terminal B cell differentiation in the pathogenesis and/or maintenance of DLBCL.
Footnotes
Address reprint requests to Wayne Tam, M.D., Ph.D., Department of Pathology & Laboratory Medicine, Weill Cornell Medical College, Starr 709, 525 East 68th Street, New York, NY 10021. E-mail: wtam@med.cornell.edu.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
Current address of H.A.: King Hussein Institute for Biotechnology and Cancer, Jordan; of M.G.: New York College of Osteopathic Medicine, New York, NY and of A.C.: Department of Pathology, Northwestern University Feinberg School of Medicine, Chicago, IL.
References
- Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- Turner CA, Jr, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. doi: 10.1016/0092-8674(94)90321-2. [DOI] [PubMed] [Google Scholar]
- Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–620. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- Kallies A, Hawkins ED, Belz GT, Metcalf D, Hommel M, Corcoran LM, Hodgkin PD, Nutt SL. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat Immunol. 2006;7:466–474. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- Chang DH, Angelin-Duclos C, Calame K. BLIMP-1: trigger for differentiation of myeloid lineage. Nat Immunol. 2000;1:169–176. doi: 10.1038/77861. [DOI] [PubMed] [Google Scholar]
- Chan YH, Chiang MF, Tsai YC, Su ST, Chen MH, Hou MS, Lin KI. Absence of the transcriptional repressor Blimp-1 in hematopoietic lineages reveals its role in dendritic cell homeostatic development and function. J Immunol. 2009;183:7039–7046. doi: 10.4049/jimmunol.0901543. [DOI] [PubMed] [Google Scholar]
- Magnusdottir E, Kalachikov S, Mizukoshi K, Savitsky D, Ishida-Yamamoto A, Panteleyev AA, Calame K. Epidermal terminal differentiation depends on B lymphocyte-induced maturation protein-1. Proc Natl Acad Sci USA. 2007;104:14988–14993. doi: 10.1073/pnas.0707323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam W, Gomez M, Chadburn A, Lee JW, Chan WC, Knowles DM. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood. 2006;107:4090–4100. doi: 10.1182/blood-2005-09-3778. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Compagno M, Houldsworth J, Monti S, Grunn A, Nandula SV, Aster JC, Murty VV, Shipp MA, Dalla-Favera R. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med. 2006;203:311–317. doi: 10.1084/jem.20052204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courts C, Montesinos-Rongen M, Brunn A, Bug S, Siemer D, Hans V, Blumcke I, Klapper W, Schaller C, Wiestler OD, Kuppers R, Siebert R, Deckert M. Recurrent inactivation of the PRDM1 gene in primary central nervous system lymphoma. J Neuropathol Exp Neurol. 2008;67:720–727. doi: 10.1097/NEN.0b013e31817dd02d. [DOI] [PubMed] [Google Scholar]
- Nie K, Gomez M, Landgraf P, Garcia JF, Liu Y, Tan LH, Chadburn A, Tuschl T, Knowles DM, Tam W. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: a potential pathogenetic lesion in Hodgkin lymphomas. Am J Pathol. 2008;173:242–252. doi: 10.2353/ajpath.2008.080009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Dahlberg JE, Tam W. MicroRNAs in tumorigenesis: a primer. Am J Pathol. 2007;171:728–738. doi: 10.2353/ajpath.2007.070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, Falini B, Banham AH, Rosenwald A, Staudt LM, Connors JM, Armitage JO, Chan WC. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allawi HT, Dahlberg JE, Olson S, Lund E, Olson M, Ma WP, Takova T, Neri BP, Lyamichev VI. Quantitation of microRNAs using a modified Invader assay. RNA. 2004;10:1153–1161. doi: 10.1261/rna.5250604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia JF, Roncador G, Sanz AI, Maestre L, Lucas E, Montes-Moreno S, Fernandez Victoria R, Martinez-Torrecuadrara JL, Marafioti T, Mason DY, Piris MA. PRDM1/BLIMP-1 expression in multiple B and T-cell lymphoma. Haematologica. 2006;91:467–474. [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Tam W, Gomez M, Nie K. Significance of PRDM1beta expression as a prognostic marker in diffuse large B-cell lymphomas. Blood. 2008;111:2488–2489. doi: 10.1182/blood-2007-09-111385. author reply 2489–2490. [DOI] [PubMed] [Google Scholar]
- Calame K. Activation-dependent induction of Blimp-1. Curr Opin Immunol. 2008;20:259–264. doi: 10.1016/j.coi.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Pal R, Janz M, Galson DL, Gries M, Li S, Johrens K, Anagnostopoulos I, Dorken B, Mapara MY, Borghesi L, Kardava L, Roodman GD, Milcarek C, Lentzsch S. C/EBPbeta regulates transcription factors critical for proliferation and survival of multiple myeloma cells. Blood. 2009;114:3890–3898. doi: 10.1182/blood-2009-01-201111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai S, Bolick SC, Maurin M, Wright KL. PU. 1 regulates positive regulatory domain I-binding factor 1/Blimp-1 transcription in lymphoma cells. J Immunol. 2009;183:5778–5787. doi: 10.4049/jimmunol.0901120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H, Thierry-Mieg D, Thierry-Mieg J, Kim HP, Oh J, Tunyaplin C, Carotta S, Donovan CE, Goldman ML, Tailor P, Ozato K, Levy DE, Nutt SL, Calame K, Leonard WJ. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 2009;31:941–952. doi: 10.1016/j.immuni.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien C, Fang CM, Huso D, Livak F, Lu R, Pitha PM. Critical role of IRF-5 in regulation of B-cell differentiation. Proc Natl Acad Sci USA. 2010;107:4664–4668. doi: 10.1073/pnas.0911193107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler HG. Guide to Leukemia-Lympohma Cell lines. Braunschweig. 2005 [Google Scholar]
- Cattoretti G, Angelin-Duclos C, Shaknovich R, Zhou H, Wang D, Alobeid B. PRDM1/Blimp-1 is expressed in human B-lymphocytes committed to the plasma cell lineage. J Pathol. 2005;206:76–86. doi: 10.1002/path.1752. [DOI] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunniff K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, Surani MA, Daley GQ. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- Basso K, Sumazin P, Morozov P, Schneider C, Maute RL, Kitagawa Y, Mandelbaum J, Haddad J, Jr, Chen CZ, Califano A, Dalla-Favera R. Identification of the human mature B cell miRNome. Immunity. 2009;30:744–752. doi: 10.1016/j.immuni.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres R, Sarosiek KA, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, Tibshirani R, Lossos IS. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. 2009;113:3754–3764. doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17:F19–F36. doi: 10.1677/ERC-09-0184. [DOI] [PubMed] [Google Scholar]
- Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, Sultmann H, Lyko F. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–1423. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Smith H, Ueno Y, Patel T. The MicroRNA let-7a modulates interleukin-6-dependent STAT-3 survival signaling in malignant human cholangiocytes. J Biol Chem. 2007;282:8256–8264. doi: 10.1074/jbc.M607712200. [DOI] [PubMed] [Google Scholar]
- Ding XC, Slack FJ, Grosshans H. The let-7 microRNA interfaces extensively with the translation machinery to regulate cell differentiation. Cell Cycle. 2008;7:3083–3090. doi: 10.4161/cc.7.19.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumaru S, Suzuki M, Yamada H, Nagino M, Takahashi T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis. 2008;29:2073–2077. doi: 10.1093/carcin/bgn187. [DOI] [PubMed] [Google Scholar]
- Zhang J, Jima DD, Jacobs C, Fischer R, Gottwein E, Huang G, Lugar PL, Lagoo AS, Rizzieri DA, Friedman DR, Weinberg JB, Lipsky PE, Dave SS. Patterns of microRNA expression characterize stages of human B cell differentiation. Blood. 2009;113:4586–4594. doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan MA, Magnusdottir E, Kuo TC, Tunyaplin C, Harper J, Arnold SJ, Calame K, Robertson EJ, Bikoff EK. Blimp-1/Prdm1 alternative promoter usage during mouse development and plasma cell differentiation. Mol Cell Biol. 2009;29:5813–5827. doi: 10.1128/MCB.00670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucci E, Onnis A, Cocco M, De Falco G, Imperatore F, Giuseppina A, Costanzo V, Cerino G, Mannucci S, Cantisani R, Nyagol J, Mwanda W, Iriso R, Owang M, Schurfeld K, Bellan C, Lazzi S, Leoncini L. B-cell differentiation in EBV-positive Burkitt lymphoma is impaired at posttranscriptional level by miRNA-altered expression. Int J Cancer. 2010;126:1316–1326. doi: 10.1002/ijc.24655. [DOI] [PubMed] [Google Scholar]
- Kallies A, Hasbold J, Tarlinton DM, Dietrich W, Corcoran LM, Hodgkin PD, Nutt SL. Plasma cell ontogeny defined by quantitative changes in blimp-1 expression. J Exp Med. 2004;200:967–977. doi: 10.1084/jem.20040973. ‘. [DOI] [PMC free article] [PubMed] [Google Scholar]