Abstract
K-RAS mutations are found in approximately 30% of lung cancers. The transcription factor Krüppel-like Factor 5 (KLF5) has been shown to mediate cellular transformation signaling events downstream of oncogenic RAS in other cancers, but a role for KLF5 in lung tumorigenesis has not been defined. We show here that knockdown of KLF5 expression significantly decreased anchorage-independent growth, but did not affect proliferation of human lung adenocarcinoma cells. Moreover, Klf5 is not required for lung tumor formation in an inducible oncogenic K-RasG12D mouse model of lung tumorigenesis, and non-small cell lung cancer patients expressing high levels of KLF5 (21/258) have a significantly better disease-specific survival than those with intermediate to no KLF5 expression. Further, KLF5 knockdown in K-RAS-mutant human lung cancer cells resulted in a fivefold increase in ATP-binding cassette, subfamily G (WHITE), member 2 (ABCG2), an anthracycline drug transporter, which lead to significantly increased resistance to doxorubicin treatment, a chemotherapeutic agent clinically used to treat lung cancer. In summary, while KLF5 is not required for oncogenic mutant K-Ras-induced lung tumorigenesis, KLF5 regulation of ABCG2 expression may be important for chemotherapeutic resistance and patient survival.
Lung cancer is the leading cause of cancer deaths throughout the world.1 Despite major advances in detection and cancer therapeutics in the past decade, the overall 5-year survival of patients with lung cancer is 15%.2 Therefore, genetic and environmental factors responsible for the susceptibility to lung cancer and therapeutic resistance need to be identified to determine more efficient ways of preventing and treating the disease.
Ras genes have been highly conserved throughout evolution in all mammalian cells, and therefore represent essential proteins for normal cellular physiology.3 Three Ras genes have been well characterized: H-Ras, K-Ras, and N-Ras. K-Ras mutations have been identified in approximately 30% of human lung cancers; however, attempts to clinically target mutant K-Ras have encountered significant problems.4,5 Ras mutants induce an exaggerated form of oncogenic signaling allowing cancer cells to experience continuous growth factor stimulation and contribute to many cell phenotypes, such as loss of contact inhibition and anchorage independence. Therefore, the presence of Ras mutations marks a poor prognosis for both early and late stage lung cancers.6 Many murine lung tumor models have failed to generate tumors that closely resemble human adenocarcinomas.7,8 However, a mouse lung tumor model has been generated in which a lox-stop-lox cassette and mutant K-RasG12D were introduced into the endogenous K-Ras locus. Cre-mediated deletion of the lox-stop-lox cassette induces K-RasG12D expression from normal cis-regulatory elements and yields pulmonary hyperplasia, adenomas, and adenocarcinomas, which mimic the human disease.9,10
Krüppel-like factor 5 (KLF5, IKLF, BTEB2) is a member of the Krüppel-like transcription factor family. KLF5 is a DNA-binding transcriptional regulator, and contains three independent C2H2 zinc fingers.11 KLF5 is expressed in multiple human tissues including small intestine, colon, prostate, kidney, pancreas, kidney, skeletal muscle, breast, and lung.12,13,14 KLF5 expression is responsive to H-Ras, Wnt-1, and ErbB2 oncogenic signaling.15,16,17 Exogenous expression of H-Ras in NIH3T3 cells leads to cellular transformation and KLF5 overexpression through the MEK/MAPK pathway.15 Inhibition of KLF5 in these cells resulted in decreased proliferation and anchorage-independent cell growth, potentially through KLF5-mediated activation of cyclin D1 expression.15 Expression of K-RasV12G in IEC-6 intestinal epithelial cells also lead to increased KLF5 expression, proliferation, and anchorage-independent cell growth that was reversed on siRNA-mediated knockdown of KLF5.18 In contrast, deletion or reduced expression of KLF5 has been associated with human prostate, breast, and familial colon cancers.14,19,20,21 Moreover, KLF5 re-expression in esophageal and breast cancer cell lines reduced cell viability and inhibited cell proliferation.14,22 Thus, KLF5 may act either as a tumor suppressor, or as an oncogene in a context-dependent manner; however, the necessity of KLF5 for Ras-mediated lung tumorigenesis in vivo has not been determined.
Solid tumors contain a minor population of cancer cells that have a high repopulation capacity.23,24 In vitro studies have shown that roughly 1 in 1000 to 5000 lung cancer cells show this high repopulation capacity, implying that not every lung cancer cell is capable of tumor initiation.25 Expression of ATP-binding cassette, subfamily G (WHITE), member 2 (ABCG2), an ATP-binding cassette transporter, permits the identification of hematopoietic stem cells by flow cytometry as a side population (SP) of cells with a distinct “low” Hoechst 33342 dye staining pattern.26,27 Moreover, the SP of a tumor may be enriched in cancer repopulating cells.25,28,29,30,31 Importantly, the ABCG2 transporter effluxes Hoechst dyes and chemotherapeutic anthracycline drugs, thus providing an intersection between the concepts of cancer stem cells and drug resistance.
To directly assess the requirement for KLF5 in lung tumorigenesis, we used in vitro and in vivo models of lung cancer in which an oncogenic allele of K-Ras was combined with manipulation of KLF5 expression. We found that KLF5 functions to support the in vitro anchorage-independent growth of Ras-mutant human lung adenocarcinoma cell lines. However, using the K-RasG12D mouse model of lung tumorigenesis, we found that Klf5 is not required for lung tumorigenesis. Moreover, KLF5 expression levels in human primary lung cancers demonstrate a positive correlation with lung cancer disease-specific survival. Furthermore, KLF5 repression of ABCG2 expression subsequently affects sensitivity to doxorubicin, an anthracycline drug, which is now used routinely in the clinic as adjuvant chemotherapy for the treatment of lung cancer. In summary, our studies demonstrate that although KLF5 is not required for Ras-mediated lung tumorigenesis in mice, KLF5 expression and target gene regulation may be relevant to chemotherapeutic resistance and patient survival.
Materials and Methods
Cell Culture
The human lung adenocarcinoma cell lines A549, H441, H23, and H460 were all obtained from the American Type Culture Collection (ATCC, Manassas, VA), and maintained in RPMI 1640 medium (Life Technologies, Bethesda, MD) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% l-glutamine. All cell lines were incubated in a humidified incubator at 37°C supplied with 5% carbon dioxide. Cells were routinely maintained in 75 cm2 tissue culture flasks (BD Biosciences Discovery Labware, San Jose, CA) and harvested when they were in the logarithmic phase of growth.
Production of Stable KLF5 Overexpressing and Knockdown Cells
The day before virus infection, 2 × 105 A549, H441, H460, and H23 cells were plated in each well of a six-well tissue culture plate. The next day, the culture medium was aspirated, cells washed, and incubated with viruses. For KLF5 knockdown experiments, lentivirus containing either VSVg-pseudotyped KLF5-specific short hairpin (sh)RNA vectors (shKLF5-A, B, and C), or nontargeting shRNA control vector (NT) that were purchased from Sigma (St. Louis, MO) were used. For KLF5 overexpression experiments, cells were instead infected with retrovirus containing either VSVg-pseudotyped MSCV γ-retroviral vector containing hemagglutinin (HA)-immunoepitope-tagged KLF5, or with empty vector serving as control. Polyclonal stable transfectants were selected using 5 μg/ml puromycin for 4 days.
Western Analysis
Nuclear extracts were generated using the Nuclear Extract Kit purchased from Active Motif (Carlsbad, CA) according to the manufacturer’s instructions. Protein concentrations were quantified by Bradford assay (Thermo Fisher Scientific, Rockford, IL). Equal amounts of total protein (35 to 40 μg per lane) were resolved on 10% polyacrylamide gels by using standard SDS-polyacrylamide gel electrophoresis (PAGE) techniques. Proteins were transferred onto a nitrocellulose membrane using a semidry transfer apparatus. Membranes were blocked in 5% nonfat milk plus 0.01% Tween 20, then incubated with anti-KLF5 (ab24331; concentration 2 μg/ml; Abcam Inc. Cambridge, MA) polyclonal antibody, followed by incubation with horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody (Calbiochem, Gibbstown, NJ) and developed using the ECL Plus Western Blotting Detection System (GE Health care Bio-Sciences Corp. Amersham, Piscataway, NJ). Membranes were stripped and reprobed for TATA-box binding protein (Abcam, Cambridge, MA) as loading control. Quantitation of band intensities were performed using Quantity One software (BioRad Laboratories, Hercules, CA) and are expressed as fold change relative to control.
Cell Proliferation Assay
The growth rates of A549, H441, H23, and H460 cells stably expressing KLF5-specific shRNA, nontargeting shRNA, KLF5 overexpression, or MSCV empty vectors were measured by colorimetric WST assay based on metabolizing tetrazolium salt (Roche, Indianapolis, IN). Briefly, 2 × 103 cells were plated into each well of a 96-well plate in triplicate. Cells were allowed to settle for 24 hours. On each day for five consecutive days, 10 μl WST was added to each well, and then the cells were incubated at 37°C for an additional 30 minutes. Optical densities were determined on a microplate reader at 490 nm. For drug sensitivity assays, Doxorubicin was added to the cells at a final concentration of 100 nmol/L, 10 nmol/L, 1 nmol/L, or 0.1 nmol/L and incubated for 24 hours. Drug resistance is represented as % viability calculated using the following formula: (absorbance of treated cells)/(absorbance of untreated cells) × 100%. Experiments were repeated thrice and graphed as average % viability ± SEM.
Anchorage-Independent Growth Assay
A549, H441, H23, and H460 cells (8 × 103 cells/well) were seeded into 0.3% Difco Agar Noble (Difco BD Biosciences, San Jose, CA) supplemented with complete culture medium. The suspension was layered over 2 ml of 0.8% agar-medium base layer in 6-well tissue culture plates (BD Biosciences Discovery Labware, San Jose, CA). After 14 days, the colonies were counted. Results are displayed as the average number of colonies from three independent experiments ± SEM.
Flow Cytometry
Side population (SP) abundance was determined similarly to the procedure described.27 Cells were trypsinized and washed twice with RPMI 1640 supplemented with 2% fetal bovine serum and 10 mmol/L HEPES (pH 7.4). For each SP analysis, ∼106 cells were resuspended in 1 ml of RPMI 1640. Hoechst 33342 (1 mg/ml in water; Sigma, St. Louis, MO) was added to a final concentration of 5 μg/ml (+/− verapamil (Sigma, St. Louis, MO) as a negative control), and the cells were incubated at 37°C in a water bath for 90 minutes and mixed by gentle vortexing every 20 minutes. At the end of the incubation, all samples were chilled on ice and transferred to microcentrifuge tubes. Cells were washed once with cold HBSS supplemented with 2% fetal bovine serum and 10 mmol/L HEPES (pH 7.4), resuspended in 1 ml of HBSS and transferred to prechilled 5 ml polystyrene tubes. To assess viability, 7AAD was added to a final concentration of 1 μg/ml for 30 minutes before running the sample on an LSRII flow cytometer (BD Biosciences, San Jose, CA). Verapamil-sensitive SP were quantified using FloJo software (Treestar, Ashland, OR).
RNA Isolation and Real-Time PCR
Total RNA was isolated with the RNeasy Mini Kit (QIAGEN, Valencia, CA), digested with DNase1 (Applied Biosystems/Ambion, Austin, TX) to remove DNA contamination, and processed using the Taqman Reverse Transcription System according to the manufacturer’s instructions (Applied Biosystems, Foster City, CA). cDNA was analyzed with probes targeting KLF5, ABCG2, and β-actin (Applied Biosystems, Foster City, CA). The expression of the glyceraldehude-3-phosphate dehydrogenase (Gapdh) gene was used for normalizing samples. The average fold expression from at least three independent samples ± SEM is shown.
Generation of K-RasG12D;Klf5+/+ and K-RasG12D;Klf5fl/fl Mice and Lung Infections with Cre-Containing Adenovirus
Klf5flox/flox mice32 were mated to K-RaslslG12D mice9 obtained from the Mouse Models of Human Cancer Consortium (NCI, Frederick, MD). The resultant experimental K-RaslslG12D;Klf5fl/fl mice (n = 5) and K-RaslslG12D;Klf5+/+ (n = 6) control mice lacking loxP sites in Klf5 were used for in vivo lung tumorigenesis studies. Adenovirus expressing Cre recombinase (AdCreM2) was produced according to Akagi et al33 to simultaneously induce the K-RasG12D oncogene expression (referred to as K-RasG12D) and recombination between the loxP sites flanking exons 2 and 3 in the Klf5 gene (referred to as Klf5fl/fl). Two- to 4-month-old mice were treated with 9.6 mg/day co-trimoxazole orally (480 mg co-trimoxazole in 250 ml drinking water, presumed intake 5 ml/day) 1 week before virus administration. Co-trimoxazole was administered to reduce perioperative mortality due to infections. One day before AdCre administration mice received 15 mg/kg cyclosporin A intraperitoneally. Mice were anesthetized with isoflurane, and intubated with AdCre virus (1 × 107 PFU). From the day of AdCre administration until 6 weeks later, mice were administered cyclosporin A orally, 20 mg/kg/day. Two K-RasG12D;Klf5fl/fl mice and one K-RasG12D;Klf5+/+ mouse were sacrificed 8 weeks postintubation for showing signs of distress, all other animals were sacrificed at 16 weeks postintubation. Animal protocols were approved by the Institutional Animal Care and Use Committee in accordance with NIH guidelines.
Mouse Lung Fixation, Tissue Collection, and Tumor Quantification
At the time of sacrifice, K-RasG12D;Klf5+/+ and K-RasG12D;Klf5fl/fl mice were perfused with 5 ml of sterile PBS through the inferior vena cava. Once the lungs were perfused to a white color, the lungs were then perfused through the trachea with 10% neutral buffered formalin (Surgipath, Richmond, IL) and the lungs were inflated to 10cm H2O with the fixative. Lungs were removed en bloc and fixed overnight, then replaced with 70% ethanol, and the tissues were processed and embedded into paraffin. Tumors were enumerated from two H&E-stained, 4-μm histological sections per mouse. Tumors were categorized as either adenoma with a defined perimeter and a size of ≥0.4 mm or adenocarcinoma displaying mitotic bodies and a size of >1 mm, in addition to the use of previously defined criteria for classification of mouse lung tumors by Nikitin et al,34 Johnson et al,10 and Jackson et al.9 The number of adenomas and adenocarcinomas per mouse were summed and averaged from the two sections and are shown graphically as tumor number per mouse. Individual areas of alveolar hyperplasia could not be accurately quantified in this study due to diffuse, and in some areas, dense inflammation and to abnormal lung parenchyma architecture, which have been described previously for these mouse models.9,32
Human Lung Cancer Tissue Array
Tissue arrays were generated from 609 cases of primary non-small cell lung cancer (NSCLC), using duplicate 0.6 mm cores.35 Of these, 460 cases had interpretable KLF5 data, as outlined in the Immunohistochemistry Materials and Methods, known cause of death, and follow-up time of greater than 30 days. There were 159 female cases, 301 male, with a median age of 64 years and median survival of 3.57 years. Patients from this historical cohort were treated before adjuvant chemotherapy (eg, anthracycline) was routinely used in the treatment of lung carcinoma. The Ras mutation status of these patients is not known. Therefore, prognosis in groups substratified based on specific treatment protocols or Ras status was not performed.
Immunohistochemistry
Immunohistochemistry was performed on 4-μm paraffin sections of K-RasG12D;Klf5+/+ and K-RasG12D;Klf5fl/fl mouse lungs prepared by the CCHMC Pathology Core Lab, and on 609 tissue biopsies on the human NSCLC tissue array.35 Endogenous peroxidases were blocked in 3% H2O2 for 20 minutes at room temperature and then in PBS with 0.1% Triton X-100 and 4% normal goat serum at 37°C for 2 hours. For KLF5 immunohistochemistry, sections were incubated first with either a commercially available KLF5 antibody (1:500 dilution; Abcam Inc. Cambridge, MA) or our own KLF5 antisera,32 and then a secondary antibody in PBS with 0.1% Triton X-100 and 4% normal goat serum at 37°C for 30 minutes. For prosurfactant protein-C (pro-SP-C) and mature surfactant protein-B (mature SP-B) staining, sections were incubated at 4°C overnight with primary rabbit polyclonal sera anti-pro-SP-C (1:3000 dilution) and mature anti-SP-B (1:1000 dilution) antibodies (Seven Hills Bioreagents, Cincinnati, OH), and then with anti-rabbit secondary antibodies. Staining was detected using the Vectastain ABC and DAB kits (Vector Labs, Burlingame, CA). Stained slides were photographed on a Zeiss Imager.A2 microscope with Zeiss Axiocam MRc5 camera (Carl Zeiss, Inc., Thornwood, NY) at ×10 magnification for KLF5 and at ×40 magnification for pro-SP-C and mature SP-B. For the NSCLC tissue array, expression of KLF5 was graded according to the following scale, for each case on the array: 0-negative, 1-weak immunoreactivity, 2-moderately intense immunoreactivity, and 3- strong immunoreactivity. Tissues that did not remain intact during the staining procedure were omitted from the scoring analysis.
Gene Expression Analysis
Total RNA was extracted from H441 cells transduced with KLF5-specific shRNA, KLF5 over expression (OE), and MSCV empty vector (NT) respectively. The cRNAs was then hybridized to Human Genome U133 Plus 2.0 Arrays (Affymetrix) according to manufacturer’s protocol. The RNA quality and quantity assessment, probe preparation, labeling, hybridization, and image scan were performed in the CCHMC Gene Expression Microarray Core Facility using standard procedure. Affymetrix Microarray Suite 5.0 was used to scan and quantitate the gene chips under default scan settings. Hybridization data were sequentially subjected to normalization, transformation, filtration, and functional classification and pathway analysis as previously described.36,37 The complete dataset can be found at Gene Expression Omnibus (GEO); Accession no. GSE16555. Data analysis was performed with BRB Array Tools software package (http://linus.nci.nih.gov/BRB-ArrayTools.html).
Chromatin Immunoprecipitation Analysis
Chromatin immunoprecipitation assay was performed in A549 cells overexpressing KLF5. 2 × 108 cells were harvested and fixed in 1% formaldehyde for 10 minutes on ice and terminated with 0.125 mol/L glycine. Cells were resuspended in cell lysis buffer (50 mmol/L Tris-HCl pH 8.1, 10 mmol/L EDTA pH 8.0, 10% glycerol, 1% SDS, 1× complete protease inhibitor) and incubated on ice for 10 minutes. Cells were then sonicated with a Sonicator 3000 cup horn (Misonix, Farmingdale, NY) on ice to generate soluble chromatin complex with DNA fragments of <1 kb length. Approximately, 1.5 ml of soluble chromatin was used per reaction for immunoprecipitation with antibody to KLF5 (ab24331, Abcam Inc. Cambridge, MA) and control mouse IgG (sc-2025, Santa Cruz Biotechnology, Santa Cruz, CA). Recovered chromatin was PCR amplified using Abcg2 (5′-AATGGCCTCTCAAAAGGTGTC-3′ and 5′-GGTTGTGGTGAGCCAAGATT-3′; product size 200 bp) or β-actin (5′ - AGCGCGGCTACAGCTTCA-3′ and 5′-CGTAGCACAGCTTCTCCTTAATGTC-3′; product size 120 bp) primer pairs. β-actin was used as a control for nonspecific enrichment.
Statistical Analyses
For statistical analyses of WST cell growth assays and soft agar anchorage-independent cell growth assays, t-test analyses were performed comparing KLF5 overexpression (KLF5oe) to empty vector control (vector) or KLF5 shRNA (A, B, or C) to nontargeting control (NT). For doxorubicin WST cell growth assays, One-way analysis of variance analyses were performed comparing KLF5 overexpression (KLF5oe), KLF5 shRNA-C (shKLF5-C), and nontargeting control (NT) for each day per treatment dose. In the case of the doxorubicin studies, the only significant differences were found between shKLF5-C and NT. For t-test and analysis of variance *P ≤ 0.05, **P ≤ 0.005, and ***P ≤ 0.0005. For clinical NSCLC tissue array, the initial 609 cases were first filtered for follow-up of >30 days and interpretable KLF5 staining, which resulted in a total of 460 cases with lung cancer including all subtypes. The overall survival of these 460 cases based on KLF5 expression level was compared by Mantel-Cox log rank analysis. For disease-specific survival analyses, the 460 filtered cases were censored for lung cancer all subtypes-specific survival totaling 258 cases, which were then compared by Mantel-Cox log rank analysis and Kaplan Meier curves generated based on KLF5 expression. For adenocarcinoma only and squamous cell carcinoma only disease-specific survival, 109 and 116 cases, respectively, were compared by Mantel-Cox log rank analysis and Kaplan Meier curves generated based on KLF5 expression. For gene microarray analyses, differentially expressed genes between shRNA versus NT, OE versus NT and shRNA versus OE in H441 cells were identified using a random-variance t-test.38 Genes were considered statistically significant if their P value was less than 0.01 and fold change greater than 1.5. The maximum proportion of false discovery rate was set to 0.1. In addition, Affymetrix “Present Call” in at least two of three replicates and coefficient of variation among replicates ≤50% were set as a requirement for gene selection.
Results
Manipulation of KLF5 Expression Significantly Alters Anchorage-Independent Growth of K-Ras Mutant Lung Cancer Cells
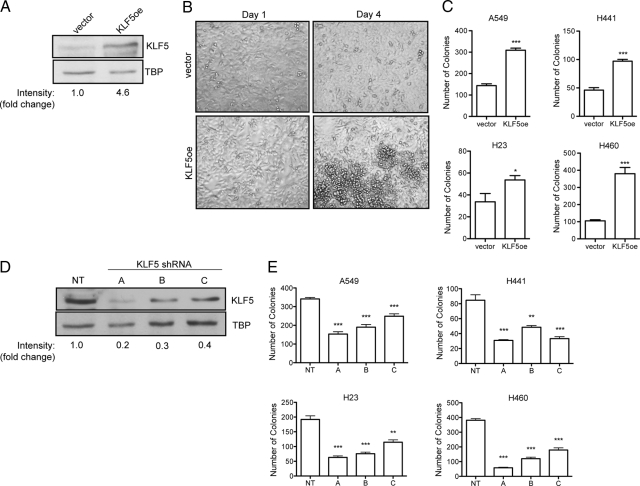
To investigate the effect of KLF5 on lung cancer, we infected the K-Ras-mutant human lung cancer cell lines A549, H441, H23, and H460 with either KLF5 overexpression retrovirus (KLF5oe) or three different KLF5-specific shRNA lentiviruses (A, B, and C). Cells were selected with puromycin to generate bulk stable cell populations with KLF5 overexpression or knockdown, and expression of KLF5 was validated by Western analysis on nuclear extracts from these cells (Figure 1, A and D, respectively). Using a growth assay based on metabolism of a tetrazolium salt (WST), we examined the growth of the cells over 5 days in culture. In all cell lines tested, neither KLF5 overexpression nor knockdown had a consistently significant effect on cell growth (Supplemental Figure S1 http://ajp.amjpathol.org and data not shown). However, on plating cells at a higher starting density, we observed that by day 4 of growth KLF5 overexpressing H441 cells continued to grow past confluency, while the empty vector control H441 cells ceased proliferation after reaching confluence maintaining a single monolayer (Figure 1B). To test whether KLF5 regulates contact-mediated cell growth, which has also been shown to be a characteristic phenotype of oncogenic mutant Ras signaling, we performed anchorage-independent cell growth assays by colony formation in soft agar. KLF5 overexpression in A549, H441, H23, and H460 lung cancer cells lead to a significant increase in the number of colonies formed in soft agar as compared with empty vector control cells (Figure 1C). Correspondingly, knockdown of KLF5 by three different shRNAs all significantly decreased the number of colonies in soft agar as compared with nontargeting shRNA controls in all lung cancer cells examined (Figure 1E). These data demonstrate that in K-Ras mutant lung cancer cells, modulation of KLF5 expression significantly alters the ability of the cells to undergo anchorage-independent cell growth in vitro.
Figure 1.
KLF5 regulates anchorage-independent growth of human lung cancer cells. A549, H441, H23, and H460 cells were stably transduced with KLF5 overexpression retrovirus (KLF5oe) or empty vector control virus (vector). A: Western analysis of KLF5 in protein lysates generated from nuclear extracts of KLF5 overexpressing or empty vector control A549 cells. Detection of TATA-binding protein (TBP) serves as loading control. KLF5 band intensities are reported as fold change relative to empty vector control. B: Representative images of KLF5 overexpressing or empty vector control H441 cells plated at high density that have reached confluence (Day 1) or three days later (Day 4). C: Average number of colonies formed in soft agar by A549, H441, H23, and H460 KLF5 overexpressing or empty vector control cells ± SEM. *P < 0.05 and ***P < 0.0005 by t-test. A549, H441, H23, and H460 cells were stably transduced with three different KLF5-specific shRNA lentiviruses (A–C) or nontargeting shRNA lentivirus control (NT). D: Western analysis of KLF5 expression in protein lysates generated from nuclear extracts of KLF5 knockdown or control A549 cells. TATA-binding protein serves as loading control. KLF5 band intensities are reported as fold change relative to nontargeting shRNA control. E: Average number of colonies formed in soft agar by A549, H441, H23, and H460 KLF5 knockdown or control cells ± SEM. **P < 0.005 and ***P < 0.0005 by t-test.
Klf5 Is Not Required for K-RasG12D-Mediated Lung Tumorigenesis
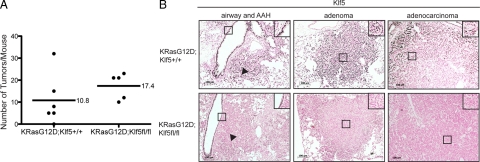
To determine the importance of Klf5 to oncogenic mutant K-Ras driven tumorigenesis in vivo, we mated mice with a K-RaslslG12D allele to mice with floxed exons 2–3 of Klf5. Subsequently, K-RaslslG12D mice (n = 6) and K-RaslslG12DKlf5fl/fl mice (n = 5) were intubated at 6 weeks of age with adenoviruses expressing the Cre recombinase to delete exons 2 and 3 of Klf5 and induce oncogenic KRasG12D expression. Mice were followed for 4 months unless they showed signs of early distress, at which point mice were sacrificed and lungs were fixed. The number of adenomas and adenocarcinomas were quantified histopathologically from at least 2 sections per mouse and averaged to generate the number of tumors per mouse represented in Figure 2A. The mean number of tumors formed in K-RasG12D;Klf5+/+ control mice was 10.8, whereas the K-RasG12D;Klf5fl/fl mice developed a mean of 17.4 tumors. Although there was a trend toward the development of more tumors in the K-RasG12D;Klf5fl/fl mice as compared with controls, this difference was not statistically significant (P = 0.28 by t-test) perhaps due to the limited number of animals in this study and variability in tumor numbers within each group. KLF5 immunohistochemical analysis of lung tissues harvested from these mice revealed strong KLF5 expression in airway epithelium, alveolar hyperplasia, and adenomas, with more moderate expression observed in the adenocarcinomas of K-RasG12D;Klf5+/+ control mice (Figure 2B). In K-RasG12D;Klf5fl/fl mice, Klf5 expression was also observed in airway epithelium, while very low to no Klf5 expression was found in areas of alveolar hyperplasia (Figure 2B arrowhead), adenomas, or adenocarcinomas (Figure 2B), which is coincident with induction of oncogenic K-RasG12D. These findings suggest that Klf5 is not required for K-RasG12D-mediated lung tumorigenesis in mice.
Figure 2.
Klf5 is not required for K-RasG12D-mediated lung tumorigenesis in mice. K-RasG12D;Klf5+/+ controls (n = 6) and K-RasG12D;Klf5fl/fl experimental (n = 5) mice were intubated with a Cre recombinase adenovirus at six weeks of age. Mice were sacrificed 8 to 16 weeks postinfection, at which point their lungs were inflation fixed and embedded in paraffin. A: The number of lung adenomas (≥0.4 mm) and adenocarcinomas (>1 mm) per mouse were enumerated histopathologically, summed, and represented as tumor number per mouse. The mean tumor number in each genotype is denoted with a straight line. B: Representative images of immunohistochemical detection of Klf5 expression in the airways, alveolar hyperplasia (AH; arrowheads), adenomas, and adenocarcinomas of K-RasG12D;Klf5+/+ and K-RasG12D;Klf5fl/fl mice are shown. Klf5 positive cells are dark brown in contrast to Klf5 negative cells and have been magnified to show their histological appearance (black boxes). Original magnification ×10. Scale bar = 100 μm.
Previously, we reported that embryonic deletion of Klf5, lead to a maturation defect in the type II cells of the lung, which specifically lacked lamellar bodies.32 It was demonstrated previously that K-RasG12D mice have normal expression of the type II cell marker pro-surfactant protein-C (proSP-C), and additionally it was hypothesized that type II cells are the cell origin of lung tumors in this model.9,39 To address whether deletion of Klf5 postnatally in mice with a K-RasG12D mutation also results in a type II cell maturation defect, we performed immunohistochemical analysis of type II cell lamellar body proteins proSP-C and mature SP-B. We found no differences in the expression of either proSP-C or mature SP-B in K-RasG12D;Klf5fl/fl lungs compared with K-RasG12D;Klf5+/+ controls (Supplemental Figure S2 http://ajp.amjpathol.org). Thus, except for differences in Klf5 expression, tumors in K-RasG12D;Klf5+/+ and K-RasG12D;Klf5fl/fl lungs appeared similar.
KLF5 Expression Predicts Disease-Specific Survival of Lung Cancer Patients
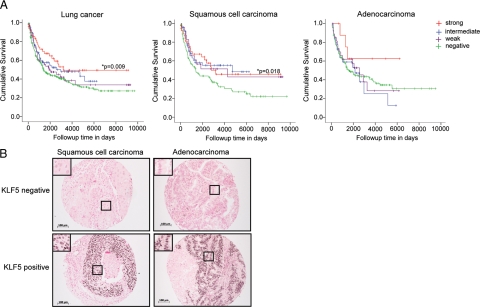
We next addressed the prognostic significance of KLF5 expression in primary human lung cancers. A human NSCLC tissue array35 was subjected to immunohistochemical analyses for KLF5 expression. Both commercially available anti-sera and our in-house antibodies specific to KLF532 revealed similar staining patterns. The overall survival of all lung cancer patients (n = 460) did not significantly correlate with KLF5 expression (P = 0.177). However, the disease-specific survival of all lung cancer patients (n = 258) was significantly better for those patients with strong KLF5 expression (P = 0.009, Figure 3A, left). In addition, the disease-specific survival of all lung cancer patients progressively worsened with decreasing levels of KLF5 expression (Figure 3A, left). The disease-specific survival of patients with squamous cell carcinoma (P = 0.018) was significantly better with strong KLF5 expression (Figure 3A, middle and right, respectively). A similar trend for adenocarcinoma was limited by the number of patients with high KLF5 expression (P = 0.476). The breakdown of patients within each KLF5 staining category are shown in Table 1 by lung cancer subclassification. Representative images of lung adenocarcinomas and squamous cell carcinomas positive and negative for KLF5 immunohistochemical staining are shown in Figure 3B. These data suggest that KLF5 expression is a novel indicator of lung cancer survival.
Figure 3.
KLF5 expression significantly correlates with better disease-specific survival of human NSCLC patients. KLF5 immunohistochemical analysis was performed on biopsies of 609 human NSCLC tissues on array slides. For disease-specific survival analyses, a total of 258 cases were compared by Mantel-Cox log rank analysis for KLF5 expression levels that had interpretable KLF5 data, follow-up information >30 days, and known cause of death. A: Kaplan-Meier disease-specific survival curves of lung cancer patients of all subtypes (left), squamous cell carcinoma (middle), and adenocarcinoma (right). P values are indicated where statistical significance was found. B: Representative images of KLF5-negative and KLF5-positive immunohistochemical staining of squamous cell carcinomas and adenocarcinomas from the NSCLC tissue array. KLF5 positive cells are dark brown in contrast to KLF5 negative cells and have been magnified to show their histological appearance (black boxes). Original magnification ×10. Scale bar = 100 μm.
Table 1.
Distribution of NSCLC Patients Represented in Kaplan Meier Disease-Specific Survival Curves Grouped by Immunohistochemical Score for KLF5 Expression
| Total cases (N) | KLF5 expression (% of Total)
|
||||
|---|---|---|---|---|---|
| Strong | Intermediate | Weak | Negative | ||
| All lung cancer | 258 | 8% | 15% | 13% | 64% |
| Squamous cell carcinoma | 116 | 16% | 18% | 10% | 56% |
| Adenocarcinoma | 109 | 3% | 14% | 14% | 70% |
| Other | 33 | - | 9% | 18% | 73% |
Identification of KLF5-Responsive Genes by Microarray Analysis
Given our findings that KLF5 regulates anchorage-independent growth of K-Ras mutant human lung cancer cells in vitro, and that KLF5 expression predicts the disease-specific survival of lung cancer patients, we wanted to further investigate potential mechanisms by which KLF5 might influence lung tumorigenesis and patient disease-specific survival. Since KLF5 is a DNA-binding transcription factor, we performed gene-expression microarray analysis on H441 cells with stable expression of KLF5-shRNA or KLF5 overexpression compared with the nontargeting shRNA control. Microarray data analysis identified 220 genes that were differentially expressed more than 1.5-fold between KLF5-overexpressing and KLF5 knockdown cells; a subset of which provided discrimination between normal and transformed lung cancer gene expression signatures (Supplemental Figure S3 http://ajp.amjpathol.org). The 20 most deregulated genes are listed in Table 2, 15 of which demonstrated at least threefold differential expression; a subset of those showed modest correlation to KLF5 expression in gene expression arrays from primary human lung cancers (Supplemental Figure S4 http://ajp.amjpathol.org). Four genes ABCG2, EED, TP63, and PROM2 were selected for validation of microarray results by TaqMan quantitative-PCR on RNA harvested from KLF5 knockdown, KLF5 overexpressing, or nontargeting control H441 and A549 cells. In agreement with the microarray results ABCG2 (Figure 4, A and C), EED, and PROM2 (Supplemental Figure S5 http://ajp.amjpathol.org) showed increased expression in the KLF5-shRNA cells and decreased expression in KLF5 overexpressing cells, whereas TP63 (Supplemental Figure S5 http://ajp.amjpathol.org) showed decreased expression in KLF5-shRNA cells and increased expression in KLF5 overexpressing cells. These data confirm the validity of the gene array results.
Table 2.
Gene Expression Microarray Analysis of H441 Cells Stably Transduced with Lentiviral Vectors Expressing KLF5-Specific shRNA, a Control Nontargeting shRNA, or a Retroviral Vector Expressing KLF5
| Gene symbol | Gene description | Affymetrix ID | Fold change |
|---|---|---|---|
| SFTPB | Surfacant, pulmonary-associated protein B | 209810 at | 0.162 |
| SFTPB | Surfacant, pulmonary-associated protein B | 37004 at | 0.176 |
| TP73L | Tumor protein p73-like (p63) | 209863 s at | 0.232 |
| TP73L | Tumor protein p73-like (p63) | 211194 s at | 0.238 |
| TP73L | Tumor protein p73-like (p63) | 207382 at | 0.282 |
| FYN | FYN oncogene related to SRC, FGR, YES | 210105 s at | 0.349 |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B (p15) | 236313 at | 0.451 |
| CCPG1 | Cell cycle progression 1 | 214152 at | 0.453 |
| EED | Embryonic ectoderm development | 210656 at | 2.577 |
| CDKN2C | Cyclin-dependent kinase inhibitor 2C (p18) | 204159 at | 2.711 |
| CDCA3 | Cell division cycle associated 3 | 223307 at | 2.745 |
| BMP5 | Bone morphogenetic protein 5 | 205430 at | 2.848 |
| SCIN | Scinderin | 1552367 a at | 2.896 |
| SCIN | Scinderin | 1552365 at | 2.927 |
| TGFB2 | Transforming growth factor, beta 2 | 228121 at | 3.005 |
| PROM2 | Prominin 2 | 1552797 s at | 3.015 |
| PKIB | Protein kinase (cAMP-dependent) inhibitor beta | 231120 × at | 3.367 |
| DAPK1 | Death-associated protein kinase 1 | 203139 at | 3.786 |
| ABCG2 | ATP-binding cassette, sub-family G (WHITE) 2 | 209735 at | 4.136 |
| ADH1A | Alcohol dehydrogenase 1A (class I) | 206262 at | 4.439 |
| VGLL1 | Vestigial like 1 (Drosophilia) | 215729 s at | 4.783 |
| BCL2A1 | BCL2-related protein A1 | 205681 at | 4.883 |
| AQP3 | Aquaporin 3 (Gill blood group) | 39248 at | 6.159 |
| DKK1 | Dickkopf homolog 1 (Xenopus laevis) | 204602 at | 8.162 |
Gene expression changes are shown as fold change of KLF5 knockdown cells relative to KLF5 overexpressing cells normalized to nontargeting control.
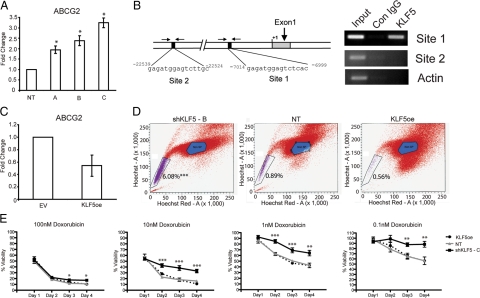
Figure 4.
KLF5 represses ABCG2 expression and controls doxorubicin sensitivity. A: Real-time quantitative gene expression analysis (TaqMan) of ABCG2 in A549 cells transduced with lentiviruses expressing KLF5-specific shRNAs (A, B, and C) or nontargeting shRNA control (NT). B: Schematic representation of the ABCG2 gene with putative KLF5 binding sites 1 and 2 (black boxes). Chromatin immunoprecipitation analysis on lysates from A549 cells overexpressing KLF5. KLF5-specific antiserum was used for immunoprecipitation of KLF5-DNA complexes, and the indicated primers flanking sites 1 and 2 (arrows) were used for identification of DNA sequences. Immunoprecipitation using IgG and amplification of β-Actin gene (actin) serve as negative controls. C: Real-time quantitative gene expression analysis (TaqMan) of ABCG2 in A549 cells transduced with retroviral vectors overexpressing KLF5 (KLF5oe) or an empty vector control (vector). D: Side population (SP) shown is purple flow cytometric analysis after Hoechst 33342 dye staining of A549 cells transduced with lentivirus expressing KLF5-specific shRNA “B” (shKLF5-B), nontargeting shRNA control (NT), or a retrovirus overexpressing KLF5 (KLF5oe). The percentages of SP cells per group are indicated. E: WST cell growth analysis of A549 cells transduced with lentivirus expressing KLF5-specific shRNA “C” (shKLF5-C), nontargeting shRNA control (NT), or a retrovirus overexpressing KLF5 (KLF5oe) treated with the indicated doses of doxorubicin. % Viability was calculated as (absorbance of treated cells)/(absorbance of untreated cells) × 100% ± SEM *P < 0.05, **P < 0.005, and ***P < 0.0005 by one-way analysis of variance.
KLF5 Represses ABCG2 Expression and Controls Anthracycline Sensitivity
Given the ability of ABCG2 to efflux chemotherapeutic agents25 we examined the ABCG2 genomic locus to find two putative KLF5 binding sites conserved between murine and human genomes (Figure 4B). Chromatin immunoprecipitation analysis revealed that KLF5 associates with Site 1, but not Site 2, upstream of the transcription start site in the ABCG2 locus in A549 cells (Figure 4B). Taken together with the gene array and quantitative-PCR data, these results suggest that KLF5 may transcriptionally repress ABCG2 expression.
To determine whether ABCG2 plays a functional role downstream of KLF5 in lung cancer cells, we first examined the ability of ABCG2 to efflux Hoechst dye 33342, as evidenced by a Hoechst-negative side population (SP) of cells measured by flow cytometric analysis of A549 cells in which KLF5 expression was manipulated. KLF5 overexpressing, KLF5-shRNA, and nontargeting control expressing cells were stained with Hoechst 33342 dye with and without verapamil treatment, which blocks the activity of Hoechst transporter. The SP gate was defined as the diminished region in the presence of verapamil (data not shown). The KLF5-shRNA cells consistently exhibited a significantly higher percentage of SP cells, 6.03%, compared with 0.89% for the nontargeting control and 0.56% in KLF5 overexpressing cells (Figure 4D and Supplemental Figure S6 http://ajp.amjpathol.org). Given that ABCG2 has been shown to efflux anthracycline chemotherapeutic drugs25 such as doxorubicin, we next examined the relative sensitivity of A549 cells (in which KLF5 expression was manipulated) to doxorubicin treatment. A549 cells expressing KLF5-shRNA “C,” KLF5 overexpressing, or nontargeting shRNA control cells were treated with increasing doses of doxorubicin, and the metabolism of tetrazolium salt (WST) was measured 24 hours later as an indication of cell survival. Drug resistance is represented as % viability calculated as (absorbance of treated cells)/(absorbance of untreated cells) × 100%. The KLF5-shRNA expressing A549 lung cancer cells consistently showed significantly increased survival from 0.1 nmol/L up to 100 nmol/L doxorubicin (Figure 4E). These data demonstrate that reducing KLF5 expression significantly decreases the sensitivity of lung cancer cells to chemotherapy with an anthracycline drug; revealing functional sequelae of KLF5-manipulated ABCG2 gene expression.
Discussion
Prior investigation has shown that KLF5 is up-regulated by oncogenic mutant Ras signaling, and functions as a downstream mediator of Ras signaling to activate expression of Ras-pathway target genes, such as cyclin D1, and to regulate cell proliferation and anchorage-independent cell growth.15,20,22,40,41 In agreement with this, we found that Klf5 is expressed in K-RasG12D mouse lung tumors, and that KLF5 expression significantly regulates the ability of K-Ras mutant human lung cancer cells to undergo anchorage-independent growth in vitro. This prompted us to use an inducible K-RasG12D mouse model of lung cancer to determine the requirement for Klf5 in K-RasG12D-mediated tumorigenesis. The deletion of exons 2–3 of Klf5, which functionally inactivates the protein and causes Klf5 expression to be undetectable, did not prevent K-RasG12D initiated lung tumors, nor did it overtly alter cyclin D1 expression (data not shown). Thus, in contrast to in vitro data using human lung cancer cell lines, Klf5 is not required for in vivo oncogenic K-RasG12D-mediated lung tumorigenesis.
Traditionally, chemotherapeutic agents have serious side effects and ideally should be given only to patients classified as high-risk. Common prognostic factors such as lymph node status and tumor size are not sufficiently accurate to identify those patients whose prognosis would not benefit from adjuvant chemotherapy, and new prognostic biomarkers are needed. Our evidence suggests that for lung cancer patients, in particular those with squamous cell carcinoma, KLF5 expression levels may be a novel indicator for disease-specific survival. In addition, our in vitro data suggests that lung cancers with high levels of KLF5 expression may be more sensitive to adjuvant chemotherapy.
We found that the disease-specific survival of human NSCLC patients with strong KLF5 expression was significantly better than those with weak or no expression, which is the opposite survival correlation for patients with Ras mutations. In addition, K-RasG12D mice with Klf5 deletion exhibited a trend toward greater tumor burden than in mice with wild-type Klf5, although this finding was not statistically significant, perhaps limited the number of animals in our current study and/or to the difficulty of distinguishing smaller hyperplastic lesions close together from the larger areas of diffuse hyperplasia. Together, these data suggest that KLF5 may act as a tumor suppressor in a subset of lung cancers. In support of this, previous studies have demonstrated down-regulation and deletion of KLF5 in both prostate and breast cancer, and KLF5 re-expression reduces cell viability and proliferation.14,19,20
Although KLF5 is typically regarded as a transcriptional activator, as with other Krüppel-like factor family members, its function is likely to be context dependent.42 We found more genes significantly up-regulated, than down-regulated, on KLF5 knockdown in H441 lung cancer cells by microarray analysis. Given that the functions of other Krüppel-like factor family members have been found to be cell type- and promoter-dependent,42 our data suggest similar complexity for KLF5 target gene regulation.
The recent concept that chemotherapeutic agents are unsuccessful due to a minority population of chemo-resistant cancer cells, which retain the ability to re-initiate the tumor, suggests that these “cancer stem cells” retain stem cell-like qualities such as self-renewal and self-preservation through expression of multidrug resistance transporters.43 Some of the KLF5-responsive genes identified in our microarray study have been previously associated with tumorigenic or stem-cell-related properties such as TP73L (TP63), EED, ABCG2, PROM2, and ADH1A, which may impact disease-specific survival of lung cancer patients.26,43,44,45,46 ABCG2, specifically, has been shown to efflux chemotherapeutic anthracycline drugs, such as doxorubicin, which leads to drug resistance.25 We showed that KLF5 expression exhibits an inverse relationship with ABCG2 expression, and KLF5 knockdown significantly increased the resistance of lung cancer cells to doxorubicin treatment. Moving forward, lung cancer patients with high KLF5 expression should be examined to determine whether they respond better to adjuvant chemotherapeutic intervention.
Acknowledgments
We acknowledge the assistance of the Cincinnati Children’s Hospital Medical Center (CCHMC) Gene Expression Microarray Core, the CCHMC Pathology Core Lab, and the CCHMC Pulmonary Biology Morphology Core. We would also like to acknowledge Dr. Susan Wert (CCHMC) for helpful discussions.
Footnotes
Address reprint requests to H. Leighton Grimes, Ph.D., 3333 Burnet Ave., MLC 7038, Cincinnati, OH 45229, E-mail lee.grimes@cchmc.org.
Supported by an American Lung Association Fellowship (H.W.), HL090156 (J.A.W.), HL079574-06S1, CA112405, CA105152 and a Barrett Cancer Center Pilot grant (H.L.G.).
S.E.M., J.R.H., and A.B. contributed equally to this work.
None of the authors disclosed any relevant financial relationships.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer Statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Pao W, Kris MG, Iafrate AJ, Ladanyi M, Janne PA, Wistuba II, Miake-Lye R, Herbst RS, Carbone DP, Johnson BE, Lynch TJ. Integration of molecular profiling into the lung cancer clinic. Clin Cancer Res. 2009;15:5317–5322. doi: 10.1158/1078-0432.CCR-09-0913. [DOI] [PubMed] [Google Scholar]
- Rodenhuis S, Slebos RJ. Clinical significance of ras oncogene activation in human lung cancer. Cancer Res. 1992;52:2665s–2669s. [PubMed] [Google Scholar]
- Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Schiller JH, Spinola M, Minna JD. New molecularly targeted therapies for lung cancer. J Clin Invest. 2007;117:2740–2750. doi: 10.1172/JCI31809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgia R, Skarin AT. Molecular abnormalities in lung cancer. J Clin Oncol. 1998;16:1207–1217. doi: 10.1200/JCO.1998.16.3.1207. [DOI] [PubMed] [Google Scholar]
- Meuwissen R, Berns A. Mouse models for human lung cancer. Genes Dev. 2005;19:643–664. doi: 10.1101/gad.1284505. [DOI] [PubMed] [Google Scholar]
- Tuveson DA, Jacks T. Modeling human lung cancer in mice: similarities and shortcomings. Oncogene. 1999;18:5318–5324. doi: 10.1038/sj.onc.1203107. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276:34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- Shi H, Zhang Z, Wang X, Liu S, Teng CT. Isolation and characterization of a gene encoding human Kruppel-like factor 5 (IKLF): binding to the CAAT/GT box of the mouse lactoferrin gene promoter. Nucleic Acids Res. 1999;27:4807–4815. doi: 10.1093/nar/27.24.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sur I, Unden AB, Toftgard R. Human Kruppel-like factor5/KLF5: synergy with NF-kappaB/Rel factors and expression in human skin and hair follicles. Eur J Cell Biol. 2002;81:323–334. doi: 10.1078/0171-9335-00257. [DOI] [PubMed] [Google Scholar]
- Chen C, Bhalala HV, Qiao H, Dong JT. A possible tumor suppressor role of the KLF5 transcription factor in human breast cancer. Oncogene. 2002;21:6567–6572. doi: 10.1038/sj.onc.1205817. [DOI] [PubMed] [Google Scholar]
- Nandan MO, Yoon HS, Zhao W, Ouko LA, Chanchevalap S, Yang VW. Kruppel-like factor 5 mediates the transforming activity of oncogenic H-Ras. Oncogene. 2004;23:3404–3413. doi: 10.1038/sj.onc.1207397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneyhill L, Pennica D. Identification of Wnt responsive genes using a murine mammary epithelial cell line model system. BMC Dev Biol. 2004;4:6. doi: 10.1186/1471-213X-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers J, Herrmann F, Rieger S, Drobyshev AL, Horsch M, Hrabe de Angelis M, Seliger B. Identification and validation of novel ERBB2 (HER2. NEU) targets including genes involved in angiogenesis. Int J Cancer. 2005;114:590–597. doi: 10.1002/ijc.20798. [DOI] [PubMed] [Google Scholar]
- Nandan MO, McConnell BB, Ghaleb AM, Bialkowska AB, Sheng H, Shao J, Babbin BA, Robine S, Yang VW. Kruppel-like factor 5 mediates cellular transformation during oncogenic KRAS-induced intestinal tumorigenesis. Gastroenterology. 2008;134:120–130. doi: 10.1053/j.gastro.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Bhalala HV, Vessella RL, Dong JT. KLF5 is frequently deleted and down-regulated but rarely mutated in prostate cancer. Prostate. 2003;55:81–88. doi: 10.1002/pros.10205. [DOI] [PubMed] [Google Scholar]
- Tong D, Czerwenka K, Heinze G, Ryffel M, Schuster E, Witt A, Leodolter S, Zeillinger R. Expression of KLF5 is a prognostic factor for disease-free survival and overall survival in patients with breast cancer. Clin Cancer Res. 2006;12:2442–2448. doi: 10.1158/1078-0432.CCR-05-0964. [DOI] [PubMed] [Google Scholar]
- Bateman NW, Tan D, Pestell RG, Black JD, Black AR. Intestinal tumor progression is associated with altered function of KLF5. J Biol Chem. 2004;279:12093–12101. doi: 10.1074/jbc.M311532200. [DOI] [PubMed] [Google Scholar]
- Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther. 2005;4:1216–1221. doi: 10.4161/cbt.4.11.2090. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA: 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67:4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- Zhou S, Schuetz JD, Bunting KD, Colapietro AM, Sampath J, Morris JJ, Lagutina I, Grosveld GC, Osawa M, Nakauchi H, Sorrentino BP. The ABC transporter Bcrp1/ABCG2 is expressed in a wide variety of stem cells and is a molecular determinant of the side-population phenotype. Nat Med. 2001;7:1028–1034. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschmann-Jax C, Foster AE, Wulf GG, Nuchtern JG, Jax TW, Gobel U, Goodell MA, Brenner MK. A distinct “side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci USA: 2004;101:14228–14233. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Setoguchi T, Taga T. Persistence of a small subpopulation of cancer stem-like cells in the C6 glioma cell line. Proc Natl Acad Sci USA: 2004;101:781–786. doi: 10.1073/pnas.0307618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuring-Buske M, Hogge DE. Hoechst 33342 efflux identifies a subpopulation of cytogenetically normal CD34(+)CD38(−) progenitor cells from patients with acute myeloid leukemia. Blood. 2001;97:3882–3889. doi: 10.1182/blood.v97.12.3882. [DOI] [PubMed] [Google Scholar]
- Wulf GG, Wang RY, Kuehnle I, Weidner D, Marini F, Brenner MK, Andreeff M, Goodell MA. A leukemic stem cell with intrinsic drug efflux capacity in acute myeloid leukemia. Blood. 2001;98:1166–1173. doi: 10.1182/blood.v98.4.1166. [DOI] [PubMed] [Google Scholar]
- Wan H, Luo F, Wert SE, Zhang L, Xu Y, Ikegami M, Maeda Y, Bell SM, Whitsett JA. Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development. 2008;135:2563–2572. doi: 10.1242/dev.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi K, Sandig V, Vooijs M, Van der Valk M, Giovannini M, Strauss M, Berns A. Cre-mediated somatic site-specific recombination in mice. Nucleic Acids Res. 1997;25:1766–1773. doi: 10.1093/nar/25.9.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin AY, Alcaraz A, Anver MR, Bronson RT, Cardiff RD, Dixon D, Fraire AE, Gabrielson EW, Gunning WT, Haines DC, Kaufman MH, Linnoila RI, Maronpot RR, Rabson AS, Reddick RL, Rehm S, Rozengurt N, Schuller HM, Shmidt EN, Travis WD, Ward JM, Jacks T. Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer Res. 2004;64:2307–2316. doi: 10.1158/0008-5472.can-03-3376. [DOI] [PubMed] [Google Scholar]
- Ionescu DN, Treaba D, Gilks CB, Leung S, Renouf D, Laskin J, Wood-Baker R, Gown AM. Nonsmall cell lung carcinoma with neuroendocrine differentiation–an entity of no clinical or prognostic significance. Am J Surg Pathol. 2007;31:26–32. doi: 10.1097/01.pas.0000213319.04919.97. [DOI] [PubMed] [Google Scholar]
- Xu Y, Liu C, Clark JC, Whitsett JA. Functional genomic responses to cystic fibrosis transmembrane conductance regulator (CFTR) and CFTR(delta508) in the lung. J Biol Chem. 2006;281:11279–11291. doi: 10.1074/jbc.M512072200. [DOI] [PubMed] [Google Scholar]
- Xu Y, Ikegami M, Wang Y, Matsuzaki Y, Whitsett JA. Gene expression and biological processes influenced by deletion of Stat3 in pulmonary type II epithelial cells. BMC Genomics. 2007;8:455. doi: 10.1186/1471-2164-8-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- Shaw AT, Meissner A, Dowdle JA, Crowley D, Magendantz M, Ouyang C, Parisi T, Rajagopal J, Blank LJ, Bronson RT, Stone JR, Tuveson DA, Jaenisch R, Jacks T. Sprouty-2 regulates oncogenic K-ras in lung development and tumorigenesis. Genes Dev. 2007;21:694–707. doi: 10.1101/gad.1526207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Benjamin MS, Sun X, Otto KB, Guo P, Dong XY, Bao Y, Zhou Z, Cheng X, Simons JW, Dong JT. KLF5 promotes cell proliferation and tumorigenesis through gene regulation and the TSU-Pr1 human bladder cancer cell line. Int J Cancer. 2006;118:1346–1355. doi: 10.1002/ijc.21533. [DOI] [PubMed] [Google Scholar]
- Ghaleb AM, Nandan MO, Chanchevalap S, Dalton WB, Hisamuddin IM, Yang VW. Kruppel-like factors 4 and 5: the yin and yang regulators of cellular proliferation. Cell Res. 2005;15:92–96. doi: 10.1038/sj.cr.7290271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. J Cell Physiol. 2001;188:143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- Donnenberg VS, Donnenberg AD. Multiple drug resistance in cancer revisited: the cancer stem cell hypothesis. J Clin Pharmacol. 2005;45:872–877. doi: 10.1177/0091270005276905. [DOI] [PubMed] [Google Scholar]
- Finlan LE, Hupp TR. p63: the phantom of the tumor suppressor. Cell Cycle. 2007;6:1062–1071. doi: 10.4161/cc.6.9.4162. [DOI] [PubMed] [Google Scholar]
- Nylander K, Vojtesek B, Nenutil R, Lindgren B, Roos G, Zhanxiang W, Sjostrom B, Dahlqvist A, Coates PJ. Differential expression of p63 isoforms in normal tissues and neoplastic cells. J Pathol. 2002;198:417–427. doi: 10.1002/path.1231. [DOI] [PubMed] [Google Scholar]
- Ura H, Usuda M, Kinoshita K, Sun C, Mori K, Akagi T, Matsuda T, Koide H, Yokota T. STAT3 and Oct-3/4 control histone modification through induction of Eed in embryonic stem cells. J Biol Chem. 2008;283:9713–9723. doi: 10.1074/jbc.M707275200. [DOI] [PubMed] [Google Scholar]