Abstract
Neuroimaging genomics is a new transdisciplinary research field, which aims to examine genetic effects on brain via integrated analyses of high throughput neuroimaging and genomic data. We report our recent work on (1) developing an imaging genomic browsing system that allows for whole genome and entire brain analyses based on visual exploration and (2) applying the system to the imaging genomic analysis of an existing MCI/AD cohort. Voxel-based morphometry is used to define imaging phenotypes. ANCOVA is employed to evaluate the effect of the interaction of genotypes and diagnosis in relation to imaging phenotypes while controlling for relevant covariates. Encouraging experimental results suggest that the proposed system has substantial potential for enabling discovery of imaging genomic associations through visual evaluation and for localizing candidate imaging regions and genomic regions for refined statistical modeling.
Index Terms: Imaging genomics, voxel-based morphometry, genome-wide association study, visual exploration
I. INTRODUCTION
Neuroimaging genomics [1], [2] has recently emerged as a transdisciplinary research field where new strategies are examined to evaluate genetic effects on brain structure and function through joint analysis of imaging and genomic data. Although genome-wide association studies have been actively performed [3], it remains a highly challenging issue to effectively relate high throughput genotyping data to large scale image data. As pointed out by Glahn et. al. [1], in prior imaging genomics studies, researchers often reduce the image data to a small number of variables (e.g., nine imaging measures used in [2]) or focus on a single SNP or gene (e.g., [4]), to limit the number of statistical tests, control Type I error, and make computation feasible. However, significant reduction in one or both data types greatly limits our capability of identifying important relationships. We report our recent work on developing an imaging genomic browsing system that allows for whole genome and entire brain analyses via visual exploration. An initial prototype of this system was reported in [5], where one-way analysis of variance (ANOVA) was used to measure the associations between brain imaging phenotypes and genotypes and was tested on a synthetic data set. In this paper, we extend this system and apply it to a real data set focusing on amnestic mild cognitive impairment (MCI) and early Alzheimer’s disease (AD) [6]. Besides ANOVA, in this work, we extend this system to include analysis of covariance (ANCOVA) so that we can not only examine the associations between imaging phenotypes and genotypes, but also study the main effects of genotypes and diagnosis and their interaction effects in relation to imaging phenotypes while controlling for covariates such as age and gender.
II. METHODS
This imaging genomic analysis was performed on an existing MCI/AD cohort [6]. Structural magnetic resonance imaging (MRI) data and genotype data were available for the cohort and subsequent imaging genomic analysis with these two types of data was performed using an imaging genomic browsing system [5]. Further information about the data and the analysis using the imaging genomic browsing system with advanced features is explained in Section II and Section III.
A. Data preprocessing
Participants of this study were selected from an existing MCI/AD cohort [6], including all the subjects in the cohort who have both imaging and genomic data available. These participants included 39 healthy older adults (HC), 36 euthymic older adults with cognitive complaints (CC), 34 older adults with amnestic MCI, and 6 adults with AD. Table I shows several participant characteristics. Structural MRI data were acquired on a 1.5 Tesla General Electric (GE) LX Horizon scanner using a T1-weighted Spoiled Gradient Recalled (SPGR) coronal series with 1.5 mm slice thickness. Voxel-based morphometry (VBM) [7] was employed for extracting gray matter (GM) maps of all participants and the SPM5 software package [8] was used for this purpose. Initial GM maps were extracted by segmenting the T1-weighted SPGR volumes after resampling them to 1 mm3 isotropic voxels. A 12-parameter model was used to spatially normalize the GM maps to the GM prior probability template. The normalized GM maps were smoothed using an isotropic spatial filter with full width half maximum of 10mm to help increase signal-to-noise ratio and account for individual differences in gyral anatomy. The smoothed normalized GM maps were used as imaging phenotypes in the subsequent analyses, where each voxel location corresponded to an imaging variable.
TABLE I.
Participant characteristics
| Age (mean±std) | Sex (M, F) | |
|---|---|---|
| HC | 71±5.1 years | 12, 27 |
| CC | 73±5.6 years | 13, 23 |
| MCI | 73±6.7 years | 19, 15 |
| AD | 71±9.2 years | 4, 2 |
| ALL | 72±6.0 years | 48, 67 |
Genotype data was acquired from a custom Affymetrix single nucleotide polymorphism (SNP) panel that included 3300 common SNPs in 1100 candidate genes selected from candidate molecular pathways for age-related memory decline [9]. These candidate genes/pathways were selected based on (1) a detailed search using Medline/PubMed and multiple databases of known or suspected genetic associations with neurological, psychiatric, neurodevelopmental and neurodegenerative disorders of central nervous system and (2) a detailed on-line and manual search for known or hypothesized genes involved in cognition and memory in humans and animal models. SNP values were coded as follows: 0 for AA, 1 for Aa, 2 for aa, and 5 for missing data. Before the imaging genomic analyses, a quality check for genotype data was performed using PLINK [3] and the results were briefly described below: Total genotyping rate was 0.99102. No individual was removed for low genotyping (missing rate per individual MIND > 0.05). 46 SNPs failed missingness test (missing rate per marker GENO > 0.1), 41 SNPs failed frequency test (minor allele frequency MAF < 0.01), and they were excluded from the analysis. All the SNPs passed Hardy-Weinberg Test (p ≤ 10−6) and none was excluded for this reason. This procedure also identified one pair of participants among 115 subjects as a sibling pair. One sibling was randomly selected and included in the analysis and the other was excluded. Thus the total number of subjects involved in the study became 114.
B. Data analysis
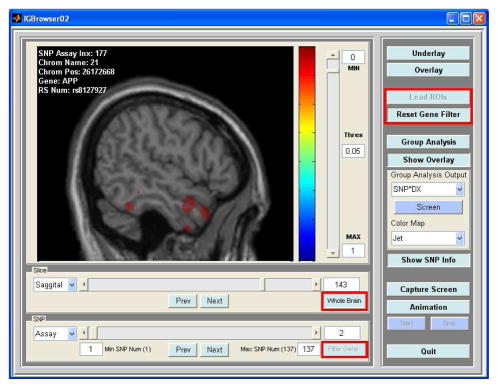
Genome-wide association studies are very computationally intensive tasks partially due to a large number of genomic variables involved. In an imaging genomic analysis, the number of imaging variables is very large as well. Therefore, the computational bottleneck becomes the major challenge that such a study faces. To expedite the procedure of this imaging genomic study, the following two features were incorporated into our imaging genomic system: selection of regions of interest (ROIs), selection of candidate genes, highlighted with red rectangles in Fig. 1. To reduce the amount of computation, instead of conducting the whole brain analysis, in this study, hippocampus, amygdala, and the entire temporal lobe in both hemispheres were chosen as ROIs because these ROIs included brain structures most affected by MCI/AD. In addition to reducing the number of imaging variables, instead of scanning 3300 SNPs, we selected a subset of SNPs known to be related to AD. These SNPs were determined based on a search on the Alzheimer Research Forum (http://www.alzforum.org/res/com/gen/alzgene/default.asp) [10]. From this search, 16 genes were found (see Table II) in common to our 1100 candidate genes and 137 SNPs were selected.
Fig. 1.
Graphical user interface of the imaging genomic browser, displaying a resultant statistical map on top of an anatomical underlay.
TABLE II.
Symbols and official full names of 16 selected genes.
| Gene Symbol | Official Full Name |
|---|---|
| ACE | angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 |
| APOE | apolipoprotein E |
| APP | amyloid beta (A4) precursor protein |
| BDNF | brain-derived neurotrophic factor |
| CHRNB2 | cholinergic receptor, nicotinic, beta 2 (neuronal) |
| CST3 | cystatin C |
| IL1B | interleukin 1 beta |
| MAPT | microtubule-associated protein tau |
| MTHFR | 5,10-methylenetetrahydrofolate reductase (NADPH) |
| NEDD9 | neural precursor cell expressed, developmentally down-regulated 9 |
| PRNP | prion protein (p27–30) |
| PSEN1 | presenilin 1 |
| PSEN2 | presenilin 2 |
| SORL1 | sortilin-related receptor, L(DLR class) A repeats-containing |
| TF | transferrin |
| TFAM | transcription factor A, mitochondrial |
This imaging genomic analysis was designed to find interaction effects of diagnosis and SNP on imaging phenotypes. The imaging phenotypes (GM maps) could be affected by other factors, such as age, gender, and intracranial volume (ICV, calculated from the FreeSurfer imaging analysis suite [11]). To remove the effect of additional factors, multi-factor ANCOVA was intended to be performed. Partially due to functional limitations of Matlab (2007b, The MathWorks, Natick, MA), multi-factor ANOVA with continuous and categorical factors was performed for each selected imaging variable instead of multi-factor ANCOVA in this study.
III. RESULTS
A. Performance of imaging genomic system
Fig. 1 shows the graphical user interface of our imaging genomic browsing system, which was used to conduct imaging genomic analyses via visual exploration. The current version of the program can perform multi-factor ANOVA with multiple categorical and continuous factors to look for main and interaction effects of factors on imaging phenotypes. In addition, it has functions to reduce search ranges in imaging and genomic domains and for users to interactively change viewing conditions such as view direction, threshold, and colormap. This system is based on Matlab, so that it can run on multiple platforms. If we run this imaging genomic system on a single machine, its performance in terms of computational time for calculating a statistical map of multi-factor ANOVA between one image and one SNP is acceptable for moderately sized data sets. With our data, it took about 2 to 4 seconds to calculate one statistical map, depending on the number of imaging variables within each image slice.
B. Statistical analysis
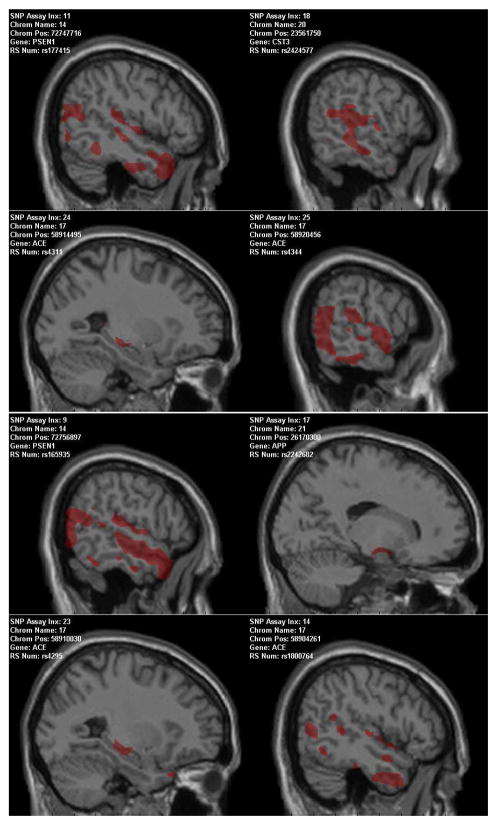
In this analysis, we focused on examining the interaction effect of genotypes and diagnosis on imaging phenotypes in order to localize ROIs in the imaging domain and candidate SNPs in the genomic domain for refined statistical modeling. We grouped all the subjects into three diagnostic groups: HC, CC, and MCIAD (i.e., MCI or AD). For each SNP, we had three genotypes: AA, Aa, and aa. Thus the interaction of genotype by diagnosis could take nine possible values. For simplicity, we defined these nine values (G1–G9) as follows: G1=AA*HC, G2=Aa*HC, G3=aa*HC, G4=AA*CC, G5=Aa*CC, G6=aa*CC, G7=AA*MCIAD, G8=Aa*MCIAD, and G9=aa*MCIAD. Given a SNP location, each subject could take one of the above nine values. Note that we only had 114 subjects but needed to divide them into nine groups for each SNP. This would result in very small groups (e.g., those containing only one or two subjects) in many cases. Clearly, performing statistical analysis on these cases would not derive meaningful results. Therefore, we added a constraint to our analysis, requiring the minimum number of subjects among these nine groups to be be equal to or greater than 5. Fig. 2 and Table III show all the results that meet this requirement. In Fig. 2, colored associations are significant at the level of p < 0.05 for visualization purpose and in each panel, at least one voxel within the selected ROIs was significantly correlated with one SNP among 137 SNPs at the level of p < 10−4. At this significance level (p < 10−4) and minimum number of subjects (≥ 5), 4 genes (ACE, APP, CST3, and PSEN1) among 16 candidate genes, listed in Table II, contained SNPs that were significantly associated with some imaging variables within the selected ROIs (hippocampus, amygdala, and the entire temporal lobe in both hemispheres).
Fig. 2.
Imaging genomic patterns.
TABLE III.
Number of subjects in each SNP by diagnosis group. Subject groups (G1–G9) were defined by SNP values and diagnoses. The number of subjects in each row is 114 or less, since the genotyping missing rates vary among these SNPs. Resultant map of each RefSNP corresponds to each panel in Fig. 2 from left to right and top to bottom.
| RefSNP | G1 | G2 | G3 | G4 | G5 | G6 | G7 | G8 | G9 |
|---|---|---|---|---|---|---|---|---|---|
| rs177415 | 7 | 23 | 8 | 6 | 14 | 15 | 9 | 18 | 11 |
| rs2424577 | 17 | 16 | 6 | 17 | 12 | 6 | 14 | 19 | 6 |
| rs4311 | 13 | 17 | 9 | 11 | 18 | 7 | 12 | 18 | 9 |
| rs4344 | 8 | 19 | 12 | 7 | 18 | 11 | 11 | 19 | 9 |
| rs165935 | 12 | 19 | 8 | 11 | 20 | 5 | 12 | 16 | 11 |
| rs2242682 | 13 | 19 | 7 | 13 | 18 | 5 | 13 | 19 | 7 |
| rs4295 | 15 | 18 | 6 | 17 | 14 | 5 | 16 | 18 | 5 |
| rs1800764 | 11 | 16 | 12 | 10 | 14 | 12 | 5 | 19 | 15 |
APP, PSEN1, and CST3 genes are known to contribute to the early-onset of Alzheimer’s disease (AD) [12], [13], [14] and ACE is associated with AD [15]. The APOE gene, which is widely established as a major genetic risk factor for the development of AD, failed genotyping on the targeted array and so was not included in this study.
IV. CONCLUSIONS
We presented our initial efforts toward developing an imaging genomic browsing system. This system was applied to an existing MCI/AD cohort and produced very encouraging results, consistent to the findings from other studies. These results support the usefulness of the imaging genomic browsing system as an analysis tool for refining results from genome-wide association studies and localizing brain regions, associated with specific genes and/or SNPs. To extend the effectiveness of the system, further refined statistical modeling will be employed for the localized imaging ROIs and candidate SNPs. In addition to confirming these results, development of new computational and algorithmic methods are under consideration to improve computational performance. The current computational performance is acceptable for moderately sized data but not fast enough to explore large-scale data. Therefore, a future plan includes further improvement of performance by developing more efficient algorithms or employing parallel computing systems.
Acknowledgments
This work was supported in part by NIA R01 AG19771, NIBIB R03 EB008674, NIA P30 AG10133, and NCI R01 CA101318 from the NIH, Foundation for the NIH, an Indiana CTSI CBR/CTR award, and grant #87884 from the Indiana Economic Development Corporation (IEDC).
Contributor Information
Sungeun Kim, Email: sk31@iupui.edu.
Li Shen, Email: shenli@iupui.edu.
Andrew J. Saykin, Email: asaykin@iupui.edu.
John D. West, Email: jdwest@iupui.edu.
References
- 1.Glahn DC, Thompson PM, Blangero J. Neuroimaging endophenotypes: Strategies for finding genes influencing brain structure and function. Hum Brain Mapp. 2007;28:488–501. doi: 10.1002/hbm.20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seshadri S, DeStefano A, Au R, Massaro J, Beiser A, et al. Genetic correlates of brain aging on MRI and cognitive test measures: a genome-wide association and linkage analysis in the framingham study. BMC Med Genet. 2007;8:S15. doi: 10.1186/1471-2350-8-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad RH, Emily MD, Daniel RW. Imaging genetics: Perspectives from studies of genetically driven variation in serotonin function and corticolimbic affective processing. Biol Psychiatry. 2006;59:888–897. doi: 10.1016/j.biopsych.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Kim S, Shen L, Saykin AJ, West JD. Data synthesis and tool development for exploring imaging genomic patterns. IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology. 2009:298–305. doi: 10.1109/CIBCB.2009.4925742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saykin AJ, Wishart HA, Rabin LA, et al. Older adults with cognitive complaints show brain atrophy similar to that of amnestic MCI. Neurology. 2006;67:834–842. doi: 10.1212/01.wnl.0000234032.77541.a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashburner J, Friston K. Voxel-based morphometry-the methods. NeuroImage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 8.Wellcome Dept. of Imaging Neuroscience. The statistical parametric mapping software package. London, UK: [accessed on March 31, 2008.]. Available at http://www.fil.ion.ucl.ac.uk/spm/ [Google Scholar]
- 9.Saykin AJ, Sloan CD, Wishart HA, Flashman LA, et al. A genome wide association study using mri and a targeted pathway array to examine medial temporal lobe morphology in older adults at-risk for alzheimer’s disease. World Cong of Psychiatric Genetics. 2007:868. [Google Scholar]
- 10.Bertram L, McQueen M, Mullin K, Blacker D, Tanzi R. [accessed on Jan 25, 2009.];Alzheimer research forum. Available at http://www.alzgene.org.
- 11.Athinoula A Martinos Center for Biomedical Imaging. [accessed on Jan 25, 2009.];FreeSurfer imaging analsysi suite. Available at http://surfer.nmr.mgh.harvard.edu/
- 12.Cruts M, Hendriks L, Van Broeckhoven C. The presenilin genes: a new gene family involved in alzheimer disease pathology. Hum Mol Genet. 1996;5:1449–1455. doi: 10.1093/hmg/5.supplement_1.1449. [DOI] [PubMed] [Google Scholar]
- 13.Ancolio K, Dumanchin C, Barelli H, Warter JM, Brice A, et al. Unusual phenotypic alteration of beta amyloid precursor protein (betaAPP) maturation by a new Val-715 → Met betaAPP-770 mutation responsible for probable early-onset alzheimer’s disease. Proc Natl Acad Sci USA. 1999;96(7):4119–4124. doi: 10.1073/pnas.96.7.4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beyer K, Lao JI, Gomez M, Riutort N, Latorre P, et al. Alzheimer’s disease and the cystatin c gene polymorphism: an association study. Neurosci Lett. 2001;23(315):17–20. doi: 10.1016/s0304-3940(01)02307-2. [DOI] [PubMed] [Google Scholar]
- 15.Narain Y, Yip A, Murphy T, Brayne C, Easton D, et al. The ACE gene and Alzheimer’s disease susceptibility. J Med Genet. 37 doi: 10.1136/jmg.37.9.695. [DOI] [PMC free article] [PubMed] [Google Scholar]