SUMMARY
We describe a mechanism of flagellar motor control by the bacterial signaling molecule c-di-GMP, which regulates several cellular behaviors. E. coli and Salmonella have multiple c-di-GMP cyclases and phosphodiesterases, yet absence of a specific phosphodiesterase YhjH impairs motility in both bacteria. yhjH mutants have elevated c-di-GMP levels and require YcgR, a c-di-GMP-binding protein, for motility inhibition. We demonstrate that YcgR interacts with the flagellar switch-complex proteins FliG and FliM, most strongly in the presence of c-di-GMP. This interaction reduces the efficiency of torque generation and induces CCW motor bias. We present a “backstop brake” model showing how both effects can result from disrupting the organization of the FliG C-terminal domain, which interacts with the stator protein MotA to generate torque. Inhibition of motility and chemotaxis may represent a strategy to prepare for sedentary existence by disfavoring migration away from a substrate on which a biofilm is to be formed.
INTRODUCTION
Cyclic or c-di-GMP has emerged as a key player in the decision between motile and sedentary forms of bacterial life (Hengge, 2009; Jenal and Malone, 2006; Romling et al., 2005; Schirmer and Jenal, 2009). c-di-GMP enhances biosynthesis of capsular and fimbrial components required for biofilm formation while inhibiting flagella and pili that allow movement (Simm et al., 2004; Wolfe and Visick 2008). Steady-state levels of c-di-GMP are thought to be maintained by diguanylate cyclases (DGCs) and phosphodiesterases (PDEs). DGC activity depends on a conserved GGDEF domain, while PDE activity requires an EAL or HD-GYP domain (Ryan et al., 2006). GGDEF and EAL domains are associated with many diverse input and output domains, suggesting that they receive a variety of signals and respond through a variety of different mechanisms. These two domains are the most abundant domains encoded in bacterial genomes, with many bacteria possessing multiple proteins with these domains (Galperin, 2005; Galperin et al., 2001). For example, Salmonella has five GGDEF proteins, eight EAL proteins, and seven proteins with both GGDEF and EAL domains (Simm et al., 2007; Solano et al., 2009). Similarly, E. coli has a large number of such proteins. Despite the existence of multiple such proteins, the association of specific phenotypes with mutation of individual proteins suggests that c-di-GMP levels may be modulated in a spatially localized manner.
Bioinformatic analysis first suggested that PilZ domains may function as c-di-GMP effectors (Amikam and Galperin 2006). Binding and mutagenesis studies of several PilZ domain proteins have confirmed this observation and demonstrated that c-di-GMP binding depends on residues in RxxxR and D/NxSxxG sequence motifs. A crystal structure of a PilZ domain–c-di-GMP complex from Vibrio cholerae shows c-di-GMP contacting seven of nine strongly conserved residues (Benach et al., 2007).
In E. coli and Salmonella, YhjH is a PDE and YcgR a PilZ domain protein that binds c-di-GMP (Ryjenkov et al., 2006; Schmidt et al., 2005). Both proteins are members of the class 3 flagellar regulon (Frye et al., 2006; Ko and Park 2000a; Wang et al., 2006). Mutation of yhjH reduces motility, while that of ycgR does not. However, ycgR suppresses the motility defect of yhjH, suggesting that in the absence of YhjH, YcgR interacts with the resultant increased levels of c-di-GMP. A yhjH mutant in E. coli was shown to have a counterclockwise (CCW) bias, suggesting a role for the YcgR - c-di-GMP in motor switching (Girgis et al., 2007). Early evidence implicating YcgR at the motor comes from the work of Ko and Park, who found that absence of H-NS results in paralyzed flagella, a defect that could be suppressed either by a mutation in ycgR or by providing yhjH in multiple copies (Ko and Park 2000a). Since H-NS was shown to interact with FliG (Donato and Kawula, 1998; Marykwas et al., 1996), a protein that interacts with both the proton-conducting protein MotA that energizes rotation and the rotor protein FliM that controls the direction of rotation, it was proposed that the YcgR–c-di-GMP complex might interfere with motor rotation by interfering with the proper association of the Mot proteins with FliG (Wolfe and Visick 2008).
The flagellar motor consists of a rotating part (the rotor) and a membrane-embedded, nonrotating part (the stator). The rotor is formed from about 25 copies of FliG, 34 copies of FliM, and more than 100 copies of FliN, together forming an assembly called the C-ring or switch complex (Francis et al., 1992; Yamaguchi et al., 1986). The switch complex is located at the bottom of the basal body and functions in flagellar assembly, rotation, and clockwise/counterclockwise (CW/CCW) directional control. The stator is formed from the membrane proteins MotA and MotB. These proteins function to conduct ions across the membrane and harness ion flow to rotation (reviewed in Blair, 2003). Rotation depends upon electrostatic interactions between sites in the stator protein MotA and the rotor protein FliG (Zhou et al., 1998a). The direction of rotation is controlled by the switch complex, which changes to the CW state in response to the signaling protein phospho-CheY (CheY~P) (Welch et al., 1993).
Here we present evidence that YcgR interacts with the flagellar rotor proteins FliM and FliG in a c-di-GMP-dependent manner. In E. coli, regions of protein-protein interaction were mapped using mutations, and the effects of YcgR on flagellar protein organization were probed by targeted crosslinking. In Salmonella, these were followed by localization of a fluorescent YcgR-GFP fusion to flagellar basal bodies. We show that overexpression of wild-type YcgR, but not the YcgRR118D mutant protein, alters the organization of the C-terminal domain of FliG, the rotor component most directly involved in generation of torque. The results are discussed in the framework of a model in which c-di-GMP-bound YcgR alters the orientation of the FliG C-terminal domain so that the rotor-stator interface is disrupted and the motor is biased toward CCW rotation.
RESULTS
Cyclic-di-GMP Acts through YcgR to Inhibit Swimming and Swarming
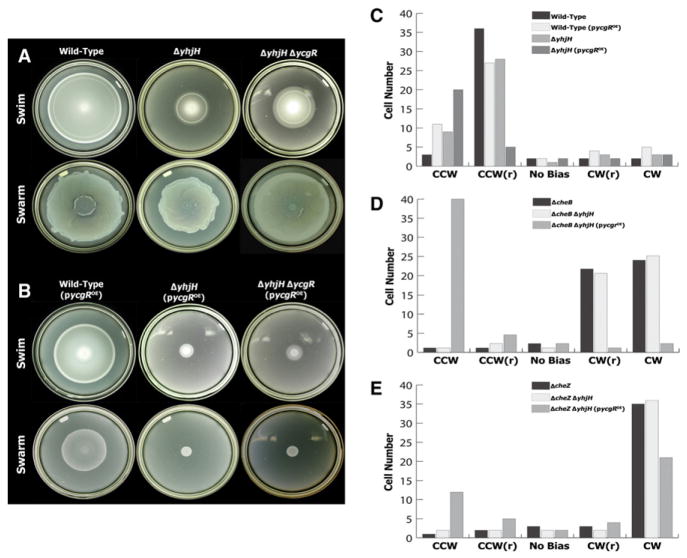
As has been noted previously, deletion of the c-di-GMP phosphodiesterase yhjH impairs swimming motility in Salmonella, and motility of the ΔyhjH strain is substantially rescued by deletion of the PilZ-domain protein YcgR (Ryjenkov et al., 2006) (Figure 1A). The inhibitory effect of the yhjH deletion on both swimming and swarming was made stronger by overexpression of YcgR from a plasmid (Figure 1B). Similar results have been obtained in experiments with Escherichia coli mutants (Ko and Park, 2000a; Ryjenkov et al., 2006) and cyclic-di-GMP was shown to bind directly to the purified YcgR protein of E. coli (Ryjenkov et al., 2006). Together, these observations indicate that cyclic-di-GMP acts through YcgR to inhibit motility and that this regulatory mechanism operates in both Salmonella and E. coli.
Figure 1. Swimming, Swarming, and Flagellar Rotational Bias of Salmonella Strains with Altered c-di-GMP Levels.
(A) Motility of wild-type, ΔyhjH, and ΔyhjHΔycgR strains. Swim plates contain 0.3% agar and swarm plates 0.6% agar in addition to 0.5% glucose. Plates were incubated at 37°C for 6–8 hr.
(B) Motility of strains shown in (A) transformed with a plasmid directing overexpression (OE) of YcgR. Plasmid gene expression was induced with 0.2% arabinose.
(C–E) Histograms display flagellar rotation bias of tethered cells in the absence of chemotactic stimuli. Forty-four tethered cells were observed for 60 s each as described in the Experimental Procedures. Cells were classified into five categories (from left to right): exclusively CCW, CCW biased with reversals (r), frequent reversals with no bias, CW biased with reversals, and exclusively CW.
Motility Inhibition Is Due to Reduced Motor Speed and Increased CCW Bias
We hypothesized that the motility inhibition in ΔyhjH cells might be caused by a loss of chemotactic function, because nonchemotactic mutants of Salmonella or E. coli cannot swarm (Burkart et al., 1998; Harshey and Matsuyama, 1994; Mariconda et al., 2006; Wang et al., 2005) and a ΔyhjH mutant of E. coli was reported to have an aberrantly CCW motor bias (Girgis et al., 2007). To look more directly at motor performance in the ΔyhjH and YcgR-overexpressing (YcgROE) cells, we used a tethered-cell assay to measure motor speed and bias (Figure 1C). In a wild-type population, most cells rotated predominantly CCW but also displayed some intervals of CW rotation. When the yhjH gene was deleted, or when YcgR was overexpressed, the number of cells showing CW intervals was decreased, with a corresponding increase in cells turning exclusively CCW. Effects of yhjH deletion and YcgR overexpression were additive; most cells of the ΔyhjH/YcgROE strain turned exclusively CCW. Motor speeds were also reduced in the mutant strains. Rotation rates for the tethered cells (mean ± SD, revs/min) were the following: wild-type, 146 ± 21; YcgROE, 132 ± 24; ΔyhjH, 138 ± 20, and ΔyhjH/YcgROE, 97 ± 25. Thus, under conditions of elevated c-di-GMP and YcgR, motor torque was reduced by about 30%.
Motor bias is normally set by the chemotactic signaling protein CheY~P, which interacts with the flagellar switch to increase the probability of CW rotation. CheY~P level is governed by the relative activities of the kinase CheA and the phosphatase CheZ. CheA kinase activity is modulated by the chemoreceptors, which are in turn regulated by both the binding of chemoeffector and the level of methylation at specific glutamyl residues (Hazelbauer et al., 2008). Receptor methylation level is determined by the activities of the methyltransferase CheR and the methylesterase CheB, and is the basis of sensory adaptation: CheA transfers some of its phosphoryl groups to CheB, activating the methylesterase and bringing the receptors back to their “kinase-off” signaling state. The absence of either the methylesterase CheB or the phosphatase CheZ elevates the level of CheY~P in the cell, either by inducing the methylated, “kinase-on” state of the receptors or by slowing the dephosphorylation of CheY~P. Both ΔcheB and ΔcheZ strains display a strong CW motor bias. The effect of YcgR on motor switching was most clearly evident in these CW-biased backgrounds; deletion of yhjH coupled with overexpression of YcgR caused many of the ΔcheB and ΔcheZ cells to switch from CW to CCW bias (Figures 1D and 1E).
YcgR Interacts with Proteins of the Rotor
The direction of motor rotation is controlled by a “switch complex” on the rotor formed from the proteins FliG, FliM, and FliN (Berg, 2003). Torque is produced at the interface between FliG and the membrane protein MotA, which is a component of the stator (Lloyd et al., 1996; Zhou et al., 1998a). To identify the target(s) of YcgR in the motor, we used pull-down assays to examine the binding of YcgR to the switch-complex proteins and to a cytoplasmic domain of MotA that is known to form the site of interaction with the rotor (Yakushi et al., 2006; Zhou et al., 1998a). The experiments used a fusion of GST (glutathione S-transferase) to the amino terminus of YcgR and were done using proteins of E. coli, which are very similar in sequence to those of Salmonella. Experiments were conducted in the presence of 5 μM c-di-GMP. Neither MotAC (a domain comprising residues 70–170 and so including residues that interact with the rotor) nor FliN was coisolated with the GST-YcgR fusion in this assay. The FliG and FliM proteins were reproducibly coisolated with the GST-YcgR fusion at levels significantly above GST-only controls (Figure 2A). In the experiments with FliM, some of the FliM was found in a low-speed pellet, consistent with the reported tendency of this protein to aggregate when expressed separately from the other switch-complex proteins (Mathews et al., 1998). Aggregation was decreased by coexpression of FliN, and because no direct YcgR-FliN interaction was detected, we included FliN in subsequent binding experiments with FliM. In experiments with both FliM and FliN present, FliM was coisolated with GST-YcgR at relatively high levels, and FliN was coisolated in smaller amounts, consistent with its known binding to FliM (Mathews et al., 1998) (data not shown).
Figure 2. YcgR Interaction with Flagellar Rotor Proteins and YcgR-GFP Localization in Cells.
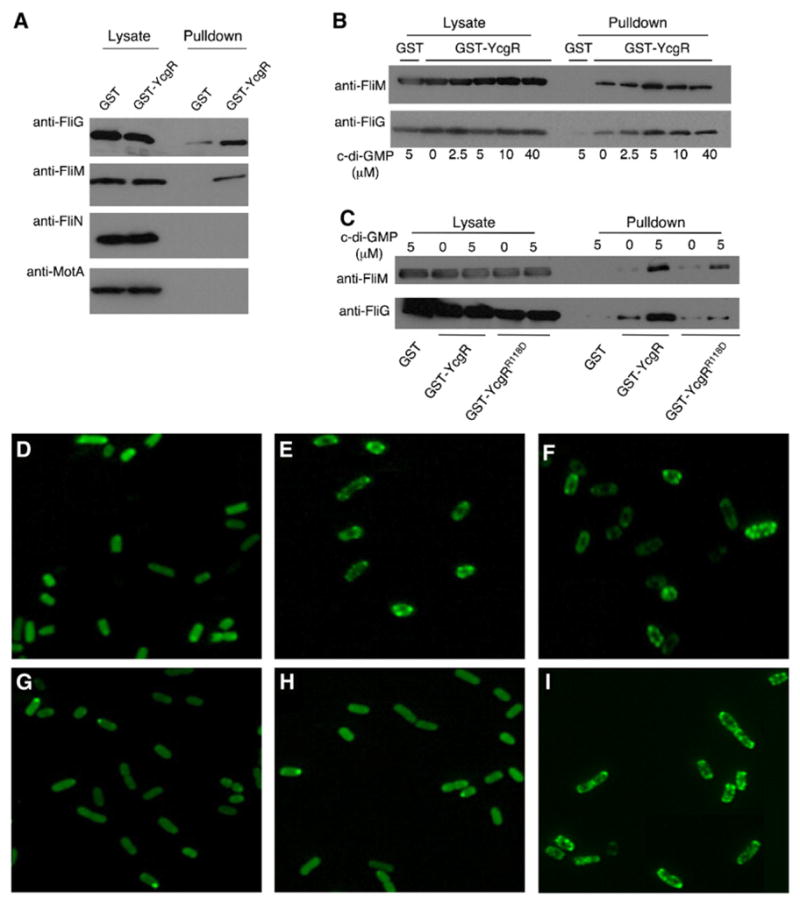
(A) Coisolation of YcgR with FliG and FliM but not with FliN or MotAC (a cytoplasmic domain of MotA). GST-YcgR and the flagellar proteins were expressed in cells of E. coli strain RP3087 (ΔflhDC). Cell lysates were incubated with Sepharose beads and 5 μM c-di-GMP. Pull-down assays were carried out as described in the Experimental Procedures. Lysate controls show protein levels in samples prior to treatment with beads.
(B) c-di-GMP dependence of YcgR binding to FliM and FliG. Samples for pull-down assays were supplemented with c-di-GMP at the concentrations indicated.
(C) Induction by c-di-GMP of binding of the mutant GST-YcgRR118D protein to FliM (top), and weakened binding of GST- YcgRR118D to FliG (bottom).
(D–I) Localization of YcgR-GFP in various Salmonella strains. Strains expressing either GFP or YcgR-GFP were grown in the presence of arabinose and analyzed by fluorescence microscopy as described in the Experimental Procedures. (D) Wild-type (pgfpOE). (E) Wild-type (pycgROE). (F) ΔyhjH (pycgROE). (G) ΔyhjH ΔfliM (pycgROE). (H) ΔyhjH ΔfliG (pycgROE). (I) ΔyhjH ΔmotA (pycgROE).
Effect of c-di-GMP on YcgR Binding to FliM and FliG
Because c-di-GMP binds to YcgR and is important for the YcgR-mediated motility inhibition, we next measured the binding of YcgR to FliM and FliG in the presence of various concentrations of c-di-GMP. YcgR showed measurable binding to both in the absence of any externally added effector, and this binding was increased by roughly 2-fold when c-di-GMP was added to 5 μM or higher (Figure 2B). Subsequent binding experiments were done in the presence of 5 μM c-di-GMP.
In experiments in E. coli, the mutation R118D in YcgR was shown to prevent c-di-GMP binding to the purified protein and to eliminate the motility impairment in the yhjH deletion strain (Ryjenkov et al., 2006). To test binding of this mutant protein to the switch proteins, we introduced the R118D mutation into the GST-YcgR fusion construct and performed the pull-down experiment as before. Binding of the YcgRR118D protein to FliM was weakened but was not prevented. Surprisingly, binding of the YcgRR118D protein to FliM was stimulated by c-di-GMP despite the reported inability of this protein to bind the effector (Figure 2C). Binding of the YcgRR118D protein to FliG was much weaker than that of wild-type YcgR, both in the presence and absence c-di-GMP (Figure 2C).
Localization of YcgR-GFP to the Flagellar Basal Body
To determine whether the interaction between YcgR and the switch proteins occurs in cells, we constructed a GFP fusion to the C terminus of Salmonella YcgR and examined the localization of the protein. The fusion protein behaved similarly to wild-type YcgR in causing motility impairment when overexpressed (data not shown). The YcgR-GFP construct displayed a punctate fluorescence in cells, unlike the diffuse fluorescence of GFP alone (Figure 2; compare Figures 2D and 2E). Punctate localization was more distinct in a ΔyhjH strain (Figure 2F) than in wild-type and was retained in a strain lacking all seven chemoreceptors (see Figure S1 available online). Puncta were eliminated in cells lacking either FliM (Figure 2G) or FliG (Figure 2H) but were still present in a strain lacking MotA (Figure 2I; an E. coli motA null strain was also punctate, as shown in Figure S1), confirming that localization is via the C-ring.
Surface on FliM Involved in Binding YcgR
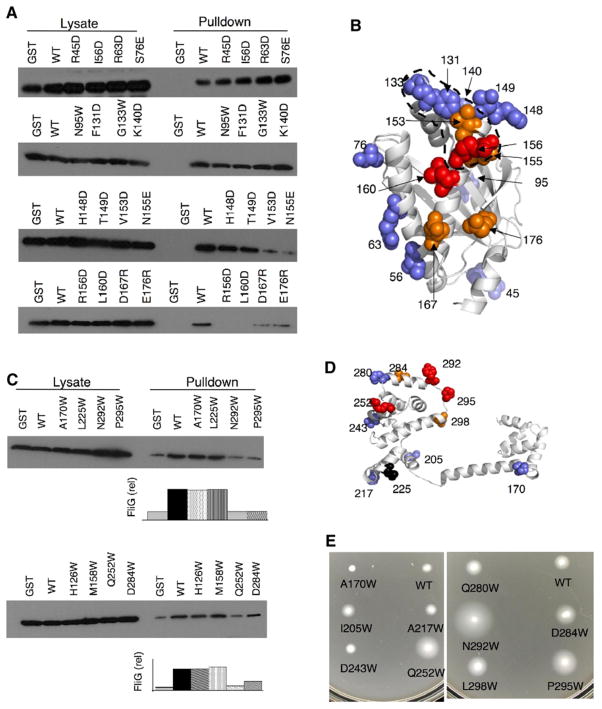
To identify the regions of FliM most important for its interaction with YcgR, we used the pull-down assay and a collection of nonconservative FliM mutations in E. coli that sample various regions of the protein surface. Binding to YcgR was eliminated by the FliM mutations R156D and L160E and was weakened by the replacements V153D, N155E, D167R, and E176R (Figure 3A). These residues lie near each other on a surface region of FliM that overlaps the binding site for FliG (Figure 3B). Binding to YcgR was not measurably weakened by the FliM mutations R45D, I56D, R63D, S76E, N95W, F131D, G133W, K140D, H148D, or T149D (Figures 3A and 3B).
Figure 3. Mapping of YcgR-Interaction Sites on FliM and FliG in E. coli.
(A) Coisolation of FliM and its mutant variants with GST-YcgR. The experiment was done in the presence of 5 μM c-di-GMP. FliN was also expressed in the cells to prevent aggregation of FliM (see the text for details).
(B) Binding results with mutants, mapped onto the FliM structure. Red coloring indicates complete elimination of YcgR binding; orange, weakened binding; blue, binding similar to wild-type. The region on FliM involved in its interaction with FliG is shown by the black dotted line (K.P. and D.F.B., unpublished data).
(C) Coisolation of FliG and its mutant variants with GST-YcgR. The experiment was done in the presence of 5 μM c-di-GMP.
(D) Binding results with mutants, mapped onto the FliG structure (with colors as in B). A mutation that gave nonflagellate phenotype (225W) (which was not included in the swimming assays) is indicated by the black sphere.
(E) Effects of FliG mutations on the motility impairment caused by elevated levels of c-di-GMP. Mutant FliG proteins were expressed from plasmids in a ΔyhjHΔfliG strain. Fresh transformants were picked onto swim plates containing tryptone and 0.27% agar and incubated at 32°C for 10 hr.
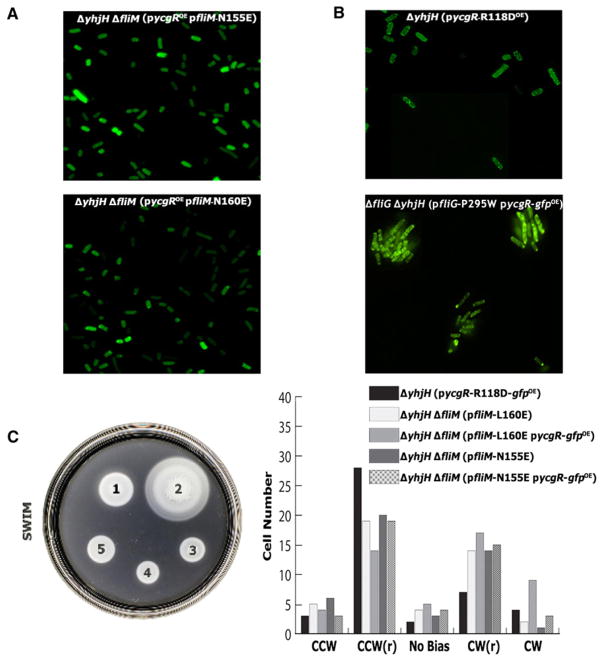
To determine the importance of this binding site in cells, we examined localization of YcgR-GFP in Salmonella cells containing the N155E, L160E, D167R, and E176R mutations in FliM. Puncta were eliminated by the N155E and L160E FliM mutations, which lie near the center of the region identified in the pull-down experiments (Figure 4A). Localization was not affected by the D167R and E176R mutations that lie nearer the edge of the YcgR-binding region (data not shown). The YcgRR118D variant was also examined and showed normal punctate localization (Figure 4B), consistent with the retention of FliM binding observed in this mutant (Figure 2C). The R118D mutation was reported to eliminate the YcgR-linked motility impairment in E. coli (Ryjenkov et al., 2006). To determine whether this is also the case in Salmonella, we used the soft agar swimming and tethered-cell rotation assays to examine motility of Salmonella cells overexpressing the YcgRR118D protein. Consistent with the previous reports in E. coli, the mutation relieved the motility impairment in both assays (Figure 4C). Thus, the YcgRR118D protein is able to bind FliM and localize to the basal body but does not affect motor bias or speed.
Figure 4. YcgR Localization and Phenotypic Analysis of Mutants in Salmonella.
(A) Diffuse localization of YcgR-GFP in cells with FliM mutations N160E and N155E.
(B) Normal punctate localization of YcgR in the YcgR R118D mutant and the FliG P295W mutant.
(C) Swimming motility (left) and rotational bias (right) of mutant strains. Strains on the swim plate are as follows: (1) ΔyhjH/pycgr-gfpOE; (2) ΔyhjH/pycgr(R118D)-gfpOE; (3) ΔyhjHΔfliM/pfliM(L160E), pycgr-gfpOE; (4) ΔyhjHΔfliM/pfliM,pycgr-gfpOE; (5) ΔyhjHΔfliM/pfliM(N155E),pycgr-gfpOE.
If the motility inhibition in the ΔyhjH strain requires binding of effector-stimulated YcgR to FliM, then FliM mutants defective in the YcgR interaction might show improved motility in the ΔyhjH background. To test this, plasmids expressing YcgR-interaction-defective FliM mutants (N155E, L160D, D167R, and E176R) were transformed into a ΔyhjHΔfliM Salmonella strain, and rates of migration on soft agar plates were measured (the V153D and R156D mutations were not included because they disrupt binding of FliM to FliG and cause a nonflagellate phenotype). The N155E and L160D mutations caused a reproducible but fairly small improvement in motility in the ΔyhjH background (Figure 4C, left; compare 3 and 5 with 4). These mutants by themselves had an altered motor bias, with a significant CW(r) fraction (Figure 4C, right). This bias remained unaltered in the presence of increased YcgR and may account for their smaller swim colony diameter. In parallel experiments in E. coli, similarly mild effects were observed (data not shown) but could be made stronger by further optimization of the experiment (described below).
FliG Mutations Relieve Motility Inhibition and Weaken the YcgR Interaction
Since FliG also interacted with YcgR in the pull-down assay, FliG mutants with defects in the YcgR interaction might relieve the motor from YcgR-mediated inhibition and show improved motility. To examine this, plasmids expressing a collection of E. coli FliG proteins with Trp replacements in various surface positions (obtained in a previous study; Brown et al., 2007) were transformed into a ΔyhjHΔfliG strain, and motility was examined on soft agar plates. Three Trp replacements in the C-terminal domain of FliG (Q252W, N292W, and P295W) showed significant motility improvement in the ΔyhjH background, while two others (D284W and L298W) showed smaller but reproducible improvements (Figure 3E). The motility inhibition was not relieved by FliG mutations at positions 170, 205, 217, 218, 243, or 280 (Figure 3; position 218 mutant not shown). Next, the mutant FliG proteins were tested in pull-down experiments to see if the YcgR-FliG binding was affected. Binding to YcgR was weakened significantly by the Q252W, N292W, and P295W mutations in FliG and was somewhat diminished by the D284W mutation (Figure 3C). The binding to YcgR was not affected by mutation of residues 126, 158, or 170 in the middle domain of FliG, or residue 225 in the hydrophobic patch of the C-terminal domain (Figures 3C and 3D). Residues 292 and 295 lie on the “top” of the domain near the charged ridge that interacts with the stator (Lloyd and Blair, 1997; Yakushi et al., 2006; Zhou et al., 1998a, 1998b), and residue 252 is on the side of the domain, adjacent to the charged ridge (Figures 3D). To test the importance of the FliG binding for YcgR localization to the basal body, we examined the distribution of YcgR-GFP in Salmonella cells containing the P295W mutant FliG. Localization appeared not to be affected (Figure 4B).
Because YcgR interacts with both FliM and FliG, we hypothesized that the presence of a suitable mutation in FliG might reveal otherwise-subtle effects of FliM mutations. To test this proposal, we remeasured the effects of selected FliM mutations on the YcgR-mediated motility inhibition with a mutation also present in FliG (N292W) to decrease the affinity for YcgR. In the presence of this FliG mutation, the FliM mutations N155E and D160R both gave substantial relief from the motility inhibition (motility plates shown in Figure S2).
Point Mutations in YcgR Affecting Its Interactions with FliM and FliG
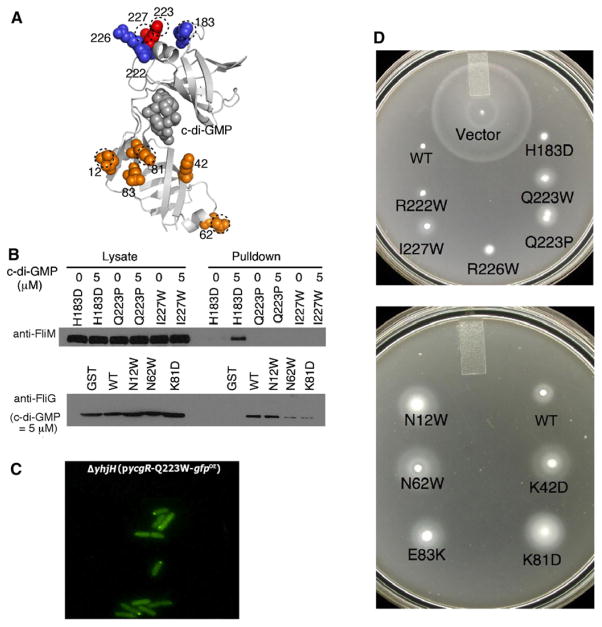
We next sought to identify the regions of YcgR protein important for its interactions with FliM and FliG. YcgR is composed of two domains, an N-terminal PilZN domain (sometimes called the YcgR domain) whose function is not precisely known, and a C-terminal domain that has been associated with the binding of c-di-GMP (Christen et al., 2007; Merighi et al., 2007; Pratt et al., 2007; Ryjenkov et al., 2006). A crystal structure is known for a YcgR-like protein of Vibrio cholerae (VCA0042/PlzD), bound to its effector c-di-GMP (Benach et al., 2007). Using this structure and a sequence alignment with E. coli YcgR, a homology model for the E. coli protein was constructed (Figure 5A). The structure consists of two β barrel domains with c-di-GMP bound in a region between the domains where it could regulate their relative positions or orientation. Additional features include an α helix followed by an antiparallel β hairpin near the C terminus (at the top of the molecule in the orientation shown). Residues in this α helix are relatively well conserved. Further, in the case of the YcgR-like protein DrgA of Caulobacter crescentus the C-terminal domain has been suggested to be involved in interaction with downstream target(s) (Christen et al., 2007). We hypothesized that the α helix in YcgRC might form the site of interaction with FliM and accordingly targeted it for mutagenesis.
Figure 5. Effects of YcgR Mutations on Motility and Binding.
(A) Homology model of E. coli YcgR, based on the crystal structure of the Vibrio cholerae protein VCA0042 (Protein Data Bank ID code 2RDE). YcgR mutations characterized in this study are indicated. Mutations that relieved the motility inhibition are colored red (in the C-terminal domain) or orange (in the N-terminal domain); mutations that did not relieve the motility inhibition are colored blue. Mutations characterized further in pull-down assays with FliM and FliG are indicated by dotted lines.
(B) Effects of YcgR mutations on the binding to FliM and FliG in E. coli.
(C) Diffuse localization of the Q223W YcgR mutant in Salmonella.
(D) Effects of YcgR mutations on the motility impairment caused by YcgR overexpression. Mutant YcgR proteins were overexpressed from plasmids in the ΔyhjHΔycgR background in E. coli by addition of 2.5 μM Na-salicylate. Soft agar plates were incubated at 32°C for 10 hr. The upper and lower plates show, respectively, the behaviors of C- and N-terminal domain mutants.
Mutations were made at five positions in YcgR, four on the α helix (R222W, Q223W and Q223P, R226W, and I227W) and one at a nearby position on the surface (H183D) (Figure 5A). The mutant ycgR genes were fused to sequences encoding an HA tag to facilitate detection of the proteins. Control blots using anti-HA antibody showed that the mutant proteins were expressed at levels similar to HA-tagged wild-type YcgR (Figure S3). The mutant YcgR proteins were expressed from plasmids in the ΔyhjHΔycgR strain to measure their effects on motility. Proteins with the H183D, R222W, or R226W replacements caused motility impairments similar to wild-type. The Q223W, Q223P, and I227W mutations gave substantial relief from the motility impairment, suggesting a role for these residues in interaction with the motor (Figure 5D).
To measure the effects of the YcgR mutations on the binding to FliM, the Q223P and I227W mutations, and the H183D replacement as a negative control, were transferred into the GST-YcgR-HA fusion protein and used in pull-down experiments. The GST-YcgR protein with the H183D mutation bound FliM as strongly as wild-type, whereas the Q223P and I227W mutants showed no binding even in the presence of 5 μM c-di-GMP (Figure 5B). The Q223W mutation was then introduced into the YcgR-GFP fusion construct to examine effects on localization. Punctate localization was eliminated in the Q223W YcgR mutant (Figure 5C). Finally, the tethered-cell assay was used to measure motor performance in ΔyhjH cells overexpressing the Q223W mutant protein. Motor speed and bias were normal (motor speeed 141 ± 27; bias data provided in Figure S4). We conclude that the α helix near the C terminus of YcgR is important for the binding of YcgR to the flagellar switch and the resulting alteration of motor function.
YcgR proteins with mutations in the α helix were also tested in pull-down assays with FliG but were found to have no effect on the YcgR-FliG interaction (Figure S5A). We hypothesized that binding to FliG might occur through the N-terminal domain of YcgR and accordingly mutagenized several moderately well-conserved surface residues in this domain. The mutant proteins were expressed in a ΔfliGΔyhjH strain, and motility was measured in soft agar plates. Of five mutations studied, three (K42D, N62W, and K81D) showed a substantial motility improvement in the ΔyhjH background, and two others (N12W and E83K) showed mild improvements (Figure 5D). To determine whether the motility improvement was due to a weakened interaction with FliG, three of the YcgR mutations (N12W, N62W, and K81D) were transferred into the GST-YcgR fusion construct and used in a pull-down experiment. Binding to FliG was weakened in the N62W and K82W mutants but was not affected in the N12W mutant (Figure 5B). Binding to FliM was normal for all three of these YcgR mutant proteins (Figure S5). Together with the motility enhancements (Figure 5D), these results implicate the N-terminal domain of YcgR in binding to FliG.
Improved Motility upon Overexpression of Stator Components
Because the motility defect in the ΔyhjH strain is due at least in part to CCW motor bias, we hypothesized that overexpression of the CW-signaling protein CheY might reverse this bias defect and improve motility of the ΔyhjH strain. However, overexpression of CheY alone did not improve motility of the ΔyhjH null strain (Figure 6A). In a study of Δhns mutants of E. coli, Ko and Park found that certain defects associated with the rotor can be suppressed by overexpression of the stator proteins (Ko and Park, 2000b). To see if this is true of the ΔyhjH mutant, we introduced plasmids expressing MotA, MotB, or MotA and MotB together and retested motility in the soft agar assay. Motility of the ΔyhjH mutant was markedly improved by overexpression of MotA, either by itself or with MotB (Figure 6A). In the strains overexpressing MotA, migration in soft agar was enhanced further by overexpression of CheY (Figure 6A). These findings are in accordance with the proposal that YcgR affects both motor bias and torque generation. Further, they suggest that the defect involves a rearrangement at the rotor-stator interface.
Figure 6. Effects of Overexpressing Stator Proteins, and CheY, in ΔyhjH E. coli Cells.
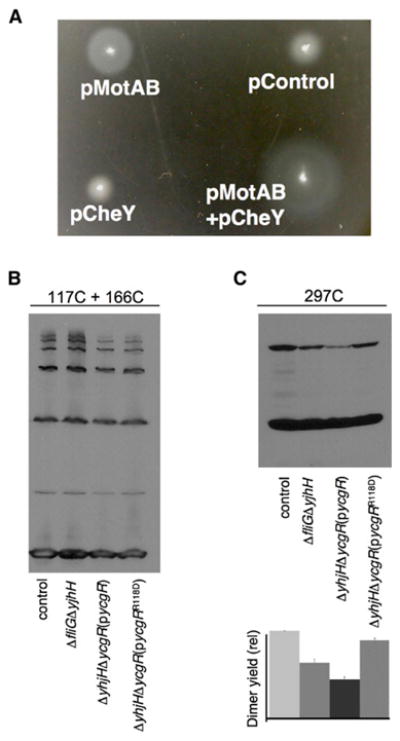
(A) Motility of ΔyhjH cells transformed with plasmids expressing the stator proteins MotA and MotB, the CW-signaling protein CheY, or all three proteins. Parent vectors pRR48 and pKG116 were used as controls. Mot proteins were expressed from the trp promoter, with induction by 100 μM indole acrylic acid. CheY expression was induced using 500 μM arabinose. Fresh transformants were picked onto a soft agar tryptone plate and incubated at 32°C for 10 hr. (B and C) Effects of yhjH deletion and ycgR overexpression on FliG domain organization. Iodine was used to induce crosslinking of FliG proteins with Cys replacements at positions 117 and 166 in the middle domain (B) or position 297 in the C-terminal domain (C). YcgR was wild-type or with the mutation R118D, as indicated. Protein expression was induced with 50 μM IPTG (YcgR) or 10 μM sodium salicylate (FliG). FliG proteins were HA tagged, and products of crosslinking were visualized on anti-HA immunoblots. Relative crosslinking yields for the position 297 experiments are shown at the bottom of (C). In (C), error bars indicate standard variation in three experimental repeats.
Effect of YcgR on Organization of the FliG C-Terminal Domain
The C-terminal domain of FliG is at the interface with the stator and contributes directly to generation of torque (Lloyd et al., 1996; Zhou et al., 1998a). In a current structural model of the switch complex, FliG is at the “top” (the membrane-proximal part) of the C-ring, to allow interactions with the stator, while the major part of FliM (the middle domain, FliMM) is located just below FliGC and positions this domain in its proper relationship to the stator (Yakushi et al., 2006; Zhou et al., 1998a). We previously examined the organization of FliG in the rotor using targeted disulfide cross-linking, identifying cysteine residues capable of forming FliG-FliG crosslinks in the wild-type motor (Lowder et al., 2005). To examine the effects of YcgR upon the organization of FliG, we examined patterns of FliG-FliG crosslinking in the ΔyhjH mutant and in the ΔyhjH mutant overexpressing YcgR. To facilitate detection of the proteins, an HA tag was fused to the FliG C terminus. Disulfide crosslinking was induced with iodine, and products were examined on immunoblots using anti-HA antibody. The HA-fusion construct was functional and, when expressed in a wild-type background, gave crosslinking results similar to those reported previously (data not shown).
Cys residues at positions 117 and 166 in the middle domain of FliG were previously shown to crosslink efficiently to yield dimers and larger multimers (Lowder et al., 2005). The crosslinking occurred in flagellate cells, but not in a nonflagellate ΔflhDC mutant, indicating that it occurs between protein subunits in the flagellum (rather than by collision in the cytosol). Patterns of crosslinking in the 117/166 double-Cys mutant were not substantially affected either by the deletion of yhjH or by overexpression of YcgR (Figure 6B). Thus, YcgR does not appear to affect the organization of the FliG middle domain.
A FliG protein with a Cys replacement of residue 297 in the C-terminal domain was shown previously to crosslink efficiently into dimers, again only when the protein was present in assembled motors (Lowder et al., 2005). Crosslinking in a single-Cys mutant presumably requires significant motion of the domain because equivalent positions in adjacent subunits should be separated by about 4 nm (assuming uniform spacing) (Thomas et al., 2006). The yield of the 297-297 crosslink was somewhat decreased in the ΔyhjH mutant and was decreased further (to about 50% of wild-type) in the mutant that also overexpressed YcgR (Figure 6C). Thus, the binding of YcgR to the motor alters the orientation and/or motional freedom of the FliG C-terminal domain. The effect was prevented by the R118D mutation in YcgR (Figure 6C) and so appears to depend on normal interactions with c-di-GMP.
DISCUSSION
In E. coli and Salmonella, YcgR is dedicated to c-di-GMP-dependent regulation of flagellum-based motility and does not appear to influence other c-di-GMP-associated phenotypes (Ryjenkov et al., 2006). YcgR is composed of two domains: an N-terminal domain of unknown function and a C-terminal PilZ domain. The PilZ domain binds c-di-GMP in vitro with an affinity that is high enough to allow it to respond to small changes in intracellular c-di-GMP levels. This domain organization is conserved in a large number of proteobacteria (Amikam and Galperin 2006). Therefore, the function of YcgR in motility control is likely to be conserved. In this work, we show that elevated c-di-GMP and YcgR slow motor speed and inhibit chemotaxis by inhibiting CW rotation. We provide evidence that YcgR, when bound to c-di-GMP, interacts strongly with the flagellar motor proteins FliG and FliM. We present a model showing how both effects can result from disrupting the organization of the C-terminal domain of FliG, altering the rotor-stator interface in a way that reduces the efficiency of torque generation and induces a CCW motor bias.
“Backstop Brake” Model for Torque Reduction and Promotion of CCW Bias by YcgR
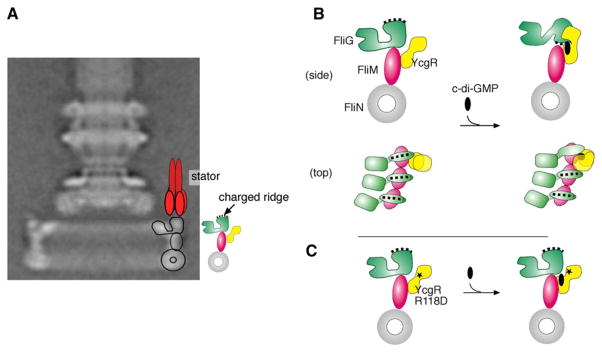
By use of mutations, we identified the region of FliM involved in the binding to YcgR and found that this binding surface overlaps the region shown previously to interact with FliG (Figure 3). This suggests that binding of YcgR could weaken the FliM-FliG inter-action, possibly displacing FliGC and allowing it to adopt an alternative conformation. Physiological and binding experiments with FliG mutants show, furthermore, that YcgR interacts directly with FliGC. Several Trp replacements in FliGC relieved the YcgR-mediated motility inhibition, and when some of these were tested further in pull-down experiments they showed reduced binding to YcgR (Figure 3). Most such FliG mutations occur near the charge-bearing ridge that interacts with the stator (Lloyd et al., 1996; Yakushi et al., 2006; Zhou et al., 1998a). Determinants for YcgR binding thus lie near the “top” of the FliGC domain, i.e., in the region that must point toward the stator in the functioning motor. In our current working model for the switch-complex structure, the charged ridge of FliGC is quite distant from the YcgR-binding region on FliM (the distance between FliG-295 and FliM-160, for example, is approximately 55 Å). Based on the structure of the YcgR homolog of Vibrio (Benach et al., 2007), the YcgR molecule would be unable to span this distance. Accordingly, we propose that binding of c-di-GMP-loaded YcgR to the motor disrupts the interaction between FliM and FliGC and causes FliGC to be reoriented with its charged ridge downward to interact with YcgR. A conserved Gly-Gly linker at the base of the FliGC domain, which has been proposed to function as a hinge during CW/CCW switching (Brown et al., 2002), could function in this context to allow this reorientation of the domain (Figure 7).
Figure 7. Model for YcgR-Mediated Motility Control.
(A) Overall disposition of stator and rotor proteins. A single stator complex (of approximately 11 present in the motor) is shown in red. Proteins of the rotor are shown in outline on the basal-body C-ring and are also shown to the side: FliG (green), FliM (majenta), and FliN (gray). YcgR (yellow) is shown attached to FliM. The charge-bearing ridge indicated on the C-terminal domain of FliG forms the site of interaction with the stator. The electron micrograph was reproduced with permission from the American Society for Microbiology (Brown et al., 2007).
(B) Hypothesis for c-di-GMP-induced rearrangements. Binding of c-di-GMP to YcgR induces movement of the YcgR N-terminal domain, which allows it to bind to the C-terminal domain of FliG. Interaction with YcgR causes the FliG C-terminal domain to reorient so that the charged ridge is no longer able to interact with the stator. YcgRN might also be expected to constrain the movement of the neighboring FliGC domain (top view), preventing movements needed for direction switching.
(C) Hypothesis for the effect of the R118D mutation in YcgRR118D (indicated by *). The R118D mutant protein retains the ability to FliM. When bound to FliM, it can also bind c-di-GMP, but this interaction is altered so that it no longer undergoes the c-di-GMP-induced domain rearrangement and does not interact with FliGC to impair motility.
Torque generation in the motor is known to depend upon the proper relationship between FliGC and MotA (Garza et al., 1996; Yakushi et al., 2006; Zhou et al., 1998a), and so the hypothesized domain movement should reduce the torque of the motor, roughly in proportion to the number of domains that are flipped. Further, given the dimensions of the Vibrio YcgR-homolog (Benach et al., 2007) and the FliGC domain (Brown et al., 2002), and assuming a 4 nm spacing between proteins in the outer C-ring (Thomas et al., 2006; Young et al., 2003), a YcgR molecule interacting with one FliGC domain could easily inhibit the movements of neighboring domains, even those that remain unflipped. Thus, even the FliGC domains that remain correctly oriented for torque generation could be prevented from undergoing movements needed for direction switching, accounting for the CCW bias of the YcgR-inhibited motors. YcgR could thus function as a “backstop brake,” both slowing the motor and inhibiting preferentially its rotation in one direction.
The model can account also for the effects of stator overexpression. Installation of the stator complexes is thought to depend in part on the FliGC-MotA interactions, which are mostly attractive. Loss of the rotor-stator interactions, as occurs when FliGC reorients, should then reduce the average number of stator complexes present in the motor. Overexpression of MotA (or MotA and MotB together) would counter this effect, increasing the number of stator units present and (because such stabilizing effects must be mutual) favoring the normal FliGC conformation. Alternatively, the Mot-overexpression effects might reflect the occurrence of a direct interaction between YcgR and MotA. While our results do not rule out any YcgR-MotA interaction, they suggest that if such an interaction occurs it is weaker than the binding to FliM and FliG (Figure 2A). It is important to point out, however, that YcgR-GFP localization is completely eliminated in single point mutants of FliM (N155E and N160E) (Figure 4A), while localization remains unaffected in a motA null strain (Figure 2I and Figure S1). Thus, YcgR-dependent motility control is primarily through interaction with the switch complex.
The YcgRR118D mutant was reported to be completely defective in c-di-GMP binding and does not impair motility when expressed in the ΔyhjH strain (Ryjenkov et al., 2006). We found, however, that the YcgRR118D protein retains the ability to bind FliM (Figure 2C), and that this binding is responsive to c-di-GMP in the same micromolar concentration range as wild-type YcgR (Figure 2). This indicates that in the binding experiment here, with FliM present, the YcgRR118D protein retains some c-di-GMP binding ability not observed with the purified protein (Ryjenkov et al., 2006). This is not unexpected: if the c-di-GMP-bound form of YcgR binds more tightly to FliM, then it must also be true that the FliM-bound form of YcgR binds more tightly to c-di-GMP, and the binding of c-di-GMP in this case may no longer be sensitive to mutation of Arg118. The R118D mutation clearly does alter YcgR function, eliminating both the motility impairment (Figure 4) and the FliGC domain reorganization (Figure 6C) and weakening the binding to FliG (Figure 2C). These results can be accounted for if the mutation affects the rearranged state of the motor, in which YcgR interacts with FliGC to displace this domain from its normal position. Based on structure of the Vibrio YcgR homolog, Arg118 lies in a hinge region between the two domains of YcgR, where it might influence the relative position of the domains. Accordingly, we propose that Arg118 is important in modulating the conformational change that occurs upon binding c-di-GMP, and that while it is also important for c-di-GMP binding affinity in the purified protein (Ryjenkov et al., 2006), this may not be the case in vivo.
Motility Control by Other YcgR-like Proteins
c-di-GMP-mediated motility control is not restricted to enterics, and can be mediated through alterations in flagellar gene expression, assembly, or function (Wolfe and Visick, 2008). The PilZ domain proteins YcgR in E. coli/Samonella and DgrA in Caulobacter crescentus are the only known examples of ci-d-GMP-binding proteins that do not affect expression of flagellar genes but directly interfere with the function of fully assembled flagella (Christen et al., 2007; Girgis et al., 2007). In Caulobacter, c-di-GMP levels control flagellar function by two different mechanisms acting through the protein FliL (Christen et al., 2007). FliL is required for flagellar rotation in Caulobacter. Elevated c-di-GMP levels, when coupled with elevated expression of DgrA, apparently inhibit FliL synthesis and consequently paralyze the flagella. FliL is also required for the developmentally programmed ejection of the flagellum during the swimmer-to-stalk cell transition in this dimorphic bacterium. Flagellar ejection is dependent on the response regulator PleD, whose C-terminal output domain has DGC activity (Paul et al., 2004). In C. crescentus, DgrA-mediated downregulation of flagellar activity is probably an early step in the attachment of a swarmer cell to a surface, and is followed by flagellar-to-stalked cell pole differentiation. In E. coli and Salmonella, FliL is important for surface swarming, but not for swimming (Attmannspacher et al., 2008). However, the YcgR phenotype is independent of FliL in Salmonella, and overexpression of FliL did not improve motility in the E. coli ΔyhjH null strain (our unpublished data), suggesting that FliL is not a major target of YcgR action in these species.
Logic of Regulating Motility after a Flagellum Is Fully Assembled
Regulating motility by controlling the function of the fully assembled flagellum has a distinct advantage over transcriptional control of flagellar gene expression. In Bacillus subtilis, a related mechanism has been demonstrated for the protein EpsE, which functions as a “clutch” by interacting with the C-terminal domain of FliG to disengage the rotor from the stator (Blair et al., 2008). The biological function of the clutch in B. subtilis is intimately associated with biofilm formation, because epsE is encoded within an operon devoted to biosynthesis of extracellular poly-saccharides (EPS). In both the EpsE case and the YcgR mechanism proposed here, motor control is more rapid, and more readily reversible, than mechanisms based on gene expression or assembly. Cells that have been partially immobilized by the action of YcgR and c-di-GMP presumably remain ready to move if the biofilm disperses.
While the targets of YcgR and EpsE are similar, involving proteins of the rotor in both cases, their modes of action show some important differences. Unlike EpsE, YcgR does not appear to disengage the rotor completely from the stator but leaves most rotor elements in position to interact with the stator, though with reduced ability to switch. This difference might reflect the different stages at which the proteins act: Eps functions after the commitment to make a biofilm has already been made and the bacteria are likely to be assured of a suitable substrate for biofilm development, whereas YcgR could facilitate an earlier step in surface attachment. Inhibition of tumbling together with the reduction in motor torque could provide cells with a longer window of opportunity to interact with the surface, setting up the platform for early stages of biofilm formation. Inhibition of chemotaxis has been reported to reduce surface hydration and inhibit swarming in Salmonella and E. coli (Mariconda et al., 2006; Wang et al., 2005). P. aeruginosa also adjusts its motility during the transition to surface living, but using yet another strategy: in this species, c-di-GMP accumulation enhances biofilm formation by increasing EPS synthesis while simultaneously inhibiting chemoreceptor methylation to decrease the frequency of flagellar reversals (Caiazza et al., 2007; Kuchma et al., 2007). When circumstances call for less-vigorous motility, flagellar function can evidently be modulated by a variety of molecular mechanisms that leave the flagellar structure intact and so capable, presumably, of subsequent revival.
EXPERIMENTAL PROCEDURES
Strains, Growth Conditions, Mutagenesis, and Motility Assays
Strains, growth conditions, mutagenesis, and motility assays are described in the Supplemental Experimental Procedures and Table S1.
Cell Tethering Assay
Tethering was performed as described previously (Mariconda et al., 2006). Bacteria were observed through phase-contrast microscopy and recorded on an external Sony video recording device. Average rotation speed per minute was calculated using a stopwatch as described (Attmannspacher et al., 2008).
Fluorescence Microscopy
Cells containing GFP-tagged fusion proteins were grown in the presence of 0.005% L-arabinose as described previously (Sourjik and Berg, 2000). Cells were visualized through a Nikon Eclipse 50i upright microscope using a B-2E/C filter. Fluorescent images were obtained using an attached Nikon digital camera. Images were archived using Nis-Elements D 3.0 software and subsequently cleaned and colorized using Adobe Photoshop CS3.
Binding Assays
Binding of FliG, FliM, FliN, and MotAC to YcgR was measured using a pull-down assay with GST fusions to HA-tagged YcgR. Proteins were expressed separately in two strains, using plasmid pHT100 (Tang et al., 1996) derivatives to express the GST fusions to YcgR-HA (or its variants) and pHT53 (Tang et al., 1996) to express FliG (or its variant), pDB72 (Tang et al., 1996) to express FliM (or its variants), pHT39 (Lloyd et al., 1996) to express FliN, and pSB1 (this study) to express the cytoplasmic domain of MotA (MotAC; comprising residues 70–170). For most experiments, FliN was coexpressed with FliM because FliM alone is prone to aggregation and did not accumulate readily in cells (Mathews et al., 1998). Control experiments used GST only, expressed from plasmid pHT100. Most binding experiments used strain RP3098, a ΔflhDC mutant that expresses no flagellar genes from the chromosome (Tang et al., 1996).
Cells were cultured overnight at 32°C in 40 ml TB containing appropriate antibiotics and 400 μM IPTG for expressing GST (pHT100), FliG (pHT53), FliN (pHT39), and MotAC (pSB1). FliM and its mutant variants (pDB72) and FliN (pKP41) were cultured at the same condition containing appropriate antibiotics. IPTG (40 μM) was used to induce expression of FliM and 10 μM Na-salicylate to induce expression of FliN. Cells expressing the GST-YcgR-HA fusion or its mutant variants were cultured as described by Ryjenkov et al. (2006) and were harvested and resuspended in lysozyme-containing phosphate-buffered saline (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, 5 mM EDTA, 0.2 mM APMSF [4-amidinophenylmethanesulfonyl fluoride], and 0.1% CHAPS [3-(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate). Following a 1 hr incubation on ice, the cells were further disrupted by sonication, debris was pelleted (16,000 g, 40 min, 4°C), and 50 μl of the supernatant was saved for use in estimating the amount of FliG, FliM, FliN, and MotAC present before addition of affinity beads. The rest (~1 ml) was transferred to a clean tube, mixed with 150 μl of a 50% slurry of glutathione Sepharose 4B (Pharmacia) prepared according to the manufacturer’s directions, and incubated for 1 hr at room temperature along with cyclic-di-GMP (Sigma) and with gentle rotation to allow binding. The Sepharose beads were then pelleted by a 1 min microcentrifuge spin, washed with 1 ml of phosphate-buffered saline, and pelleted again by a brief spin. FliG was washed slightly differently with phosphate-buffered saline containing 1% BSA and 0.1% Triton-X. The beads were then incubated with 50 μl of elution buffer (50 mM reduced glutathione in 50 mM Tris-HCl [pH 8.0]) for 10 min at room temperature with gentle rotation to release the GST-YcgR-HA and associated proteins. Beads were then pelleted and the supernatant was collected for analysis by SDS-PAGE and immunoblotting using corresponding antibodies.
The expression of GST-YcgR-HA and its variants were also examined by anti-HA antibody (Covance, USA); these experiments used strain MS1280 (Table S1) and the culturing procedures described by Ryjenkov et al. (2006).
Crosslinking
Crosslinking experiments were carried out as described by Lowder et al. (2005). Products of crosslinking were examined on immunoblots using anti-HA antibody (Covance, USA).
SDS-PAGE and Immunoblotting
Protein samples were separated on 12% SDS-PAGE minigels and transferred to nitrocellulose using a semidry transfer apparatus (Bio-Rad). Rabbit poly-clonal antibodies against FliG, FliM, FliN, and MotA were prepared as described previously (Kim et al., 2008; Tang et al., 1995; Tang and Blair, 1995) and were used at 1500-fold dilution. YcgR-HA tagged proteins were detected using mouse anti-HA antibody at 1000-fold dilution (Covance, USA). Bands were visualized using the Super Signal West Picoluminol system (Pierce) and X-ray film.
Supplementary Material
Acknowledgments
We gratefully acknowledge Chankyu Park for his comments and helpful discussion in the early stages of this study. We thank J.S. Parkinson, Chankyu Park, and Tim Wang for strains; Duncan Brunstetter for technical assistance; and Matthew Ramsey and Aimee Wessel for help in the use of the Whiteley lab microscope. This work was supported by grants from the Welch foundation (F-1351 to R.M.H.) and the U.S. National Institutes of Health (NIH) (R01GM64664 to D.F.B. and GM57400 to R.M.H.) and a Dale A. Stringfellow Graduate Fellowship in Microbiology to K.P.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, one table, and Supplemental References and can be found with this article online at doi:10.1016/j.molcel.2010.03.001.
References
- Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- Attmannspacher U, Scharf BE, Harshey RM. FliL is essential for swarming: motor rotation in absence of FliL fractures the flagellar rod in swarmer cells of Salmonella enterica. Mol Microbiol. 2008;68:328–341. doi: 10.1111/j.1365-2958.2008.06170.x. [DOI] [PubMed] [Google Scholar]
- Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007;26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- Blair DF. Flagellar movement driven by proton translocation. FEBS Lett. 2003;545:86–95. doi: 10.1016/s0014-5793(03)00397-1. [DOI] [PubMed] [Google Scholar]
- Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science. 2008;320:1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- Brown PN, Hill CP, Blair DF. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 2002;21:3225–3234. doi: 10.1093/emboj/cdf332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PN, Terrazas M, Paul K, Blair DF. Mutational analysis of the flagellar protein FliG: Sites of interaction with FliM and implications for organization of the switch complex. J Bacteriol. 2007;189:305–312. doi: 10.1128/JB.01281-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart M, Toguchi A, Harshey RM. The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2568–2573. doi: 10.1073/pnas.95.5.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazza NC, Merritt JH, Brothers KM, O’Toole GA. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:3603–3612. doi: 10.1128/JB.01685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen M, Christen B, Allan MG, Folcher M, Jeno P, Grzesiek S, Jenal U. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc Natl Acad Sci USA. 2007;104:4112–4117. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato GM, Kawula TH. Enhanced binding of altered H-NS protein to flagellar rotor protein FliG causes increased flagellar rotational speed and hypermotility in Escherichia coli. J Biol Chem. 1998;273:24030–24036. doi: 10.1074/jbc.273.37.24030. [DOI] [PubMed] [Google Scholar]
- Francis NR, Irikura VM, Yamaguchi S, DeRosier DJ, Macnab RM. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc Natl Acad Sci USA. 1992;89:6304–6308. doi: 10.1073/pnas.89.14.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, McClelland M, Hughes KT. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:2233–2243. doi: 10.1128/JB.188.6.2233-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 2005;5:35. doi: 10.1186/1471-2180-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- Garza AG, Biran R, Wohlschlegel J, Manson MD. Mutations in motB suppressible by changes in stator or rotor components of the bacterial flagellar motor. J Mol Biol. 1996;258:270–285. doi: 10.1006/jmbi.1996.0249. [DOI] [PubMed] [Google Scholar]
- Girgis HS, Liu Y, Ryu WS, Tavazoie S. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 2007;3:1644–1660. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harshey RM, Matsuyama T. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc Natl Acad Sci USA. 1994;91:8631–8635. doi: 10.1073/pnas.91.18.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7:263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu Rev Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- Kim EA, Price-Carter M, Carlquist WC, Blair DF. Membrane segment organization in the stator complex of the flagellar motor: implications for proton flow and proton-induced conformational change. Biochemistry. 2008;47:11332–11339. doi: 10.1021/bi801347a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Park C. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J Mol Biol. 2000a;303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- Ko M, Park C. H-NS-dependent regulation of flagellar synthesis is mediated by a LysR family protein. J Biol Chem. 2000b;182:4670–4672. doi: 10.1128/jb.182.16.4670-4672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O’Toole GA. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J Bacteriol. 2007;189:8165–8178. doi: 10.1128/JB.00586-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd SA, Blair DF. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J Mol Biol. 1997;266:733–744. doi: 10.1006/jmbi.1996.0836. [DOI] [PubMed] [Google Scholar]
- Lloyd SA, Tang H, Wang X, Billings S, Blair DF. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J Bacteriol. 1996;178:223–231. doi: 10.1128/jb.178.1.223-231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowder BJ, Duyvesteyn MD, Blair DF. FliG subunit arrangement in the flagellar motor probed by targeted cross-linking. J Bacteriol. 2005;187:5640–5647. doi: 10.1128/JB.187.16.5640-5647.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariconda S, Wang Q, Harshey RM. A mechanical role for the chemotaxis system in swarming motility. Mol Microbiol. 2006;60:1590–1602. doi: 10.1111/j.1365-2958.2006.05208.x. [DOI] [PubMed] [Google Scholar]
- Marykwas DL, Schmidt SA, Berg HC. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J Mol Biol. 1996;256:564–576. doi: 10.1006/jmbi.1996.0109. [DOI] [PubMed] [Google Scholar]
- Mathews MAA, Tang HL, Blair DF. Domain analysis of the FliM protein of Escherichia coli. J Bacteriol. 1998;180:5580–5590. doi: 10.1128/jb.180.21.5580-5590.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65:876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 2004;18:715–727. doi: 10.1101/gad.289504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt JT, Tamayo R, Tischler AD, Camilli A. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J Biol Chem. 2007;282:12860–12870. doi: 10.1074/jbc.M611593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Gomelsky M, Galperin MY. C-di-GMP: the dawning of a novel bacterial signalling system. Mol Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Fouhy Y, Lucey JF, Dow JM. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J Bacteriol. 2006;188:8327–8334. doi: 10.1128/JB.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryjenkov DA, Simm R, Romling U, Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J Biol Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- Schirmer T, Jenal U. Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol. 2009;7:724–735. doi: 10.1038/nrmicro2203. [DOI] [PubMed] [Google Scholar]
- Schmidt AJ, Ryjenkov DA, Gomelsky M. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J Bacteriol. 2005;187:4774–4781. doi: 10.1128/JB.187.14.4774-4781.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Simm R, Lusch A, Kader A, Andersson M, Romling U. Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar Typhimurium. J Bacteriol. 2007;189:3613–3623. doi: 10.1128/JB.01719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano C, Garcia B, Latasa C, Toledo-Arana A, Zorraquino V, Valle J, Casals J, Pedroso E, Lasa I. Genetic reductionist approach for dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proc Natl Acad Sci USA. 2009;106:7997–8002. doi: 10.1073/pnas.0812573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- Tang H, Blair DF. Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J Bacteriol. 1995;177:3485–3495. doi: 10.1128/jb.177.12.3485-3495.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Billings S, Wang X, Sharp L, Blair DF. Regulated underexpression and overexpression of the FliN protein of Escherichia coli and evidence for an interaction between FliN and FliM in the flagellar motor. J Bacteriol. 1995;177:3496–3503. doi: 10.1128/jb.177.12.3496-3503.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Braun TF, Blair DF. Motility protein complexes in the bacterial flagellar motor. J Mol Biol. 1996;261:209–221. doi: 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]
- Thomas DR, Francis NR, Xu C, DeRosier DJ. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:7039–7048. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Suzuki A, Mariconda S, Porwollik S, Harshey RM. Sensing wetness: a new role for the bacterial flagellum. EMBO J. 2005;24:2034–2042. doi: 10.1038/sj.emboj.7600668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Mariconda S, Suzuki A, McClelland M, Harshey RM. Uncovering a large set of genes that affect surface motility in Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:7981–7984. doi: 10.1128/JB.00852-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M, Oosawa K, Aizawa S, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ, Visick KL. Get the message out: cyclic-Di-GMP regulates multiple levels of flagellum-based motility. J Bacteriol. 2008;190:463–475. doi: 10.1128/JB.01418-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakushi T, Yang J, Fukuoka H, Homma M, Blair DF. Roles of charged residues of rotor and stator in flagellar rotation: comparative study using H+-driven and N+-driven motors in Escherichia coli. 2006;188:1466–1472. doi: 10.1128/JB.188.4.1466-1472.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Aizawa SI, Kihara M, Isomura M, Jones CJ, Macnab RM. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986;168:1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young HS, Dang H, Lai Y, DeRosier DJ, Khan S. Variable symmetry in Salmonella typhimurium flagellar motors. Biophys J. 2003;84:571–577. doi: 10.1016/S0006-3495(03)74877-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Lloyd SA, Blair DF. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1998a;95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol. 1998b;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.