Abstract
The aryl hydrocarbon receptor (AhR) participates in the differentiation of mouse regulatory T cells (Treg cells) and interleukin 17 (IL-17)-producing helper T cells (TH17 cells), but its role in human T cell differentiation is unknown. We investigated the role of AhR in the differentiation of human induced Treg cells (iTreg cells). We found that AhR activation promoted the differentiation of CD4+Foxp3− T cells, which produce IL-10 and control responder T cells through granzyme B. However, activation of AhR in the presence of transforming growth factor-β1 induced Foxp3+ iTreg cells, which suppress responder T cells through the ectonucleoside triphosphate diphosphohydrolase CD39. The induction of functional Foxp3+ iTreg cells required coordinated action of the transcriptional regulators Smad1 and Aiolos. Thus, AhR is a potential target through which functional iTreg cells could be induced in human autoimmune disorders.
In healthy people, the immune response is controlled by several subsets of regulatory T cells (Treg cells) that are generated in the thymus (natural Treg) and also in the periphery in response to various tolerogenic stimuli (induced Treg cells (iTreg cells)1. One of these subsets is a population of CD4+ T cells characterized by expression of the transcription factor Foxp3 (A002750)1. In mice, Foxp3 is a specific marker for Treg cells, and forced expression of Foxp3 (refs. 2,3) or its induction with transforming growth factor-β1 (TGF-β1)4 promotes the differentiation of functional Foxp3+ Treg cells. In humans, however, Foxp3 expression is not always linked to regulatory function: activated T cells transiently express Foxp3 (refs. 5,6), and neither forced overexpression of Foxp3 (ref. 7) nor its induction with TGF-β1 (ref. 8) results in the differentiation of suppressive Foxp3+ Treg cells. Thus, additional signals beyond those controlled by Foxp3 are required for the generation of human functional Foxp3+ Treg cells.
An additional subset of CD4+ Treg cells are the Foxp3−IL-10+ T cells, called type 1 regulatory T cells (Tr1 cells), initially generated after repeated cycles of in vitro stimulation in the presence of interleukin 10 (IL-10)9. Additional regimens, such as stimulation in the presence of IL-27, the addition of antibody to CD46 (anti-CD46), treatment with dexamethasone and vitamin D, or the administration of rapamycin and IL-10 can also promote the differentiation of Tr1 cells10,11. Tr1 cells are important in immune homeostasis and the control of graft-versus-host disease10, but the signaling pathways that control their differentiation, especially in humans, are largely unknown.
Mice that lack functional Foxp3+ Treg cells develop severe auto-immunity2, and Treg cell deficits have been described in several human autoimmune diseases12. Because Treg cells have positive effects in experimental models of autoimmunity, their induction is viewed as a promising approach for the treatment of human autoimmune disorders. Several methods have been reported to differentiate and expand human Foxp3+ iTreg cell populations in vitro, but their ability to produce substantial numbers of functional cells in a consistent manner is limited13. It is therefore important to characterize the pathways that control the generation of functional human Foxp3+ iTreg cells.
The ligand-activated transcription factor aryl hydrocarbon receptor (AhR (A000229)) controls the differentiation of mouse Treg cells and IL-17-producing helper T cells (TH17 cells) in vitro and in vivo14–20. Activation of AhR by its high-affinity ligand TCDD (2,3,7,8-tetrachlorodibenzo-p-dioxin) in vivo results in the induction of CD4+CD25+Foxp3+ Treg cells14. These cells are functional and suppress the development of experimental autoimmune encephalo-myelitis14, experimental autoimmune uveoretinitis18 and spontaneous autoimmune diabetes21. However, it is not known whether AhR signaling can be exploited to promote the differentiation of functional human iTreg cells.
To address that question, we investigated the effect of AhR ligands on the differentiation of human Tr1 and Foxp3+ iTreg cells. We found that AhR activation induced human Tr1-like cells that suppressed responder T cells by a granzyme B–dependent mechanism. In addition, AhR activation in the presence of TGF-β1 induced the differentiation of functional human Foxp3+ iTreg cells that suppressed responder T cells through the ectonucleoside triphosphate diphosphohydrolase CD39. The induction of functional Foxp3+ iTreg cells by the concurrent activation of TGF-β1 and AhR signaling was mediated, at least partially, by the transcription factors Smad1 and Aiolos. Thus, our data suggest that AhR might be an important target for the generation of various types of Treg cells in humans and that nontoxic AhR ligands could provide new drug candidates for the induction of Treg cells in vivo and for the management of autoimmune diseases.
RESULTS
AhR activation induces T cells that produce IL-10
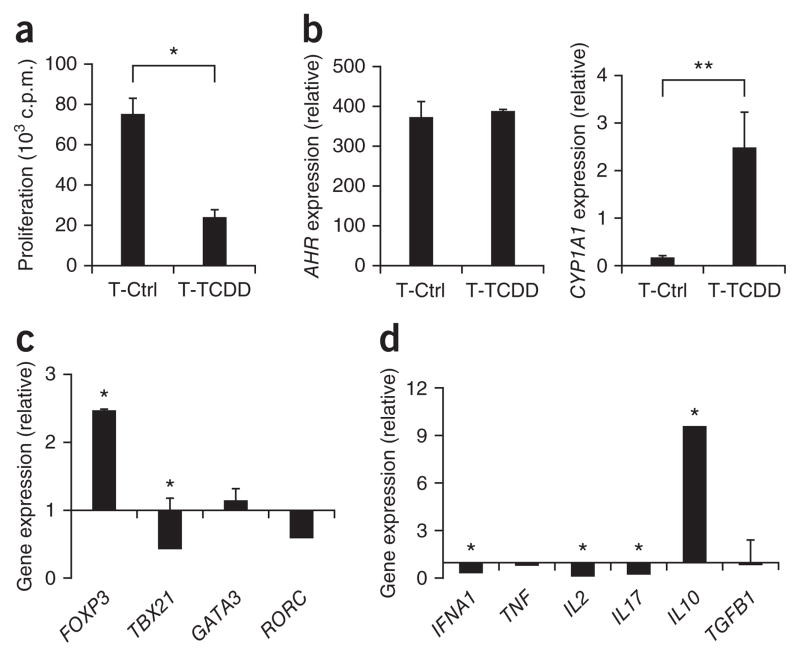
AhR participates in the differentiation of mouse Foxp3+ Treg cells14–18. To investigate whether AhR contributes to the differentiation of human Treg cells, we isolated naive CD4+ T cells from peripheral blood mononuclear cells obtained from healthy donors and activated them with anti-CD3, anti-CD28 and IL-2 with or without the AhR ligand TCDD (Supplementary Fig. 1). Naive T cells differentiated in the presence of TCDD showed a much lower proliferative response after restimulation (Fig. 1a). Moreover, T cell activation in the presence of TCDD upregulated expression of the AhR target gene CYP1A1 (which encodes a cytochrome p450 protein)22 but did not modify the expression of AHR, as measured by quantitative real-time PCR (Fig. 1b).
Figure 1.
AhR activation induces T cells that produce IL-10. (a) Proliferative response of human naive CD4+ T cells activated for 6 d with plate-bound anti-CD3 and anti-CD28, with (T-TCDD) or without (T-Ctrl) TCDD, then restimulated with bead-conjugated anti-CD3 and anti-CD28. (b–d) Real-time PCR analysis of the expression of AHR and the AhR target CYP1A1 (b), FOXP3, TBX21, GATA3 and RORC (c) and cytokines (d) on differentiated T cells (only TCDD-treated cells in c,d); results are presented relative to the expression of GAPDH (encoding glyceraldehyde phosphate dehydrogenase; b–d) and as the ratio of expression in T-TCDD cells to that in T-Ctrl cells (c,d). *P < 0.05 and **P < 0.01, compared with T-Ctrl (Student’s t-test). Data are representative of five experiments (a; mean + s.d. of triplicate wells) or represent one of three to five independent experiments (b–d; mean + s.d. of duplicates).
To characterize the phenotype of the human T cells treated with TCDD, we analyzed expression of the lineage-specific transcription factor genes FOXP3 (Treg cells), GATA3 (T helper type 2 (TH2) cells), TBX21 (TH1 cells) and RORC (TH17 cells). Activation of naive T cells in the presence of TCDD led to significant upregulation of FOXP3 expression (Fig. 1c). However, we did not detect upregulation of Foxp3 by flow cytometry (Supplementary Fig. 2). We detected no change in the expression of GATA3 or RORC, but there was significantly lower TBX21 expression after treatment with TCDD (Fig. 1c). In addition, TCDD-treated T cells expressed significantly more IL10 concomitant with significantly lower expression of IFNA1 (encoding interferon-γ), IL2 and IL17A than that of untreated T cells (Fig. 1d). We found no difference in the expression of TNF or TGFB1 (Fig. 1d). We obtained similar results when we activated human naive CD4+ T cells in the presence of the AhR ligand FICZ (6-formylindolo[3,2-b]carbazole; data not shown).
AhR transactivates the IL10 promoter
The transcription factor c-Maf (A003947) controls the synthesis of IL-10 by mouse T cells23–25. To further characterize the IL-10-producing T cells induced by AhR activation, we studied the expression of MAF (which encodes c-Maf) by quantitative real-time PCR. We found that MAF was expressed by TCDD-treated and IL-27 induced Tr1 cells, but its expression was not significantly higher in those cells than in control cells (Supplementary Fig. 3).
To investigate whether the effect of TCDD on IL10 expression was mediated by AhR, we knocked down AhR expression with small interfering RNA (siRNA). After 6 d, siRNA decreased the expression of AHR mRNA by 75% (Supplementary Fig. 4a). Correspondingly, the knockdown of AHR suppressed the induction of IL10 by TCDD (Supplementary Fig. 4a). Thus, AhR controls IL10 expression by TCDD-treated T cells.
Given the importance of AhR for the expression of IL10 by TCDD-treated T cells, we hypothesized that AhR transactivates the human IL10 promoter. Moreover, given the expression of MAF by TCDD-treated cells (Supplementary Fig. 3) and its reported interaction with the mouse Il10 promoter25, we investigated the role of c-Maf in regulating human IL10 expression. We used bioinformatics analysis to identify a potential AhR-binding site (xenobiotic response element (XRE)) that partially overlaps with a c-Maf-recognition element (MARE) in the IL10 promoter (Supplementary Fig. 4b).
To determine whether AhR can bind the XRE in the IL10 promoter, we used electrophoretic mobility-shift assay to study the interaction between an oligonucleotide containing the XRE and in vitro–translated AhR protein in complex with the AhR nuclear translocator. In vitro–translated AhR–AhR nuclear translocator complexes bound the XRE in the IL10 promoter, and this interaction was significantly lower after the addition of an excess of a competitor oligonucleotide containing an XRE found in CYP1A1 (Supplementary Fig. 4c). To investigate whether AhR interacts with the XRE in the IL10 promoter in TCDD-treated cells, we used chromatin immunoprecipitation (ChIP) assays. AhR bound substantially to the IL10 promoter region that contained the XRE in TCDD-treated T cells but not in control T cells (Supplementary Fig. 4d). We also detected a substantial interaction between c-Maf and the MARE in the IL10 promoter in TCDD-treated cells (Supplementary Fig. 4d). Thus, AhR and c-Maf interact with the XRE and MARE, respectively, in the IL10 promoter in TCDD-treated cells.
To analyze the functional relevance of the binding of AhR and c-Maf to the IL10 promoter, we did reporter assays using a construct containing the firefly luciferase gene under the control of the human IL10 promoter26. AhR and c-Maf separately transactivated the IL10 promoter; this transactivation was greater after activation of transfected Jurkat cells with anti-CD3 and anti-CD28 (Supplementary Fig. 4e). Notably, cotransfection with plasmids encoding both AHR and MAF resulted in an additive transactivation of IL10 (Supplementary Fig. 4e). Similarly, forced overexpression of AHR in naive human CD4+ T cells followed by treatment with TCDD triggered IL10 expression, which was further upregulated by forced coexpression of MAF (Supplementary Fig. 4f). In summary, these data suggest that AhR and c-Maf act together to control the transcription of IL10.
It has been shown that c-Maf interacts physically with other transcription factors whose responsive elements are located close to MARE motifs in target genes27. Therefore, we investigated the interaction between AhR and c-Maf in coimmunoprecipitation experiments with constructs encoding AhR tagged with hemagglutinin and c-Maf tagged with the red fluorescent protein mCherry. We found that AhR and c-Maf precipitated together when we used anti-hemagglutinin or anti-mCherry but not when we used an isotype-matched control antibody (Supplementary Fig. 4g). These data suggest that AhR interacts with c-Maf to control the transcriptional activity of the IL10 promoter.
AhR activation induces human Tr1-like cells
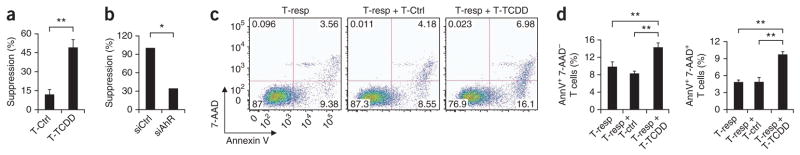
The production of IL-10 and the lower proliferative response of TCDD-treated T cells resembled the phenotype of Tr1 cells, which have in vitro suppressive activity9. We therefore analyzed the suppressive activity of TCDD-treated T cells in coculture assays. Human TCDD-treated T cells showed suppressive activity in vitro (Fig. 2a). This suppressive activity was controlled by AhR, as it was abrogated when we knocked down AHR expression with a specific siRNA (Fig. 2b). To further characterize the suppressive activity of the IL-10-producing T cells induced by AhR activation, we resorted TCDD-treated T cells, washed them extensively and cultured them together with responder T cells stained with annexin V and 7-amino-actinomycin D (7-AAD) after 3 d of coincubation. Coincubation of responder T cells with TCDD-treated T cells resulted in a greater frequency of annexin V–positive, 7-AAD− (early apoptotic) cells and annexin V–positive, 7-AAD+ (late apoptotic and/or dead) cells (Fig. 2c,d), which suggested that suppressive TCDD-treated T cells trigger apoptosis in responder T cells.
Figure 2.
AhR activation induces human Tr1-like cells. (a) Suppressive activity of human naive CD4+ T cells activated for 6 d with plate-bound anti-CD3 and soluble anti-CD28, with or without TCDD. (b) Effect of AHR knockdown on the suppressive activity of TCDD-treated T cells transduced with non–target-specific control siRNA (siCtrl) or AHR-specific siRNA (siAhR). (c) Flow cytometry of annexin V and 7-AAD in responder human CD4+ T cells (T-resp) coincubated with T cells with or without TCDD treatment. Numbers in quadrants indicate percent cells in each. (d) Frequency of annexin V–positive (AnnV+), 7-AAD− T cells (left) and annexin V–positive, 7-AAD+ T cells (right) in the cocultures in c. *P < 0.05 and **P < 0.01, compared with T-Ctrl (Student’s t-test). Data are representative of five experiments (a; mean + s.d. of triplicate wells), two experiments (b), three to eight independent experiments (c) or three independent experiments (d; mean + s.d.).
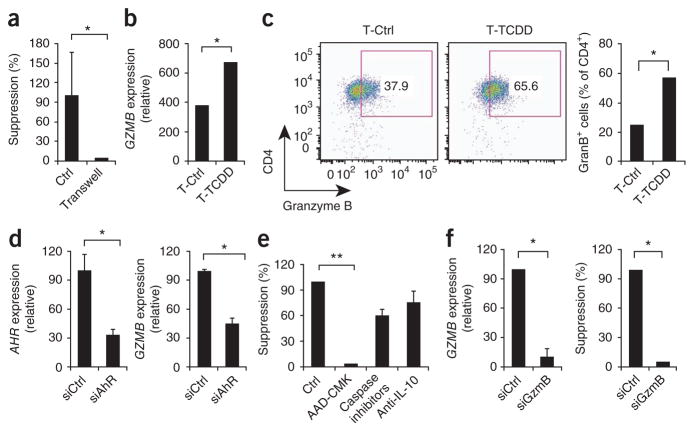
Treg cells use several mechanisms to control the activity of effector T cells28. Using a Transwell system, we found that the suppressive activity of TCDD-treated cells required cell contact (Fig. 3a). Granzyme-triggered apoptosis can mediate the suppressive activity of Tr1 cells in a cell contact–dependent manner29, so we used quantitative real-time PCR to analyze the expression of GZMA (encoding granzyme A) and GZMB (encoding granzyme B) in TCDD-treated T cells. We found significantly higher GZMB expression in TCDD-treated T cells (Fig. 3b) but no significant change in the expression of GZMA (data not shown). We also found significantly higher granzyme B expression when we used flow cytometry to analyze TCDD-treated T cells (Fig. 3c). To investigate whether the induction of GZMB by TCDD was mediated by AhR, we knocked down AHR expression with siRNA (Fig. 3d). Knockdown of AHR significantly decreased the induction of GZMB expression by TCDD (Fig. 3d). Thus, AhR activation directly or indirectly upregulates the expression of GZMB.
Figure 3.
The suppressive activity of Tr1-like cells induced by AhR activation is mediated by granzyme B. (a) Suppressive activity of human naive CD4+ T cells activated with plate-bound anti-CD3 and soluble anti-CD28, with TCDD, and incubated in contact with responder T cells (Ctrl) or with a Transwell (Transwell). (b) Quantitative real-time PCR analysis of GZMB expression on T cells with or without TCDD, presented relative to GAPDH expression. (c) Flow cytometry of granzyme B expression on T cells with or without TCDD. Numbers in outlined areas (left) indicate percent CD4+ granzyme B–positive (GranB+) cells. (d) Expression of AHR (left) and GZMB (right) in TCDD-treated cells transduced with nonspecific control or AHR-specific siRNA, presented relative to GAPDH expression. (e) Suppressive activity of TCDD-treated T cells left unstimulated or treated with the granzyme B inhibitor AAD-CMK, caspase inhibitors or neutralizing anti-IL-10. (f) GZMB expression (left) and the suppressive activity (right) of TCDD-treated T cells transduced with control siRNA or GNZB-specific siRNA (siGnzB). *P < 0.05 and **P < 0.01, compared with T-Ctrl (b,c), siCtrl (d,f) or no treatment (e; Student’s t-test). Data are representative of two experiments (a,d,f; mean + s.d. of triplicate wells in a and mean + s.d. in d,f), four experiments (b,c; mean + s.d. of duplicates (b) or mean (right, c) or three experiments (e; mean + s.d.).
To assess the relevance of granzyme B for the suppressive activity of TCDD-treated T cells, we used the granzyme B inhibitor AAD-CMK (benzyloxycarbonyl-Ala-Ala-Asp-chloromethylketone). AAD-CMK abrogated the suppressive activity of TCDD-treated T cells (Fig. 3e). However, consistent with published reports30, inhibition of caspase-3 activity had no significant effect on the suppressive activity (Fig. 3e). Moreover, knockdown of GZMB expression with specific siRNA abrogated the suppressive activity of TCDD-treated T cells in vitro (Fig. 3f). Neutralizing antibodies to other known suppressive molecules, such as IL-10, TGF-β, Fas or FasL, had no significant effect on the suppressive activity of TCDD-treated T cells (data not shown). Together these data show that activation of AhR induces Tr1-like cells that control responder T cells in a granzyme B–dependent manner.
AhR activation plus TGF-β1 induce FOXP3+ T cells
TGF-β1 has an important role in the differentiation, maintenance and function of Treg cells31. TGF-β1 promotes the differentiation of functional Foxp3+ Treg cells in the mouse4; however, although naive human T cells activated in the presence of TGF-β1 express Foxp3, they are not endowed with suppressive activity8. Given the reported effects of AhR14–16,18,21 and TGF-β1 (ref. 4) on mouse Foxp3+ Treg cells, we investigated the combined effect of TGF-β1 and TCDD-mediated activation of AhR on naive human T cells.
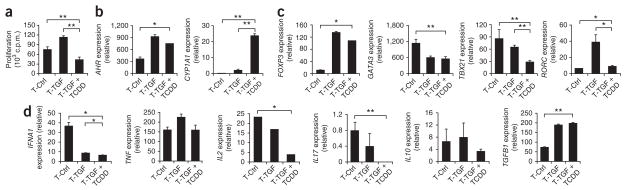
Naive T cells activated in the presence of TGF-β1 and TCDD showed a significantly smaller proliferative response after being restimulated with beads coated with anti-CD3 and anti-CD28 than did control or TGF-β1-treated T cells (Fig. 4a). Furthermore, activation of T cells in the presence of TGF-β1 or of TGF-β1 plus TCDD resulted in significant upregulation of AHR expression (Fig. 4b, left). The upregulation of AHR expression by TGF-β1 led to only a little induction of the AhR-controlled gene CYP1A1, but we achieved higher CYP1A1 expression after treating cells with both TGF-β1 and TCDD (Fig. 4b, right).
Figure 4.
AhR activation plus TGF-β1 induces Foxp3+ T cells. (a) Proliferative response of human naive CD4+ T cells activated for 6 d with plate-bound anti-CD3 and soluble anti-CD28 alone (T-Ctrl) or together with TGF-β1 alone (T-TGF) or TGF-β1 plus TCDD (T-TGF + TCDD) and restimulated with bead-conjugated anti-CD3 and anti-CD28. (b–d) Expression of AHR and CYP1A1 (b), FOXP3, GATA3, TBX21 and RORC (c) and cytokines (d) on cells treated as described in a; results are presented relative to GAPDH expression. *P < 0.05 and **P < 0.01, compared with T-Ctrl or T-TGF (Student’s t-test). Data are representative of five experiments (a; mean + s.d. of triplicate wells) or three to five experiments (b–d; mean + s.d. of duplicates).
We then analyzed the expression of lineage-specific transcription factors in T cells treated with TCDD and TGF-β1. The upregulation of FOXP3 was similar in naive human T cells treated with TGF-β1 and in those treated with TGF-β1 plus TCDD (Fig. 4c). This upregulation of FOXP3 expression was 50 times greater than the upregulation that followed treatment with TCDD alone (Figs. 1c and 4c) and we also detected it by flow cytometry (Supplementary Figs. 2 and 5). Treatment with TGF-β1 plus TCDD, but not with TGF-β1 alone, resulted in significant downregulation of the expression of GATA3 and TBX21 (Fig. 4c). Moreover, treatment with TGF-β1 plus TCDD suppressed the induction of RORC expression triggered by TGF-β1 alone (Fig. 4c). In T cells treated with TGF-β1 plus TCDD, the expression of TGFB1 was upregulated (Fig. 4d) and the expression of IFNA1, IL2 and IL17 was downregulated, but that of IL10 and TNF did not change (Fig. 4d).
TCDD plus TGF-β1 induce functional Foxp3+ Treg cells
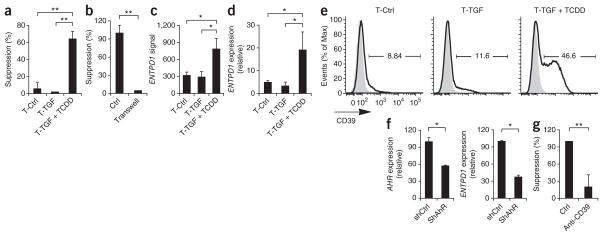
Naive human T cells activated in the presence of TGF-β1 express Foxp3 but do not have suppressive activity8. However, T cells treated with TGF-β1 plus TCDD were suppressive in vitro (Fig. 5a), and their suppressive activity was greater than that of T cells treated with TCDD alone (Figs. 2a and 5a). The suppressive activity of T cells treated with TGF-β1 and TCDD was mediated by a cell contact–dependent mechanism (Fig. 5b) independently of granzyme B (Supplementary Fig. 6).
Figure 5.
AhR activation plus TGF-β1 induces functional human Foxp3+ Treg cells. (a) Suppressive activity of human naive CD4+ T cells activated with plate-bound anti-CD3 and anti-CD28 alone or together with TGF-β1 alone or TGF-β1 plus TCDD (as in Fig. 4a). (b) Suppressive activity of T cells activated with plate-bound anti-CD3 and anti-CD28 alone together with TGF-β1 plus TCDD, incubated in contact with responder T cells (control (Ctrl)) or in a Transwell system (Transwell). (c,d) Gene microarray analysis (c) and quantitative PCR analysis (d) of ENTPD1 expression on cells treated as described in a; results in d are presented relative to GAPDH expression. (e) Flow cytometry of CD39 (ENTPD1) expression (dark lines) on cells treated as described in a; gray filled histograms, isotype-matched control antibody. Numbers above bracketed lines indicate percent CD39+ cells. (f) Expression of AHR (left) and ENTPD1 (right) in T cells as described in a transduced with nonspecific control or AHR-specific siRNA, presented relative to GAPDH expression. (g) Suppressive activity of cells activated as described in b and treated with control antibody or anti-CD39. *P < 0.05 and **P < 0.01, compared with T-Ctrl or T-TGF (a,c,d), shCtrl (f,g) or no Transwell (b; Student’s t-test). Data are representative of five (a,d), two (b,f) or three (e) experiments (mean and s.d. in a–d) or three (c) or five (g) independent experiments (mean and s.d. of all).
We used gene microarray to analyze the transcriptional profile of T cells treated with TGFβ-1 and TCDD to identify the mechanism of suppression used by human Treg cells induced by AhR stimulation in presence of TGF-β1. We found significantly higher expression of ENTPD1 (encoding CD39) in T cells treated with TGF-β plus TCDD (Fig. 5c). We obtained similar results when we analyzed ENTPD1 expression in an independent set of samples by quantitative real-time PCR (Fig. 5d) and by flow cytometry (Fig. 5e). To investigate whether the induction of ENTPD1 was mediated by AhR, we knocked down AHR expression with short hairpin RNA (shRNA). After 6 d, AHR expression was 45% lower (Fig. 5f). The knockdown of AHR resulted in significantly lower ENTPD1 expression in T cells differentiated with TGF-β1 plus TCDD (Fig. 5f). Thus, AhR activation in the presence of TGF-β1 controls the expression of ENTPD1.
CD39 (ENTPD1) hydrolyzes ATP and mediates the suppressive activity of murine and human Foxp3+ Treg cells32. Neutralizing antibodies to CD39 abrogated the suppressive activity of T cells treated with TGF-β plus TCDD (Fig. 5g). Together these results show that activation of AhR in the presence of TGF-β induces functional human Foxp3+ Treg cells that suppress effector T cells by a CD39-dependent mechanism.
AhR activation plus TGF-β1 induces Smad1 and Aiolos
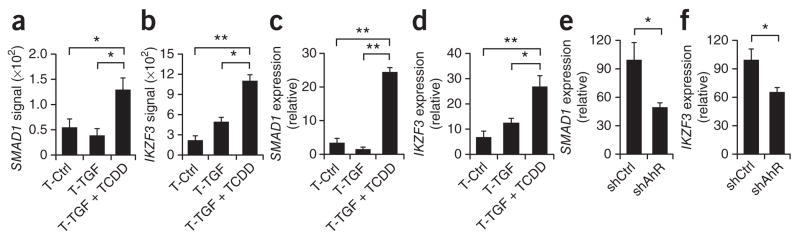
Naive human T cells activated in the presence of TGF-β1 express Foxp3 but do not have suppressive activity8. On the basis of the finding that suppressive human Foxp3+ Treg cells can be induced by TGF-β1 plus TCDD, we hypothesized that AhR activation might induce additional transcription factors needed for the induction of functional Foxp3+ Treg cells. Gene microarray showed that expression of the transcription factors Smad1 and Aiolos (encoded by IKZF3) was significantly upregulated in T cells treated with TGF-β1 plus TCDD (Fig. 6a,b). We confirmed that upregulation by quantitative real-time PCR in an independent set of samples (Fig. 6c,d). To investigate whether the induction of Smad1 and Aiolos was mediated by AhR, we knocked down AHR expression with shRNA. Knockdown of AHR resulted in significantly lower expression of SMAD1 and IKZF3 in T cells differentiated with TGF-β1 plus TCDD (Fig. 6e,f). Thus, AhR activation in the presence of TGF-β1 upregulates the expression of Smad1 and Aiolos.
Figure 6.
AhR activation plus TGF-β1 induce the expression of Smad1 and Aiolos. (a,b) Gene microarray analysis of the expression of SMAD1 (a) and IKZF3 (Aiolos; b) by human naive CD4+ T cells activated for 6 d with plate-bound anti-CD3 and soluble anti-CD28 alone or together with TGF-β1 alone or TGF-β1 plus TCDD (as in Fig. 4a). (c,d) Quantitative real-time PCR analysis of the expression of SMAD1 (c) and IKZF3 (d) on cells treated as described in a,b, presented relative to GAPDH expression. (e,f) Expression of SMAD1 (e) and IKZF3 (f) by human naive CD4+ T cells activated for 6 d with plate-bound anti-CD3 and soluble anti-CD28 together with TGF-β1 plus TCDD and transduced with nonspecific control or AHR-specific shRNA, presented relative to GAPDH expression. *P < 0.05 and **P < 0.01, compared with T-Ctrl or T-TGF (a–d) or shCtrl (e,f; Student’s t-test). Data are representative of three independent experiments (a,b; mean and s.d. of all) or five (c,d) or two (e,f) experiments (mean and s.d.).
Smad1 regulates Foxp3 enhancer activity
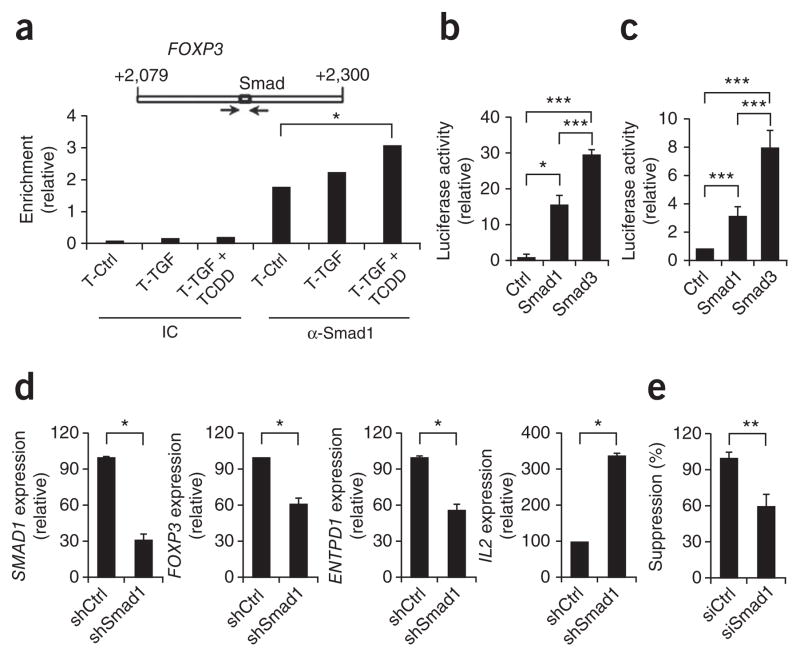
Smad1 belongs to a family of transcription factors involved in TGF-β signaling. Smad3 binds to an enhancer in the region of positions +2079 to +2198 of human FOXP3 and thus controls its expression33. Because Smad3 and Smad1 recognize similar DNA-binding motifs34, we investigated the binding of Smad1 to the enhancer in this region (+2079 to +2198) in FOXP3 in human T cells treated with TGF-β1 and TCDD. Using ChIP, we found that the interaction between Smad1 and the FOXP3 enhancer located in this region was upregulated in T cells treated with TGF-β1 and TCDD compared with that of control T cells (Fig. 7a).
Figure 7.
Smad1 regulates FOXP3 enhancer activity. (a) Smad-binding site in the FOXP3 enhancer (top) and ChIP analysis (below) of the interaction of Smad1 with that binding site in human naive CD4+ T cells activated with plate-bound anti-CD3 and soluble anti-CD28 alone or together with TGF-β1 alone or TGF-β1 plus TCDD (as in Fig. 4a), precipitated with isotype-matched control antibody (IC) or anti-Smad1 (α-Smad1); results are presented as enrichment relative to input chromatin. (b) Luciferase activity in EL4 cells transfected with a reporter containing six copies of the Smad-binding motif in the FOXP3 enhancer together with empty control vector (Ctrl) or vector encoding Smad1 or Smad3, and activated in the presence of TGF-β1 (ref. 33); results are presented relative to renilla luciferase. (c) Luciferase activity in EL4 cells transfected with a reporter for the FOXP3 enhancer shown in a33, together with empty control vector or vector encoding Smad1 or Smad3, and activated in the presence of TGF-β1; results are presented relative to renilla luciferase. (d) Expression of SMAD1, FOXP3, ENTPD1 and IL2 by human naive CD4+ T cells activated with plate-bound anti-CD3 and soluble anti-CD28 together with TGF-β1 plus TCDD and transduced with nonspecific control shRNA (shCtrl) or SMAD1-specific shRNA (shSmad1), presented relative to GAPDH expression. (e) Suppressive activity of cells activated and transduced as described in d. *P < 0.05, **P < 0.01 and ***P < 0.001, compared with T-Ctrl (a), Ctrl or Smad1 (b,c), shCtrl (d) or siCtrl (e; Student’s t-test). Data are representative of three (a–c) or two (d,e) experiments (mean and s.d.).
To investigate the functional relevance of the binding of Smad1 to the enhancer we transfected the EL4 mouse lymphoma cell line with a reporter system in which six copies of the Smad-binding motif in the Foxp3 enhancer in the region of positions +2079 to +2198 control the expression of firefly luciferase33 (Supplementary Fig. 7a). As reported before33 luciferase activity was induced after cotransfection with a vector encoding Smad3 (Fig. 7b). Moreover, luciferase activity was also induced after cotransfection with a construct encoding Smad1 (Fig. 7b). We obtained similar results when we investigated the ability of constructs encoding Smad1 or Smad3 to activate a luciferase reporter controlled by the Foxp3 enhancer in this region33 (Fig. 7c and Supplementary Fig. 7b). Thus, stimulation with TGF-β1 concurrent with activation of AhR results in the induction of Smad1, which binds and activates the Foxp3 enhancer in the region of positions +2079 to +2198.
To investigate the effect of the binding and activation of the FOXP3 enhancer in that region by Smad1 on FOXP3 expression, we knocked down SMAD1 expression with lentivirus-delivered shRNA in naive human T cells activated in the presence of anti-CD3, anti-CD28 and IL-2 with TGF-β1 and TCDD. The expression of SMAD1 mRNA was 80% lower after treatment with the Smad1-specific shRNA (Fig. 7d). Correspondingly, the knockdown of SMAD1 led to significantly lower expression of FOXP3 and ENTPD1 by Foxp3+ Treg cells induced with TGF-β1 and TCDD. Conversely, IL2 expression was significantly upregulated after knockdown of SMAD1 (Fig. 7d). Moreover, the knockdown of SMAD1 led to significantly lower suppressive activity of Foxp3+ Treg cells induced with TGF-β1 plus TCDD (Fig. 7e). Together these data show that Smad1 controls the expression of FOXP3 and the suppressive activity in Foxp3+ Treg cells that have been differentiated with TCDD and TGF-β1.
Aiolos-Foxp3 interaction silences IL2 expression
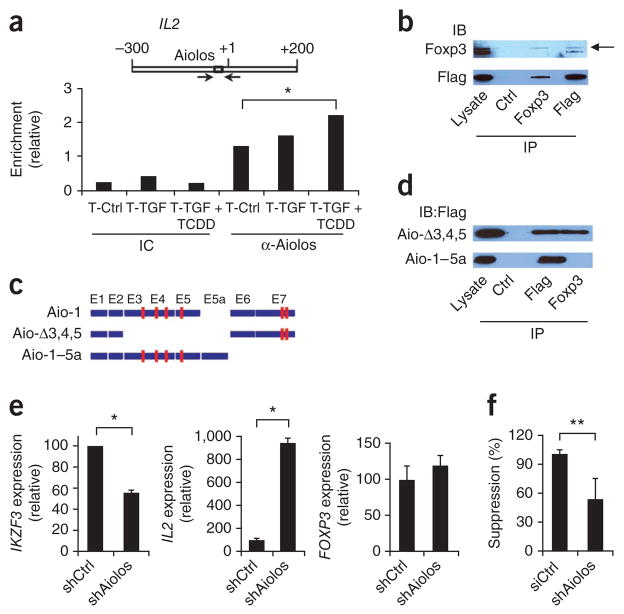
Aiolos is a transcription factor of the Ikaros family; members of this family are important in the development of hematopoietic cells35. Eos, another member of the Ikaros family, forms a complex with Foxp3 and mediates the silencing of target genes such as Il2 in mouse Foxp3+ Treg cells36. DNA target sequences bound by members of the Ikaros family are highly conserved37, and Eos interacts with the Il2 promoter36, so we used ChIP assays to investigate whether Aiolos binds the IL2 promoter in human T cells activated with TGF-β1 and TCDD. The binding of Aiolos to the Eos-binding site in the IL2 promoter was greater in naive T cells activated in the presence of TGF-β1 and TCDD (Fig. 8a).
Figure 8.
Aiolos interacts with Foxp3 to silence IL2 expression. (a) Aiolos-binding site in the IL2 promoter (top) and ChIP analysis (below) of the interaction between Aiolos and that binding site in the IL2 promoter in human naive CD4+ T cells activated for 6 d with plate-bound anti-CD3 and soluble anti-CD28 alone or together with TGF-β1 alone or TGF-β1 plus TCDD (as in Fig. 4a), precipitated with isotype-matched control antibody (IC) or anti-Aiolos (α-Aiolos). (b) Physical interaction between Aiolos and Foxp3 in 293 human embryonic kidney cells transfected with constructs encoding Foxp3 and Flag-tagged Aiolos and lysed 24 h later, followed by immunoprecipitation (IP) with isotype-matched control antibody (Ctrl), anti-Foxp3 or anti-Flag, and analysis by immunoblot (IB) with anti-Foxp3 or anti-Flag. Arrow indicates Foxp3. Lysate, immunoblot analysis before immunoprecipitation. (c) Aiolos isoforms. Red boxes indicate zinc-finger motifs; E1–E7 indicate exons 1–7. (d) Immunoassay of 293 cells transfected with constructs encoding Foxp3 or Flag-tagged isoforms of Aiolos and lysed 48 h later, followed by immunoprecipitation with isotype-matched control antibody, anti-Foxp3 or anti-Flag, and analysis by immunoblot with anti-Flag. (e) Expression of IKZF3, IL2 and FOXP3 by human naive CD4+ T cells activated with plate-bound anti-CD3 and soluble anti-CD28 together with TGF-β1 plus TCDD and transduced with nonspecific control shRNA (shCtrl) or IKZF3-specific shRNA (shAiolos), presented relative to GAPDH expression. (f) Suppressive activity of cells activated and transduced as described in e. *P < 0.05 and **P < 0.01, compared with T-Ctrl (a), shCtrl (e) or siCtrl (f; Student’s t-test). Data are representative of three (a,b,d) or two (e,f) experiments (mean and s.d. in a,e,f).
Eos and Foxp3 form a protein complex that represses the expression of target genes36. To investigate whether Aiolos and Foxp3 can interact physically, we did coimmunoprecipitation studies with constructs encoding Foxp3 and Flag-tagged Aiolos. Anti-Foxp3 and anti-Aiolos precipitated a protein complex containing Foxp3 and Aiolos, but control IgG did not (Fig. 8b). Thus, Foxp3 and Aiolos physically associate with each other.
Aiolos can form homodimers and can also form complexes with other proteins through interactions mediated by zinc fingers in its C-terminal domain37. There are also four more zinc fingers in the N-terminal portion of the protein. To determine whether the interaction of Aiolos with Foxp3 was mediated by its C-terminal domain, we did coimmunoprecipitation studies with a vector encoding the Aio-1–5a isoform of Aiolos, which lacks the two C-terminal zinc fingers involved in homo- and heterodimerization38 (Fig. 8c). As a control, we used the Aio-Δ3,4,5 isoform, which lacks all the zinc fingers at the N terminus but retains the C-terminal dimerization domain (Fig. 8c). Like full-length Aiolos, Aio-Δ3,4,5 interacted with Foxp3 (Fig. 8d). However, the deletion of the C-terminal domain in Aio-1–5a disrupted its interaction with Foxp3 (Fig. 8d). Therefore, like Eos, Aiolos forms a complex with Foxp3 through interactions mediated by its C-terminal zinc fingers.
Eos participates in the repression of Il2 expression by Foxp3 (ref. 36). Naive T cells activated in the presence of TGF-β1 plus TCDD had lower IL2 expression (Fig. 4d) and concomitant induction of Aiolos (Fig. 6b,d). To investigate the role of Aiolos in the repression of IL2 expression by Foxp3, we did knockdown experiments with naive human T cells activated in the presence of anti-CD3, anti-CD28 and IL-2 with TGF-β1 and TCDD using lentivirus-delivered shRNA specific for IKZF3. Treatment with shRNA resulted in significantly lower expression of IKZF3 (Fig. 8e) and higher IL2 expression in T cells treated with TGF-β1 plus TCDD (Fig. 8e). Foxp3 expression is independent of Eos36. Accordingly, the knockdown of IKZF3 did not result in significant changes in FOXP3 expression (Fig. 8e). However, the knockdown of IKZF3 led to significantly lower suppressive activity of Foxp3+ Treg cells induced with TGF-β1 plus TCDD (Fig. 8f). Together these data suggest that Foxp3 forms a complex with Aiolos to repress target genes and control the suppressive activity of Foxp3+ Treg cells that have been differentiated with TCDD and TGF-β1.
DISCUSSION
Here we have shown that activation of AhR promoted the differentiation of functional human Tr1 or Foxp3+ iTreg cells in vitro, depending on the cytokine context. Activation of CD4+ T cells in the presence of AhR ligands promoted the differentiation of Foxp3− IL-10-producing T cells that controlled responder T cells through granzyme B. However, activation of AhR in the presence of TGF-β1 induced Foxp3+ Treg cells that suppressed responder T cells through CD39. The induction of functional Foxp3+ Treg cells by the concurrent activation of TGF-β1 and AhR signaling was mediated at least in part by the transcription factors Smad1 and Aiolos. Smad1 regulated the FOXP3 enhancer located in positions +2079 to +2198, and Aiolos formed a complex with Foxp3 to silence IL2 expression. Thus, in different cytokine milieus, AhR activation can promote the differentiation of human suppressive Tr1 or Foxp3+ Treg cells or proinflammatory TH17 cells20.
Activation of naive human CD4+ T cells in the presence of AhR ligands resulted in the differentiation of suppressive Tr1-like cells. Tr1 cells are Foxp3− Treg cells that produce IL-10 (ref. 9), and in different in vitro and in vivo scenarios they also produce TGF-β1, IL-5 and interferon-γ and have transient low expression of Foxp3 (refs. 10,11). The phenotypic diversity of Tr1 cells and the disparate settings that promote their differentiation10,11 suggest that there are several lineages of CD4+IL-10+ Treg cells. Accordingly, transcription factors such as Sp1, Sp3, C/EBP-β, IRF1, STAT3 and c-MAF can activate the Il10 promoter24. In particular, c-MAF has been proposed to be a universal transcription factor that regulates the production of IL-10 by T cells24. Our data, together with those of Apetoh et al. published in this issue of Nature Immunology39, show that AhR interacts with c-Maf to control the transcription of IL10.
The production of IL-10 does not necessarily result in a suppressive phenotype; proinflammatory IL-9-producing helper T cells, for example, have high expression of IL-10 (ref. 40). We found that the Tr1-like cells induced by AhR activation suppressed effector T cells by a granzyme B–dependent mechanism independently of IL-10. TCDD-induced mouse Tr1-like cells also express granzyme B41. However, we do not believe that AhR alone or in combination with c-Maf acts as a lineage-specification transcription factor for Tr1 cells. AhR is expressed by many lineages in addition to Tr1 cells, such as Foxp3+ Treg cells14,16 and TH17 cells14,16,20. Similarly, c-Maf is expressed not only by Tr1 cells but also by TH1 cells24 and TH17 cells25. Instead, our findings, along with those of Apoteh et al.39, suggest that AhR acts in synergy with c-Maf and other transcription factors to control part of the transcriptional program linked to Tr1 cell differentiation.
In humans, neither forced Foxp3 overexpression7 nor Foxp3 expression triggered by TGF-β1 alone8 results in the differentiation of suppressive Foxp3+ Treg cells, which indicates that additional signals beyond those controlled by Foxp3 are required for the generation of functional Foxp3+ Treg cells. We found that the differentiation of suppressive Foxp3+ Treg cells by the concurrent activation of AhR and TGF-β1 signaling required the combined activities of Smad1 and Aiolos. A conserved noncoding sequence (CNS-1) in Foxp3 controls the differentiation of Foxp3+ iTreg cells in gut-associated lymphoid tissue42. The CNS-1 region in FOXP3 contains a functional Smad-binding site that interacts with Smad3 and drives FOXP3 expression upon activation with TGF-β1 (ref. 33). We found that T cell activation in the presence of TGF-β1 plus TCDD induced the expression of Smad1, which interacted with the Smad-binding motif in CNS-1 to promote FOXP3 expression. It has been proposed that stable Foxp3 expression is achieved when the transcription factors NFAT, Smad, CREB and c-Rel form an enhanceosome that transactivates FOXP3 (ref. 43). In this model, various members of the Smad or NFAT family can be incorporated into the enhanceosome that drives FOXP3 expression43. Our data suggest that in the Foxp3+ iTreg cells induced in vitro by the concomitant activation of TGF-β1 and AhR signaling, Smad1 alone or in combination with Smad3 and/or Smad4 interacts with CNS-1 to activate FOXP3 expression.
Proteins of the Ikaros family share a DNA-binding domain that recognizes sequences containing the GGGA core motif37. Accordingly, we found that Aiolos and Eos bound the same DNA motif in IL2 to inhibit its expression. Members of the Ikaros family control gene expression by anchoring protein complexes that regulate chromatin remodeling and histone deacetylation in target genes44. In addition, proteins in the Ikaros family can also repress gene expression by mechanisms independent of chromatin remodeling and histone deacetylation45. Together our data suggest that Aiolos interacts with Foxp3 to silence the transcriptional program of effector T cells and further promote the differentiation of functional Foxp3+ Treg cells.
It has been suggested that Tr1 cells and Foxp3+ iTreg cells constitute alternative fates of T cell differentiation whose immunoregulatory function might be partially redundant in gut-associated lymphoid tissue42. Here we have reported that depending on the cytokine milieu, AhR activation promoted the differentiation of human Tr1 or Foxp3+ Treg cells. Thus, it is conceivable that AhR ligands provided by the diet46 or the intestinal flora47 influence the differentiation of Tr1, Foxp3+ iTreg and TH17 cells in vivo. Moreover, our data suggest that AhR ligands can be used to promote iTreg cell differentiation in vitro, which could be exploited to generate functional iTreg cells in vitro for adoptive-transfer regimes aimed at reestablishing immune tolerance. Alternatively, nontoxic AhR ligands could constitute potential new drugs for the therapeutic induction of Treg cells in vivo and the management of autoimmune disorders.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/natureimmunology/.
Supplementary Material
Acknowledgments
We thank V. Kuchroo (Center for Neurologic Diseases, Brigham and Women’s Hospital, Harvard Medical School) for discussions and the IL10 promoter reporter construct; Y. Tone and M. Tone (Cedars-Sinai Medical Center) for reporter constructs for FOXP3 +2079 to +2198; and E. Ballestar (Bellvitge Biomedical Research Institute) for vectors encoding Aiolos and its isoforms. Supported by the US National Institutes of Health (AI435801 and NS38037 to H.L.W. and 1K99AI075285 to F.J.Q.), the National Multiple Sclerosis Society (RG4151A12 to H.L.W. and RG4111A1 to F.J.Q.) and the Harvard Medical School Office for Diversity and Community Partnership (to F.J.Q.).
Footnotes
Accession codes. UCSD-Nature Signaling Gateway (http://www.signaling-gateway.org): A002750, A000229 and A003947.
Note: Supplementary information is available on the Nature Immunology website.
AUTHOR CONTRIBUTIONS
R.G., D.K., E.J.B., M.N. and A.L. did experiments; D.K. sorted cells by flow cytometry; B.D. provided advice; R.G., D.K., E.J.B., H.L.W. and F.J.Q. analyzed data; R.G. and F.J.Q. wrote the manuscript; and H.L.W. and F.J.Q. supervised the study and edited the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Sakaguchi S. Naturally arising CD4+ regulatory T cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- 2.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 3.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 4.Chen W, et al. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allan SE, et al. Activation-induced FOXP3 in human T effector cells does not suppress proliferation or cytokine production. Int Immunol. 2007;19:345–354. doi: 10.1093/intimm/dxm014. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Ioan-Facsinay A, van der Voort EI, Huizinga TW, Toes RE. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur J Immunol. 2007;37:129–138. doi: 10.1002/eji.200636435. [DOI] [PubMed] [Google Scholar]
- 7.Allan SE, et al. The role of 2 FOXP3 isoforms in the generation of human CD4+ Tregs. J Clin Invest. 2005;115:3276–3284. doi: 10.1172/JCI24685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran DQ, Ramsey H, Shevach EM. Induction of FOXP3 expression in naive human CD4+FOXP3 T cells by T-cell receptor stimulation is transforming growth factor-β dependent but does not confer a regulatory phenotype. Blood. 2007;110:2983–2990. doi: 10.1182/blood-2007-06-094656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groux H, et al. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389:737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 10.Roncarolo MG, et al. Interleukin-10-secreting type 1 regulatory T cells in rodents and humans. Immunol Rev. 2006;212:28–50. doi: 10.1111/j.0105-2896.2006.00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 12.Baecher-Allan C, Hafler DA. Human regulatory T cells and their role in autoimmune disease. Immunol Rev. 2006;212:203–216. doi: 10.1111/j.0105-2896.2006.00417.x. [DOI] [PubMed] [Google Scholar]
- 13.Tran DQ, Shevach EM. Therapeutic potential of FOXP3+ regulatory T cells and their interactions with dendritic cells. Hum Immunol. 2009;70:294–299. doi: 10.1016/j.humimm.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quintana FJ, et al. Control of Treg and TH17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;23:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 15.Hauben E, et al. Activation of the aryl hydrocarbon receptor promotes allograft-specific tolerance through direct and dendritic cell-mediated effects on regulatory T cells. Blood. 2008;112:1214–1222. doi: 10.1182/blood-2007-08-109843. [DOI] [PubMed] [Google Scholar]
- 16.Kimura A, Naka T, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Natl Acad Sci USA. 2008;105:9721–9726. doi: 10.1073/pnas.0804231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel CF, Goth SR, Dong B, Pessah IN, Matsumura F. Aryl hydrocarbon receptor signaling mediates expression of indoleamine 2,3-dioxygenase. Biochem Biophys Res Commun. 2008;375:331–335. doi: 10.1016/j.bbrc.2008.07.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, et al. Suppression of experimental autoimmune uveoretinitis by inducing differentiation of regulatory T cells via activation of aryl hydrocarbon receptor. Invest Ophthalmol Vis Sci. 2010;51:2109–2117. doi: 10.1167/iovs.09-3993. [DOI] [PubMed] [Google Scholar]
- 19.Veldhoen M, Hirota K, Christensen J, O’Garra A, Stockinger B. Natural agonists for aryl hydrocarbon receptor in culture medium are essential for optimal differentiation of Th17 T cells. J Exp Med. 2009;206:43–49. doi: 10.1084/jem.20081438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veldhoen M, et al. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 2008;453:106–109. doi: 10.1038/nature06881. [DOI] [PubMed] [Google Scholar]
- 21.Kerkvliet NI, et al. Activation of aryl hydrocarbon receptor by TCDD prevents diabetes in NOD mice and increases Foxp3+ T cells in pancreatic lymph nodes. Immunotherapy. 2009;1:539–547. doi: 10.2217/imt.09.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones PB, Galeazzi DR, Fisher JM, Whitlock JP., Jr Control of cytochrome P1–450 gene expression by dioxin. Science. 1985;227:1499–1502. doi: 10.1126/science.3856321. [DOI] [PubMed] [Google Scholar]
- 23.Pot C, et al. Cutting edge: IL-27 induces the transcription factor c-Maf, cytokine IL-21, and the costimulatory receptor ICOS that coordinately act together to promote differentiation of IL-10-producing Tr1 cells. J Immunol. 2009;183:797–801. doi: 10.4049/jimmunol.0901233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saraiva M, et al. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity. 2009;31:209–219. doi: 10.1016/j.immuni.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu J, et al. c-Maf regulates IL-10 expression during Th17 polarization. J Immunol. 2009;182:6226–6236. doi: 10.4049/jimmunol.0900123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JI, Ho IC, Grusby MJ, Glimcher LH. The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity. 1999;10:745–751. doi: 10.1016/s1074-7613(00)80073-4. [DOI] [PubMed] [Google Scholar]
- 27.Planque N, et al. Interaction of Maf transcription factors with Pax-6 results in synergistic activation of the glucagon promoter. J Biol Chem. 2001;276:35751–35760. doi: 10.1074/jbc.M104523200. [DOI] [PubMed] [Google Scholar]
- 28.Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grossman WJ, et al. Human T regulatory cells can use the perforin pathway to cause autologous target cell death. Immunity. 2004;21:589–601. doi: 10.1016/j.immuni.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Thomas DA, Du C, Xu M, Wang X, Ley TJ. DFF45/ICAD can be directly processed by granzyme B during the induction of apoptosis. Immunity. 2000;12:621–632. doi: 10.1016/s1074-7613(00)80213-7. [DOI] [PubMed] [Google Scholar]
- 31.Rubtsov YP, Rudensky AY. TGF-β signalling in control of T-cell-mediated self-reactivity. Nat Rev Immunol. 2007;7:443–453. doi: 10.1038/nri2095. [DOI] [PubMed] [Google Scholar]
- 32.Bynoe MS, Viret C. Foxp3+CD4+ T cell-mediated immunosuppression involves extracellular nucleotide catabolism. Trends Immunol. 2008;29:99–102. doi: 10.1016/j.it.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Tone Y, et al. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 34.Massague J, Seoane J, Wotton D. Smad transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 35.Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol. 2002;2:162–174. doi: 10.1038/nri747. [DOI] [PubMed] [Google Scholar]
- 36.Pan F, et al. Eos mediates Foxp3-dependent gene silencing in CD4+ regulatory T cells. Science. 2009;325:1142–1146. doi: 10.1126/science.1176077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morgan B, et al. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16:2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caballero R, et al. Combinatorial effects of splice variants modulate function of Aiolos. J Cell Sci. 2007;120:2619–2630. doi: 10.1242/jcs.007344. [DOI] [PubMed] [Google Scholar]
- 39.Apetoh L, et al. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat Immunol. doi: 10.1038/ni.1912. advance online publication 1 August 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dardalhon V, et al. IL-4 inhibits TGF-β-induced Foxp3+ T cells and, together with TGF-β, generates IL-9+IL-10+Foxp3− effector T cells. Nat Immunol. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marshall NB, Vorachek WR, Steppan LB, Mourich DV, Kerkvliet NI. Functional characterization and gene expression analysis of CD4+CD25+ regulatory T cells generated in mice treated with 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Immunol. 2008;181:2382–2391. doi: 10.4049/jimmunol.181.4.2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng Y, et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruan Q, et al. Development of Foxp3+ regulatory t cells is driven by the c-Rel enhanceosome. Immunity. 2009;31:932–940. doi: 10.1016/j.immuni.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim J, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 45.Thompson EC, et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 2007;26:335–344. doi: 10.1016/j.immuni.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 46.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 47.Perdew GH, Babbs CF. Production of Ah receptor ligands in rat fecal suspensions containing tryptophan or indole-3-carbinol. Nutr Cancer. 1991;16:209–218. doi: 10.1080/01635589109514159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.