Abstract
We have developed a method for dissecting single neurons from the nematode Ascaris suum, in order to determine their peptide content by mass spectrometry (MS). In this paper, we use MALDI-TOF MS and tandem MS to enumerate and sequence the peptides present in the two neurons, ALA and RID, that comprise the dorsal ganglion. We compare the peptide content determined by MS with the results of immunocytochemistry and in situ hybridization of previously isolated peptides AF2, AF8, and six peptides encoded by the afp-1 transcript. We find complete agreement among the three techniques, which validates single neuron MS as a method for peptide localization. We also discovered and sequenced six novel peptides in the ALA neuron. Cloning of cDNAs and database searching of Genomic Survey Sequences showed that transcript afp-12 encodes peptide AF36 (VPSAADMMIRFamide) and afp-13 encodes AF19 (AEGLSSPLIRFamide), AF34 (DSKLMDPLIRFamide), AF35 (DPQQRIVTDETVLRFamide), and three nonamidated peptides (PepTT, PepTL, and PepGE). We have found no similarities with reported peptide expression in the nematode Caenorhabditis elegans. This method promises to be ideally suited for determining the peptide content of each of the 298 neurons in the nervous system of this nematode.
Keywords: Neuropeptide, MALDI-TOF, nematode, Ascaris suum, single neuron, de novo sequencing
Peptides have long been known to be important intercellular signaling molecules in a wide variety of species. Their role as hormones was established many years ago, yet new peptide hormones continue to be discovered (e.g., kisspeptin (1)). In the nervous system, peptides can act as primary neurotransmitters (2). More commonly, they act as neuromodulators, affecting multiple aspects of neuronal activity. Their effects include modulation of the ion channels involved in action potential propagation and synaptic transmission and modulation of the molecular machinery of transmitter release (3). In most well-studied systems, the number of peptide signaling molecules is impressively large, and as with peptide hormones, new neuropeptides are still being discovered. The details of their activity and sites of action are intricate and particular, so a full description of the role of peptides in a neuronal circuit can be complex, but it must be complete if we are to understand how neuronal circuits work.
The nervous system of the parasitic nematode Ascaris suum contains only 298 neurons (4). This numerical simplicity makes it an attractive system for the study of the role of peptides in neuronal function. We have already shown in A. suum that neuropeptides are numerous, widespread, and varied and that they have potent effects on muscle and subsets of neurons (5−8). Most of the peptides we have identified in A. suum are FMRFamide-like peptides (FLPs), and are named AF peptides (Ascaris FMRFamide-like peptides). The transcripts and the genes that encode the AF peptides are named afp’s (Ascaris FMRFamide-like precursor proteins).
In the related, free-living nematode Caenorhabditis elegans, three large families of peptides have been predicted by genome searches or sequenced directly (9−12): FLPs encoded by 34 flp genes, NLPs (neuropeptide-like proteins) encoded by 42 nlp genes, and insulin-like peptides encoded by 39 ins genes (10). In all cases, the sequences suggest that in nematodes the formation of neuropeptides is similar to that in other organisms: the gene encodes a precursor protein that is directed by an N-terminal signal peptide to the secretory pathway, where it is cleaved by specific protease processing enzymes and in many cases modified by other enzymes, for example, those responsible for C-terminal amidation (13,14). Since there are several processing proteases with differing specificity and with unknown cellular localization, it is difficult to predict the structure of processed peptides from precursor protein sequences. We therefore emphasize direct sequencing of the peptides themselves. In this paper, we describe a method that allows us to localize peptides, including several newly discovered ones, to single identified neurons in A. suum.
The rate at which we are discovering new peptides is accelerating, implying that there are many more yet to be discovered. For each peptide, it is essential to determine the molecular structure in order to synthesize the amount needed to investigate its biological activity. In addition, we need to determine its cellular expression pattern. In other systems, single cell dissection has been combined with mass spectrometry to allow simultaneous peptide characterization and localization (15−21). In A. suum, the neurons are tightly enwrapped with non-neural tissue (hypodermis), and this impeded our previous attempts at single cell isolation. In this paper, we show that in A. suum it is possible to dissect single neurons. We subjected each neuron to matrix assisted laser desorption/ionization−time of flight mass spectrometry (MALDI−TOF MS) for peptide detection and tandem MS (MS/MS) for sequence determination. MALDI-TOF MS is a sensitive technique that was previously used on dissected A. suum ganglia (7,22,23). It detected all previously identified AF peptides, as well as many unknown peptides; six new peptide sequences were reported. Although this method provides a great improvement in peptide discovery and localization, we are concerned about its adequacy for determining the complete peptide inventory of each neuron. In particular, some peptides that are present in a cell might not produce a measurable peak in an MS spectrum due to ion suppression or low probability of ionization (24).
As proof-of-concept, we chose to study the simplest ganglion in this organism, the dorsal ganglion (DG), which includes only two neurons, ALA and RID (25). By MS, we detected different subsets of peptides in each neuron. We validated the method by demonstrating that the peptide content revealed in each cell by MS corroborates the peptide expression previously established by other techniques, namely, immunocytochemistry (ICC) using specific antipeptide antibodies and in situ hybridization (ISH) with riboprobes against transcripts that encode known peptides (26). In addition, we sequenced six novel peptides from these neurons. This technique has been successfully extended to other ganglia, containing many more neurons: this will be reported elsewhere.
A comparison of cell-specific expression of neuropeptides in A. suum to that in the well-studied nematode Caenorhabditis elegans is particularly interesting, given that comparison of the morphology of the nervous systems shows that C. elegans is a faithful miniature version of A. suum. These nematodes are estimated to have diverged about 550 million years ago (27), yet their neuroanatomy is so similar that the same neurons are recognizable and are often given the same name in the two organisms. However, when we compare our results in A. suum with those reported in C. elegans(28), the cellular expression of peptides is noticeably different in the two organisms.
Results and Discussion
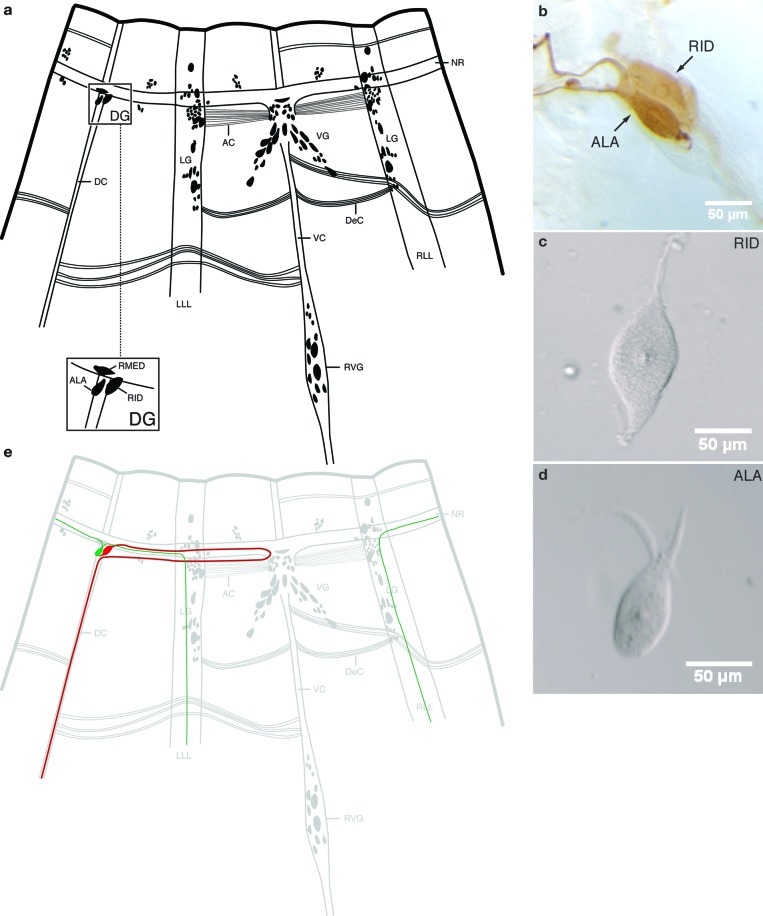
A thorough knowledge of the chemical identity and cellular localization of neuropeptides is essential in understanding the neuronal control of behavior. We report here a cell-by-cell MALDI-MS peptidomic map of the smallest ganglion in A. suum. The two neurons of the DG, ALA and RID, were manually dissected with tungsten needles from collagenase-treated worms under a dissecting microscope. The morphology of these neurons is distinctive (Figure 1) and allows each of them to be individually identified in most preparations (26), both before and after dissection. ALA has two processes that branch from the anterior portion of the cell body, project into the left or right side of the nerve ring, and then send a process down each lateral line to the tail. RID, by contrast, is a bipolar neuron. Its anterior process travels in the nerve ring to the ventral midline where, just posterior to the ring, it gives rise to a process that runs back around the body to the dorsal cord (in nematodes this is termed a dorsoventral commissure); this branch of RID then projects posteriorly along the dorsal nerve cord. The posterior process from its cell body extends along the dorsal cord for a very short distance (Figure 1e).
Figure 1.
The neurons of the dorsal ganglion (DG). (a) Diagram of the head ganglia of A. suum, modified from Goldschmidt (63). The worm has been split near the dorsal axis and opened flat. Neuronal cell bodies and commissural processes are shown. The nerve ring (NR), ventral ganglion (VG), dorsal ganglion (DG), lateral ganglia (LG), and retrovesicular ganglion (RVG) are indicated. LLL, left lateral line; RLL, right lateral line; VC, ventral nerve cord; DC, dorsal nerve cord; DeC, deirid commissures; AC, amphidial commissures. The inset shows higher magnification of the identified neurons ALA and RID in the DG. Neuron RMED lies within the NR, not the DG. (b) Immunological staining (ICC) of DG in a whole mount of the head region using anti-FMRFa antibody (25), which stains both cells of the DG. (c) Dissected unstained individual RID. (d) Dissected unstained individual ALA. (e) Diagram showing the cell bodies and neural processes of ALA (green) and RID (red).
Peptide Content of ALA and RID Neurons
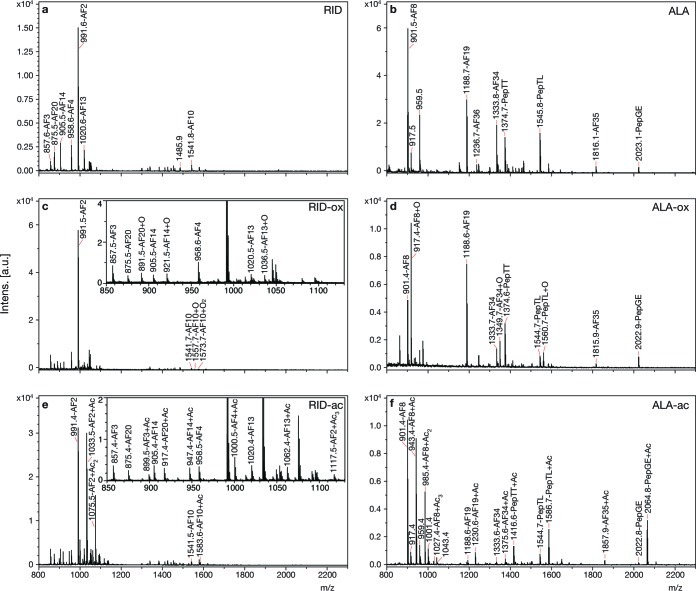
Previous results from ISH showed that ALA expresses afp-3 (encoding AF8) and RID expresses both afp-1 (encoding AF3, AF4, AF10, AF13, AF14, and AF20) and afp-4 (encoding AF2) (26). In this paper, we report the use of MS both to confirm the cellular expression of these peptides and to discover novel peptides expressed in these cells. Nine ALA neurons and 11 RID neurons were examined. ALA and RID exhibited distinct peptide profiles that included peaks with the mass-to-charge ratio (m/z) of the peptides encoded by afp-1, afp-3, and afp-4, as well as several other peptides (Figure 2a,b, Table 1; see also Supplementary Table S1, Supporting Information). The spectra were simpler and more reproducible than those from dissected ganglia (23). The most intense peaks were always present, but minor peaks sometimes varied from sample to sample (Supplementary Table S1a, Supporting Information). AF2 (m/z 991.5) in RID and AF8 (m/z 901.5) in ALA were always the most intense peaks present. The six afp-1-encoded peptides were relatively minor peaks in the RID profile (m/z 857.5, 875.5, 905.5, 958.5, 1020.5, and 1541.7). In one preparation, all of the afp-1 peptides were absent and in two more preparations AF10 (m/z 1541.7) was not detected. Preliminary confirmation that the peaks from RID and ALA were indeed the peptides encoded by afp-1, afp-3, and afp-4 was obtained using two types of chemical modification. First, on-target acetylation of free amines with acetic anhydride adds 42 Da for the N-terminus and 42 Da for each lysine; incomplete acetylation of tyrosines also has been detected. Second, oxidation of methionines with methylene blue adds 16 Da for each methionine. Although methylene blue was initially used to aid visualization of neurons for dissection, rather than for oxidation, its ability to oxidize is known (29). Oxidation of methionine by methylene blue is not as robust as oxidation by H2O223 and allows some unoxidized peptide to remain for direct comparison within the same spectrum. Modification by acetylation or oxidation is variable, probably because uniform exposure of the modifying agents to the peptides is difficult to obtain in these very small biological samples. However, evidence for the expected modifications was seen in almost all cases (Figure 2c−f). Peaks corresponding to the afp-1, afp-3, and afp-4 peptides were sequenced by MS/MS, which confirmed the identity of these peptides (Figure 3; Supplementary Figure S2, Supporting Information). Since MS/MS does not distinguish between leucine and isoleucine, gene sequences were used to resolve the leucine/isoleucine ambiguities.
Figure 2.
Neuropeptides from single DG neurons in native and chemically modified forms. (a) Mass spectrum from an individual RID with no chemical modifications. (b) Spectrum from an individual ALA with no chemical modifications. (c) Spectrum from an individual RID stained with methylene blue, which oxidizes methionines and adds 16 mass units. The inset is an expansion of the small peaks between 850 and 1130 m/z. (d) Spectrum from an individual ALA stained with methylene blue. (e) Spectrum from an individual RID treated with acetic anhydride, which acetylates free amines (N-terminus, lysine) and to a lesser extent tyrosine and adds 42 mass units. The inset is an expansion of the small peaks between 850 and 1130 m/z. (f) Spectrum from an individual ALA treated with acetic anhydride. X axis, m/z, is mass-to-charge ratio. Y axis is intensity of MS signal in arbitrary units, a.u.
Table 1. Comparison of Peptides Localized within Cells of the DG by MALDI-MS, ISH, and ICC.
| neuron | gene | peptide name | peptide sequence | obsd [M + H] | ISHa | ICC |
|---|---|---|---|---|---|---|
| RID | afp-1 | AF3 | AVPGVLRFa | 857.5 | + | +b |
| AF4 | GDVPGVLRFa | 958.5 | ||||
| AF10 | GFGDEMSMPGVLRFa | 1541.7 | ||||
| AF13 | SDMPGVLRFa | 1020.5 | ||||
| AF14 | SMPGVLRFa | 905.5 | ||||
| AF20 | GMPGVLRFa | 875.5 | ||||
| afp-4 | AF2 | KHEYLRFa | 991.5 | + | NDd | |
| ALA | afp-3 | AF8 | KSAYMRFa | 901.5 | + | NDd |
| afp-12 | AF36 | VPSAADMMIRFa | 1236.6 | NDd | + | |
| afp-13 | AF19 | AEGLSSPLIRFa | 1188.7 | NDd | +c | |
| AF34 | DSKLMDPLIRFa | 1333.7 | ||||
| AF35 | DPQQRIVTDETVLRFa | 1816.0 | ||||
| PepTT | TPPEEDLLGRFT | 1374.7 | ||||
| PepTL | TNIMGENRLNRNL | 1544.8 | ||||
| PepGE | GNSYRSFFNPNEYTVVE | 2022.9 |
ISH results are from previous work (26).
ICC of the afp-1 peptides was performed with an anti-PGVLRFa antibody (Sithigorngul and Stretton, unpublished).
ICC of the afp-13 peptides was represented by staining with an anti-AF19 antibody (see Figure 7b). Newly sequenced peptides are in bold.
ND = Not done.
Figure 3.
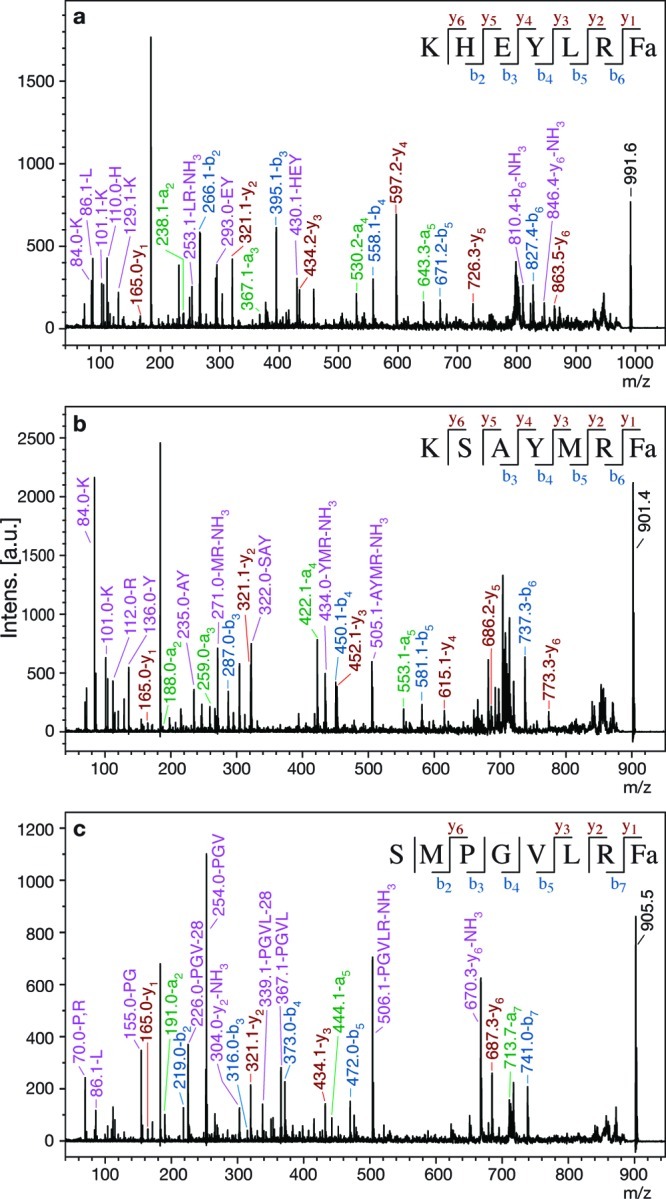
MS/MS spectra of peptides with known expression in DG. Peaks representing a (green), b (blue), y (red), and high-intensity internal fragment (purple) ions are labeled, and b and y ions are summarized in the sequence at the top of each spectrum. (a) MS/MS spectrum from RID of AF2 (afp-4), m/z 991.6. (b) MS/MS spectrum from ALA of AF8 (afp-3), m/z 901.4. (c) MS/MS spectrum from RID of AF14 (afp-1), m/z 905.5.
The relatively low intensity of the peaks for the peptides derived from the AFP-1 precursor protein is probably an effect of ion suppression (24) of the six peptides expressed on this precursor. Other methods suggest that the expression of afp-1 in RID is robust. ICC with an antibody that recognizes the C-terminal −PGVLRFamide in these peptides stains RID strongly (Sithigorngul and Stretton; unpublished). A riboprobe based on a sequence from afp-1 produces a strong ISH signal in RID (26). The interpretation of other minor peaks in the spectra is uncertain. Many of these smaller peaks vary in intensity from spectrum to spectrum and may be absent in individual preparations. Among the low intensity and variable peaks in ALA were members of a family of peptides encoded by afp-11, including AF11 (data unpublished). An antibody raised against AF11 recognized ALA in some preparations (Viola, I., personal communication). Variability in levels of peptide expression among individual A. suum would not be surprising, since they are not isogenic and their individual history, including their interactions with their swine host and its drug treatments, is unknown. There are also potential sources of variability from sample preparation, for example, effects of collagenase digestion and dissection. Differences in the interactions between matrix and the sample on the MS target plate could also contribute to variability, but we attempt to minimize such effects by using small drops of matrix and aiming the laser beam directly at or very near the cell body on the target.
Inter-preparation variability was considered previously when assessing the cellular localization of peptides or peptide-encoding transcripts by ICC and ISH (26). Preparation-to-preparation variability was seen with both of these techniques. Significantly, as with the MS data, there was also a consensus set of strong signals, and variability was seen only with low-level signals. None of these three techniques is quantitative. It is therefore prudent to use at least one of the ancillary techniques alongside MS to determine the peptide content of an individual neuron. The use of MS is primary because it yields sequence information on which the generation of antibodies or riboprobes is based.
All peptides found in MS of single DG cells were also found in previous experiments using MS on whole dorsal ganglia from A. suum(23). We find that the sum of the peptide profiles of ALA and RID is less complex than the profile obtained from the whole DG, and we suspect that this is due to contamination in the whole ganglia from neighboring neural tissue, for example fragments of the nerve ring or dorsal nerve cord.
Studying whole ganglia allows the rapid localization of many peptides to a particular ganglion, but it is only single cell MS that can determine the specific cellular localization of a peptide. This is a significant technological advance toward our aim of mapping the peptide content of each cell in the A. suum nervous system.
De Novo Sequencing of Novel Peptides
afp-12 Encoded Peptides
The MS spectra from ALA contain a minor peak of m/z 1236.6 (Figure 2b; Supplementary Table S1, Supporting Information). MS/MS analysis of this peak yielded the partial sequence DMM(L/I)RFa (leucine and isoleucine are isobaric and therefore cannot be distinguished by MS/MS (Figure 4a). A direct search of an A. suum Genome Survey Sequence (GSS) database (Mitreva, M., personal communication) was performed with both DMMLRFG and DMMIRFG, including a C-terminal G as the precursor to a C-terminal amide. The search yielded a translated amino acid sequence, flanked with basic amino acid cleavage sites and containing a C-terminal G, which when fully processed would produce the peptide VPSAADMMIRFa (AF36) with the molecular mass of 1236.6 Da (Figure 4b); this also resolved the L/I ambiguity. Upon further inspection of the MS/MS spectrum, additional b and y ions that were not used to create the original partial sequence were identified and provided further confirmation of the peptide sequence. In addition, synthetic AF36 was subjected to MS/MS and produced a similar fragmentation pattern to the MS/MS of native AF36 (Supplementary Figure S4a, Supporting Information).
Figure 4.
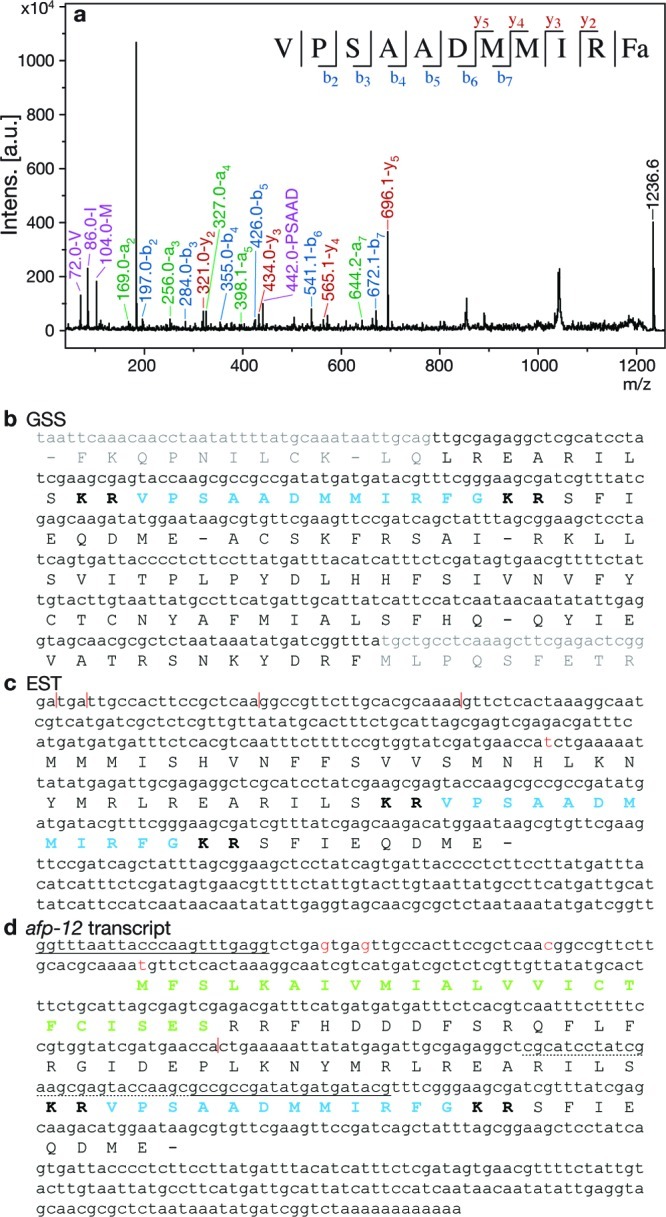
AF36 and afp-12. (a) MS/MS spectrum from ALA of AF36, m/z 1236.6. Peaks representing a (green), b (blue), y (red), and high-intensity internal fragment (purple) ions are labeled, and b and y ions are summarized in the sequence at the top of the spectrum. (b) Sequence from GSS database encoding AF36. (c) Sequence from EST library encoding AF36 (CB040272 (30)). (d) Cloned afp-12 transcript deduced by unique primers and 3′ RACE (HM125966). Gray indicates suspected intronic region. Blue indicates encoded peptide. Bold indicates basic cleavage sites. Green indicates signal peptide. Sequences used to design primers for PCR have a solid underline; the 5′ primer was SL1. Nucleotides with a dotted underline were used for the primer for 3′ RACE. In panels c and d red bases and vertical lines indicate insertions/deletions between the two sequences.
A search of the A. suum EST library produced the same peptide sequence within a predicted precursor protein that does not include a signal peptide (Figure 4c, CB040272 (30)). Comparison of the EST sequence with the full transcript (afp-12), which we obtained by PCR and 3′ RACE (Figure 4d, HM125966), showed one base insertion and four single base deletions, so we ascribe the lack of a signal peptide in the sequence predicted from the EST to phase shifts introduced by sequencing errors. The transcript that we obtained encoded a predicted precursor protein that includes an initiating methionine and a signal peptide. The 5′ end of this transcript includes SL1, a trans-spliced leader sequence found in 80% of A. suum transcripts (31). The one copy of AF36 appears to be the only peptide encoded by afp-12.
The localization of AF36 in ALA was confirmed by ICC with an affinity-purified polyclonal antibody against AF36 (Figure 5a). Staining was blocked by preincubating the antibody with 40 μg/mL AF36 (data not shown).
Figure 5.
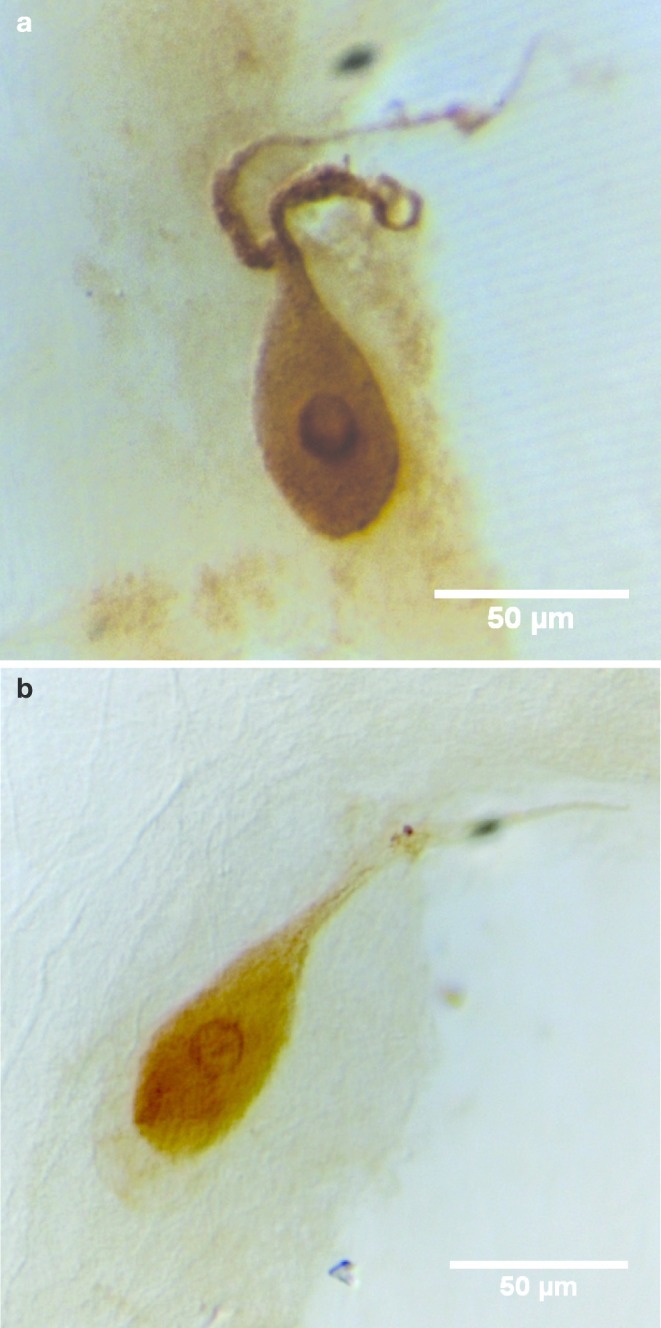
Confirmation by ICC of peptides localized in ALA. (a) ICC with anti-AF36 antibody. (b) ICC with anti-AF19 antibody.
afp-13 Encoded Peptides
Also present in the MS spectra from ALA were peaks with m/z 1333.7 and 1374.5 (Figure 2b; Supplementary Table S1, Supporting Information). MS/MS analysis yielded the partial sequences DP(L/I)(L/I)RFa and TPPEED(L/I)(L/I), respectively (Figure 6a,b). These sequences were used to direct searches of the A. suum GSS database, using all possible combinations of L and I, and extending the first sequence to DP(L/I)(L/I)RFG. The first search identified a sequence DPLIRFG (GSS 2, Figure 7b) that was part of a translated amino acid sequence flanked by two dibasic amino acid sequences, which would predict the processed peptide DSKLMDPLIRFa (AF34), which has a molecular mass of 1333.7. A second search yielded GSS 3, which included the predicted processed peptide TPPEEDLLGRFT (PepTT), which was flanked with basic amino acid cleavage sites (Figure 7c). The mass of this peptide is 1374.5. Further confirmation was obtained from MS/MS of synthetic AF34 and PepTT. Spectra were similar to those from the native peptide (Supplementary Figure S4b,c, Supporting Information). In particular, the lability of bonds C-terminal to aspartate residues was diagnostic.
Figure 6.
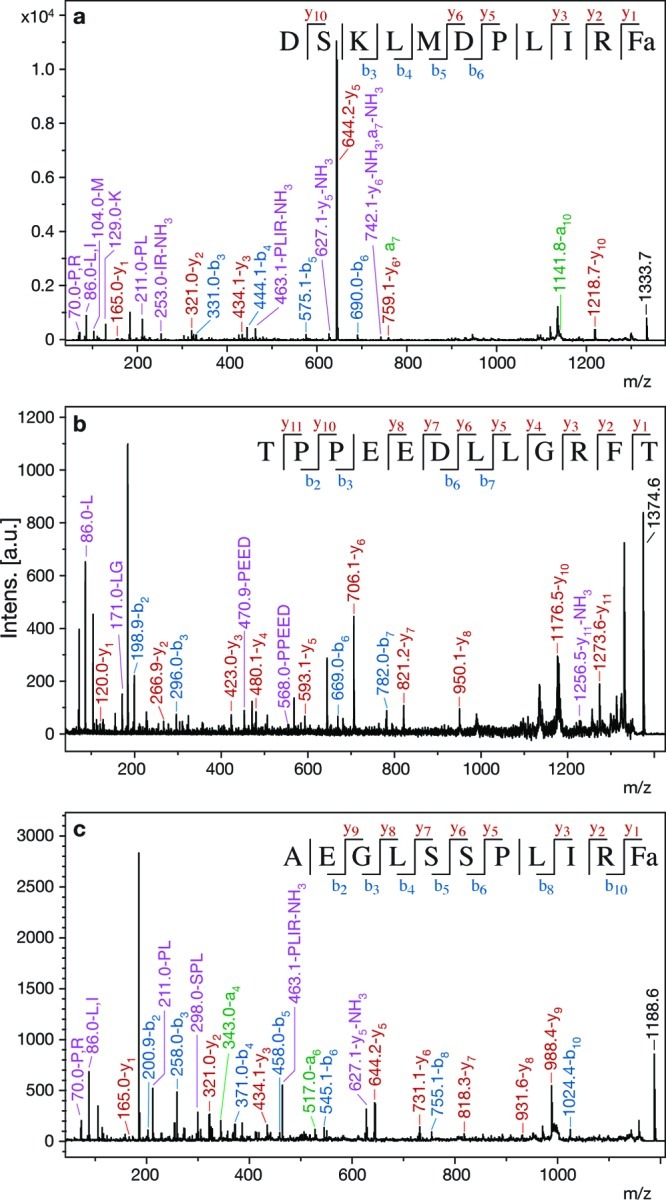
MS/MS of peptides encoded by afp-13. Peaks representing a (green), b (blue), y (red), and high-intensity internal fragment (purple) ions are labeled, and b and y ions are summarized in the sequence at the top of each spectrum. (a) MS/MS spectrum from ALA of AF34, m/z 1333.7. (b) MS/MS spectrum from ALA of PepTT, m/z 1374.6. (c) MS/MS spectrum from ALA of AF19, m/z 1188.6.
Figure 7.
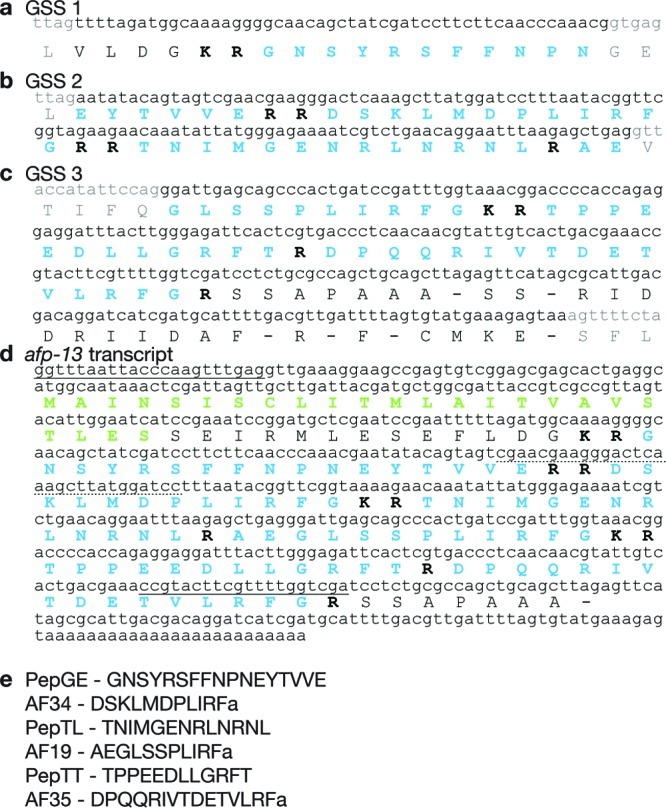
Nucleotide sequence and deduced amino acid sequence of afp-13: (a) Sequence from GSS database (GSS 1) encoding a portion of PepGE. (b) GSS 2 encoding AF34, PepTL, a small portion of AF19, and the remaining portion of PepGE. (c) GSS 3 encoding PepTT, AF35, and the remaining portion of AF19. (d) Cloned afp-13 transcript deduced by unique primers and 3′ RACE (HM125967). (e) Processed peptides from afp-13. Gray indicates predicted intronic region. Blue indicates encoded peptide. Bold indicates basic cleavage sites. Green indicates signal peptide. Sequences used to design primers for PCR have a solid underline; the 5′ primer was SL1. Nucleotides with a dotted underline were used for the primer for 3′ RACE.
Interestingly, in the translated amino acid sequence of GSS 3, N-terminal to PepTT, was a predicted amidated peptide GLSSPLIRFa, which is the C-terminal sequence of AF19 (AEGLSSPLIRFa, 1188.7 Da), a previously known A. suum peptide with an unknown transcript (6). A peak with m/z 1188.7 is present in ALA (Figure 2b; Supplementary Table S1, Supporting Information), and MS/MS confirms that it is AF19 (Figure 6c). GSS 2 and 3 have predicted splice sites such that the 3′ region of GSS 2 would provide the N-terminal AE of peptide AF19. GSS 3 also encodes a predicted amidated peptide DPQQRIVTDETVLRFa (AF35, mass 1815.9). GSS 2 also includes predicted peptide TNIMGENRLNRNL (PepTL, mass 1544.4). Peaks with these masses were also present in ALA neurons (Figure 2b; Supplementary Table S1, Supporting Information) and were confirmed by MS/MS (Supplementary Figure S3a,b, Supporting Information). Comparison of MS/MS of synthetic AF35 and PepTL with MS/MS of native peptides also confirmed the sequences (Supplementary Figure S4c,d, Supporting Information).
The predicted splicing of GSS 2 and 3 was investigated by PCR using SL1 and a unique second sequence as primers. The amplified sequence (Figure 7d, HM125967) showed that the predicted transcript segments were indeed spliced together. In addition, the 5′ end of the mRNA predicts another splicing event, to GSS 1 (Figure 7a), yielding the predicted peptide GNSYRSFFNPNEYTVVE (PepGE, mass 2022.9). A peak with the m/z of PepGE is present in ALA (Figure 2b; Supplementary Table S1, Supporting Information), and we have partial sequence confirmation by MS/MS of the acetylated peptide (Supplementary Figure S3c, Supporting Information). The 3′ region of the transcript was found by 3′ RACE, and the full length transcript (afp-13) is shown in Figure 7d.
Affinity-purified polyclonal antibodies against AF19 confirmed peptide localization in ALA (Figure 5b). Immunolocalization of AF19 in ALA was only seen with fixation with glutaraldehyde and not with formaldehyde. This reinforces the need to use multiple techniques to determine peptide localization. Preincubating the antibody with 40 μg/mL AF19 blocked staining of ALA (data not shown).
Peptide Discovery
The majority of previously characterized A. suum peptides were discovered by large-scale chemical separation followed by Edman degradation (32−34). The original process required accumulation of 60 000 worms over the course of 2 years, followed by seven steps of high-performance liquid chromatography (HPLC) to obtain the chemically pure sample required for Edman degradation. In most cases, subsequent comparison of the isolated sequences with predicted cleaved and processed sequences from translated cDNAs gave perfect matches. However, in addition to being laborious and time-consuming, this method does not yield any information about cellular localization and exposes the peptides to possible modifications or degradation. This was specifically a problem with the characterization of AF17, which was later found by cloning and MS to contain an N-terminal aspartic acid that was removed during acid/methanol extraction (35; Yew, J., personal communication).
In C. elegans, the large majority of peptides were discovered by prediction from the genome (9,10). This method (currently limited in A. suum) does not necessarily predict the mature form of the peptide, since the specificity and cellular localization of the processing enzymes are not known accurately. For example, cleavage of the precursor protein at conventional di- or monobasic amino acid sites or at unconventional cleavage sites is not completely predictable. More recently, many of the predicted peptides in C. elegans have been confirmed by liquid chromatography/MS (11,12), and indeed this technique has also shown that some of the original predictions were incorrect, for example, some of the products of the flp-1 and flp-4 genes.
By direct MALDI-TOF MS of whole dorsal, ventral, lateral, or retrovesicular ganglia, or the nerve ring, this laboratory sequenced six novel peptides (7,22), the same number of novel peptides we report here from just two cells. The whole-ganglia MS experiments were performed before the availability of the GSSs, which could have enabled the discovery of many more peptides by whole ganglion MS. Although powerful, whole ganglion MS may also be limited by the complexity of the spectra from so many cells, increasing the probability of problems from ion suppression (24). Higher complexity also increases the probability that two different peptides with identical masses will be present. In simpler spectra, the probability of peaks including two peptides of identical or very similar m/z is much reduced. The m/z window for peptides taken for MS/MS is several daltons wide, so complex MS spectra increase the likelihood of sampling multiple peptides. This is important because interpretation of MS/MS of multiple peptides is extremely difficult.
Peptide Localization
Previously, peptide localization in A. suum was ascertained by ICC, ISH, and whole ganglia MS. Each technique, although valuable, has limitations. Whole ganglia MS, although it yielded a great deal of information, was less reproducible; in addition there was possible contamination from the nerve ring or nerve cords. ISH detects the peptide-encoding transcript and not the peptide itself, yielding no direct information about which mature peptides are present. The strength of ICC is that this method detects the peptide product and therefore stains the entire cell including its processes. This allows for more robust cellular identification, since the morphology of the cell processes is an important feature for neuronal identification. In contrast, ISH stains only the cell body, which can make cell identification more difficult (26). A limitation of ICC is that it requires the production of a highly specific antibody to a short amino acid sequence that often has sequence overlap with other neuropeptides. Although all of our antibodies are rigorously tested for specificity, it is difficult to exclude cross-reactivity with other endogenous peptides, the majority of which are presently unknown. On the other hand, in ISH there is little doubt about the specificity of the probe. Another limitation of ICC is that variations in fixation techniques can greatly affect staining patterns, as occurred with the ICC results for AF19. Both ICC and ISH require prior knowledge of sequences, either of the peptide itself for raising antibodies or of transcript sequences for making riboprobes. This problem is overcome by the use of MS, where no prior sequence knowledge is needed.
Is the Peptide Map Accurate and Complete?
It is important to question whether our list of the peptide content of individual neurons is complete. MALDI-TOF MS is not a quantitative technique, and the ability to detect a peptide depends both on the complexity of the mixture, where ion suppression may be a problem, and on the chemical nature of each peptide, which affects the probability that it will accept charge during the ionization process (24). The simplification of the spectra of single cells, in comparison with dissected ganglia, is striking, and this greatly facilitates the scoring of lower intensity peaks. On the other hand, it is noticeable that all the peptides we detected in both whole ganglia and single-cell MS include at least one basic amino acid (arginine, lysine, or histidine); most of these peptides are FLPs (FMRFamide-like peptides), with C-terminal −RFa, although other peptides, cleaved from precusor proteins encoding FLPs, have been detected. Are there other families of peptides present? In C. elegans, many NLPs have been predicted that do not contain basic amino acids (10). Some NLPs have been predicted in A. suum(36). In future studies of single neurons in A. suum, it will be interesting to see whether any NLPs, especially those with no basic amino acids, can be detected.
Alternatives to Single-Cell Dissection
One limitation of single-cell MALDI-MS is that the throughput is low due to the high level of manual skill required for dissection. There has been interest in imaging mass spectrometry (IMS) and laser capture microdissection (LCM) coupled with MALDI-MS (37−39), as alternatives. IMS creates an image of peptide or protein expression across a tissue section. Although this is a powerful and relatively high-throughput technique, resolution and analyte migration are problematic (40). In addition, even with cell-sized resolution, it is difficult to identify the specific cell that is being investigated. By contrast, LCM utilizes a laser to precisely dissect tissues. Attempts to use LCM on A. suum neurons were not successful (Yew, J., personal communication).
Comparison to Other Nematodes
The well-studied nematode C. elegans is anatomically remarkably similar to A. suum. Although dramatically different in size, the underlying structure of the nervous system is so similar that the same cell names can be used in both organisms (41−43). Expression of small molecular weight neurotransmitters such as GABA and ACh is also highly similar (44−48), although there is a clear example of a difference in expression in 2 out of 26 GABA-expressing neurons (48,49). Cellular expression of peptides, however, appears to be quite different (Table 2). This is especially remarkable since the structure of the peptides is very similar, and in many cases identical, in both species. Thus far, the only method widely employed for peptide localization in C. elegans has been the use of GFP constructs (28,50), a method currently unavailable in A. suum. It is not yet clear whether the upstream 5′ sequences used in the GFP constructs include a complete set of the sites that control peptide-gene expression in these cells (51). Thus, comparisons between A. suum and C. elegans must be made with caution. Nevertheless, based on available data, peptide expression in the same identified neurons in these two organisms is completely different. There are three pairs of genes we can directly compare: flp-6/afp-3, flp-13/afp-13, and flp-18/afp-1 (flp-24, corresponding to afp-12, has not been tested by GFP constructs). Of these three, which are expressed in the A. suum DG, none showed expression in the DG of C. elegans. Surprisingly, one of the constructs important for our comparison, flp-14 (AF2), failed to express in any cells even though the peptide has been isolated from C. elegans(52,53). Of the two flp genes expressed in the DG of C. elegans, one (flp-7) has no known analog in A. suum. For the other (flp-2), analogous peptide products (AF37, AF38) have been sequenced from a cell in another ganglion, although the mRNA has not yet been cloned from A. suum (unpublished observation). Although a peak with the m/z of AF37 is present in RID, subsequent MS/MS of the peptide showed it to be AF13, an isobaric peptide (Supplementary Figure S2d, Supporting Information). Cellular localization of nlp gene expression in C. elegans revealed no expression of any known NLPs in ALA or RID (50).
Table 2. Comparison of Analogous Peptides Localized within the DG in A. suum by MALDI-TOF MS and C. elegans by GFP Constructs (28).
|
A. suum |
C. elegans |
||||||
|---|---|---|---|---|---|---|---|
| gene | peptide name | peptide sequence | DGa | gene | peptide name | peptide sequence | DGa |
| afp-1 | AF3 | AVPGVLRFa | RID | flp-18 | FLP-18-1 | (DFD)GAMPGVLRFa | NONEb |
| AF4 | GDVPGVLRFa | FLP-18-2 | EMPGVLRFa | ||||
| AF10 | GFGDEMSMPGVLRFa | FLP-18-3 | (SYFDEKK)SVPGVLRFa | ||||
| AF13 | SDMPGVLRFa | FLP-18-4 | EIPGVLRFa | ||||
| AF14 | SMPGVLRFa | FLP-18-5 | SEVPGVLRFa | ||||
| AF20 | GMPGVLRFa | FLP-18-6 | DVPGVLRFa | ||||
| afp-4 | AF2 | KHEYLRFa | RID | flp-14 | FLP-14 | KHEYLRFa | NAc |
| afp-3 | AF8 | KSAYMRFa | ALA | flp-6 | FLP-6-1 | KSAYMRFa | NONEb |
| afp-12 | AF36 | VPSAADMMIRFa | ALA | flp-24 | FLP-24 | VPSAGDMMVRFa | NAc |
| afp-13 | AF19 | AEGLSSPLIRFa | ALA | flp-13 | FLP-13-1 | AMDSPLIRFa | NONEb |
| AF34 | DSKLMDPLIRFa | FLP-13-2 | AADGAPLIRFa | ||||
| AF35 | DPQQRIVTDETVLRFa | FLP-13-3 | APEASPFIRFa | ||||
| PepTT | TPPEEDLLGRFT | FLP-13-4 | ASPSAPLIRFa | ||||
| PepTL | TNIMGENRLNRNL | FLP-13-5 | SPSAVPLIRFa | ||||
| PepGE | GNSYRSFFNPNEYTWE | FLP-13-6 | ASSAPLIRFa | ||||
| FLP-13-7 | SAAAPLIRFa | ||||||
| AF37 | FRGEPIRFa | NONEb | flp-2 | FLP-2-1 | SPREPIRFa | RID | |
| AF38 | AQREPIRFa | FLP-2-2 | LRGEPIRFa | ||||
| NAc | flp-7 | FLP-7-1 | SPMQRSSMVRFa | ALA | |||
| FLP-7-2 | TPMQRSSMVRFa | ||||||
| FLP-7-3 | SPMERSAMVRFa | ||||||
| FLP-7-4 | SPMDRSKMVRFa | ||||||
Dorsal ganglion.
NONE = No evidence of peptide.
NA = Not applicable.
The peptides present in the A. suum DG are highly conserved throughout the nematode phylum. Searches of the nematode EST libraries find 20 species of nematodes expressing flp-14/afp-4-related transcripts, 16 expressing flp-6/afp-3-related transcripts, 16 expressing flp-13/afp-13-related transcripts, 15 expressing flp-18/afp-1-related transcripts, and 7 expressing flp-24/afp-12-related transcripts (30). Although the peptides encoded by these transcripts are probably present in all these nematodes, there is only fragmentary supportive evidence from direct studies of the peptides themselves (54). The study of peptide cellular localization in other nematodes is fairly limited. ISH has been performed in the nematode Globodera pallida to localize the G. pallida transcripts of AF2, AF8, and the afp-1-related peptides (55). The reported localization did not match the expression we report in A. suum, although cellular identification is difficult in G. pallida. These peptide expression differences give further weight to the idea that, despite great morphological similarities between species, one factor that determines species-specific behavior is the different patterns of cellular expression of identical or closely related neuropeptides.
Conclusions
We have reported here a significant technological improvement for peptide localization and discovery of neuropeptides in nematodes. Direct MALDI-TOF MS of single dissected identified neurons of the DG confirmed known expression and identified six novel peptides. Comparison with reported C. elegans expression found no similarities. Overall, this is an important first step toward the use of this method to describe the peptide expression in the entire A. suum nervous system.
Methods
Animals
A. suum were harvested from the small intestines of pigs at a regional slaughterhouse. The worms were maintained for up to 3 days at 37 °C in phosphate-buffered saline (PBS; 8.5 mM sodium phosphate, 150 mM sodium chloride, pH 7.4).
Sample Preparation for Mass Spectrometry
Adult female A. suum were injected with 2 mg/mL collagenase (Sigma, St. Louis, MO) in Ascaris saline (4 mM sodium chloride, 125 mM sodium acetate, 24.5 mM potassium chloride, 5.9 mM calcium chloride, 4.9 mM magnesium chloride, and 5 mM 3-(N-morpholino)propanesulfonic acid (MOPS) buffer, pH 6.8). After 1.5 h at 37 °C, the head was removed and rinsed with water to remove dissociated muscle cells. The head region was then cut longitudinally between the lateral line and the dorsal cord and pinned flat on a Sylgard-lined Petri dish filled with 170 mM ammonium acetate (56). Electrolytically sharpened tungsten needles were used to manually dissociate the desired cell from the hypodermis and the other neuron in the dorsal ganglion. In some experiments, cells were stained with methylene blue to aid cell identification: prior to dissection, the preparation was exposed to 16 mM methylene blue in EtOH. After 1−2 min, the methylene blue dye was removed by rinsing 2−3 times with 170 mM ammonium acetate. Dissection then proceeded as described above.
Cells were transferred with tungsten needles to a Bruker MTP ground stainless steel target plate. Single cells were rinsed on-target with 0.5 μL of isopropanol (57) and allowed to dry. The matrix solution was prepared by vortexing solid α-cyano-4-hydroxycinnamic acid (CHCA; Sigma, St. Louis, MO) with 5% acetic acid for a minute. After removal of the acetic acid, 50 μL of water and 50 μL of acetonitrile were added to the solid, and the mixture was vortexed for 90 s yielding a saturated solution of CHCA with few Na+ ions present. Using a Nanoliter Cool Wave Syringe II (58), matrix was applied robotically to the sample 3 times in 60−100 nL amounts and allowed to dry after each application.
MS Acquisition
A Bruker Ultraflex III MALDI-TOF/TOF MS (Bruker Daltonics, Billerica, MA) equipped with a Smartbeam laser, a reflectron, and a LIFT cell was used to obtain MS and MS/MS spectra. Compass v. 1.2 software was used to control the instrument. Due to the delicacy of the sample, all spectra were obtained from 50 laser shots per acquisition instead of the default 200 shots. MS spectra were obtained in positive ion reflector mode, with an m/z range of 450−4000 Da. Poly(propylene glycol) (PPG) 1000 or poly(ethylene glycol) (PEG) 1500 in methanol was used for external calibration.
Acetylation of Cells
Prior to application of CHCA, 0.5 μL of 3:1 methanol/acetic anhydride was applied to each cell and allowed to dry. After the solution evaporated, the cell was covered with matrix as described above.
Oxidation of Cells
In cells exposed to methylene blue, methionine residues were oxidized to the sulfoxide with a gain of 16 Da (See Sample Preparation for Mass Spectrometry).
Assignment of Peaks and Interpretation of Mass Spectra
All spectra were analyzed using Bruker Daltonics flexAnalysis 3.0 software. For all MS spectra, masses were assigned automatically by the software. All MS/MS data underwent background subtraction and smoothing prior to automatic assignment of masses. Occasionally, additional peaks were assigned a mass manually. Peaks were considered to be above noise when the intensity of the peak was at least twice that of the baseline.
A peak was initially identified as a known peptide if the observed m/z value was within ±0.2 Da of a previously sequenced A. suum peptide (6,7,22,23,32−34). Molecular masses were calculated by Prospector MS-Product (http://prospector.ucsf.edu). Assignments were confirmed by MS/MS and chemical modifications (acetylation and oxidation). To identify novel peptides, de novo sequencing was performed by hand. Further verification was obtained by chemical modification of the cell, by comparing the MS/MS spectra of synthetic peptides to cellular spectra, or by tBLASTn searches and cloning of gene sequences.
Database Searches
All putative peptide sequence assignments were searched against a catalog of 447 546 A. suum GSSs (Mitreva, M., personal communication). Since the peptide sequences are short a local tBLASTn search was used with a word size of 2, an E value of 20 000, and a PAM30 matrix. Search results were translated using the ExPASy Translate tool (http://au.expasy.org/tools/dna.html) and visually examined for possible peptide cleavage sites, as well as a C-terminal glycine for amidated peptides. Possible splice sites were predicted by GenScan (http://genes.mit.edu/GENSCAN.html59) or were detected directly by PCR cloning of the mRNA sequence and comparison with genomic sequences. Signal peptides were predicted by SignalP (http://www.cbs.dtu.dk/services/SignalP/60).
Peptide Synthesis
Synthetic peptides were synthesized by the University of Wisconsin-Madison Biotechnology Center.
RNA Isolation
A. suum total RNA was extracted from 20−30 flash-frozen A. suum heads using silica spin columns (ClonTech, Palo Alto, CA, RNA II Nucleospin Kit).
cDNA Preparation for Polymerase Chain Reaction (PCR)
First-strand cDNA was generated from the isolated RNA by using the following reaction in a 10 μL volume: 100 ng of total RNA, 0.5 μL of dNTPs (10 mM each of dATP, dCTP, dGTP, and dTTP), 0.5 μL of oligo(dT)12−18 (0.5 M), 1 μL of 10× RT buffer (200 mM Tris-HCl, pH 8.4, 500 mM KCl; Invitrogen, La Jolla, CA), 2 μL of 25 mM MgCl2, 1 μL of 100 mM dithiothreitol (DTT), 0.5 μL of RNase inhibitor (40 U/μL), and 1 μL of Superscript II reverse transcriptase (50 U/μL; Invitrogen). After incubation for 50 min at 42 °C, the reaction was terminated by incubation at 70 °C for 15 min.
Polymerase Chain Reaction (PCR)
PCR reactions were carried out with the splice leader sequence (SL1) forward primer (5′-GGTTTAATTACCCAAGTTTGAG-3′) and the reverse primers afp-12rev (5′-CGTATCATCATATCGGCGGC-3′) or afp-13rev (5′-TCGACCAAACGAAGTACGG-3′). In a 50 μL volume, the following were mixed: 2 μL of A. suum cDNA or 2 μL of cDNA reaction solution with no reverse transcriptase as a control, 5 μL of 10× PCR Gold buffer (Applied Biosystems, Foster City, CA), 2−8 μL of 25 mM MgCl2, 1 μL each of 10 mM dNTPs, 1 μL of each primer (20 μM), and 0.25 μL of AmpliTaq Gold (Applied Biosystems; 5 U/μL). The PCR conditions were as follows: 95 °C for 3 min, followed by 40 cycles of 94 °C for 30 s, 58 °C for 30 s, 72 °C for 1 min. The PCR products were purified (Qiaquick Gel Purification, Qiagen, Chatsworth, CA) and cloned into Escherichia coli cells (TOPO TA Cloning, Invitrogen). Automated sequencing of the purified plasmid DNA by capillary electrophoreses was carried out by the DNA Sequencing service at the University of Wisconsin-Madison Biotechnology Center.
3′ RACE
3′ RACE was performed with a SMARTer RACE cDNA amplification kit (Clontech, Mountain View, CA) according to the manufacturer’s instructions with either the primer afp-12gsp (5′-CGCATCCTATCGAAGCGAGTACCAAGC-3′) or afp-13gsp (5′-CGAACGAAGGGACTCAAAGCTTATGGATCC-3′). The products were purified, cloned, and sequenced as described above.
Antibody Generation and Affinity Purification
Synthetic AF36 was conjugated with glutaraldehyde to keyhole limpet hemocyanin (KLH; Pierce, Rockford, IL) and then used to raise rabbit polyclonal antibodies (Panigen, Blanchardville, WI). Anti-AF19 antibodies were raised in the same manner (61). Polyclonal antibodies from rabbit serum were affinity-purified using Affigel-10 (Bio-Rad, Hercules, CA) according to the manufacturer’s instructions. Retained antibodies were eluted with Actisep (Sterogene, Carlsbad, CA). The antibody solution was dialyzed against PBS then concentrated using a molecular weight cutoff filter (Centricon Ultracel 30K filters, Millipore, Bedford, MA). The specificity of the antibody against each of the peptides was tested on a dot-ELISA spotted with peptides AF1−36, PepNY, PepTT, and PepTL as described previously (62).
Whole Mount Immunocytochemistry
Whole mounts were prepared by fixing collagenase-treated female A. suum heads overnight at 4 °C in 1% paraformaldehyde and 0.5% glutaraldehyde in PBS. After washing with PBS, the preparations were treated with 1% ethanolamine-HCl, pH 9, incubated at room temperature for 30 min, rinsed with PBS, and then washed extensively with P1 (0.05% NP-40 and 0.1% BSA in PBS44). For blocking experiments, prior to treatment of the tissue, the antibodies were incubated with 40 μg/mL AF36 or AF19 overnight at 4 °C with gentle rocking. The antibody to AF36 was diluted 1:5000. The antibody to AF19 was diluted 1:500. Both antibodies were diluted with P1+ (P1 with 10% calf serum and 1% Triton X-100) and applied for 16 h at 4 °C with gentle rocking. The heads were washed several times with P1 then incubated with the horseradish peroxidase (HRP)-labeled secondary antibody (goat antirabbit IgG) at 1:500 in P1+. Following overnight incubation at 4 °C, the heads were washed as before. Peroxidase activity was revealed by incubation with 0.03% 3,3′-diaminobenzidine 4-HCl (Sigma) and 0.006% H2O2 in PBS. The heads were dehydrated in a graded ethanol series, cleared in xylene, and mounted on glass slides in Permount (Fisher Scientific).
Acknowledgments
We are grateful to Makedonka Mitreva at Washington University for access to her large GSS database, Ivan Chevere-Colon for the use of his AF19-antibody, and Bill Feeny for his significant help with the figures. We also thank Philippa Claude, Catharine Reinitz, and India Viola for critically reviewing the manuscript.
Abbreviations
AF, Ascaris FMRFamide-like peptide; afp, Ascaris FMRFamide-like precursor protein gene; DG, dorsal ganglion; EST, expressed sequence tag; flp, FMRFamide-like peptide gene; ICC, immunocytochemistry; ISH, in situ hybridization; MALDI-TOF, matrix-assisted laser desorption/ionization−time of flight; MS, mass spectrometry; m/z, mass-to-charge ratio; nlp, neuropeptide-like protein gene; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; RACE, rapid amplification of cDNA ends.
Supporting Information Available
Peak lists and comparisons of peaks present in ALA and RID, MS/MS of additional native peptides, and MS/MS of synthetic peptides. This material is available free of charge via the Internet at http://pubs.acs.org.
J.J. was responsible for MS, bioinformatics, data analysis, molecular biology, and manuscript preparation. K.A. was responsible for dissections. C.K. was responsible for molecular biology and ICC. J.K. was responsible for molecular biology and dot-ELISAs. M.V. was responsible for MS. A.S. was responsible for manuscript preparation and data analysis.
This research was supported by the National Institutes of Health (Grants AI15429, NRSA T32 GM007507, and NCRR/SIG S10RR024601).
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Kotani M.; Detheux M.; Vandenbogaerde A.; Communi D.; Vanderwinden J. M.; Le Poul E.; Brezillon S.; Tyldesley R.; Suarez-Huerta N.; Vandeput F.; Blanpain C.; Schiffmann S. N.; Vassart G.; Parmentier M. (2001) The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J. Biol. Chem. 276, 34631–34636. [DOI] [PubMed] [Google Scholar]
- Cottrell G. A. (1997) The first peptide-gated ion channel. J. Exp. Biol. 200, 2377–2386. [DOI] [PubMed] [Google Scholar]
- Marder E.; Bucher D. (2007) Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu. Rev. Physiol. 69, 291–316. [DOI] [PubMed] [Google Scholar]
- Stretton A.; Donmoyer J.; Davis R.; Meade J.; Cowden C.; Sithigorngul P. (1992) Motor behavior and motor nervous system function in the nematode Ascaris suum. J. Parasitol. 78, 206–214. [PubMed] [Google Scholar]
- Davis R. E.; Stretton A. O. (2001) Structure-activity relationships of 18 endogenous neuropeptides on the motor nervous system of the nematode Ascaris suum. Peptides 22, 7–23. [DOI] [PubMed] [Google Scholar]
- Davis R. E.; Stretton A. O. (1996) The motornervous system of Ascaris: Electrophysiology and anatomy of the neurons and their control by neuromodulators. Parasitology 113(Suppl), S97–117. [DOI] [PubMed] [Google Scholar]
- Yew J. Y.; Davis R.; Dikler S.; Nanda J.; Reinders B.; Stretton A. O. (2007) Peptide products of the afp-6 gene of the nematode Ascaris suum have different biological actions. J. Comp. Neurol. 502, 872–882. [DOI] [PubMed] [Google Scholar]
- Reinitz C. A.; Herfel H. G.; Messinger L. A.; Stretton A. O. (2000) Changes in locomotory behavior and cAMP produced in Ascaris suum by neuropeptides from Ascaris suum or Caenorhabditis elegans. Mol. Biochem. Parasitol. 111, 185–197. [DOI] [PubMed] [Google Scholar]
- Li C.; Nelson L. S.; Kim K.; Nathoo A.; Hart A. C. (1999) Neuropeptide gene families in the nematode Caenorhabditis elegans. Ann. N.Y. Acad. Sci. 897, 239–252. [DOI] [PubMed] [Google Scholar]
- Li C.; Kim K. (2008) Neuropeptides. WormBook 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson S. J.; Clynen E.; Baggerman G.; De Loof A.; Schoofs L. (2005) Discovering neuropeptides in Caenorhabditis elegans by two dimensional liquid chromatography and mass spectrometry. Biochem. Biophys. Res. Commun. 335, 76–86. [DOI] [PubMed] [Google Scholar]
- Husson S. J.; Landuyt B.; Nys T.; Baggerman G.; Boonen K.; Clynen E.; Lindemans M.; Janssen T.; Schoofs L. (2009) Comparative peptidomics of Caenorhabditis elegans versus C. briggsae by LC-MALDI-TOF MS. Peptides 30, 449–457. [DOI] [PubMed] [Google Scholar]
- Kreil G. (1984) Occurrence, detection, and biosynthesis of carboxy-terminal amides. Methods Enzymol. 106, 218–223. [DOI] [PubMed] [Google Scholar]
- Bradbury A. F.; Smyth D. G. (1987) Biosynthesis of the C-terminal amide in peptide hormones. Biosci. Rep. 7, 907–916. [DOI] [PubMed] [Google Scholar]
- van Veelen P.; Jiménez C.; Li K.; Wildering W.; Geraerts W.; Tjaden U.; van der Greef J. (1993) Direct peptide profiling of single neurons by matrix-assisted laser desorption-ionization mass spectrometry. Org. Mass Spectrom. 28, 1542–1546. [DOI] [PubMed] [Google Scholar]
- Garden R. W.; Moroz L. L.; Moroz T. P.; Shippy S. A.; Sweedler J. V. (1996) Excess salt removal with matrix rinsing: Direct peptide profiling of neurons from marine invertebrates using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Mass Spectrom. 31, 1126–1130. [DOI] [PubMed] [Google Scholar]
- Redeker V.; Toullec J. Y.; Vinh J.; Rossier J.; Soyez D. (1998) Combination of peptide profiling by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and immunodetection on single glands or cells. Anal. Chem. 70, 1805–1811. [DOI] [PubMed] [Google Scholar]
- Neupert S.; Predel R. (2005) Mass spectrometric analysis of single identified neurons of an insect. Biochem. Biophys. Res. Commun. 327, 640–645. [DOI] [PubMed] [Google Scholar]
- Neupert S.; Predel R.; Russell W. K.; Davies R.; Pietrantonio P. V.; Nachman R. J. (2005) Identification of tick periviscerokinin, the first neurohormone of Ixodidae: Single cell analysis by means of MALDI-TOF/TOF mass spectrometry. Biochem. Biophys. Res. Commun. 338, 1860–1864. [DOI] [PubMed] [Google Scholar]
- Rubakhin S. S.; Churchill J. D.; Greenough W. T.; Sweedler J. V. (2006) Profiling signaling peptides in single mammalian cells using mass spectrometry. Anal. Chem. 78, 7267–7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert S.; Johard H. A.; Nassel D. R.; Predel R. (2007) Single-cell peptidomics of Drosophila melanogaster neurons identified by Gal4-driven fluorescence. Anal. Chem. 79, 3690–3694. [DOI] [PubMed] [Google Scholar]
- Yew J. Y.; Dikler S.; Stretton A. O. (2003) De novo sequencing of novel neuropeptides directly from Ascaris suum tissue using matrix-assisted laser desorption/ionization time-of-flight/time-of-flight. Rapid Commun. Mass Spectrom. 17, 2693–2698. [DOI] [PubMed] [Google Scholar]
- Yew J. Y.; Kutz K. K.; Dikler S.; Messinger L.; Li L.; Stretton A. O. (2005) Mass spectrometric map of neuropeptide expression in Ascaris suum. J. Comp. Neurol. 488, 396–413. [DOI] [PubMed] [Google Scholar]
- Annesley T. M. (2003) Ion suppression in mass spectrometry. Clin. Chem. 49, 1041–1044. [DOI] [PubMed] [Google Scholar]
- Cowden C.; Sithigorngul P.; Brackley P.; Guastella J.; Stretton A. O. (1993) Localization and differential expression of FMRFamide-like immunoreactivity in the nematode Ascaris suum. J. Comp. Neurol. 333, 455–468. [DOI] [PubMed] [Google Scholar]
- Nanda J. C.; Stretton A. O. (2010) In situ hybridization of neuropeptide-encoding transcripts afp-1, afp-3, and afp-4 in neurons of the nematode Ascaris suum. J. Comp. Neurol. 518, 896–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanfleteren J. R.; Van de Peer Y.; Blaxter M. L.; Tweedie S. A.; Trotman C.; Lu L.; Van Hauwaert M. L.; Moens L. (1994) Molecular genealogy of some nematode taxa as based on cytochrome c and globin amino acid sequences. Mol. Phylogenet. Evol. 3, 92–101. [DOI] [PubMed] [Google Scholar]
- Kim K.; Li C. (2004) Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J. Comp. Neurol. 475, 540–550. [DOI] [PubMed] [Google Scholar]
- Estevam M. L.; Nascimento O. R.; Baptista M. S.; Di Mascio P.; Prado F. M.; Faljoni-Alario A.; Zucchi Mdo R.; Nantes I. L. (2004) Changes in the spin state and reactivity of cytochrome C induced by photochemically generated singlet oxygen and free radicals. J. Biol. Chem. 279, 39214–39222. [DOI] [PubMed] [Google Scholar]
- McVeigh P.; Leech S.; Mair G. R.; Marks N. J.; Geary T. G.; Maule A. G. (2005) Analysis of FMRFamide-like peptide (FLP) diversity in phylum Nematoda. Int. J. Parasitol. 35, 1043–1060. [DOI] [PubMed] [Google Scholar]
- Maroney P. A.; Denker J. A.; Darzynkiewicz E.; Laneve R.; Nilsen T. W. (1995) Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: A role for spliced leader addition in translational efficiency. RNA 1, 714–723. [PMC free article] [PubMed] [Google Scholar]
- Cowden C.; Stretton A. O.; Davis R. E. (1989) AF1, a sequenced bioactive neuropeptide isolated from the nematode Ascaris suum. Neuron 2, 1465–1473. [DOI] [PubMed] [Google Scholar]
- Cowden C.; Stretton A. O. (1993) AF2, an Ascaris neuropeptide: Isolation, sequence, and bioactivity. Peptides 14, 423–430. [DOI] [PubMed] [Google Scholar]
- Cowden C.; Stretton A. O. (1995) Eight novel FMRFamide-like neuropeptides isolated from the nematode Ascaris suum. Peptides 16, 491–500. [DOI] [PubMed] [Google Scholar]
- Nanda J. (2004) Molecular biological analysis of neuropeptide gene transcripts from the nematode, Ascaris suum, Ph.D. Thesis, University of Wisconsin-Madison, Madison. [Google Scholar]
- McVeigh P.; Alexander-Bowman S.; Veal E.; Mousley A.; Marks N. J.; Maule A. G. (2008) Neuropeptide-like protein diversity in phylum Nematoda. Int. J. Parasitol. 38, 1493–1503. [DOI] [PubMed] [Google Scholar]
- Stoeckli M.; Chaurand P.; Hallahan D. E.; Caprioli R. M. (2001) Imaging mass spectrometry: A new technology for the analysis of protein expression in mammalian tissues. Nat. Med. 7, 493–496. [DOI] [PubMed] [Google Scholar]
- DeKeyser S. S.; Kutz-Naber K. K.; Schmidt J. J.; Barrett-Wilt G. A.; Li L. (2007) Imaging mass spectrometry of neuropeptides in decapod crustacean neuronal tissues. J. Proteome Res. 6, 1782–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B. J.; Caprioli R. M.; Sanders M. E.; Jensen R. A. (2002) Direct analysis of laser capture microdissected cells by MALDI mass spectrometry. J. Am. Soc. Mass Spectrom. 13, 1292–1297. [DOI] [PubMed] [Google Scholar]
- Hummon A. B.; Amare A.; Sweedler J. V. (2006) Discovering new invertebrate neuropeptides using mass spectrometry. Mass Spectrom. Rev. 25, 77–98. [DOI] [PubMed] [Google Scholar]
- Stretton A. O.; Fishpool R. M.; Southgate E.; Donmoyer J. E.; Walrond J. P.; Moses J. E.; Kass I. S. (1978) Structure and physiological activity of the motoneurons of the nematode Ascaris. Proc. Natl. Acad. Sci. U.S.A. 75, 3493–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. G.; Southgate E.; Thomson J. N.; Brenner S. (1986) The structure of the nervous system of the nematode C. elegans. Philos. Trans. R. Soc. London, Ser. B: Biol. Sci. 314, 1–340. [DOI] [PubMed] [Google Scholar]
- Sithigorngul P.; Stretton A. O.; Cowden C. (1990) Neuropeptide diversity in Ascaris: An immunocytochemical study. J. Comp. Neurol. 294, 362–376. [DOI] [PubMed] [Google Scholar]
- Johnson C. D.; Stretton A. O. (1987) GABA-immunoreactivity in inhibitory motor neurons of the nematode Ascaris. J. Neurosci. 7, 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. D.; Reinitz C. A.; Sithigorngul P.; Stretton A. O. (1996) Neuronal localization of serotonin in the nematode Ascaris suum. J. Comp. Neurol. 367, 352–360. [DOI] [PubMed] [Google Scholar]
- Johnson C. D.; Stretton A. O. (1985) Localization of choline acetyltransferase within identified motoneurons of the nematode Ascaris. J. Neurosci. 5, 1984–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand J. B. (2007) Acetylcholine. WormBook 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuske K.; Beg A. A.; Jorgensen E. M. (2004) The GABA nervous system in C. elegans. Trends Neurosci. 27, 407–414. [DOI] [PubMed] [Google Scholar]
- Guastella J.; Johnson C. D.; Stretton A. O. (1991) GABA-immunoreactive neurons in the nematode Ascaris. J. Comp. Neurol. 307, 584–597. [DOI] [PubMed] [Google Scholar]
- Nathoo A. N.; Moeller R. A.; Westlund B. A.; Hart A. C. (2001) Identification of neuropeptide-like protein gene families in Caenorhabditis elegans and other species. Proc. Natl. Acad. Sci. U.S.A. 98, 14000–14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger J. F.; Lorch A.; Sleumer M. C.; Zapf R.; Jones S. J.; Marra M. A.; Holt R. A.; Moerman D. G.; Hobert O. (2007) The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 21, 1653–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson S. J.; Clynen E.; Baggerman G.; Janssen T.; Schoofs L. (2006) Defective processing of neuropeptide precursors in Caenorhabditis elegans lacking proprotein convertase 2 (KPC-2/EGL-3): Mutant analysis by mass spectrometry. J. Neurochem. 98, 1999–2012. [DOI] [PubMed] [Google Scholar]
- Husson S. J.; Janssen T.; Baggerman G.; Bogert B.; Kahn-Kirby A. H.; Ashrafi K.; Schoofs L. (2007) Impaired processing of FLP and NLP peptides in carboxypeptidase E (EGL-21)-deficient Caenorhabditis elegans as analyzed by mass spectrometry. J. Neurochem. 102, 246–260. [DOI] [PubMed] [Google Scholar]
- Geary T. G.; Price D. A.; Bowman J. W.; Winterrowd C. A.; Mackenzie C. D.; Garrison R. D.; Williams J. F.; Friedman A. R. (1992) Two FMRFamide-like peptides from the free-living nematode Panagrellus redivivus. Peptides 13, 209–214. [DOI] [PubMed] [Google Scholar]
- Kimber M. J.; Fleming C. C.; Prior A.; Jones J. T.; Halton D. W.; Maule A. G. (2002) Localisation of Globodera pallida FMRFamide-related peptide encoding genes using in situ hybridisation. Int. J. Parasitol. 32, 1095–1105. [DOI] [PubMed] [Google Scholar]
- Berman E. S.; Fortson S. L.; Checchi K. D.; Wu L.; Felton J. S.; Wu K. J.; Kulp K. S. (2008) Preparation of single cells for imaging/profiling mass spectrometry. J. Am. Soc. Mass Spectrom. 19, 1230–1236. [DOI] [PubMed] [Google Scholar]
- Schwartz S. A.; Reyzer M. L.; Caprioli R. M. (2003) Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass Spectrom. 38, 699–708. [DOI] [PubMed] [Google Scholar]
- Hilker B.; Clifford K. J.; Sauter A. D. Jr.; Sauter A. D. 3rd; Harmon J. P. (2009) The measurement of charge for induction-based fluidic MALDI dispense event and nanoliter volume verification in real time. J. Am. Soc. Mass Spectrom. 20, 1064–1067. [DOI] [PubMed] [Google Scholar]
- Burge C.; Karlin S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- Bendtsen J. D.; Nielsen H.; von Heijne G.; Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340, 783–795. [DOI] [PubMed] [Google Scholar]
- Chevere-Colon I. (2001) Neuropeptide study in Ascaris by immunocytochemical and molecular biology methods, Ph.D. Thesis, University of Wisconsin-Madison, Madison. [Google Scholar]
- Sithigorngul P.; Stretton A. O.; Cowden C. (1991) A versatile dot-ELISA method with femtomole sensitivity for detecting small peptides. J. Immunol. Methods 141, 23–32. [DOI] [PubMed] [Google Scholar]
- Goldschmidt R. (1908) Das nervensystem von Ascaris lumbricoides und megalocephala. Ein Versuch in den Aufbau eines einfacken Nervensystem einzudringen. Z. Wiss. Zool. 90, 73–136. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.