Abstract
The extent to which variation in chromatin structure and transcription factor binding may influence gene expression, and thus underlie or contribute to variation in phenotype, is unknown. To address this question, we cataloged both individual-to-individual variation and differences between homologous chromosomes within the same individual (allele-specific variation) in chromatin structure and transcription factor binding in lymphoblastoid cells derived from individuals of geographically diverse ancestry. Ten percent of active chromatin sites were individual-specific; a similar proportion were allele-specific. Both individual-specific and allele-specific sites were commonly transmitted from parent to child, which suggests that they are heritable features of the human genome. Our study shows that heritable chromatin status and transcription factor binding differ as a result of genetic variation and may underlie phenotypic variation in humans.
Control of gene transcription is believed to be important in determining organismal phenotype and fitness. Variations in genomic DNA, such as single-nucleotide polymorphisms (SNPs), insertions, or deletions (indels), may act singly or in combination to influence gene regulation (1, 2). These heritable variations have been thought to affect the binding of sequence-specific transcription factors or to affect the physical conformation of packaged DNA, namely chromatin. Humans typically harbor two copies (alleles) of every gene, and recent studies show that for between 10% and 22% of human genes, the two copies are regulated differently—for example, one copy may be transcribed while the other is not (3). Such allele-specific expression can be created in part by underlying biological processes such as imprinting, but little is known about other molecular determinants of allele-specific gene regulation in humans or to what extent these events are genetically determined, given that variation in gene regulation can also be caused by nongenetic phenomena including epigenetic, environmental, or stochastic effects (4–6). To aid in our understanding of the molecular basis of allele-specific gene regulation and the separate but related topic of phenotypic variation between individuals, we have cataloged allele-specific and individual-specific variation in transcription factor binding and chromatin structure.
To assay individual variation and how it relates to the allele-specific behavior of chromatin, we used deoxyribonuclease I hypersensitive (DNase I HS) site mapping, which broadly identifies regulatory DNA elements such as promoters, enhancers, silencers, and insulators (7, 8). We also performed chromatin immunoprecipitation (ChIP) for elements associated with the CCCTC-binding factor (CTCF), a multifunctional transcriptional and chromatin regulator (9–12). The combination of these two different methods, DNase I HS mapping and CTCF ChIP, allowed us to independently validate our results. Assays were performed on cell lines from one CEU (CEPH Utah reference family; residents with ancestry from northern and western Europe) family (both parents and their daughter) and one YRI (Yoruba from Ibadan, Nigeria) family (both parents and their daughter) in the 1000 Genomes Project (13). The study design therefore features four unrelated adults (the parents) and two children who are directly related to one pair of adults but unrelated to the other pair or each other (Fig. 1A). This design allows us to dissect individual- and allele-specific information in the context of these families, and thereby to determine heritability and the contribution from genetic or epigenetic processes. Previous studies have identified very few individual-specific sites and have not explored their heritability (14).
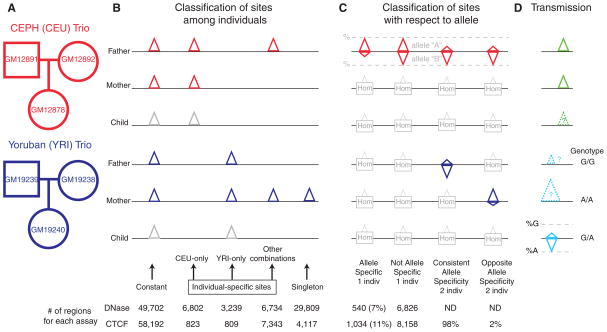
Fig. 1.
(A) Cell lines from CEU and YRI parent-child trios. (B) Classification of DNase I HS or CTCF binding sites among individuals. Constant sites are those occurring in all four parents. CEU- and YRI-only sites occurred in both parents within only one population. Sites occurring in one individual (singletons) or in other combinations were also noted (table S2). Sites in children were not used in this initial classification (gray). (C) Sites that are allele-specific (skewed toward one allele) in one individual, not allele-specific in one individual, consistent allele-specific in two individuals, and opposite allele-specific in two individuals. Homozygous (Hom) individuals for an allele are not informative. (D) Transmission tests show that CEU- or YRI-only sites are more likely in children from the same population (green; see also Fig. 2), and allele-specific sites in children correspond to signal intensities in parents who are homozygous for different alleles (turquoise; see also Fig. 3). Numbers and percentages of all categories are indicated at the bottom. The orientation of the triangles indicates the two alleles that are assayed; triangle sizes indicate differences in signal strength (in terms of number of sequence reads).
We generated DNase-seq and CTCF ChIP-seq (deep sequencing) data from two independent cell growths for each cell line (Fig. 1 and fig. S1) (13). Sites were classified as “constant” (present in all four unrelated parents), “individual-specific” (present in at least two of the parents and absent in the other two parents), or “singletons” (present in just one individual) (Fig. 1B, Fig. 2, A and B, fig. S2, and table S1). Global analysis of the 10,041 (DNase) or 1632 (CTCF) individual-specific sites specific to one set of parents compared to the other showed that the children’s signals at those sites were closer to their own parents than to that of the unrelated family (Fig. 2, C and D). Given the large number of individual sites tested, this result shows that these chromatin signals are heritable. However, this analysis alone cannot distinguish among genetic, epigenetic, or other causes for inheritance. The high degree of concordance at the 54,621 sites identified by both assays also supports the heritability of binding-level specificity (fig. S3).
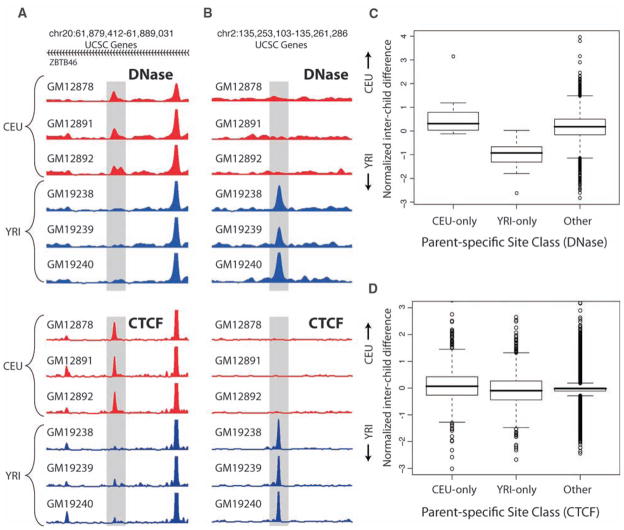
Fig. 2.
Individual-specific chromatin transmission. (A) Example of CEU-only individual-specific DNase I HS and CTCF sites (shaded areas). (B) Example of YRI-only individual-specific sites. (C and D) Genome-wide individual-specific DNase I HS sites (C) and CTCF sites (D) were categorized as CEU-only, YRI-only, or other combinations. The standard box plots of the relative normalized interchild differences for these categories show that the child signal was significantly closer to the parental sites from its own population (P < 10−15 for DNase I HS, P < 10−8 for CTCF; Wilcoxon rank-sum test). Numbers at top of (A) and (B) are chromosome numbers followed by start-stop coordinates from the UCSC Genome Browser. In (A) the indicated sites occur in the ZBTB46 gene, whose direction of transcription is right to left (as indicated by arrowheads).
We next examined the correlation of individual variation in these chromatin sites with variation in gene expression. The presence of an individual-specific DNase I HS site near the transcription start site of a gene was positively correlated with expression of that gene in that individual, relative to genes that were farther away (fig. S4, A and C). Individual-specific CTCF sites were associated with both activation and repression of nearby genes, suggesting a more complex relationship to gene expression (fig. S4, B and D).
The use of high-throughput sequencing allowed us to assess allele-specific chromatin signals by detecting preferential recovery of sequence reads containing one allele over the other when there was an underlying heterozygous SNP in the individual. When aligning our sequences containing such a mixture of alleles at a given heterozygous SNP to the reference human genome sequence, we found a marked preference for alignment of sequence reads containing the allele that also happened to be represented in the reference sequence (fig. S5). After correcting for this technical bias (13), we assessed the true allele specificity of each heterozygous SNP sequenced at sufficient depth for each assay, and found that 7% of DNase I HS sites and 11% of CTCF sites have significant allele specificity after multiple testing correction (Fig. 1C).
Although allele-specific sites occurred on all chromosomes, the X chromosome was particularly enriched for such sites. This would be expected if DNase I HS and CTCF binding on the two X chromosomes is unequal in females, provided that one of the two X chromosomes is preferentially inactivated in the cell population (fig. S6, A and B). Indeed, we established that X inactivation patterns were nonrandom in the cell lines studied, and that the paternal X was preferentially inactivated in 90% of cells in each cell line from both daughters (fig. S7A). Most X-chromosome allele-specific CTCF sites showed a bias toward the active maternal X (fig. S7B), thus demonstrating that allelic imbalance in CTCF binding is generally associated with epigenetic silencing in X inactivation. We found several sites at which CTCF bound equally to the inactive and active X alleles or preferentially bound the allele on the inactive X. These could represent CTCF binding in regions escaping inactivation, or sites involved in or otherwise reflecting epigenetic changes associated with dosage compensation (9).
To establish that the allele-specific CTCF binding biases were not an artifact, we tested four allele-specific and five non–allele-specific CTCF sites using matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry (MALDI-TOF MS) (fig. S8A and table S4) (15). Each of the four allele-specific sites showed a significantly higher proportion of the enriched allele (fig. S8B), although the absolute levels of enrichment were lower as assayed by MALDI-TOF MS than by ChIP-seq. In contrast, none of the five non–allele-specific ChIP-seq CTCF sites showed significant bias by MALDI-TOF MS (fig. S8B and table S5).
Chromatin signals could be individual-specific or allele-specific as a result of nongenetic factors, such as environmental, epigenetic, or stochastic differences between individuals (4, 5). If allele-specific chromatin structure has a direct genetic basis, the relationship between a specific allele and the chromatin signal should be maintained between individuals. When we considered the 10,364 shared heterozygous sites present in two or more individuals, if two individuals showed significant allele-specific CTCF binding, it was nearly always toward the same allele (Fig. 3, A and B). We next examined the prevalence of an autosomal imprinting-like process for generating allele specificity. Because the male and female parental alleles are randomly distributed with respect to any genetic haplotype, one would expect that if a site were subjected to a parent-of-origin imprinting-like process, half of such sites would have reversed allele specificity in unrelated individuals with the same heterozygous sites. However, only about 2% of interindividual pairs showed significantly opposite behavior (Fig. 1C) (13). This shows that an autosomal imprinting-like mechanism is not a major contributor to allelic bias, at least for CTCF binding.
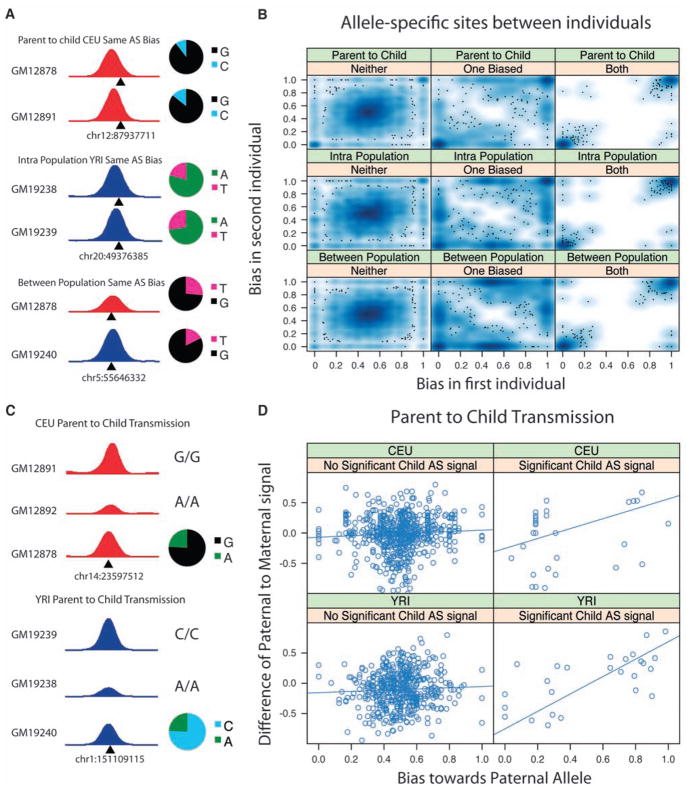
Fig. 3.
Comparison of allele-specific sites between individuals. (A) Each subpanel shows a different allele-specific site in two individuals in the indicated category. The overlapping SNP is indicated below. Adjoining pie charts show concordant allelic bias within the ChIP-seq reads for each site. (B) Allele-specific CTCF site correlations, as shown by smoothed scatterplots (13) of biases between any two individuals (parent-child, intrapopulation, and between-population) where the bias was significant in neither, one, or both individuals. Because of the large number of sites, the density of sites is shown by shades of blue, with outlying sites to this density shown as points. In each pairwise comparison, the bias was predominantly correlated (lower left and upper right of each plot). (C) Allele-specific CTCF sites in a child where both parents are homozygous, showing transmissibility. Peak heights indicate relative binding strength in the parents. The parent homozygous for the allele that was overrepresented in the child has a stronger signal than the other parent. (D) Heterozygous CTCF sites in children where both parents were homozygous. Child sites were classified as allele-specific (right) or not (left). CTCF signal differences between parents were compared to each of the children. Zero on x axis represents 100% maternal bias; 1 represents 100% paternal bias.
Using the parent-child structure of our study, we could also examine the relationship between allele-specific information present in the children and individual-specific information in the parents. Unlike the earlier transmission test of individual-specific sites (Fig. 2), this comparison specifically assesses a genetic mechanism for generating allele specificity. At the 62 CTCF sites where there was a significant allele-specific signal in the child and where one parent was homozygous for one allele and the other parent homozygous for the other (Fig. 1D), the allele bound most strongly by CTCF in the child was most often (65%) the allele carried by the parent who showed the greatest level of CTCF binding, and the extent of parental differential CTCF binding was correlated to the extent of the child’s allele specificity (P = 6.6 × 10−5, Spearman’s correlation) (Fig. 3, C and D). These results suggest a heritable genetic rather than an epigenetic basis for a large proportion of the allele-specific binding of CTCF. There was a strong tendency for the same allele to be preferred in both the CTCF and DNase I HS assays when both could be measured (fig. S9). It is thus likely that DNase I HS sites are also correlated between individuals and are transmissible from parent to child.
SNPs underlying the allele-specific sites could directly affect transcription factor binding and chromatin. Alternatively, these SNPs could merely be markers for other cis polymorphisms such as indels that we did not incorporate into our reconstructed reference genomes. We therefore examined whether SNPs themselves disrupted the CTCF binding motif, and whether the effect of any disruption was consistent with the observed effect on CTCF binding (13). At sites where CTCF showed allele-specific binding, the motif score tended to be higher for the favored allele, whereas at sites lacking differences in CTCF binding, motif scores were similar (fig. S10). Moreover, strongly conserved positions in the motif were more likely to harbor allele-specific SNPs (Fig. 4). Thus, SNPs underlying many allele-specific binding sites are likely to directly affect the binding of CTCF, further suggesting that there is a genetic basis for allele-specific binding.
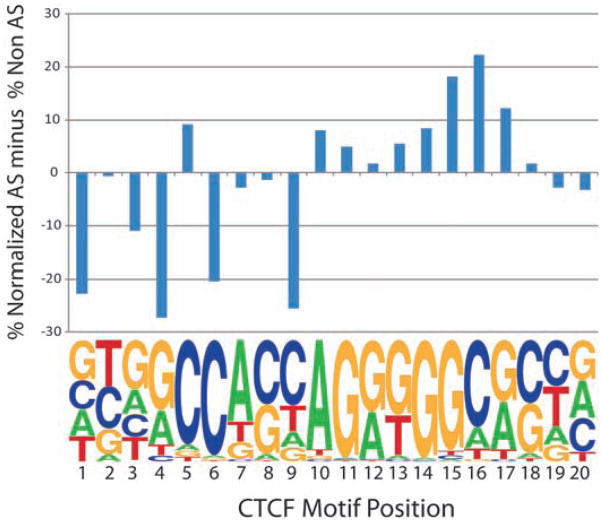
Fig. 4.
Representation of allele-specific and non–allele-specific SNPs across the CTCF binding motif (17). The y axis indicates the difference between the two as a percentage of normalized total SNPs. Higher bars indicate an increased representation of allele-specific SNPs relative to other positions, which tends to occur at conserved positions.
Our results suggest a strong genetic component for allele-specific differences at the level of transcription factor binding and chromatin structure. In addition to the genetic effects, we expect that some individual-specific differences may be due to nongenetic or epigenetic differences between individuals, such as DNA methylation, which could vary without regard to the underlying genotype. Our results are not consistent with widespread random allelic inactivation in lymphoblastoid lines (16), and they place limits on the extent of an imprinting-like process affecting transcription factor binding and chromatin structure. Chromatin structure is thought to be an important reservoir of epigenetic information as well as part of the means by which genetic and epigenetic changes affect phenotypes. Because we can now reliably measure individual differences in chromatin structure, our data may have implications for the identification and characterization of common noncoding polymorphisms associated with disease risk.
Supplementary Material
Acknowledgments
We thank T. Severson, F. Ye, and L. Bukovnik at the Duke IGSP Sequencing Core Facility and K. Moon and S. Luo at Illumina for sequencing; S. Tenenbaum and S. Chittur at the State University of New York, Albany, for expression data; J. Lucas and Z. Zhang at Duke for gene expression analysis; the Texas Advanced Computing Center for computational infrastructure; and the 1000 Genomes Project for genotypes. E.B. has been a paid consultant to EagleGenomics, UK. Supported by National Human Genome Research Institute (NHGRI) ENCODE Consortium grant U54 HG004563-03 and NHGRI grant Z01 HG000024. Raw data from this paper are available at Gene Expression Omnibus (GSE15805 and GSE19622) and at the University of California, Santa Cruz (UCSC) Genome Browser.
Footnotes
Supporting Online Material
References and Notes
- 1.Cheung VG, Spielman RS. Nat Rev Genet. 2009;10:595. doi: 10.1038/nrg2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morley M, et al. Nature. 2004;430:743. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang K, et al. Nat Methods. 2009;6:613. doi: 10.1038/nmeth.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatchwell E, Greally JM. Trends Genet. 2007;23:588. doi: 10.1016/j.tig.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Montgomery SB, Dermitzakis ET. Hum Mol Genet. 2009;18:R211. doi: 10.1093/hmg/ddp400. [DOI] [PubMed] [Google Scholar]
- 6.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 7.Gross DS, Garrard WT. Annu Rev Biochem. 1988;57:159. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 8.Wu C, Bingham PM, Livak KJ, Holmgren R, Elgin SC. Cell. 1979;16:797. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- 9.Filippova GN. Curr Top Dev Biol. 2007;80:337. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- 10.Lewis A, Murrell A. Curr Biol. 2004;14:R284. doi: 10.1016/j.cub.2004.03.026. [DOI] [PubMed] [Google Scholar]
- 11.Lewis A, Reik W. Cytogenet Genome Res. 2006;113:81. doi: 10.1159/000090818. [DOI] [PubMed] [Google Scholar]
- 12.Phillips JE, Corces VG. Cell. 2009;137:1194. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.See supporting material on Science Online.
- 14.Kadota M, et al. PLoS Genet. 2007;3:e81. doi: 10.1371/journal.pgen.0030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohlke KL, et al. Proc Natl Acad Sci USA. 2002;99:16928. doi: 10.1073/pnas.262661399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plagnol V, et al. PLoS ONE. 2008;3:e2966. doi: 10.1371/journal.pone.0002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim TH, et al. Cell. 2007;128:1231. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.