Summary
Cells from Fanconi anemia (FA) patients are extremely sensitive to DNA inter-strand crosslinking (ICL) agents, but the molecular basis of the hypersensitivity remains to be explored. FANCM (FA complementation group M), and its binding partner, FAAP24, anchor the multi-subunit FA core complex to chromatin after DNA damage and may contribute to ICL-specific cellular response. Here we show that the FANCM/FAAP24 complex is specifically required for the recruitment of replication protein A (RPA) to ICL-stalled replication forks. ICL-induced RPA foci formation requires the DNA binding activity of FAAP24 but not the DNA translocase activity of FANCM. Furthermore, FANCM/FAAP24-dependent RPA foci formation is required for efficient ATR-mediated checkpoint activation in response to ICL. Therefore, we propose that FANCM/FAAP24 plays a role in ICL-induced checkpoint activation through regulating RPA recruiment at ICL-stalled replication forks.
Keywords: Fanconi anemia, DNA inter-strand crosslink (ICL), FANCM/FAAP24, RPA, ICL-induced checkpoint response
INTRODUCTION
Fanconi Anemia (FA) is a recessive genetic disorder characterized by congenital abnormalities, chromosome instability, and cancer susceptibility (D’Andrea and Grompe, 2003; Moldovan and D’Andrea, 2009; Wang, 2007). The thirteen known Fanconi Anemia (FA) proteins cooperate in DNA inter-strand crosslink (ICL) repair. Eight of the FA proteins (A, B, C, E, F, G, L, M) comprise a multisubunit ubiquitin E3 ligase complex, referred to as the FA core complex (Kennedy and D’Andrea, 2005). In response to DNA damage during DNA replication, the FA core complex monoubiquitinates and activates FANCD2 and FANCI (Garcia-Higuera et al., 2001; Smogorzewska et al., 2007). The activated FANCD2/FANCI complex then interacts with downstream FA proteins (FANCD1, FANCJ, FANCN) in the process of homologous recombination (HR) repair (D’Andrea and Grompe, 2003; Moldovan and D’Andrea, 2009; Wang, 2007).
Cells from FA patients are extremely sensitive to ICL-inducing agents, such as mitomycin C (MMC) and cisplatin, and this hypersensitivity is the hallmark feature of FA patients. However, FA cells, in general, show normal resistance or mild sensitivity to other DNA damaging agents such as hydroxyurea (HU), ultraviolet (UV) and ionizing radiation (IR), although camptothecin (CPT) sensitivity has been reported in one FA complementation group deficient cells (Auerbach, 2009; Singh et al., 2009). The molecular basis underlying the ICL hypersensitivity of FA cells is one of the critical unanswered questions in the field. There are at least two possible mechanisms for this ICL hypersensitivity. First, ICL repair may require monoubiquitinated FANCD2, which is absent from cells from most FA subtypes. Second, ICL damaged DNA may generate a unique structure, which needs to be specifically recognized by FA proteins.
FANCM is a component of FA core complex and it contains a conserved helicase domain and associated ATP-dependent DNA translocase activity (Ciccia et al., 2008; Meetei et al., 2005; Mosedale et al., 2005). FAAP24 was identified as a FA-associated protein (FAAP) that is required for FANCD2 monoubiquitination (Ciccia et al., 2007). FAAP24 exhibits structure-specific DNA binding activity for DNA replication fork-like structures (Ciccia et al., 2007), suggesting that it may play an important role in the recognition of DNA damage during S phase. FANCM interacts with FAAP24 and the FANCM/FAAP24 complex constitutively localizes on chromatin, which allows nuclear FA core complex to access to the DNA damage sites (Kim et al., 2008). Recent studies have shown that FANCM and its interacting proteins, such as FAAP24, MHF1, and MHF2, play a role in controlling the processing and stabilization of stalled forks (Luke-Glaser et al., 2010; Schwab et al., 2010; Singh et al., 2010; Yan et al., 2010). Furthermore, FANCM/FAAP24 regulates the ATR-CHK1 checkpoint signaling through the interaction with the checkpoint protein HCLK2 independently of the FA core complex (Collis et al., 2008).
Critical to checkpoint activation after replication stress is the generation of local single-stranded DNA (ssDNA). Locally accumulated ssDNA is rapidly coated by the replication protein A (RPA) complex (RPA70, RPA32, RPA14). RPA subsequently binds ATRIP, resulting in ATR recruitment and checkpoint activation (Zou and Elledge, 2003). The accumulation of a long stretch of ssDNA results from the uncoupling of DNA helicase and DNA polymerase at the stalled replication fork (Byun et al., 2005). Importantly, the length of ssDNA and the amount of coated RPA determine the potency of the ATR-mediated checkpoint signaling (MacDougall et al., 2007; Zou and Elledge, 2003).
In the current study, we examined the role of FANCM/FAAP24 in the DNA damage recognition and checkpoint activation following different types of replication stress, including MMC, HU, UV, and CPT. The FANCM/FAAP24 complex was required for RPA foci formation in response to ICL but not other genotoxic stress. ICL-induced RPA foci formation required the DNA binding activity of FAAP24 but not the DNA translocase activity of FANCM. Furthermore, FAAP24-depleted cells showed impaired chromatin association of ATR/ATRIP, resulting in the defects in ICL-induced checkpoint activation. Moreover, by using an in vitro DNA binding assay, we demonstrated that FANCM/FAAP24 promotes efficient RPA binding to ICL DNA. Taken together, we propose that FANCM/FAAP24 can form a unique structure with RPA at an ICL-blocked replication fork and is required for ICL-induced RPA foci formation and checkpoint activation.
RESULTS
ICL-blocked replication forks generate a low level of ssDNA
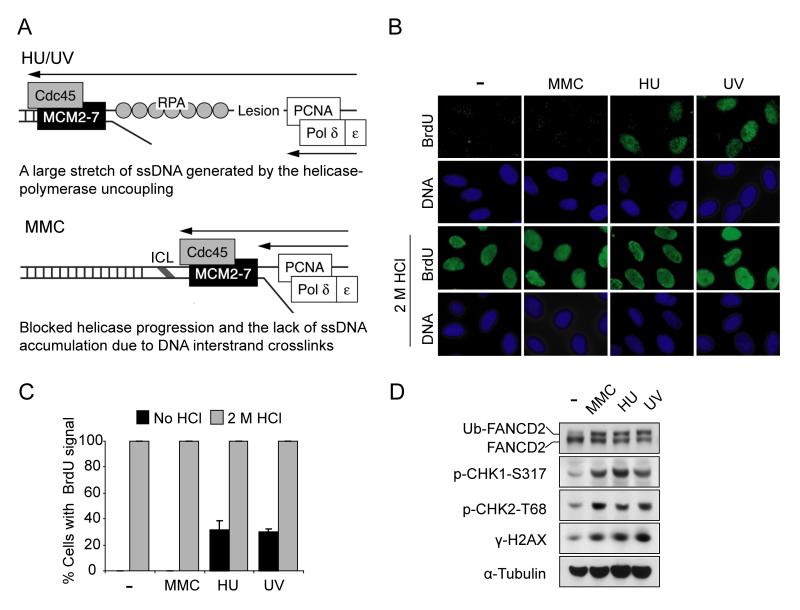
Previous studies indicate that replication forks blocked by HU or UV generate a long stretch of ssDNA due to the uncoupling between the replicative helicase, MCM and DNA polymerase (Cortez, 2005). Compared to HU and UV, an ICL-blocked replication fork may fail to generate a sufficient amount of ssDNA since both helicase and DNA polymerase are blocked by an ICL (Figure 1A). To confirm and extend this model, we initially analyzed ssDNA levels following different kinds of DNA damaging agents (Figure 1B). ssDNA was detected in situ using an anti-BrdU antibody (Raderschall et al., 1999). As predicted, HU and UV treatment caused extensive ssDNA generation, as shown by an increase in BrdU foci. In contrast, ssDNA after exposure to the DNA crosslinker, MMC, at least under these experimental conditions, was only weakly detected, although BrdU was equally incorporated into DNA (as shown under DNA-denaturing conditions) (Figure 1B and 1C). The doses of MMC, HU and UV applied are to reduce cell survival to similar extents (data not shown) and to induce similar levels of FANCD2 monoubiquitination and H2AX phosphorylation (γ-H2AX) (Figure 1D). Time-course and dose-response experiments, using higher concentrations and longer treatment with MMC, did not increase the ssDNA amount (data not shown). Intriguingly, despite the lack of ssDNA generation after MMC treatment, ATR-mediated checkpoint signaling, as indicated by CHK1 phosphorylation (CHK1-S317), was efficiently activated (Figure 1D). Therefore, we hypothesize that cells may have an alternative mechanism for ATR activation in response to ICL, which is independent of large regions of ssDNA.
Figure 1. ICL fails to generate ssDNA at stalled replication forks.
(A) A schematic model comparing stalled DNA replication forks caused by ICL and HU/UV. (B) Representative images showing less ssDNA generated by MMC treatment compared to HU or UV treatments. HeLa cells were labeled with 20 μM BrdU for 36 hr, then were treated with MMC (1 μM, 6 hr), HU (2 mM, 6 hr) or UV (30 J/m2, 6 hr post irradiation), respectively. ssDNA was detected using anti-BrdU antibody without denaturation. The BrdU intensity under the denaturing condition (2 M HCl) shows total BrdU incorporation. (C) Quantification of ssDNA generation shown in (B). The percentage of BrdU-positive cells was determined by counting at least 500 cells from each sample. Data are represented as mean ± SD from three independent experiments. (D) Immunoblot showing similar activation of DNA damage response in the conditions described in (B).
FANCM/FAAP24 is required for ICL-induced RPA foci assembly and phosphorylation
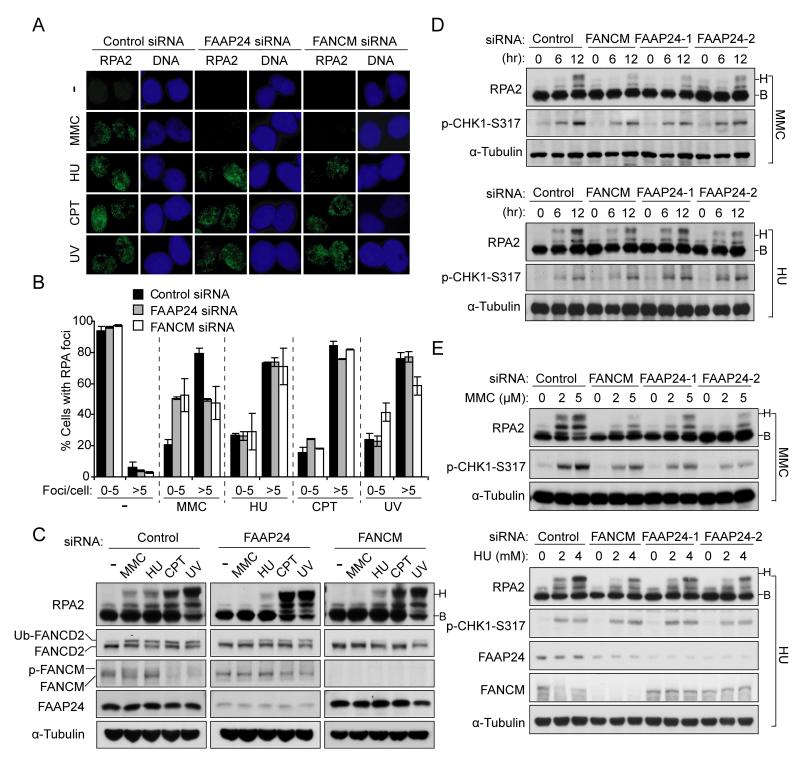
The RPA complex is a ssDNA binding complex that is involved in DNA replication, repair, and recombination (Zou et al., 2006). A diverse range of DNA lesions can generate extended regions of ssDNA-bound RPA (Cortez, 2005; Paulsen and Cimprich, 2007). The RPA nuclear foci formation at DNA damage sites is required for the recruitment of ATR, resulting in the activation of replication checkpoint (Zou and Elledge, 2003). Previous studies have indicated that FANCM and FAAP24 depletion increases RPA foci in the absence of DNA damaging agents, due to spontaneous DNA damage (Collis et al., 2008). However, we found that depletion of either FANCM or FAAP24 reduced RPA foci formation after MMC treatment, but did not significantly affect RPA foci assembly following other DNA damaging agents, including HU, CPT and UV (Figure 2A and 2B). This result indicates that FANCM/FAAP24 complex is preferentially required for RPA foci formation following ICL-inducing agents such as MMC. Disruption of FA core complex by FANCA depletion, in which the FANCM/FAAP24 complex still persists (Kim et al., 2008), had only a slight effect on ICL-induced RPA foci formation (Figure S1A-S1C). This suggests that the FA core complex may be dispensable for ICL-induced RPA foci formation, although it may still be required for optimal checkpoint activation.
Figure 2. FANCM and FAAP24 are selectively required for MMC-induced RPA foci formation and RPA2 phosphorylation.
(A) Representative images showing the requirement of FANCM and FAAP24 for MMC-induced RPA foci assembly but not other replication stress. HeLa cells were treated with MMC (1 μM, 6 hr), HU (2 mM, 6 hr), CPT (500 nM, 6 hr) or UV (30 J/m2, 6 hr post irradiation) respectively at 48 hr after transfection with either FANCM or FAAP24 siRNAs. RPA foci were detected using anti-RPA2 antibody. (B) Quantification for RPA foci shown in (A). The percentage of cells containing RPA foci was determined by counting at least 200 cells from each sample. Data are represented as mean ± SD from three independent experiments. (C) Immunoblot showing that depletion of either FANCM or FAAP24 more severely reduced RPA2 phosphorylation following MMC treatment. HeLa cells were treated as described in (A). Different mobility of RPA2 caused by phosphorylation is indicated as “B” (baseline) and “H” (hyperphosphorylated), respectively. (D) Time-course of RPA2 phosphorylation in FAAP24- or FANCM-depleted cells. HeLa cells transfected with indicated siRNAs were harvested at 0, 6, or 12 hr post treatment with MMC (1 μM) or HU (2 mM). (E) Dose-response of RPA2 phosphorylation in FAAP24- or FANCM-depleted cells. HeLa cells transfected with siRNAs were treated with MMC or HU at indicated concentrations for 8 hr.
In response to DNA damage, RPA accumulates at sites of DNA damage and undergoes DNA damage-dependent phosphorylation on numerous sites of RPA2 (Anantha et al., 2007; Binz et al., 2004; Blackwell et al., 1996; Liu et al., 2005). ICL-induced sequential phosphorylation of RPA2 is largely dependent on ATR activity (Figure S1D). We next examined RPA2 phosphorylation following different types of DNA damaging agents. Consistent with the data for RPA foci formation (Figure 2A and 2B), depletion of either FANCM or FAAP24 largely reduced MMC-induced RPA2 phosphorylation, but barely affected RPA2 phosphorylation following CPT or UV treatment (Figure 2C). Consistent with the known function of FANCM/FAAP24 in ATR-mediated checkpoint after HU treatment (Collis et al., 2008), HU-induced RPA2 phosphorylation was noticeably reduced by FAAP24 or FANCM depletion (Figure 2C-2E). The substantial reduction of MMC-induced RPA2 phosphorylation in either FANCM or FAAP24 depleted cells was also confirmed by different doses and at different time points (Figure 2D and 2E). Taken together, these results indicate that ICL-induced RPA foci formation and RPA2 phosphorylation are correlated, and both events depend on FANCM and FAAP24.
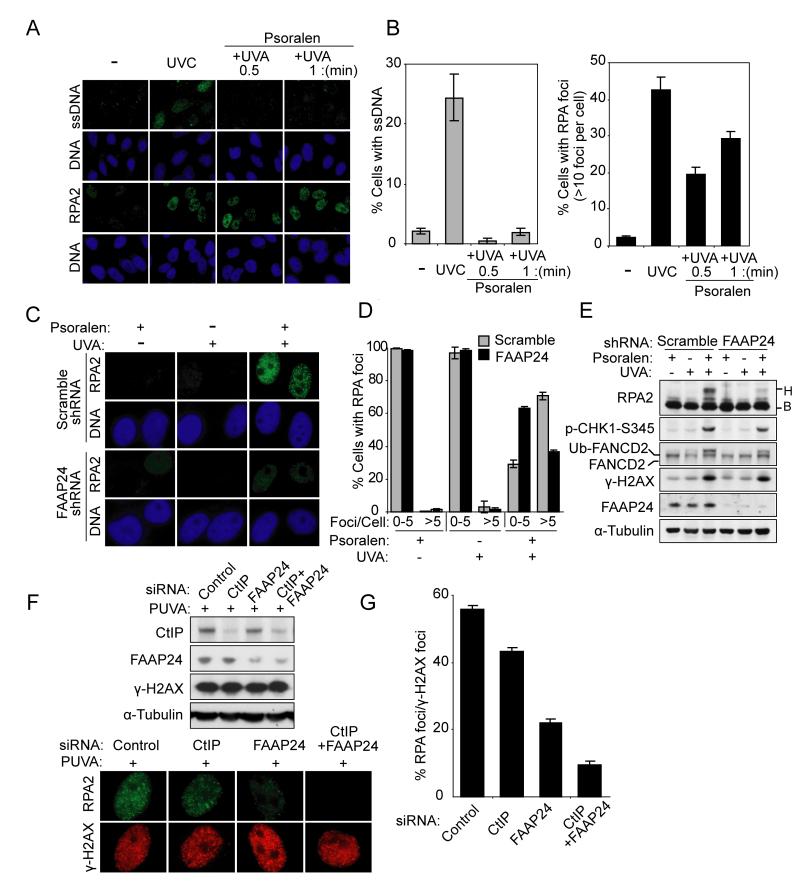
To further confirm that FANCM/FAAP24 is selectively required for ICL-induced RPA foci formation and RPA2 phosphorylation, we used psoralen plus UV-A (PUVA) to induce ICL. Cells treated with increasing doses of PUVA showed undetectable levels of ssDNA but formed discernible RPA foci in a dose-dependent manner (Figure 3A and 3B). Furthermore, consistent with data from MMC treatment (Figure 2), FAAP24 depletion reduced RPA foci formation and RPA2 phosphorylation following PUVA treatment (Figure 3C-3E). These results support our hypothesis that FANCM/FAAP24 may play a key role in ICL-induced DNA damage response through regulating RPA foci formation.
Figure 3. FANCM and FAAP24 are required for RPA foci formation and RPA2 phosphorylation following PUVA-induced ICL formation.
(A) Representative images of ssDNA staining and RPA foci showing that PUVA treatment induced RPA foci but not ssDNA. ssDNA and RPA foci were detected using anti-BrdU antibody or anti-RPA2 antibody 4 hr after PUVA treatment in HeLa cells. The immunostaining intensities are compared to those of UVC treatment. (B) Quantification of ssDNA staining and RPA foci shown in (A). The percentages of cells containing ssDNA (left panel) or cells containing RPA foci (right panel) were determined by counting at least 200 cells from each sample. Data are represented as mean ± SD from three independent experiments. (C) Representative images showing FAAP24-dependent RPA foci formation after PUVA treatment. RPA foci were detected in HeLa cells with scramble shRNA or FAAP24 shRNA 4 hr after PUVA treatment. (D) Quantification of RPA foci shown in (C). The percentage of cells containing RPA foci was determined by counting at least 200 cells from each sample. Data are represented as mean ± SD from three independent experiments. (E) Immunoblot showing reduced RPA2 phosphorylation in FAAP24-depleted cells. Cells were treated as described in (C). Different mobility of RPA2 caused by phosphorylation is indicated as “B” (baseline) and “H” (hyperphosphorylated), respectively. (F) Representative images showing that CtIP depletion resulted in a mild reduction in RPA foci compared to the depletion of FAAP24. HeLa cells were treated with PUVA at 48 hr after transfection with indicated siRNAs. Immunoblot showing the depletion efficiency of indicated proteins (upper panel). After co-staining with RPA and γ-H2AX, RPA foci formation was compared in CtIP-, FAAP24-, or CtIP- and FAAP24-depleted cells (lower panel). (G) Quantification of RPA foci formation shown in (F). The percentage of cells containing RPA foci among γ-H2AX-positive cells was determined by counting at least 200 γ-H2AX-positive cells from each sample. Data are represented as mean ± SD from three independent experiments.
ICL-induced DNA damage also generates DNA double-strand breaks (DSBs), where RPA can be recruited following DNA end resection. To determine the impact of DNA end resection on ICL-induced RPA foci formation, CtIP, which is required for DNA end resection (Sartori et al., 2007), was depleted by siRNA (Figure 3F). Consistent with previous work (Sartori et al., 2007), depletion of CtIP largely abolished CPT-induced RPA foci formation (Figure S3). Following PUVA treatment, CtIP depletion resulted in a mild reduction in RPA foci compared to the depletion of FAAP24 (Figure 3F and 3G). Co-depletion of CtIP and FAAP24 further reduced the proportion of cells displaying RPA foci. These results indicate that FANCM/FAAP24 plays a major role in ICL-induced RPA foci formation, which is independent of the pathway of DSBs resection.
FANCM/FAAP24 is required for ICL-induced checkpoint response
The impairment of RPA recruitment resulting from FAAP24 depletion may lead to the defects in ICL-induced ATR recruitment to chromatin and subsequent checkpoint activation. Indeed, we observed reduced chromatin association of ATR and ATRIP in FAAP24-depleted cells following MMC treatment (Figure S4A). We next examined the impact of FAAP24 in ICL-induced checkpoint response by monitoring the abundance of CDC25A, which is rapidly degraded upon the activation of G2/M checkpoint (Bartek et al., 2004). In FAAP24 short hairpin RNA (shRNA) stable cells, degradation of CDC25A was defective after MMC treatment compared to scramble shRNA cells (Figure S4B). We also analyzed the phosphorylation of Histone H3 as a measure of mitotic entry (Figure S4C and S4D). Scramble and FANCA shRNA cells showed an intact G2/M checkpoint response. However, in contrast to this, knockdown of FAAP24 specifically disrupted the MMC-induced, but not the UV-induced, checkpoint response (Figure S4C and S4D). These results therefore suggest that FANCM/FAAP24 plays an important role in ICL-induced checkpoint activation.
The DNA translocase activity of FANCM is dispensable for ICL-induced RPA foci formation but required for optimal checkpoint activation
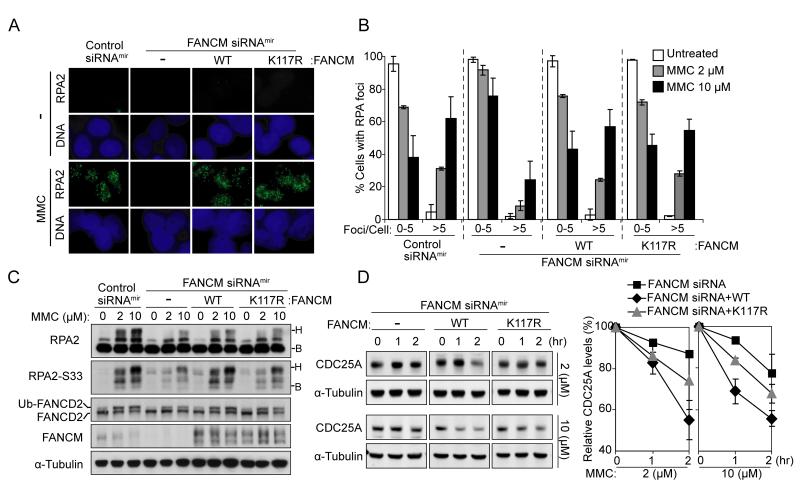
Our results thus suggest that FANCM/FAAP24 activates the ICL checkpoint by promoting RPA recruitment to ICL-stalled replication forks. Previous studies have shown that FANCM possesses ATP-dependent DNA translocase activity in vitro (Gari et al., 2008; Meetei et al., 2005; Mosedale et al., 2005), which may expose ssDNA regions by opening DNA helix. We therefore tested whether this activity could contribute to FANCM/FAAP24-dependent RPA recruitment (Figure 4A and 4B). We examined RPA foci formation in FANCM-depleted FANCM siRNAmir cells (Deans and West, 2009) complemented with either wild-type FANCM or ATPase mutant K117R FANCM. The defect in RPA foci formation in FANCM-depleted cells was rescued by the expression of either wild-type FANCM or K117R FANCM, indicating that ICL-induced RPA foci formation does not require the DNA translocase activity of FANCM (Figure 4A and 4B). However, expression of K117R FANCM did not efficiently recover RPA2 phosphorylation compared with wild-type FANCM (Figure 4C). These data indicate the K117R FANCM-corrected cells exhibit a checkpoint defect despite their intact RPA foci formation.
Figure 4. The DNA translocase activity of FANCM is dispensable for ICL-induced RPA foci assembly.
(A) Representative images showing that K117R FANCM rescued the defective RPA foci formation in FANCM-depleted cells. Non-targeting control siRNAmir, FANCM siRNAmir, and wild-type FANCM or K117R FANCM-corrected FANCM siRNAmir HEK293 Flip-in cells were treated with MMC (10 μM) for 12 hr. RPA foci were detected using anti-RPA2 antibody. (B) Quantification of RPA foci induced by 2 and 10 μM MMC treatment as described in (A). The percentage of cells containing RPA foci was determined by counting at least 200 cells from each sample. Data are represented as mean ± SD from three independent experiments. (C) Immunoblot comparing the defects in RPA2 phosphorylation in the condition as described in (A). Different mobility of RPA2 caused by phosphorylation is indicated as “B” (baseline) and “H” (hyperphosphorylated), respectively. (D) Immunoblot (left panel) and quantification (right panel) for the levels of CDC25A showing that K117R FANCM-corrected FANCM siRNAmir cells had a moderate defect in checkpoint activation. Cells were treated with MMC and harvested at indicated time points. The level of CDC25A was quantified and normalized with that of α-Tubulin. Data are represented as mean ± SD from three independent experiments.
We next examined the ICL-induced checkpoint responses in these cells (Figure 4D). FANCM-depleted cells failed to efficiently degrade CDC25A following MMC treatment, consistent with the results from FAAP24-depleted cells (Figure S4B). Wild-type FANCM-corrected cells recovered the efficient CDC25A degradation in response to MMC treatment, while K117R FANCM-expressing cells showed a moderate defect in CDC25A degradation. These results further suggest that the DNA translocase activity of FANCM is dispensable for RPA foci formation, but is required for efficient checkpoint activation downstream of RPA foci formation.
The DNA binding activity of FAAP24 is required for ICL-induced RPA foci assembly
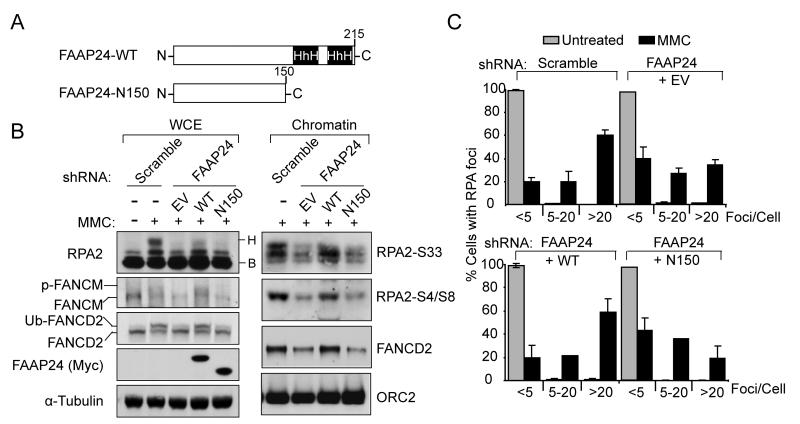
Given that DNA translocase activity of FANCM was dispensable for ICL-induced RPA foci formation, we next tested whether DNA binding activity of FAAP24 is required for RPA foci formation. FAAP24 contains two tandem helix-hairpin-helix (HhH) DNA binding motifs at its carboxyl terminus (Figure 5A), which are necessary for structure-specific DNA binding activity and interaction with FANCM (Ciccia et al., 2007; Ciccia et al., 2008). Expression of wild-type FAAP24 recovered the levels of FANCM and FANCD2 monoubiquitination in FAAP24 shRNA cells (Figure 5B). In contrast, a C-terminal truncated mutant of FAAP24, lacking the (HhH)2 domains, failed to correct these defects (Figure 5B). Furthermore, correction with wild-type FAAP24 recovered RPA foci formation and RPA2 phosphorylation (Figure 5B and 5C), while C-terminal truncated mutant of FAAP24 was defective in these functions. These data thus indicate that DNA binding activity of FAAP24 is critical for FANCM/FAAP24-mediated RPA foci formation and RPA2 phosphorylation.
Figure 5. DNA binding activity of FAAP24 is required for ICL-induced RPA foci formation.
(A) A scheme showing full-length and C-terminal truncated FAAP24. (B) Immunoblot showing wild-type FAAP24 (WT) but not C-terminal truncated FAAP24 mutant (N150) rescued defective RPA2 phosphorylation in FAAP24-depleted HeLa cells. FAAP24 shRNA cells, transfected with empty-vector (EV), Myc-tagged WT or N150 FAAP24, were treated with 1 μM of MMC for 10 hr. Whole cell extracts (WCE) and chromatin enriched extracts (Chromatin) were analyzed by immunoblotting using indicated antibodies. (C) Quantification showing recovered RPA foci by WT but not N150 FAAP24. Cells as described in (B) were treated with MMC 1 μM for 6 hr. RPA foci were detected using anti-RPA2 antibody. The percentage of cells containing RPA foci was determined by counting at least 200 cells from each sample. Data are represented as mean ± SD from three independent experiments.
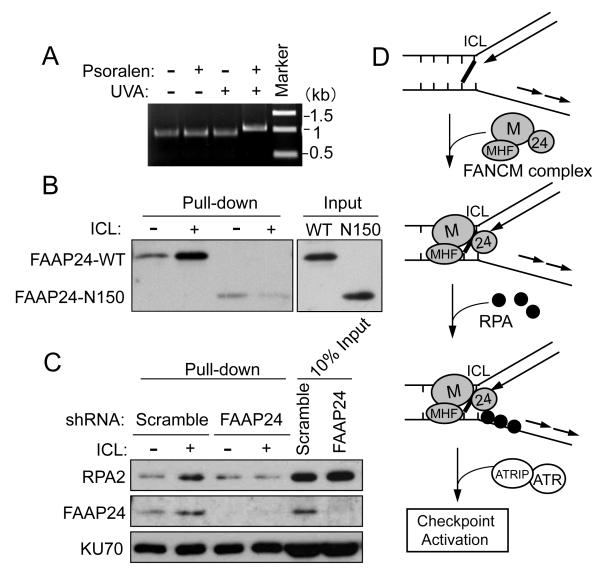
To further investigate the role of FANCM/FAAP24 in ICL-induced RPA recruitment, we employed a DNA pull-down assay to detect RPA binding to ICL DNA in vitro. For this study, we used biotin-labeled double-stranded DNA (dsDNA) as a DNA template instead of stalled replication fork structures. Due to its high affinity to ssDNA in vitro, RPA may bind to ssDNA regions at fork structures even in the absence of FANCM/FAAP24. To circumvent this potential problem, we developed a system using ICL-containing dsDNA for the RPA binding assay, where RPA can bind to dsDNA in an ICL-dependent manner. Using this system, FANCM/FAAP24-dependent RPA binding to ICL DNA was examined. ICL formation on DNA was induced by PUVA treatment in vitro and confirmed by gel electrophoresis (Figure 6A). ICL DNA migrated more slowly than DNA without ICL on a denaturing gel. The recombinant His-tagged wild-type FAAP24 bound preferentially to ICL DNA, which is consistent with previous work (Shen et al., 2009) (Figure 6B). In contrast, the binding of C-terminal truncated FAAP24 to ICL DNA was not increased, indicating that the C-terminal (HhH)2 domains of FAAP24 are critical for the structure-specific DNA binding activity of FAAP24. We next examined the RPA binding to ICL DNA in the nuclear extracts derived from scramble or FAAP24 shRNA cells (Figure 6C). Increased binding affinity of both FAAP24 and RPA to ICL DNA was observed in scramble shRNA cells. In contrast, in the absence of FAAP24, ICL-dependent RPA binding was not detected. We also tested the stability of DNA templates in nuclear extracts and there was no detectable level of DNA processing (Figure S5A).
Figure 6. RPA binding to ICL DNA is dependent on FAAP24.
(A) Migration of ICL DNA on a denaturing gel. (B) Immunoblot showing that FAAP24 wild-type (WT) but not the C-terminal truncated FAAP24 mutant (N150) binds to ICL DNA. Biotinylated control DNA or ICL-DNA (100 ng) were attached to streptavidin-coated beads and incubated with purified His-tagged WT FAAP24 or N150 FAAP24. (C) Immunoblot showing that RPA loading to ICL DNA was decreased in FAAP24-depleted nuclear extracts. Nuclear extracts (100 μg) derived from HeLa scramble or FAAP24 shRNA cells were incubated with biotinylated control DNA or ICL-DNA (100 ng) respectively. The DNA-bound FAAP24, RPA2 and KU70 were detected. (D) Proposed working model for the role of FANCM/FAAP24 in the ICL-induced checkpoint response.
Taken together, these results suggest that FANCM/FAAP24, presumably with MHF proteins, forms a unique structure with RPA at the stalled replication fork and promotes efficient RPA binding, which may in turn recruit other checkpoint proteins and initiate ATR-mediated checkpoint signaling (Figure 6D and Figure S5B).
DISCUSSION
Molecular basis of ICL hypersensitivity
A wide variety of DNA damaging agents can activate the FA core complex dependent monoubiquitination of FANCD2, including MMC, IR, UV, and HU (Andreassen et al., 2004; Garcia-Higuera et al., 2001). However, FA cells are hypersensitive to ICL agents compared to other DNA damage agents and the molecular mechanism underlying this ICL hypersensitivity remains unclear. Here we show that FANCM, and its binding partner FAAP24, may be critical determinants of this crosslinker hypersensitivity. The FANCM/FAAP24 complex appears to have an affinity for ICL-stalled replication forks. FANCM/FAAP24 can recruit RPA to these sites, even in the absence of a long stretch of ssDNA. Once RPA is efficiently recruited, it can further recruit ATR and ATRIP (Figure 6D).
Interestingly, in FANCA shRNA cells, depletion of FANCA only slightly affected ICL-induced RPA foci compared to the depletion of FAAP24 (Figure S1A-S1C), suggesting that the FA core complex may not be involved in ICL-induced RPA foci formation. However, it has been shown that xFANCL-depleted Xenopus extracts have a defect in RPA binding on chromatin (Ben-Yehoyada et al., 2009). While this could reflect differences in depletion efficiencies or species, one possibility of the discrepancy is that xFANCL depletion may co-deplete xFANCM or xFAAP24 in Xenopus extracts. It is also possible that xFANCL may function in RPA recruitment independent of FA core complex.
Given the fact that FA cells, including FANCA deficient cells, exhibit partial intra-S phase checkpoint defects in response to ICL (Centurion et al., 2000; Sala-Trepat et al., 2000), it is possible that the FA core complex plays a role in the efficient S-phase checkpoint activation downstream of RPA foci formation.
The function of FANCM/FAAP24 in the ICL checkpoint response
FANCM/FAAP24 also plays a more general role in the replication checkpoint response (Collis et al., 2008). Collis et al showed that FANCM and FAAP24 are components of a protein complex containing HCLK2, which is involved in ATR-mediated checkpoint response after replication stress. We tested whether reduced ATR activity, resulting from depletion of either FANCM or FAAP24, was the cause of the defect in RPA foci assembly. We compared the effects of ATR depletion or FAAP24 depletion on RPA foci assembly. ATR depletion did not decrease MMC or HU-induced RPA foci formation (Figure S2A). In contrast, FAAP24 depletion specifically disrupted MMC-induced RPA foci formation, indicating that the decrease in RPA foci assembly by FAAP24 depletion occurs in an ATR-independent manner. We also tested RPA foci assembly of a RPA2 phospho-mimetic mutant (RPA2-D4) and a RPA2 phospho-deficient mutant (RPA2-A9) (Vassin et al., 2004) after FANCM/FAAP24 depletion (Figure S2B). MMC-induced RPA2 foci formation was similarly reduced in FAAP24-depleted cells expressing either RPA2 phospho-mimetic mutant (Figure S2C and S2D) or RPA2 phospho-deficient mutant (Figure S2E and S2F). These results further confirmed that the defect in RPA foci assembly by FANCM/FAAP24 depletion is independent of RPA phosphorylation by ATR.
The checkpoint mediator protein TopBP1 binds and activates the ATR/ATRIP complex (Burrows and Elledge, 2008; Kumagai et al., 2006). More recently, TopBP1 has been shown to interact with BACH1/FANCJ and function in the DNA replication checkpoint (Gong et al., 2010). Following MMC treatment, the presence of TopBP1 on chromatin fractions did not differ from those of control cells (Figure S4A). However, another study reported that FANCM-deficient DT40 cells fail to retain TopBP1 on chromatin after CPT treatment (Schwab et al., 2010). The discrepancies in these studies could be explained, at least in part, by the differences in cell type, DNA damaging agent, or depletion efficiencies of the proteins. Based on our observation, we assume that FANCM/FAAP24-mediated ATR activation after ICL damage, is unlikely due to the promotion of chromatin association of TopBP1.
Proposed working model for FANCM/FAAP24-dependent RPA recruitment
There are several models for FANCM/FAAP24 mediated RPA recruitment. First, FANCM/FAAP24 may form a specific structure at the ICL-stalled replication forks, where FANCM/FAAP24 binding may expose small regions of ssDNA, so that RPA can gain access to the ICL lesions. Second, FANCM/FAAP24 may directly bind to RPA. Although neither FANCM nor FAAP24 contains RPA2 interaction motifs, similar to those found in TIPIN, XPA, UNG2, and RAD52 (Mer et al., 2000; Unsal-Kacmaz et al., 2007), previous work has shown that the yeast homologue of FANCM, mph1, interacts with RPA through its C-terminal motif (Banerjee et al., 2008). Third, FANCM/FAAP24 may indirectly bind to RPA through interaction with other DNA repair proteins. FANCM was originally immunoisolated as a member of the BRAFT (for BLM, RPA, FA, and Topo III3B1) complex (Meetei et al., 2003). Indeed, recent reports have shown that FANCM associates with BLM, which is known to interact with RPA (Brosh et al., 2000; Deans and West, 2009; Meetei et al., 2005).
In summary, in addition to the general role of FANCM/FAAP24 in DNA replication checkpoint, we found that FANCM/FAAP24 participates early in ICL-induced DNA damage response through regulating RPA foci formation at ICL-stalled replication forks (Figure 6D and S5B). This function of FANCM/FAAP24 may contribute, at least in part, to the ICL-specific cellular response and implicate an important role for FA proteins in ICL DNA damage response.
Experimental Procedures
Cell Culture and constructs
HeLa and HEK293 Flip-in cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen) supplemented with 15% fetal bovine serum. FAAP24 and FANCA shRNA stable HeLa cells were generated based on lentiviral pLKO-puro vector (Open Biosystem). shRNA resistant cDNAs for wild-type and C-terminal truncated FAAP24 mutant were cloned into pcDNA3.1(+) Myc vector. Site-directed mutagenesis was performed following the instructions of QuickChange protocol (Stratagene). Plasmid transfection was performed using Lipofectamine 2000 (Invitrogen). All the siRNAs were purchased from Qiagen and transfected using Lipofectamine™ RNAimax (Invitrogen). Information regarding siRNA sequences can be found in the supplemental information.
Antibodies
Anti-FAAP24 and anti-FANCM antibodies were described previously (Kim et al., 2008). Other antibodies used in this study are as follows: anti-RPA2 antibody (EMD biosciences); anti-ATM, anti-ATRIP, anti-phospho-RPA2-S33, and anti-phospho-RPA2-S4/S8 antibodies (Bethyl Laboratories); anti-FANCD2, anti-CtIP and anti-CDC25A antibodies (Santa Cruz Biotechnology); anti-KU70 antibody (NeoMarkers); anti-phospho-CHK1-S317, anti-phospho-CHK1-S345, anit-phospho-CHK2-T68, anti-ATR, Alexa Fluor 488 conjugated phospho-Histone H3 (S10) antibodies (Cell signaling Technology); anti-γ-H2AX antibody (Upstate); anti-ORC2 antibody (BD Pharmingen); and anti-α-Tubulin antibody (Sigma).
PUVA treatment
Cells were washed with PBS and incubated in PBS containing 100 ng/ml of 8-methoxypsoralen (Sigma) for 15 min. After incubation, cells were irradiated with UV-A in the presence of 8-methoxypsoralen.
Detection of ss DNA with BrdU incorporation
ssDNA generated by DNA damage was detected as described previously with minor modifications (Raderschall et al., 1999). Briefly, cells were grown for 36 h in culture medium containing 20 μM BrdU and then cultured without BrdU overnight before treatment with different damaging agents. Cells were fixed with cold methanol, rinsed with acetone and then incubated with Anti-BrdU antibody (BD Biosciences) without DNA denaturation. The total amount of incorporated BrdU was determined under denatured condition using 2 M HCl treatment.
Immunofluorescence
Cells were pre-extracted with extraction buffer (0.5% Triton-X100 in 200 mM HEPES at pH 7.4, 50 mM NaCl, 3 mM MgCl2 and 300 mM sucrose) on ice for 5 min prior to fixation with 4% paraformaldehyde. RPA foci were detected using anti-RPA2 antibody and visualized using Alexa Fluor 488-conjugated secondary antibody. The quantification of cells with RPA foci was performed by counting the number of cells with RPA foci. At least 200 cells were counted from each sample. Data are represented as mean ± SD from three independent experiments.
Preparation of ICL DNA in vitro
Biotinylated 1 kb DNA fragments were incubated with 20 μg/ml 8-methoxypsoralen at room temperature for 30 min prior to UV-A irradiation. The efficiency of crosslinking was confirmed on denaturing alkaline agarose gel. 1 kb DNA fragments were prepared by PCR amplification from pME18S plasmid and the locations of PCR primers were randomly set.
DNA Binding Assay
DNA binding assays and preparation of nuclear extract were performed as described (Shiotani and Zou, 2009) with modifications. The biotinylated DNA fragments were attached to streptavidin-coated magnetic beads according to manufacturer’s instructions (Dynal). The beads coated with DNA were incubated with purified wild-type FAAP24 or C-terminal truncated FAAP24 mutant with nuclear extracts in the binding buffer [10 mM Tris-HCl, (pH 7.5), 100 mM NaCl, 10% glycerol, 0.01% NP-40, and 10 μg/ml bovine serum albumin]. After a 30-min incubation at room temperature, beads were retrieved and washed 2 times with the binding buffer. The proteins bound to beads were denatured in the SDS sample buffer, separated on SDS-PAGE, and analyzed by immunoblotting.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Andrew J. Deans and Stephen C. West for providing the FANCM siRNAmir, wild-type FANCM- and K117R FANCM-corrected FANCM siRNAmir HEK293 Flip-in cells and James A. Borowiec for the Myc-RPA2-WT, Myc-RPA2-D4 and Myc-RPA2-A9 plasmids. We thank Patricia A. Stuckert and Jason T. Tsichlis for technical assistance. We also thank Dipanjan Chowdhury, Clark C. Chen, Johannes Walter, Brendan D. Price and the members of the D’Andrea laboratory for helpful discussions. K.Y. is a Harvard University Presidential Scholar. This work was supported by NIH grants PO1CA092584, RO1DK43889, and RO1HL52725 to A.D.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anantha RW, Vassin VM, Borowiec JA. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J. Biol. Chem. 2007;282:35910–35923. doi: 10.1074/jbc.M704645200. [DOI] [PubMed] [Google Scholar]
- Andreassen PR, D’Andrea AD, Taniguchi T. ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev. 2004;18:1958–1963. doi: 10.1101/gad.1196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbach AD. Fanconi anemia and its diagnosis. Mutat. Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Smith S, Oum JH, Liaw HJ, Hwang JY, Sikdar N, Motegi A, Lee SE, Myung K. Mph1p promotes gross chromosomal rearrangement through partial inhibition of homologous recombination. J. Cell Biol. 2008;181:1083–1093. doi: 10.1083/jcb.200711146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell. Biol. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- Ben-Yehoyada M, Wang LC, Kozekov ID, Rizzo CJ, Gottesman ME, Gautier J. Checkpoint signaling from a single DNA interstrand crosslink. Mol. Cell. 2009;35:704–715. doi: 10.1016/j.molcel.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz SK, Sheehan AM, Wold MS. Replication protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- Blackwell LJ, Borowiec JA, Mastrangelo IA. Single-stranded-DNA binding alters human replication protein A structure and facilitates interaction with DNA-dependent protein kinase. Mol. Cell. Biol. 1996;16:4798–4807. doi: 10.1128/mcb.16.9.4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM, Jr., Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J. Biol. Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- Burrows AE, Elledge SJ. How ATR turns on: TopBP1 goes on ATRIP with ATR. Genes Dev. 2008;22:1416–1421. doi: 10.1101/gad.1685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centurion SA, Kuo HR, Lambert WC. Damage-resistant DNA synthesis in Fanconi anemia cells treated with a DNA cross-linking agent. Exp. Cell Res. 2000;260:216–221. doi: 10.1006/excr.2000.4995. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, de Winter JP, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Ciccia A, McDonald N, West SC. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 2008;77:259–287. doi: 10.1146/annurev.biochem.77.070306.102408. [DOI] [PubMed] [Google Scholar]
- Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol. Cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Cortez D. Unwind and slow down: checkpoint activation by helicase and polymerase uncoupling. Genes Dev. 2005;19:1007–1012. doi: 10.1101/gad.1316905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- Deans AJ, West SC. FANCM Connects the Genome Instability Disorders Bloom’s Syndrome and Fanconi Anemia. Mol. Cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D’Andrea AD. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- Gong Z, Kim JE, Leung CC, Glover JN, Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol. Cell. 2010;37:438–446. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy RD, D’Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- Kim JM, Kee Y, Gurtan A, D’Andrea AD. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111:5215–5222. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kvaratskhelia M, Hess S, Qu Y, Zou Y. Modulation of replication protein A function by its hyperphosphorylation-induced conformational change involving DNA binding domain B. J. Biol. Chem. 2005;280:32775–32783. doi: 10.1074/jbc.M505705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke-Glaser S, Luke B, Grossi S, Constantinou A. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J. 2010;29:795–805. doi: 10.1038/emboj.2009.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 2003;23:3417–3426. doi: 10.1128/MCB.23.10.3417-3426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mer G, Bochkarev A, Chazin WJ, Edwards AM. Three-dimensional structure and function of replication protein A. Cold Spring Harb. Symp. Quant. Biol. 2000;65:193–200. doi: 10.1101/sqb.2000.65.193. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, D’Andrea AD. How the fanconi anemia pathway guards the genome. Annu. Rev. Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale G, Niedzwiedz W, Alpi A, Perrina F, Pereira-Leal JB, Johnson M, Langevin F, Pace P, Patel KJ. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat. Struct. Mol. Biol. 2005;12:763–771. doi: 10.1038/nsmb981. [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Raderschall E, Golub EI, Haaf T. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. U. S. A. 1999;96:1921–1926. doi: 10.1073/pnas.96.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Trepat M, Rouillard D, Escarceller M, Laquerbe A, Moustacchi E, Papadopoulo D. Arrest of S-phase progression is impaired in Fanconi anemia cells. Exp. Cell Res. 2000;260:208–215. doi: 10.1006/excr.2000.4994. [DOI] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab RA, Blackford AN, Niedzwiedz W. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J. 2010;29:806–818. doi: 10.1038/emboj.2009.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Do H, Li Y, Chung WH, Tomasz M, de Winter JP, Xia B, Elledge SJ, Wang W, Li L. Recruitment of fanconi anemia and breast cancer proteins to DNA damage sites is differentially governed by replication. Mol. Cell. 2009;35:716–723. doi: 10.1016/j.molcel.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani B, Zou L. Single-stranded DNA orchestrates an ATM-to-ATR switch at DNA breaks. Mol. Cell. 2009;33:547–558. doi: 10.1016/j.molcel.2009.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Bakker ST, Agarwal S, Jansen M, Grassman E, Godthelp BC, Ali AM, Du CH, Rooimans MA, Fan Q, et al. Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood. 2009;114:174–180. doi: 10.1182/blood-2009-02-207811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Saro D, Ali AM, Zheng XF, Du CH, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol. Cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D’Andrea AD, Elledge SJ. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol. Cell. Biol. 2007;27:3131–3142. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassin VM, Wold MS, Borowiec JA. Replication protein A (RPA) phosphorylation prevents RPA association with replication centers. Mol. Cell. Biol. 2004;24:1930–1943. doi: 10.1128/MCB.24.5.1930-1943.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- Yan Z, Delannoy M, Ling C, Daee D, Osman F, Muniandy PA, Shen X, Oostra AB, Du H, Steltenpool J, et al. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol. Cell. 2010;37:865–878. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell. Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.