Abstract
Scrub typhus is a rickettsial disease transmitted to humans through the bite of chigger mites infected with Orientia tsutsugamushi, and is an endemic disease in Taiwan. To elucidate the molecular epidemiology of O. tsutsugamushi, the complete open reading frame of the 56-kDa type-specific antigen gene sequence of strains isolated from scrub typhus patients were determined and analyzed. A total of 116 isolates of O. tsutsugamushi were successfully isolated from patients infected in diverse geographic origins including Taiwan and three offshore islets, Kinmen, Matsu, and Penghu between May 2006 and December 2007. Sequence analysis revealed that 22 distinct sequence types could be identified that were broadly distributed in different clusters of the phylogenetic tree. Most of the isolates belong to Karp, Kawasaki, and Kuroki genotypes and are closely related to strains from Thailand, Japan, and Korea, whereas unique isolates different from other countries were also found in Taiwan. Distinct seasonal distributions were found in different sequence types. Some sequence types caused disease in the cold season, whereas others caused disease in the warm season.
Introduction
Scrub typhus is acute febrile illnesses caused by the obligate intracellular bacterium Orientia tsutsugamushi after the bite of an infected larval-stage trombiculid mite (chigger).1 Symptoms of scrub typhus include fever, headache, rash, cough, and myalgias. Severe manifestations may include pneumonitis, meningitis, encephalitis, disseminated intravascular coagulation, and multiorgan failure. The case fatality rates can go up to 50%, if not treated appropriately.2–4 The disease can be treated effectively with antibiotics such as doxycycline, azithromycin, rifampin, and chloramphenicol. No effective vaccine is currently available.
Scrub typhus is endemic in Asia, northern Australia, and the Western Pacific regions.5,6 Scrub typhus is the most important acute febrile illness associated with the major rickettsioses and has been designated as a reportable communicable disease in Taiwan since 1955. The number of laboratory confirmed cases of scrub typhus from 2004 to 2008 were 369, 462, 384, 510, and 492, respectively, in Taiwan (http://www.cdc.gov.tw/en).
The phenotypic variation identified by polyclonal or monoclonal antibodies of the O. tsutsugamushi isolates depend on the antigenicity of a 56-kDa type-specific antigen (TSA), which is located on the outer membrane surface of the bacteria.7–10 This immunodominant 56-kDa TSA gene of O. tsutsugamushi has been the target for phylogenetic analysis because of its sequence variation.11–14 In this study, we reported the isolation of 116 O. tsutsugamushi strains from scrub typhus patients infected in various regions of Taiwan including three offshore islands, Kinmen, Matsu, and Penghu between May 2006 and December 2007. Phylogenetic analysis of O. tsutsugamushi isolates on the basis of the sequences of a complete open reading frame (ORF) of 56-kDa TSA genes was conducted to determine the genetic relatedness of strains with previous isolates in Taiwan and strains from other countries.
Materials and Methods
Human blood samples.
Scrub typhus is a notifiable infectious disease in Taiwan. Blood samples from patients with suspected scrub typhus were collected and sent to the Taiwan Centers for Disease Control (Taiwan CDC) for laboratory diagnosis. Samples were considered positive for scrub typhus with a positive real-time polymerase chain reaction (PCR) test or the detection of O. tsutsugamushi-specific antibodies by indirect immunofluroescent assay (IFA) (≧ 4-fold increase of O. tsutsugamushi-specific immunoglobulin M (IgM) or IgG antibody in paired sera). Bacterial isolation was performed for PCR-positive samples. Blood samples used in this study were derived from confirmed cases submitted to the Taiwan CDC from May 2006 to December 2007.
Cell culture and O. tsutsugamushi isolation.
Peripheral blood mononuclear cells (PBMC) collected from acute-phase blood samples of scrub typhus patients were used for isolation of O. tsutsugamushi. Bacterial isolation in cell culture was performed using the centrifugation shell vial technique as described previously.15,16 Briefly, O. tsutsugamushi were propagated in L929 mouse fibroblast cells (ATCC CCL-1, NCTC Clone 929) at 32°C in a 5% CO2 incubator for 10 to 14 days and then detected by IFA using O. tsutsugamushi-specific antibody. Each positive shell vial was harvested and inoculated into a T-25 flask containing a monolayer of confluent L929 cells. After 14–20 days of incubation, bacteria-infected L929 cells were scraped and frozen at −80°C.
PCR amplification and DNA sequencing.
Bacterial DNA was extracted from O. tsutsugamushi-infected L929 cells using the QIAamp DNA blood Mini Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer's instructions and stored at −80°C. Primers used for PCR and nucleotide sequencing in this study are shown in Table 1. Primers OT-1F and OT-2134r were used for PCR to amplify a complete open reading frame of 56-kDa TSA gene of O. tsutsugamushi. All primers were used for DNA sequencing. The PCR amplification was performed in 50-μL volumes using the QIAGEN Taq PCR Core Kit (QIAGEN) according to the manufacturer's protocol. The reaction mixture contained 0.2 μM each of the primer, 0.5 mM of deoxyribonucleotide triphosphate mixture, and 2.5 U of Taq DNA polymerase (QIAGEN). Specific fragments were amplified with one cycle of 3 minutes at 94°C, 35 cycles of 30 seconds at 94°C, 45 seconds at 55°C, 1 minute at 72°C, and one cycle of 10 minutes at 72°C. After electrophoresis, gels were stained with ethidium bromide, and amplicons were visualized with an ultraviolet transilluminator. The PCR products were purified using the QIAquick Gel Extraction Kit (QIAGEN), and DNA sequencing was carried out using the ABI Prism 3700 DNA sequencer (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. The PCR products were sequenced twice in each direction.
Table 1.
Primers used for polymerase chain reaction (PCR) and DNA sequencing of Orientia tsutsugamushi
| Primer | Sequence (5′ → 3′) |
|---|---|
| OT-1F | GTT AGT GTG GCT AAA TAA TTA G |
| OT-1FN | GTT TAG AAT GGT TAC CAC |
| OT-1R | GGC ATT ATA GTA GGC TGA GG |
| OT-2F | AGC AGA GCT AGG GGT TAT GT |
| OT-2R | GTA TCT GTT CGA CAG ATG CAC |
| OT-3F | CTC CTG TCA AAG TAT TAA GTG |
| OT-3R | GTC AGC ATA GAG TTT AAC TTG |
| OT-4F | AGA TCT TGT TAA ATT GCA GCG T |
| OT-1889r | TGA ACT TGA TAA TGC AGA AA |
| OT-2134r | CTT TGC TCT GGT TTA GTT GTA |
Phylogenetic analysis.
The complete ORF of 56-kDa TSA gene of O. tsutsugamushi isolates were determined and aligned with global sequences, and retrieved from GenBank for genetic analysis. Sequence analysis was performed with the Lasergene software packages (DNASTAR Inc., Madison, WI). The nucleotide sequences were aligned using Clustal W software. Phylogenetic and molecular analyses were conducted using MEGA version 417 or PHYLIP version 3.66.18 Phylogenetic trees were obtained from DNA sequences by using the neighbor-joining and maximum likelihood methods and genetic distances were calculated using the Kimura two-parameter distance algorithm. One thousand bootstrap replicates were performed to estimate the node reliability of the phylogenetic trees obtained by the two methods. Bootstrap support values above 75 are considered significant.
Nucleotide sequence accession numbers.
The GenBank nucleotide sequence accession numbers for the nucleotide sequence data generated in this study are GQ332742 to GQ332763. Accession numbers of the gene sequences used in this study were indicated within the figure.
Results
Characteristics of O. tsutsugamushi strains isolated from scrub typhus patients in Taiwan.
Figure 1 shows the Taiwan map and geographic locations of O. tsutsugamushi strains isolated in this study. Analysis of the 56-kDa TSA gene sequence of 116 O. tsutsugamushi isolates obtained in this study showed that these isolates could be grouped into 22 distinct sequence types (TW-1 to TW-22), according to their sequence homologies. Sequence similarities were > 99.9% within each of the sequence type. Table 2 shows the 22 sequence types, the representative isolate of each of the sequence types, and pairwise nucleotide sequence similarities with strains isolated from other countries. Most of the sequence types are closely related to strains from Thailand, whereas others are similar to strains from Japan, Korea, and China.
Figure 1.
A map indicating geographic areas in Taiwan from which Orientia tsutsugamushi strains were isolated.
Table 2.
The sequence type, representative strains, and phylogenetic closest foreign strains of Orientia tsutsugamushi
| Sequence type | Representative isolate | Length of ORF of 56kD-TSA gene | Isolation site of representative isolate | Isolation date (month/year) of representative isolate | Pairwise nucleotide sequence similarity (%) to phylogenetically closest foreign O. tsutsugamushi strain from NCBI | GenBank accession no. | Genotype |
|---|---|---|---|---|---|---|---|
| TW-1 | KM0605a | 1608 | Kinmen Island | 05/2006 | 98.3% to UT150 (EF213086), Thailand | GQ332742 | Karp |
| TW-2 | TY0610a | 1605 | Taoyuan County | 10/2006 | 97.4% to UT336 (EF213089), Thailand | GQ332743 | Karp |
| TW-3 | TP0607a | 1605 | Taipei County | 07/2006 | 97.5% to Karp (M33004), New Guinea | GQ332744 | Karp |
| TW-4 | TP0708a | 1608 | Taipei County | 08/2007 | 96.0% to UT336 (EF213089), Thailand | GQ332745 | Karp |
| TW-5 | KM0607h | 1632 | Kinmen Island | 07/2006 | 95.8% to UT176 (EF213081), Thailand | GQ332746 | Karp |
| TW-6 | KHC0609c | 1608 | Kaohsiung City | 09/2006 | 97.7% to UT176 (EF213081), Thailand | GQ332747 | Karp |
| TW-7 | KHC0606a | 1608 | Kaohsiung City | 06/2006 | 96.4% to yeo-joo (AF430144), Korea | GQ332748 | Karp |
| TW-8 | CH0711a | 1692 | Changhua County | 11/2007 | 96.3% to pa-joo (AF430142), Korea | GQ332749 | Karp |
| TW-9 | TPC0701a | 1599 | Taipei City | 01/2007 | 99.7% to Boryong (AM494475), Korea | GQ332750 | Kuroki |
| TW-10 | KHC0704a | 1566 | Kaohsiung City | 04/2007 | 93.8% to TA763 (U80636), Thailand | GQ332751 | TA763 |
| TW-11 | NT0707a | 1584 | Nantou County | 07/2007 | 96.7% to TA763 (U80636), Thailand | GQ332752 | TA763 |
| TW-12 | TT0705a | 1593 | Taitung County | 05/2007 | 86.8% to UT302 (EF213095), Thailand | GQ332753 | TW-12 |
| TW-13 | NT0711a | 1557 | Nantou County | 11/2007 | 92.6% to Sxh951 (AF050669), China | GQ332754 | Kawasaki |
| TW-14 | TT0711a | 1551 | Taitung County | 11/2007 | 92.6% to Ikeda (AP008981), Japan | GQ332755 | Kawasaki |
| TW-15 | PT0712b | 1569 | Pingtung County | 12/2007 | 99.3% to Kawasaki (M63383), Japan | GQ332756 | Kawasaki |
| TW-16 | KHC0707a | 1572 | Kaohsiung City | 07/2007 | 97.2% to UT329 (EF213099), Thailand | GQ332757 | Kawasaki |
| TW-17 | TPC0707a | 1596 | Taipei City | 07/2007 | 97.7% to UT125 (EF213096), Thailand | GQ332758 | Kawasaki |
| TW-18 | KHC0706a | 1596 | Kaohsiung City | 06/2007 | 98.4% to UT125 (EF213096), Thailand | GQ332759 | Kawasaki |
| TW-19 | KM0606a | 1572 | Kinmen Island | 06/2006 | 97.2% to UT125 (EF213096), Thailand | GQ332760 | Kawasaki |
| TW-20 | HC0605a | 1572 | Hsinchu County | 05/2006 | 99.9% to LF-1 (AF173050), Malaysia | GQ332761 | Kato |
| TW-21 | KM0607b | 1590 | Kinmen island | 07/2006 | 98.6% to Kato (M63382), Japan | GQ332762 | Kato |
| TW-22 | KHC0606b | 1575 | Kaohsiung City | 06/2006 | 88.3% to FPW1038 (EF213087), Thailand | GQ332763 | TW-22 |
Phylogenetic analysis of O. tsutsugamushi isolated in Taiwan.
Figure 2 shows the phylogenetic analysis of 58 strains including 22 representative isolates described in this study, seven local strains from rodent species in Taiwan previously reported by Qiang and others,12 and 29 global reference strains available from GenBank. Most of the Taiwan strains isolated in this study grouped with the Karp genotype (44.8%; 52/116) including sequence type TW-1 to TW-8. The most closely related global reference strains to TW-1, TW-2, TW-4, TW-5, and TW-6 were strains isolated from Thailand. TW-8 isolates were grouped into Saitama-type strains isolated from Japan and Korea. TW-9 isolates belong to Kuroki genotype and are most closely related to Boryong and Kuroki strains from Korea and Japan, respectively. Two isolates, TW-10 and TW-11, were clustered with TA763-type strains with sequence similarity 93.8% and 96.7% to TA763 strain, respectively.
Figure 2.
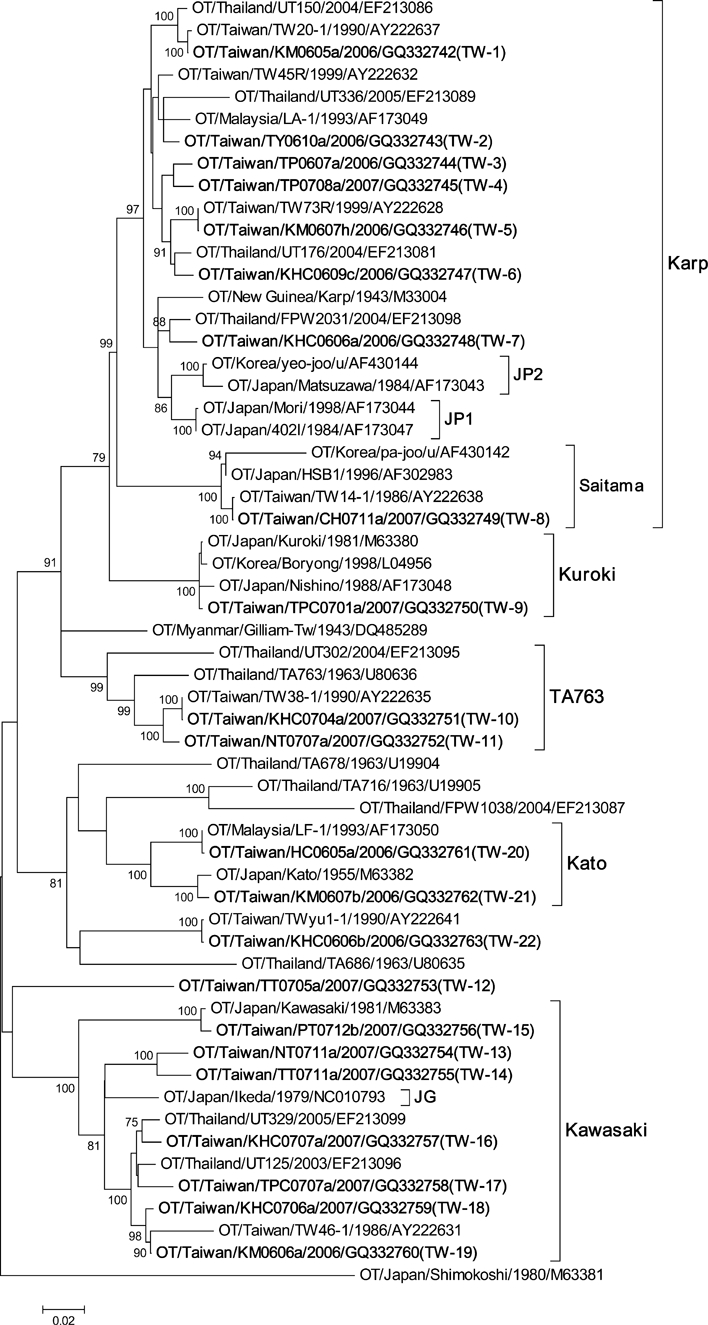
Phylogenetic tree based on the 56-kDa TSA gene ORF of 58 strains of Orientia tsutsugamushi. The tree was constructed by the neighbor-joining method. Bootstrap support values > 75 are shown. The 22 representative strains described in this study are designated in bold. Orientia tsutsugamushi strains were identified using the nomenclature of OT/country/strain name/year of isolation/GenBank accession no.; u = unknown.
TW-12 isolate was unique in that greater sequence variation was found compared with the other global reference strains. Strains of TW-13 to TW-19 belong to Kawasaki genotype. TW-13 and TW-14 are most closely related to global reference strains from China and Thailand, respectively. TW-15 is very similar to the Kawasaki strain from Japan, whereas TW-16 to TW-19 isolates are most closely related to strains from Thailand. TW-20 and TW-21 belong to Kato genotype and are most closely related to strains from Malaysia and Japan, respectively. Sequences of TW-22 isolates were unique and distinct from strains isolated from other countries.
Geographic distribution of O. tsutsugamushi strains.
Figure 1 and Table 3 show the geographic location and distribution of 116 O. tsutsugamushi isolates obtained in this study. TW-1 contains the most abundant O. tsutsugamushi isolates (39 isolates) and these isolates are widely distributed in Taiwan and offshore islands, especially in Kinmen Island. Isolates of TW-2, TW-3, TW-4, and TW-17 were only found in northern Taiwan. TW-9 contains 17 isolates and these isolates were found only in the Taiwan main island. TW-19 contains 14 isolates and these isolates were distributed in the south part of Taiwan and Kinmen and Matsu Islands. TW-22 contains 19 isolates including 11 from Kaohsiung City and County and 8 from Kinmen Island.
Table 3.
Geographic distribution of sequence types
| Location | TW−1 | TW−2 | TW−3 | TW−4 | TW−5 | TW−6 | TW−7 | TW−8 | TW−9 | TW−10 | TW−11 | TW−12 | TW−13 | TW−14 | TW−15 | TW−16 | TW−17 | TW−18 | TW−19 | TW−20 | TW−21 | TW−22 | No. isolates |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Taipei City and County | 1 | 1 | 2 | 1 | 5 | ||||||||||||||||||
| Taoyuan County | 2 | 1 | 3 | 6 | |||||||||||||||||||
| Hsinchu County | 3 | 1 | 4 | ||||||||||||||||||||
| Miaoli County | 2 | 1 | 3 | ||||||||||||||||||||
| Taichung City and County | 1 | 2 | 3 | ||||||||||||||||||||
| Changhua County | 1 | 1 | |||||||||||||||||||||
| Nantou County | 4 | 1 | 1 | 6 | 12 | ||||||||||||||||||
| Kaohsiung City and County | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 11 | 23 | ||||||||||||
| Hualien County | 1 | 1 | |||||||||||||||||||||
| Pingtung County | 3 | 1 | 1 | 5 | |||||||||||||||||||
| Taitung County | 1 | 1 | 1 | 3 | |||||||||||||||||||
| Kinmen Island | 26 | 4 | 3 | 2 | 8 | 43 | |||||||||||||||||
| Matsu Island | 1 | 1 | 2 | ||||||||||||||||||||
| Penghu Island | 3 | 2 | 5 | ||||||||||||||||||||
| No. isolates | 39 | 1 | 1 | 1 | 4 | 2 | 1 | 3 | 17 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 14 | 2 | 2 | 19 | 116 |
Discussion
In this study, phylogenetic analysis of the 56-kDa TSA gene sequences shows that large genetic diversities are recognized among the O. tsutsugamushi strains isolated in Taiwan. Among 22 sequence types identified, 11 sequence types are closely related to strains isolated from Thailand, whereas other sequence types are related to strains isolated from Japan (TW-14, TW-15, and TW-21), Korea (TW-7, TW-8, and TW-9), China (TW-13), Malaysia (TW-20), and New Guinea (TW-3). On the other hand, two sequence types, TW-12 and TW-22, are unique in Taiwan and distinct from isolates found in other countries.
Qiang and others12 reported the analysis of 14 strains of O. tsutsugamushi isolated from mites and rodent species in Kinmen Island, Penghu Island, Lanyu Island, and Chengkung harbor in Taitung County between 1986 and 1999. These strains showed high varieties in sequence homologies of the 56-kDa TSA gene and were different from those distributed in other countries. In this study, we have isolated similar strains described by Qiang from blood samples of scrub typhus patients during 2006–2007. TW20-1 and TWyu8-1 strains were similar to TW-1 isolates with sequence similarities ≧ 99.8%. TW73R, TW14-1, and TW44R were similar to TW-5, TW-8, and TW-22 with sequence similarity ≧ 99.9%, respectively, whereas TW38-1 was identical to TW-10. These results indicate that many isolates obtained from mites and rodent species in Taiwan are pathogenic for humans.
More recently, Kelly and others19 analyzed 271 sequences of 56 kDa antigen gene at the global level and found that Karp-related strains (~40%) are the most common of all genotyped isolates, followed by the JG-related genotype strains (~18%), throughout the region of endemicity. Similar results were found in our study, Karp-related isolates (TW-1 to TW-7), except TW-8 (Saitama), are most common (42.4%, 49/116) and JG-related genotype (TW-13 and TW-14; TW-16 to TW-19) make up 17.2% (20/116) of all isolates. However, in contrast to the study of Kelly and others, the Kuroki-related genotype isolates (TW-9) were more abundant (14.7% versus 5.5%) in Taiwan.
It is interesting to note seasonal prevalence of different O. tsutsugamushi strains in this study. Table 4 shows the monthly distribution of scrub typhus infections of 116 cases. Notably, sequence types that caused scrub typhus disease in the cold season (November to January), including TW-8, TW-9, TW-13, TW-14, and TW-15, are closely related to the strains isolated in Japan, Korea, and China, whereas most of the sequence types that transmitted in the warm season (April to October) are closely related to the strains isolated from Thailand. As in Southeast Asia and Western Pacific, scrub typhus also occurs virtually all year around in Taiwan. The larval chigger of Leptotrombidium deliense is the principal vector of scrub typhus and is present throughout the year, with a peak occurring in summer in Taiwan.20,21 These mites are resistant to high humidity and warm temperature. In addition, species other than the main vector species have also been collected in Kinmen and Matsu Islands, such as Leptotrombidium scutellare and Leptotrombidium pallidum. Whether different vector species were responsible for the changes of seasonal prevalence of scrub typhus disease remained to be studied. Our results provide genetic evidence that a great diversity of O. tsutsugamushi pathogenic strains existed in Taiwan, which is located in the endemic center of the geographic triangle of scrub typhus.
Table 4.
Monthly distribution of Orientia tsutsugamushi sequence types in Taiwan
| Month | TW−1 | TW−2 | TW−3 | TW−4 | TW−5 | TW−6 | TW−7 | TW−8 | TW−9 | TW−10 | TW−11 | TW−12 | TW−13 | TW−14 | TW−15 | TW−16 | TW−17 | TW−18 | TW−19 | TW−20 | TW−21 | TW−22 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| January | • | |||||||||||||||||||||
| February | ||||||||||||||||||||||
| March | ||||||||||||||||||||||
| April | • | |||||||||||||||||||||
| May | • | • | • | • | • | • | ||||||||||||||||
| June | • | • | • | • | • | |||||||||||||||||
| July | • | • | • | • | • | • | • | • | • | |||||||||||||
| August | • | • | • | • | • | |||||||||||||||||
| September | • | • | • | • | • | |||||||||||||||||
| October | • | • | • | • | • | |||||||||||||||||
| November | • | • | • | • | • | • | ||||||||||||||||
| December | • | • |
The 56-kDa TSA is a type-specific protein of O. tsutsugamushi and contains hypervariable regions. Analysis of genetic relationship of Orientia strains using DNA sequences of 56-kDa TSA gene may have exaggerated the differences of the evolution of Orientia. Analysis of the sequence of other genes, such as 16S rRNA and groEL genes showed that sequence divergence of these genes within the Orientia genus is very limited and might not provide useful data for studies of genetic differentiation within Orientia strains. Because the whole genome of O. tsutsugamushi has been determined, some of the genes might be useful for phylogenetic study in the future. It is worth analyzing the sequence divergence of other genes including the housekeeping gene to determine the genetic relationship of Orientia strains.
Footnotes
Financial support: This study is in part supported by grant 98-0324-01-F-20 from National Research Program for Genome Medicine and by grant DOH96-DC-2010 from Centers for Disease Control, Department of Health, Taipei, Taiwan, Republic of China.
Authors' addresses: Hsiu-Ying Lu, Sheng-Kai Yu, Chia-Hsin Cheng, Jr-Shiang Yang, Chien-Ling Su, Huai-Chin Hu, Jyh-Hsiung Huang, and Pei-Yun Shu, Vector-Borne Viral and Rickettsial Diseases Laboratory, Research and Diagnostic Center, Centers for Disease Control, Department of Health, Taipei, Taiwan, R.O.C., E-mails: hsiuying@cdc.gov.tw, ysk@cdc.gov.tw, eukaryota@cdc.gov.tw, shiang23@cdc.gov.tw, sue@cdc.gov.tw, huaichinhu@cdc.gov.tw, jhhuang@cdc.gov.tw, and pyshu@cdc.gov.tw. Kun-Hsien Tsai, Graduate Institute of Epidemiology, College of Public Health, National Taiwan University, Taipei, Taiwan, R.O.C., E-mail: kunhtsai@ntu.edu.tw. Hsi-Chieh Wang, Vector Biology Laboratory, Research and Diagnostic Center, Centers for Disease Control, Department of Health, Taipei, Taiwan, R.O.C., E-mail: sjwang@cdc.gov.tw.
References
- 1.Tamura A, Ohashi N, Urakami H, Miyamura S. Classification of Rickettsia tsutsugamushi in a new genus, Orientia gen. nov., as Orientia tsutsugamushi comb. nov. Int J Syst Bacteriol. 1995;45:589–591. doi: 10.1099/00207713-45-3-589. [DOI] [PubMed] [Google Scholar]
- 2.Berman SJ, Kundin WD. Scrub typhus in South Vietnam. A study of 87 cases. Ann Intern Med. 1973;79:26–30. doi: 10.7326/0003-4819-79-1-26. [DOI] [PubMed] [Google Scholar]
- 3.Yi KS, Chong Y, Covington SC, Donahue BJ, Rothen RL, Rodriguez J, Arthur JD. Scrub typhus in Korea: importance of early clinical diagnosis in this newly recognized endemic area. Mil Med. 1993;158:269–273. [PubMed] [Google Scholar]
- 4.Watt G, Strickman D. Life-threatening scrub typhus in a traveler returning from Thailand. Clin Infect Dis. 1994;18:624–626. doi: 10.1093/clinids/18.4.624. [DOI] [PubMed] [Google Scholar]
- 5.Walker DH. Rickettsial diseases in travelers. Travel Med Infect Dis. 2003;1:35–40. doi: 10.1016/S1477-8939(03)00025-5. [DOI] [PubMed] [Google Scholar]
- 6.Watt G, Parola P. Scrub typhus and tropical rickettsioses. Curr Opin Infect Dis. 2003;16:429–436. doi: 10.1097/00001432-200310000-00009. [DOI] [PubMed] [Google Scholar]
- 7.Hanson B. Identification and partial characterization of Rickettsia tsutsugamushi major protein immunogens. Infect Immun. 1985;50:603–609. doi: 10.1128/iai.50.3.603-609.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamura A, Ohashi N, Urakami H, Takahashi K, Oyanagi M. Analysis of polypeptide composition and antigenic components of Rickettsia tsutsugamushi by polyacrylamide gel electrophoresis and immunoblotting. Infect Immun. 1985;48:671–675. doi: 10.1128/iai.48.3.671-675.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohashi N, Tamura A, Ohta M, Hayashi K. Purification and partial characterization of a type-specific antigen of Rickettsia tsutsugamushi. Infect Immun. 1989;57:1427–1431. doi: 10.1128/iai.57.5.1427-1431.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seong SY, Kim MK, Lee SM, Odgerel Z, Choi MS, Han TH, Kim IS, Kang JS, Lim BU. Neutralization epitopes on the antigenic domain II of the Orientia tsutsugamushi 56-kDa protein revealed by monoclonal antibodies. Vaccine. 2000;19:2–9. doi: 10.1016/s0264-410x(00)00167-5. [DOI] [PubMed] [Google Scholar]
- 11.Enatsu T, Urakami H, Tamura A. Phylogenetic analysis of Orientia tsutsugamushi strains based on the sequence homologies of 56-kDa type-specific antigen genes. FEMS Microbiol Lett. 1999;180:163–169. doi: 10.1111/j.1574-6968.1999.tb08791.x. [DOI] [PubMed] [Google Scholar]
- 12.Qiang Y, Tamura A, Urakami H, Makisaka Y, Koyama S, Fukuhara M, Kadosaka T. Phylogenetic characterization of Orientia tsutsugamushi isolated in Taiwan according to the sequence homologies of 56-kDa type-specific antigen genes. Microbiol Immunol. 2003;47:577–583. doi: 10.1111/j.1348-0421.2003.tb03420.x. [DOI] [PubMed] [Google Scholar]
- 13.Kollars TM, Jr, Bodhidatta D, Phulsuksombati D, Tippayachai B, Coleman RE. Short report: variation in the 56-kD type-specific antigen gene of Orientia tsutsugamushi isolated from patients in Thailand. Am J Trop Med Hyg. 2003;68:299–300. [PubMed] [Google Scholar]
- 14.Blacksell SD, Luksameetanasan R, Kalambaheti T, Aukkanit N, Paris DH, McGready R, Nosten F, Peacock SJ, Day NP. Genetic typing of the 56-kDa type-specific antigen gene of contemporary Orientia tsutsugamushi isolates causing human scrub typhus at two sites in north-eastern and western Thailand. FEMS Immunol Med Microbiol. 2008;52:335–342. doi: 10.1111/j.1574-695X.2007.00375.x. [DOI] [PubMed] [Google Scholar]
- 15.Tsai KH, Wang HC, Chen CH, Huang JH, Lu HY, Su CL, Shu PY. Isolation and identification of a novel spotted fever group Rickettsia, strain IG-1, from Ixodes granulatus ticks collected on Orchid Island (Lanyu), Taiwan. Am J Trop Med Hyg. 2008;79:256–261. [PubMed] [Google Scholar]
- 16.Tsai KH, Lu HY, Tsai JJ, Yu SK, Huang JH, Shu PY. Human case of Rickettsia felis infection, Taiwan. Emerg Infect Dis. 2008;14:1970–1972. doi: 10.3201/eid1412.080525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Seattle, WA: Department of Genome Sciences, University of Washington; 2005. Distributed by the author. [Google Scholar]
- 19.Kelly DJ, Fuerst PA, Ching WM, Richards AL. Scrub typhus: the geographic distribution of phenotypic and genotypic variants of Orientia tsutsugamushi. Clin Infect Dis. 2009;15((Suppl 3)):S203–S230. doi: 10.1086/596576. [DOI] [PubMed] [Google Scholar]
- 20.Van Peenen PF, Lien JC, Santana FJ, See R. Correlation of chigger abundance with temperature at a hyperendemic focus of scrub typhus. J Parasitol. 1976;62:653–654. [PubMed] [Google Scholar]
- 21.Wang HC, Chung CL, Lin TH, Wang CH, Wu WJ. Studies on the vectors and pathogens of scrub typhus on murine-like animals in Kinmen County, Taiwan. Formosan Entomol. 2004;24:257–272. [Google Scholar]


