The microRNA156-targeted SQUAMOSA PROMOTER BINDING PROTEIN LIKE genes, which were reported to define an endogenous phase transition pathway, temporally control the trichome distribution on the stem and inflorescences by activating the trichome negative regulator genes TRICHOMELESS1 and TRIPTYCHON.
Abstract
The production and distribution of plant trichomes is temporally and spatially regulated. After entering into the flowering stage, Arabidopsis thaliana plants have progressively reduced numbers of trichomes on the inflorescence stem, and the floral organs are nearly glabrous. We show here that SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) genes, which define an endogenous flowering pathway and are targeted by microRNA 156 (miR156), temporally control the trichome distribution during flowering. Plants overexpressing miR156 developed ectopic trichomes on the stem and floral organs. By contrast, plants with elevated levels of SPLs produced fewer trichomes. During plant development, the increase in SPL transcript levels is coordinated with the gradual loss of trichome cells on the stem. The MYB transcription factor genes TRICHOMELESS1 (TCL1) and TRIPTYCHON (TRY) are negative regulators of trichome development. We show that SPL9 directly activates TCL1 and TRY expression through binding to their promoters and that this activation is independent of GLABROUS1 (GL1). The phytohormones cytokinin and gibberellin were reported to induce trichome formation on the stem and inflorescence via the C2H2 transcription factors GIS, GIS2, and ZFP8, which promote GL1 expression. We show that the GIS-dependent pathway does not affect the regulation of TCL1 and TRY by miR156-targeted SPLs, represented by SPL9. These results demonstrate that the miR156-regulated SPLs establish a direct link between developmental programming and trichome distribution.
INTRODUCTION
Trichomes are specialized epidermal cells that act as barriers to protect plants from herbivores, UV irradiation, and excessive transpiration (Johnson, 1975; Mauricio and Rausher, 1997). In Arabidopsis thaliana, the distribution of trichomes is spatially and temporally regulated. During the early vegetative phase, trichomes are evenly distributed on the adaxial side of juvenile rosette leaves. As trichome production initiates on the abaxial side of leaves, the plant prepares for the transition from the vegetative to the reproductive stage (Telfer et al., 1997). After entering into the reproductive stage, the number of trichomes produced on the main inflorescence stem is gradually reduced. Floral organs are nearly glabrous except for a few trichomes present on the abaxial surface of sepals.
Previous studies of Arabidopsis trichome development have focused on rosette leaves (Marks, 1997; Hülskamp et al., 1999; Larkin et al., 2003; Ishida et al., 2008). A series of genes positively regulating trichome initiation and development have been identified, including TRANSPARENT TESTA GLABRA1 (TTG1) (Galway et al., 1994; Walker et al., 1999; Bouyer et al., 2008), GLABRA1 (GL1) (Oppenheimer et al., 1991), GL3, and ENHANCER OF GLABRA3 (EGL3) (Payne et al., 2000; Szymanski et al., 2000; Zhang et al., 2003). TTG1, GL1, and GL3+EGL3 encode a WD40 protein, an R2R3 MYB transcription factor, and two basic helix-loop-helix–type transcription factors, respectively, and they form a ternary complex that initiates trichome cell development by activating GL2, which encodes a homeodomain/leucine zipper t ranscription factor (Rerie et al., 1994; Masucci et al., 1996; Schiefelbein, 2003; Pesch and Hülskamp, 2004; Ramsay and Glover, 2005). Another group of genes encode the single-repeat R3 MYB factors TRIPTYCHON (TRY), CAPRICE (CPC), ENHANCER OF TRY AND CPC1 (ETC1), ETC2, ETC3, and TRICHOMELESS1 (TCL1) (Wada et al., 1997; Esch et al., 2004; Kirik et al., 2004a, 2004b; Schellmann et al., 2002; Simon et al., 2007; Wang et al., 2007). They suppress trichome initiation in a redundant manner, although TRY, TCL1, and CPC exert a major influence on rosette leaves, inflorescences, and roots (root hairs), respectively (Wada et al., 1997; Schnittger et al., 1998; 1999; Schellmann et al., 2002; Wang et al., 2007). It was suggested that expression of some of the negative regulator genes, including TRY, CPC, ETC1, and ETC3, was completely or partially dependent on the GL1-GL3-TTG1 protein complex Morohashi et al., 2007), whereas TCL1 and ETC2 were regulated by yet unidentified mechanisms (Wang et al., 2008b). The single-repeat MYB proteins can move from the trichome into neighboring cells where they compete with GL1 for the binding site of GL3 to prevent the formation of the active protein complex, resulting in blockage of trichome initiation. This lateral inhibition mechanism explains how the trichome spacing pattern is generated in rosette leaves (Schellmann et al., 2002; Digiuni et al., 2008).
Although trichome production is gradually repressed in Arabidopsis plants after bolting, the phytohormones of gibberellins (GAs) and cytokinins can stimulate trichome initiation on the stem and inflorescence (Chien and Sussex, 1996; Telfer et al., 1997; Perazza et al., 1998; Gan et al., 2006, 2007). A group of transcription factor genes involved in phytohormone-activated trichome formation has been identified: GLABROUS INFLORESCENCE STEMS (GIS), GIS2, and ZINC FINGER PROTEIN8 (ZFP8) (Gan et al., 2006, 2007). GIS was first reported to act in a GA-responsive pathway; loss-of-function mutation of GIS decreases trichome initiation on the stem, whereas overexpression of GIS leads to excessive trichome production on most of the floral organs (Gan et al., 2006). Later, it was shown that both GIS2 and ZFP8 play important roles in mediating GA and cytokinin responses in initiating trichome development (Gan et al., 2007).
For developmentally regulated trichome patterning in the reproductive stage, TCL1 plays an important role. Loss-of-function mutants of TCL1 induce ectopic trichome development on the main inflorescence stem and pedicels (Wang et al., 2007). TCL1 has a high sequence identity to other single-repeat MYB proteins, such as TRY, CPC, ETC1, 2, and 3, and it also negatively regulates GL1 transcription (Wang et al., 2007). However, among the genes that encode single-repeat R3 MYB proteins, TCL1 is unique in that it is activated after bolting and its expression does not require the GL1-GL3-TTG1 complex (Wang et al., 2008b). Thus, dissection of TCL1 regulation is key to discovering the signal that links the developmental timing and trichome patterning at the reproductive stage.
Flowering time is precisely controlled by both endogenous and environmental cues (Baurle and Dean, 2006). Previous studies of Arabidopsis revealed four major pathways that are integrated into the flowering control network: the photoperiod, vernalization, autonomous, and the phytohormone GA pathways (Komeda, 2004; He and Amasino, 2005). Two types of positive regulators, the plant-specific transcription factor LEAFY (LFY) and members of the APETALA1 (AP1)/FRUITFULL (FUL) clade of MADS box transcription factors, are activated during phase transition and promote flowering (Litt and Irish, 2003; Benlloch et al., 2007). More recently, miR156 and its target genes, SQUAMOSA BINDING PROMOTER BINDING PROTEIN-LIKE (SPL), have been identified to define an endogenous flowering timing pathway. SPLs promote flowering by directly activating LFY and a group of MADS box genes, including SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), FUL, and AP1 (Wang et al., 2009; Yamaguchi et al., 2009).
Here, we show that the miR156-regulated SPL genes also control trichome patterning on the inflorescence stems and floral organs, a phenotype that was also described by a recent report (Shikata et al., 2009). We demonstrate that SPL transcription factors suppress trichome formation by directly activating TCL1 and TRY gene expression and show that an increase in SPL transcript levels was coordinated with the reduced trichome density on successive stem internodes, thus linking the developmental timing to trichome development.
RESULTS
SPLs Levels Are Inversely Correlated to Trichome Numbers
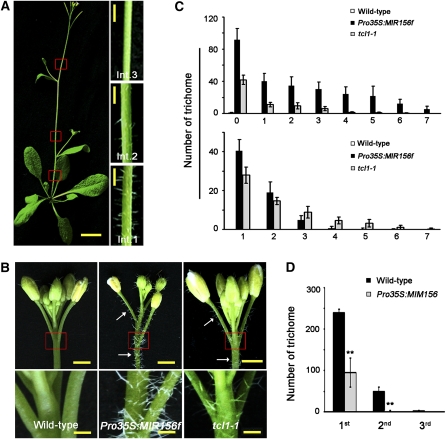
In the juvenile vegetative stage, trichomes were evenly distributed on the adaxial side of rosette leaves of Arabidopsis. After entering into the reproductive stage, when the plant was bolting, the numbers of emerging trichomes on the successive internodes of the main stem were gradually reduced. The third (counting from the bottom) and upper internodes became nearly glabrous (Figure 1A). In flowers, all other floral organs, including pedicel, were glabrous or nearly so (Figure 1B), with the exception that unbranched trichomes were found sparsely on the abaxial side of the sepal. The spatial distribution of trichomes indicates a coordination of developmental timing and trichome patterning.
Figure 1.
Regulation of Inflorescence Trichomes by miR156-Targeted SPL Genes.
(A) Spatial distribution of trichomes on aerial parts of Arabidopsis plants. Int. represents the three main stem internodes numbered from bottom to top. Red rectangles show locations of inset photographs. Bars = 1 cm in main photograph and 1 mm in insets.
(B) Trichomes on inflorescences of wild-type (left), Pro35S:MIR156f (middle), and tcl1-1 mutant (right) plants. Arrows indicate the ectopic trichomes on the main inflorescence stem and pedicels. The bottom panels represent the area marked by the red square in the top panels. Bars in top panels = 2 mm, and bars in bottom panels = 500 μm.
(C) Trichome density of wild-type, Pro35S:MIR156f, and tcl1-1 plants. Top: the main inflorescence stem internodes below (numbered 0) and above (numbered 1 to 7) the site of the first pedicel (flower); bottom: the first seven main inflorescence pedicels (numbered 1 to 7 from bottom to top). Data are given as mean ± sd (n > 15).
(D) Trichome density of the main stem of wild-type and Pro35S:MIM156 plants. 1st to 3rd represent the main stem internodes from bottom to top. Data are given as mean ± sd (n > 15) and analyzed by t test. **P ≤ 0.01, very significant difference.
Recent reports demonstrated that miR156-regulated SPLs define an endogenous flowering pathway and promote phase transition (Wang et al., 2009; Wu et al., 2009; Yamaguchi et al., 2009). To explore the relation between floral transition and trichome repression, we first examined trichome phenotypes of transgenic Pro35S:MIR156f plants, which overexpress MIR156f, an RNA encoded by one of the eight loci of MIR156 in Arabidopsis. Pro35S:MIR156f plants produced ectopic trichomes on both inflorescence stems and pedicels, a phenotype also observed with the tcl1-1 mutant (Figures 1B and 1C). We then analyzed several mutants of floral induction pathways. It has been reported that LFY and the MADS box genes of SOC1 and FUL are direct targets of SPL transcription factors (Wang et al., 2009; Yamaguchi et al., 2009); FLOWERING LOCUS T (FT) encodes a small protein that is a movable flowering signal (Kardailsky et al., 1999; Kobayashi et al., 1999), and TERMINAL FLOWER1 (TFL1) encodes an FT homolog with an opposite function (Bradley et al., 1997; Ohshima et al., 1997). These results suggest that miR156 plays an important role in determining the production of trichomes on stems and inflorescences.
In the Arabidopsis genome, 11 of the 17 SPLs are targeted by miR156 (Rhoades et al., 2002; Gandikota et al., 2007; Wang et al., 2008a; Wu et al., 2009). These SPL genes can be further classified into four clades, represented by SPL3, SPL10, SPL13, and SPL9, respectively (Guo et al., 2008; Wu et al., 2009). To check if those miR156-targeted SPL genes were responsible for the trichome phenotype observed in the Pro35S:MIR156f plants, we analyzed transgenic plants expressing a mimicry target of miR156 (Pro35S:MIM156), which resulted in elevated levels of SPLs (Franco-Zorrilla et al., 2007; Wang et al., 2008a). We found that the Pro35S:MIM156 plants indeed had a dramatic decrease of trichome densities on the stem (Figure 1D).
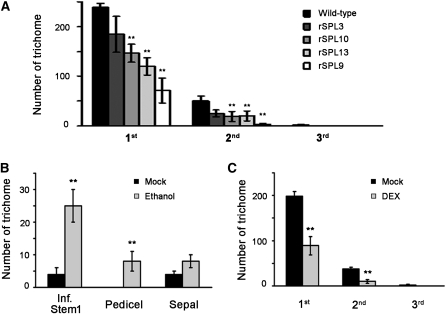
To provide more direct evidence that increased levels of SPLs were the cause of reduced trichome production, we examined transgenic plants expressing the miR156-resistant form (rSPL) of four SPL genes, Pro35S:rSPL3, ProSPL10:rSPL10, ProSPL13:rSPL13, and ProSPL9:rSPL9, which contained synonymous substitutions that changed the miR156f-pairing sequence (Wang et al., 2009). Compared with the wild type, trichome formation on the main stem of these transgenic plants was suppressed to varying degrees (Figure 2A). Together, these data suggest that the miR156-targeted SPLs are involved in trichome regulation.
Figure 2.
SPLs Mediate Temporal Control of Trichome Patterning.
(A) Trichome density of the main stems of wild-type, Pro35S:rSPL3, ProSPL10:rSPL10, ProSPL13:rSPL13, and ProSPL9:rSPL9 plants. 1st to 3rd represent the main stem internodes from bottom to top. Data are given as mean ± sd (n > 15). **P < 0.01 compared with the wild type.
(B) Trichome density of the main inflorescence stem, pedicels, and sepals of ProAlcA:MIR156f plants treated with 1% ethanol. Water was used as mock control. Inf. Stem1 represents the main inflorescence stem internode between the first two pedicels. Pedicel and sepal were from the first lower inflorescence. Data are given as mean ± sd (n > 15). **P < 0.01 compared with mock control.
(C) Trichome density of the main stem of ProSPL9:rSPL9-GR plants treated with 10 μM DEX. DMSO was used as mock control. 1st to 3rd represent the main stem internodes from bottom to top. Data are given as mean ± sd (n > 15). **P < 0.01 compared with mock control.
To further examine if SPLs were directly involved in suppressing trichome formation on the inflorescence stem and floral organs, we used an ethanol-inducible cassette of miR156 (ProAlcA:MIR156f) and a hormone-inducible cassette of SPL9, (ProSPL9:rSPL9-GR), in which the genomic fragment of SPL9 was translationally fused to the hormone binding domain of the glucocorticoid receptor. When ethanol was applied to ProAlcA:MIR156f plants, an increased number of stem trichomes was observed (Figure 2B). By contrast, treatment of ProSPL9:rSPL9-GR plants with the GR ligand of dexamethasone (DEX) resulted in reduced trichome formation on the stem (Figure 2C).
TCL1 and TRY Act Downstream of SPLs
Previous investigation showed that the single-repeat MYB transcription factors TCL1, TRY, and CPC are involved in repressing trichome development on the inflorescence stems and pedicels (Wang et al., 2007). In comparison with the tcl1-1 mutant, the Pro35S:MIR156f plants showed a similar, or even severe, phenotypic change regarding to the ectopic trichomes produced on the inflorescence (Figure 1B, Table 1). To understand the genetic interaction between SPLs and TCL1, we generated a double mutant of tcl1-1 Pro35S:MIM156. We found that tcl1-1 was epistatic to Pro35S:MIM156 as all the tcl1-1 Pro35S:MIM156 plants showed a similar trichome phenotype to tcl1-1 single mutant in terms of ectopic production of trichomes on the inflorescence stems and pedicels, while the Pro35S:MIM156 inflorescence was glabrous (Figure 3A, Table 1). These data suggest that SPLs act upstream of TCL1.
Table 1.
Trichome Production in Wild-Type, Mutant, and Transgenic Lines
| No. of Trichomes |
|||||||
| 1st Rosette Leaves | Internode |
||||||
| Genotypes | Cotyledon | 1st | 2nd | 3rd | 1st Pedicel | 1st Sepal | |
| Wild type | 0.0 ± 0.0 | 31.0 ± 1.7 | 234.0 ± 10.1b1 | 41.8 ± 20.4b1 | 1.2 ± 1.2b1 | 0.0 ± 0.0 | 6.3 ± 1.6b2 |
| tcl1-1 | 0.0 ± 0.0 | 33.1 ± 2.7 | 298.2 ± 16.6a1,b1 | 149.7 ± 20.1a1,b1 | 62.5 ± 15.1a1,b1 | 34.8 ± 5.9a1,b1 | 14.3 ± 3.0a1,b1 |
| Pro35S:MIM156 | 0.0 ± 0.0 | 27.1 ± 5.0 | 94.8 ± 8.2a1 | 2.4 ± 1.3a1 | 0.0 ± 0.0a1 | 0.0 ± 0.0 | 3.7 ± 2.1a2 |
| Pro35S:MIM156 tcl1-1 | 0.0 ± 0.0 | 28.3 ± 4.3 | 230.9 ± 15.0b1 | 86.3 ± 18.6a2,b1 | 35.1 ± 10.3a1,b1 | 15.6 ± 7.2a1,b1 | 7.2 ± 3.3b2 |
At least 10 plants for each line were analyzed. Values indicate mean ± sd.
aRelative to the corresponding wild-type line. a1P < 0.01, very significant difference; a2P < 0.05, significant difference.
bRelative to the corresponding Pro35S:MIM156 line. b1P < 0.01, very significant difference; b2P < 0.05, significant difference.
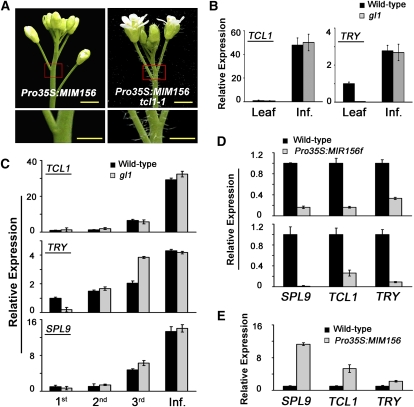
Figure 3.
SPLs Positively Regulate TCL1 and TRY Expression.
(A) Trichomes on Pro35S:MIM156 (left) and tcl1-1 Pro35S:MIM156 (right) inflorescence. The bottom panels represent the area marked by the red square in the top panels. Bars in top panels = 2 mm, and bars in bottom panels = 1 mm.
(B) Expression of TCL1 (left) and TRY (right) in seedling rosette leaves and inflorescences. Seedling (10-d-old) rosette leaves and inflorescences (Inf.) of the wild type and gl1 mutant were collected and analyzed by qRT-PCR. β-TUBULIN-2 was used as the internal standard. Error bars indicate sd of three technical replicates, and the results were consistent in three biological replicates.
(C) Expression of TCL1, TRY, and SPL9 in three successive internodes of the main stem and the inflorescence of wild-type and gl1 plants. Error bars indicate sd of three technical replicates, and the results were consistent in three biological replicates.
(D) Expression levels of SPL9, TCL1, and TRY and in the main stem (top) and developing inflorescence (bottom) of wild-type and Pro35S:MIR156f plants. Error bars indicate sd of three technical replicates, and the results were consistent in three biological replicates.
(E) Expression levels of SPL9, TCL1, and TRY in rosette leaves of 10-d-old wild-type and Pro35S:MIM156 plants. Error bars indicate sd of three technical replicates, and the results were consistent in three biological replicates.
We then examined the expression of TCL1, TRY, and CPC genes in wild-type and gl1 mutant plants. Quantitative real-time RT-PCR (qRT-PCR) analyses showed that in rosette leaves, TRY and CPC were clearly expressed in the wild type, but their expression levels were drastically reduced in the gl1 mutant, whereas the TCL1 transcript level was low in either the wild-type or the gl1 mutant seedling rosette leaves (Figure 3B). In inflorescences, however, both TCL1 and TRY were highly upregulated and the transcript levels were comparable between the wild type and the gl1 mutant; by contrast, the CPC expression level decreased in the wild-type inflorescences compared with seedling rosette leaves (see Supplemental Figure 1A online). These data suggest that TRY and TCL1 expression in inflorescences is activated and maintained by a pathway that is independent of GL1.
Since the trichome density was inversely correlated to the progress of floral transition, we compared the TCL1, TRY, CPC, and SPL9 expression levels in the main stem internodes 1 to 3 (bottom to top) and inflorescence (Figures 1A and B). SPL9 was chosen here as a representative of miR156-targeted SPL genes. Transcript levels of both TCL1 and TRY were gradually elevated from internode 1 to internode 3, and the highest level was detected from the inflorescence (Figure 3C). SPL9 exhibited a similar change of spatial expression as did TCL1 and TRY (Figure 3C). CPC, on the contrary, showed an opposite expression pattern in the stem as its transcript level was decreasing from internode 1 to 3, although the expression in inflorescence was evident (see Supplemental Figure 1B online). The gl1 mutation did not affect TCL1 expression in the main stem and the inflorescence, but it did cause an obvious reduction of TRY transcript level in the first internode of the main stem (Figure 3C). Therefore, while TCL1 expression was unrelated to GL1, TRY expression was still dependent on GL1 at the early reproductive stage, as manifested by the first internode, and this dependence was then lost along with plant development.
Because of the similar tendency of SPL9, TCL1, and TRY expression in the stem and inflorescence, we wondered whether SPL9, and possibly other SPLs too, played a role in elevating the expression level of TCL1 and TRY during floral transition. As described above, stem trichome numbers were inversely correlated to expression levels of SPL genes in transgenic plants. qRT-PCR analyses revealed that expression of both TRY and TCL1 genes was indeed downregulated in Pro35S:MIR156f plants (Figure 3D) but upregulated in ProSPL13:rSPL13, ProSPL10:rSPL10, ProSPL9:rSPL9, Pro35S:rSPL3 (see Supplemental Figure 2 online), and Pro35S:MIM156 plants (Figure 3E). However, accumulation of CPC transcripts was slightly enhanced rather than downregulated in Pro35S:MIR156f plants, and there was little change of its expression in Pro35S:MIM156 plants (see Supplemental Figure 1C online). Taken together, both genetic and gene expression data support that TCL1 and TRY, but not CPC, act downstream of SPLs in repressing trichome production in the stem and inflorescence.
SPL9 Directly Targets TCL1 and TRY Promoters
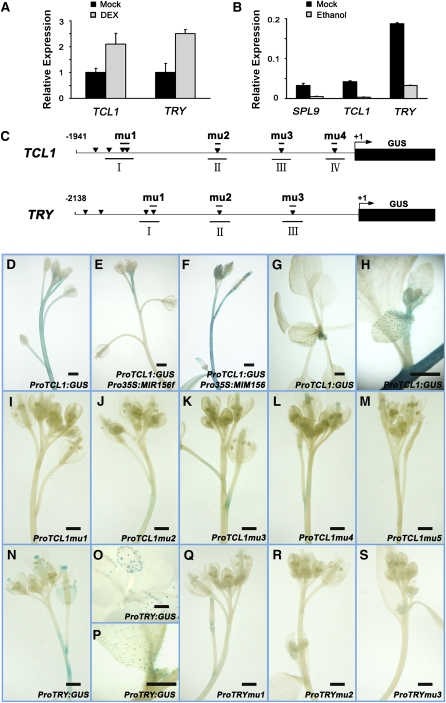
To know whether TCL1 and TRY were directly regulated by SPL transcription factors, we tested the activity of SPL9 in activating TCL1 and TRY gene expression. One-week-old ProSPL9:rSPL9-GR seedling rosette leaves were sprayed with 10 μM DEX to allow of the translocation of the rSPL9-GR fusion protein into the nucleus. A significant increase of TRY and TCL1 transcript abundance was observed after 5 h (Figure 4A), suggesting that SPL9 rapidly activated the transcription of TRY and TCL1. By contrast, downregulation of SPLs by a 6-h ethanol treatment of the 1-month-old ProAlcA:MIR156 plants almost blocked the TRY and TCL1 expression in inflorescences (Figure 4B).
Figure 4.
TCL1 and TRY as Direct Targets of SPL9.
(A) Expression of TCL1 and TRY in rosette leaves of 10-d-old ProSPL9:rSPL9-GR seedlings after a 5-h DEX treatment or a mock treatment with DMSO. Error bars indicate sd of three technical replicates, and the results were consistent in three biological replicates.
(B) Expression of SPL9, TCL1, and TRY in ProAlcA:MIR156f inflorescences after a 6-h ethanol treatment or a mock treatment with water. Error bars indicate sd of three technical replicates, and the results were consistent in three biological replicates.
(C) Schematic diagram of ProTCL1:GUS (top) and ProTRY:GUS (bottom) constructs. Top diagram: A 1941-bp 5′-upstream fragment of TCL1 was fused to GUS. Bottom diagram: A 2138-bp 5′-upstream fragment of TRY was fused to GUS. In both diagrams, triangles indicate GTAC boxes in the promoter, mu1 to mu4 represent the GTAC motifs mutated, and I to IV represent the DNA fragments used for ChIP analyses.
(D) to (F) GUS staining of inflorescences showing ProTCL1:GUS expression in wild-type (D), Pro35S:MIR156f (E), and Pro35S:MIM156 (F) backgrounds.
(G) and (H) GUS staining showing ProTCL1:GUS expression in trichomes of rosette (G) and cauline (H) leaves in the wild-type background.
(I) to (M) GUS staining of inflorescences expressing mutated versions of ProTCL1:GUS, mu1 to mu5, respectively. Note that GUS activities were greatly reduced in mu1 to mu4 and to an undetectable level in mu5 inflorescences.
(N) GUS staining of ProTRY:GUS inflorescences.
(O) and (P) GUS staining of ProTRY:GUS activities in trichomes of rosette (O) and cauline (P) leaves in the wild-type background.
(Q) to (S) GUS staining of inflorescences expressing mutated versions of ProTRY:GUS, mu1 to mu3, respectively.
Bars = 2 mm.
To view the spatial expression pattern of TCL1, we generated a reporter line by fusing the 2-kb upstream regulatory fragment of TCL1 to the coding sequence of β-glucuronidase (GUS) (ProTCL1:GUS; Figure 4C). Most of the ProTCL1:GUS transgenic plants (17/20) exhibited a strong GUS expression in the inflorescence (Figure 4D). The GUS staining was increasingly intensive in the main inflorescence stem from bottom to top, which was consistent with the expression pattern of TCL1 revealed by qRT-PCR (Figure 3C). GUS staining became paler in Pro35S:MIR156f and stronger in Pro35S:MIM156 plants, respectively (Figures 3E and 3F). In addition, we observed trichome-specific distribution of GUS activity in young rosette leaves (Figure 4G) and in cauline leaves of the high-expression lines (Figure 4H), a pattern different from the universal expression of TCL1 in the inflorescence stem epidermis, possibly due to the higher level of SPLs in the latter.
A consensus sequence, GTAC, has been identified as the core binding motif of SPLs (Klein et al., 1996; Birkenbihl et al., 2005; Kropat et al., 2005; Liang et al., 2008). There are seven GTAC motifs within the 2-kb upstream fragment of the TCL1 gene (Figure 4C). To determine the role of these GTAC motifs, we constructed four mutant versions of the TCL1 promoter (ProTCL1mu1:GUS, ProTCL1mu2:GUS, ProTCL1mu3:GUS, and ProTCL1mu4:GUS), in which the GTAC motif was changed to AAGA, respectively (Figure 4C). In addition, we made a reporter line (ProTCL1mu5:GUS) with five motifs simultaneously mutated. ProTCL1mu1:GUS, ProTCL1mu2:GUS, ProTCL1mu3:GUS, and ProTCL1mu4:GUS plants showed a reduction of GUS activity in the inflorescences to a different extend in comparison with ProTCL1:GUS (Figures 4I to 4L), whereas GUS staining was not observed in ProTCL1mu5:GUS plants (Figure 4M), indicating that these GTAC motifs are indispensable for the TCL1 promoter activity in the inflorescences.
We also scanned the TRY promoter for SPL binding site (GTAC) and constructed three single mutant versions (ProTRYmu1:GUS, ProTRYmu2:GUS, and ProTRYmu3:GUS), using the same method as for TCL1 promoter (Figure 4C). Most of the ProTRY:GUS transgenic plants showed a clear GUS staining in the inflorescence stem, floral organs (Figure 4N), and trichomes of rosette and cauline leaves (Figures 4O and 4P), whereas single mutations of the TRY promoter dramatically decrease GUS activity in the stem and flowers (Figures 4Q to 4S), indicating that the SPL binding sites are also important for TRY expression in these organs.
Because TCL1 expression level in rosette leaves were rather low (Figure 3B), GUS activities of ProTCL1:GUS were then detected in trichome cells of cauline leaves of high-expression lines (see Supplemental Figure 3A online). Mutation of single GTAC motifs in the TCL1 promoter did not severely affect its activity and trichome specificity; mutation of five GTAC cis-elements, however, decreased the promoter activity (see Supplemental Figures 3B to 3F online). For TRY promoter, mutation of single GTAC motifs did not decrease its activity or trichome specificity either (see Supplemental Figures 3G to 3J online). These data demonstrate that the cis-elements recognized by SPL transcription factors are less important for TCL1 and TRY expression in leaves than in inflorescences.
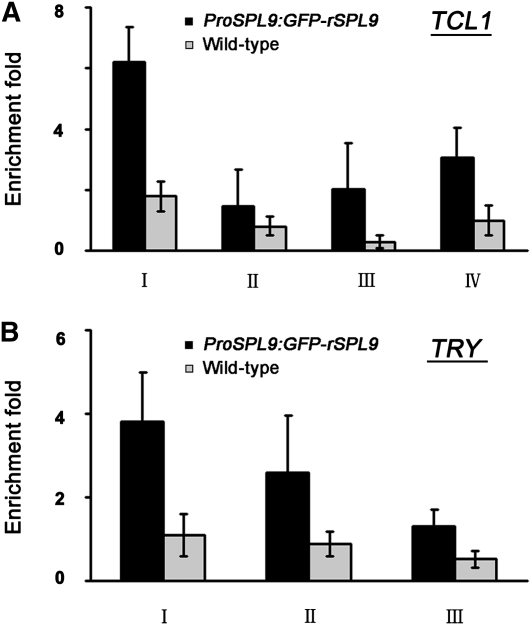
To further demonstrate that SPL9 could bind directly to the promoters of TCL1 and TRY, we performed chromatin immunoprecipitation (ChIP) assay. We used transgenic lines in which a miR156-resistant form of SPL9 was translationally fused to the green fluorescent protein (GFP) coding region (ProSPL9:GFP-rSPL9). The ProSPL9:GFP-rSPL9 plants had a similar phenotype as Pro35S:MIM156 with fewer trichomes on the stem internodes (Figure 2A) compared with the wild type. We chose four fragments spanning the GTAC motifs from TCL1 promoter (Figure 3C). Fragments I, III, and IV were enriched after ChIP with GFP antibody in the transgenic ProSPL9:GFP-rSPL9 compared with the wild-type plants (Figure 5A). We also detected the enrichment of the TRY promoter fragments (Figures 4C and 5B). These data further support that TCL1 and TRY are direct targets of SPL9.
Figure 5.
Binding of SPL9 to TCL1 and TRY Promoters.
ChIP enrichment of TCL1 (A) and TRY (B) promoter regions bound by GFP-SPL9. Rosette leaves of the 3-week-old ProSPL9:GFP-rSPL9 and wild-type plants were used. The DNA fragments were quantitatively analyzed by PCR, and the β-TUBULIN-2 promoter was used as a reference. Error bars indicate sd of three quantitative PCR replicates, and the results were consistent in three precipitations.
Expression of SPL9, TCL1, and TRY Is Not Dependent on GISs
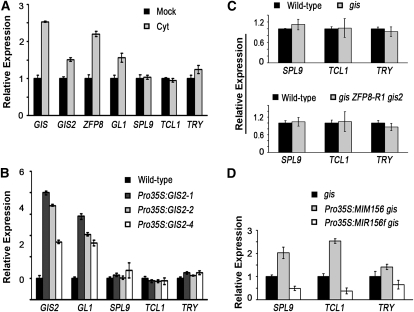
The putative C2H2 transcription factors GIS, GIS2, and ZFP8 mediate another a phytohormone-dependent pathway in which GAs and cytokinins induce trichome formation on the stem and inflorescence. It has been proposed that these proteins are involved in positive regulation of GL1 (Gan et al., 2006, 2007). We tested whether there is a close relation between GIS-dependent trichome production and SPL-dependent regulation of the negative regulators of trichome initiation. First, we treated the wild-type plants with cytokinin 6-benzylaminopurine (6-BA), which resulted in a clear increase in the expression levels of GIS, GIS2, and ZFP8, although the induction magnitudes varied (Figure 6A). The GL1 expression level was also elevated in the 6-BA–treated plants (Figure 6A), consistent with the previous report (Gan et al., 2007). If SPL9, TCL1, and TRY were involved in this hormone-induced trichome pathway, we would expect a repression of these genes in the 6-BA–treated plants. However, none of them were downregulated after the hormone treatment (Figure 6A). We also treated the plants with GA3 and a similar induction pattern was observed, although the induction effect was not as great as that by 6-BA in our experimental conditions.
Figure 6.
Expression of SPL9, TCL1, and TRY in Plants with Altered Levels of GISs.
(A) qRT-PCR analysis of the expression of GISs, SPL9, and trichome regulatory genes (GL1, TCL1, and TRY) in inflorescences of wild-type plants after 6-BA treatment. Error bars indicate sd of three technical replicates, and the results were consistent in two biological replicates. Cyt, 6-benzylaminopurine.
(B) Expression of SPL9, TCL1, and TRY in inflorescences of Pro35S:GIS2 plants. Error bars indicate sd of three biological replicates.
(C) Expression of SPL9, TCL1, and TRY in inflorescences of gis single and gis ZFP8-R1 gis2 triple mutant plants. Error bars indicate sd of three biological replicates.
(D) Expression of SPL9, TCL1, and TRY in the first pair of rosette leaves of Pro35S:MIM156 and Pro35S:MIR156f in the gis mutant background. Error bars indicate sd of three biological replicates.
Because overexpression of GIS or GIS2 produced a similar phenotype as did the hormone treatments with regard to ectopic trichome production in the stem and inflorescence, we analyzed the changes of gene expression in Pro35S:GIS2 plants. We found that, while the GL1 transcript level highly correlated with that of GIS2, as previously reported (Gan et al., 2007), the transcript levels of SPL9, TCL1, and TRY were not affected by GIS2 overexpression (Figure 6B). Furthermore, the SPL9, TCL1, and TRY transcript levels were similar in the wild type, gis, and the gis zfp8-R1 gis2 (Gan et al., 2006, 2007) triple mutant plants (Figure 6C). To further examine whether the regulation of TCL1 and TRY by SPL transcription factors requires GIS, we introduced Pro35S:MIM156 and Pro35S:MIR156f into the gis mutant, respectively. Following the changed levels of SPL9, the TCL1 and TRY transcript levels were elevated in the former and decreased in the latter (Figure 6D). Consistent with the change of transcript levels, trichome numbers on the first mainstem internode of the gis Pro35S:MIM156 plants were decreased significantly, and overexpression of MIR156f in gis mutant led to ectopic trichomes on inflorescences (see Supplemental Figure 4 and Supplemental Table 1 online). Thus, the regulation of TCL1 and TRY by SPLs was not impaired by the gis mutation. These data suggest that the GIS-dependent pathway does not play a major role in regulating SPL and the trichome negative regulator genes of TCL1 and TRY.
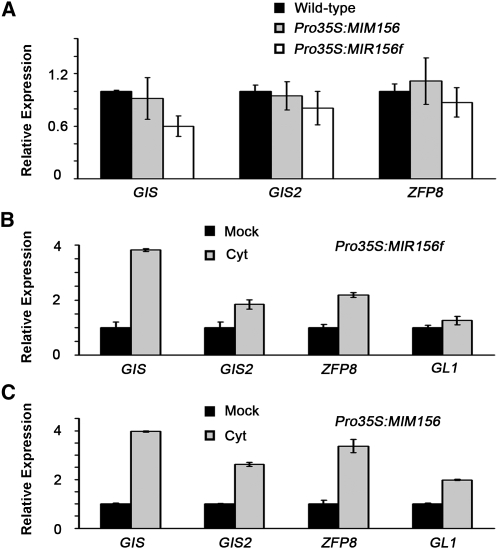
We then examined the effects of altered levels of SPLs on GIS expression. In plants with increased levels of SPLs due to Pro35S:MIM156 expression, expression levels of the GIS genes were largely unchanged in comparison with those in the wild-type plants. On the other hand, in Pro35S:MIR156f plants in which SPLs were repressed and trichomes were formed on the stem and inflorescence, expression of the GIS genes was either unchanged or even slightly downregulated (Figure 7A). Finally, we applied 6-BA to the Pro35S:MIM156 and Pro35S:MIR156f plants. We found that GIS, GIS2, and ZFP8 genes were clearly upregulated in both Pro35S:MIR156f and Pro35S:MIM156 plants after treatments (Figures 7B and 7C), as observed with the wild-type plants (Figure 6A). GL1 gene was also upregulated by 6-BA treatment of the Pro35S:MIM156 plants. In Pro35S:MIR156f plants, GL1 expression was induced only slightly (Figure 7B), possibly due to a high basal level. Thus, altered levels of SPLs did not interfere with 6-BA induction of GISs genes, and GL1 gene induction was not affected, at least not in plants with an elevated level of SPLs.
Figure 7.
Expression of GIS, GIS2, and ZFP8 in Pro35S:MIM156 and Pro35S:MIR156f Plants.
(A) Expression of GIS, GIS2, and ZFP8 in inflorescences of the Pro35S:MIM156 and Pro35S:MIR156f plants. Error bars indicate sd of three biological replicates.
(B) and (C) Expression of GIS, GIS2, and ZFP8 in inflorescences of Pro35S:MIR156f (B) and Pro35S:MIM156 (C) plants after 6-BA treatment. Error bars indicate sd of three technical replicates, and the results were consistent in two biological replicates. Cyt, 6-benzylaminopurine.
Together, these data demonstrate that while the GIS-dependent pathway promotes trichome initiation through the positive regulator gene GL1, the miR156-targeted SPLs repress trichome formation by upregulating the negative regulator genes TCL1 and TRY. It is likely that the two pathways act in parallel in determining trichome formation in the stem and inflorescences.
DISCUSSION
The epidermis is the outermost layer covering plant aerial organs. During colonization of land, plants evolved many typical epidermal features. One of the specialized cell types of aerial epidermis is the trichome, which generally forms a barrier to protect the plant. However, the excess of trichomes on floral organs may offer a few disadvantages to cleistogamous plants, in which self-pollination occurs before flower opening. It was reported that the rate of natural self-pollination was negatively correlated with the distance from the stigma to anther (Shimizu and Ichimura, 2002). This could be an explanation why the carpel and stamen of such cleistogamous plants as Arabidopsis and Pisum sativum do not bear branched trichomes. The conical-papillate cells form another type of epidermal structures, which are found only on petals in the majority of entomophilous (insect-pollinated) flowers, and they allow the pollinators to grip flowers (Martin and Glover, 2007). From these points of view, it is reasonable to assume that plants couple epidermal cell differentiation to floral transition.
There could be alternate ways of transducing phase transition signals to trichome repression in the stem and inflorescence at the gene expression level: developmental factors could downregulate GL1 or other positive regulator genes of trichome initiation, or the developmental factors could upregulate negative regulators of the trichome pathway. Previous identification of the TCL1 gene and phenotypic analysis of the tcl1-1 try double mutant plants supported the latter possibility: both negative regulators of TCL1 and TRY are required for progressive repression of trichome formation in the reproductive stage (Wang et al., 2007, 2008b).
Recently, Shikata et al. (2009) reported reduced trichome initiation on cauline leaves and floral organs in plants expressing chimeric repressors of SPL10, SPL11, and SPL12. In this investigation, we demonstrated that it is the miR156-regulated SPLs that activate the TCL1 and TRY expression. Our results establish a direct link between transition to flowering stage and trichome patterning and explain the gradual loss of trichome formation on the stem internodes and floral organs of Arabidopsis. First, expression of miR156/SPLs is age regulated and tightly coupled to the transition from the vegetative to the reproductive stage. The miR156 levels decrease as the plant ages. In short days, miR156 levels were the highest in young seedling rosette leaves and subsequently declined over several weeks. On the contrary, the expression levels of miR156 target genes, including SPL9, were inversely related to miR156 expression with a gradual increase during reproductive stage (Schmid et al., 2005; Wang et al., 2009). This temporal expression pattern fits the timing of trichome repression during plant development. Secondly, the miR156-regulated SPL transcription factors are master regulators of Arabidopsis phase transition and one of the endogenous flowering pathways. They directly activate expression of transcription factor genes of LFY, FUL, AP1, and SOC1, which control the timing of flowering (Wang et al., 2009; Yamaguchi et al., 2009). Our transgenic plant and ChIP analyses data revealed that SPL9, and likely other SPLs, positively regulates TCL1 and TRY gene expression through binding to and activating their promoters. Thus, the key factors of promoting flowering also directly specify certain epidermal features of inflorescence stems and floral organs. Nevertheless, the results presented here do not exclude the possibility that other flowering factors also participate in controlling trichome formation in specific floral organs.
In the lateral inhibition model proposed for rosette leaf trichome pattering, the negative regulator genes, such as TRY, are activated in trichome cells by the GL1-GL3-TTG1 protein complex and moved to neighboring cells to repress trichome initiation (Schellmann et al., 2002; Digiuni et al., 2008). Consistent with GL1, the TRY expression in rosette leaves is also trichome specific (Schellmann et al., 2002; Esch et al., 2003). However, this model requires the presence of trichomes as a precondition to prevent trichome initiation and does not explain the glabrous phenotype of such aerial organs as petal, pedicel, and upper internodes of inflorescence stem. Activation of TRY and TCL1 by age-related SPLs provides a solution to this paradox. At the basal part of stem, TRY expression is still influenced by GL1, but this effect becomes negligible along with the increasing levels of SPLs during shoot growth. In agreement with the expression patterns of SPL genes (Ito et al., 2004), promoter activities of TRY and TCL1 are widely distributed in at least certain regions of epidermal cells of the inflorescence stems and floral organs, with higher activities in the upper internodes despite the absence of trichomes. We propose that, after plants entering into the reproductive stage, the progressive elevation of SPL levels causes a general activation of TRY and TCL1 genes in stem and floral epidermal cells, resulting in increasingly higher ratios of negative (such as TCL1 and TRY) to positive (mainly GL1) regulators and the gradual lose of trichome formation.
Interestingly, overexpression of SPL genes promoted, rather than inhibited, trichome formation on abaxial surface of leaves, likely due to an accelerated transition to adult phase (Cardon et al., 1997; Wang et al., 2009; Wu et al., 2009). Thus, suppression of trichome formation by SPLs seems not to be universal, and the factors controlling trichome initiation on leaf abxial surfaces remain to be identified. In addition to TRY and TCL1, Arabidopsis has at least four more single-repeat R3 MYB factors that function as negative regulators of trichome development. CPC, for example, is involved in suppression of trichome formation and flavonoid biosynthesis, in addition to its role in specifying root hairs (Wada et al., 1997; Broun, 2005; Wang et al., 2008b; Zhu et al., 2009). Our data presented herein revealed that CPC is progressively downregulated in stems during plant development, and its expression is not regulated by SPLs. Therefore, these negative regulator genes must be differentially regulated in different organs during development, emphasizing that the regulatory network controlling trichome patterning requires further investigation.
Previous investigations demonstrated that promotion of trichome formation by phytohomones of GAs and cytokinins is mediated by GIS and related transcription factors of GIS2 and ZFP8. Unlike SPLs that activate the trichome negative regulator genes, the GIS-dependent pathway upregulates the positive regulator gene GL1 (Perazza et al., 1998; Gan et al., 2006, 2007). Our analysis of gis mutants and the transgenic plants with altered SPL levels suggested that GIS factors were not involved in the SPL-dependent regulation of trichome negative regulator genes of TCL1 and TRY, neither did the SPLs play a major role in regulating GIS and related genes. Collectively, data of previous reports and data presented herein suggest that the miR156/SPL- and the GIS-dependent pathways function in parallel in regulating trichome regulator complex in the main stem and inflorescences. However, at present, a possible interaction of the two pathways at an early step cannot be excluded, since both are involved regulating phase transition, shoot maturation, and flowering, besides trichome initiation. Further genetic and biochemical investigations are needed to answer how the two pathways are integrated into plant development programming.
In addition to the endogenous hormone-mediated flowering signals, environmental conditions also influence trichome development. It was reported that the trichome density is closely related to temperature, light, soil moisture, wounding, and other factors in various plant species (Wellso and Hoxie, 1982; Gianfagna et al., 1992; Pérez-Estrada et al., 2000; Yoshida et al., 2009). It will be interesting to examine the regulation of trichome production by these exogenous and endogenous cues and their crosstalk with the miR156/SPLs pathway.
METHODS
Plant Materials and Primers
Plants of Arabidopsis thaliana, ecotype Columbia (Columbia-0) were grown at 22°C in long days (16 h light/8 h dark). The trichome numbers were counted, and data were given as mean ± sd and were analyzed by t test.
Primers used in PCR are listed in Supplemental Table 2 online.
Constructs and Plant Transformation
To make ProTCL1:GUS and ProTRY:GUS constructs, the promoters of TCL1 and TRY (~2 kb) were PCR amplified using Pyrobest DNA polymerase (TaKaRa) and individually fused into the GUS coding region. Promoter mutations were created by two-round PCR, using primers as shown in Supplemental Table 2 online.
For ProAlcA:MIR156f, a genomic fragment of MIR156f was cloned between AlcA promoter and OCS terminator. The resulting fragment was inserted into pMLBart vector, which harbors a Pro35S:AlcR cassette (Roslan et al., 2001).
Constructs were delivered into Agrobacterium tumefaciens strain GV3101 (pMP90). Arabidopsis plants were transformed using a flower-dip method (Clough and Bent, 1998). Transgenic seedlings were selected with 40 μg/mL kanamycin (for pCAMBIA2300) on plates or 0.1% glufosinate (BASTA, for pCAMBIA3300) in soil. For each construct at least 25 T1 seedlings were analyzed.
Plant Treatments
For DEX induction, 1-week-old ProSPL9:rSPL9-GR seedling rosette leaves were sprayed twice a week with 10 μM DEX (Sigma-Aldrich) or DMSO (mock control) solutions until the plants started bolting. For ethanol treatment, ProAlcA:MIR156f plants were grown in soil until flowered. The plants were then sprayed with 1% ethanol or water (mock control) twice a week.
For measuring the effect of transient downregulation of miR156-regulated SPLs on TCL1 and TRY expression, ProAlcA:MIR156f plants (n > 18) were grown in soil until the inflorescence stem had reached a length of 2 to 3 cm. The plants were then sprayed with 1% ethanol or water (mock control), and the shoots were harvested after 6 h. For transient upregulation of SPL9, 1-week-old ProSPL9:rSPL9-GR seedling rosette leaves were sprayed with either 10 μM DEX or 10 μM DMSO (control). The plants were harvested 5 h after treatments, followed by RNA extraction.
Hormone treatments were performed as described (Gan et al., 2007). Briefly, the cytokinin 6-BA (Sigma-Aldrich) and the gibberellin GA3 (Sigma-Aldrich) were used, and the plants were grown in pots at 22°C in long-day conditions (16 h light/8 h dark). Until the first five to six leaves had emerged, the pots were soaked in 6-BA (50 μM) or GA3 (100 μM) for 4 h, respectively, once every 2 d. After shoots had reached 6 to 8 cm in length, the plants were treated for another 4 h with the same hormone, and the inflorescences were harvested for RNA extraction.
Expression Analyses
Total RNAs were extracted with Trizol reagent (Invitrogen), from which 1 μg was used for reverse transcription in a 20-μL reaction system with the RNA PCR (AMV) kit (Promega). Quantitative real-time RT-PCR was performed with SYBR-Green PCR Mastermix (Takara), and amplification was real-time monitored on a Mastercycler ep RealPlex2 (Eppendorf). The level of transcript abundance relative to reference gene (termed ΔCT) was determined according to the function ΔCT = CT (test gene) − CT (reference gene). To compare untreated and treated expression levels, the function ΔΔCT was first determined using the equation ΔΔ CT = Δ CT (treatment) − Δ CT (control) where control represented mock-treated plants. The induction ratio of treatment/control was then calculated by the equation 2−ΔΔ CT. The primer sequences are listed in Supplemental Table 2 online.
GUS activity was detected by staining plant tissues at 37°C in 0.1 M sodium phosphate buffer, pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 0.5 mM potassium ferrocyanide, 0.5 mM potassium ferricyanide, and 0.5 mg/mL X-glucuronide. Subsequent washes with 70% ethanol were performed to remove chlorophyll and enhance contrast. At least six independent transformants for each constructs were selected.
ChIP
ChIP experiments were performed according to published protocols (Wang et al., 2002, 2009). About 3 g tissues of the 3-week-old ProSPL9:GFP-rSPL9 plants were harvested. After fixation, the material was washed and ground under liquid nitrogen, the resultant powder was resuspended in extraction buffer (0.4 M sucrose, 10 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 5 mM mercaptoethanol, 0.1 mM PMSF, and 1× protease inhibitor [Roche]) and lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% deoxycholate sodium, and 0.1% SDS), successively, followed by sonification (Output 3, 6 × 10 s). One-third of the solution was saved as input total DNA without precipitation. An anti-GFP antibody (Abmart) was added to another one-third and the remaining one-third was precipitated in parallel with anti-HA (Sigma-Aldrich) as a negative control. The resulting DNA samples were purified using a PCR purification kit (Qiagen). The relative concentrations of the DNA fragments were analyzed by quantitative PCR, using the β-TUBULIN2 gene promoter as the reference. Relative enrichment fold was calculated first by normalizing the amount of a target DNA fragment against the TUBULIN2 promoter fragment (1153 to 1309 bp upstream of the translation start codon) and then by normalizing the value for anti-GFP against the value for anti-HA.
Accession Numbers
Arabidopsis Genome Initiative gene identifiers are as follows: SPL9 (At2g42200), SPL3 (At2g33810), SPL10 (AT1G27370), SPL13 (At5g50670), MIR156f (AT5G26147), TFL1 (At5g03840), FUL (At5g60910), SOC1 (At2g45660), LFY (At5g61850), FT (At1g65480), GL1 (AT3G27920), TCL1 (At2g30432), TRY (At5G53200), GIS (At3g58070), GIS2 (At5g06650), ZFP8 (At2g41940), and β-TUBULIN-2 (At5g62690). Mutants and transgenic plants of gl1 (SALK_039478), try (SALK_029760), tcl1-1 (SALK_055460), Pro35S:MIR156f, Pro35S:MIM156, soc1-6 (SALK_138131), ful-7 (SALK_033647), lfy-12, ft-10, tfl1-1, ProSPL13:rSPL13, ProSPL10:rSPL10, Pro35S:rSPL3, ProSPL9:rSPL9, ProSPL9:rSPL9-GR, ProSPL9:GFP-rSPL9, gis (GABI _423G08), gis2 (NASC_N119489), and gis zfp-R1 gis2 were as described previously (Wang et al., 2004, 2007, 2008a, 2009; Gan et al., 2006, 2007).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Expression Pattern of CPC in Wild-Type, gl1, and Transgenic Plants.
Supplemental Figure 2. Overexpression of SPLs Upregulates TCL1 and TRY Expression.
Supplemental Figure 3. GUS Staining of Leaves of the ProTCL1:GUS and ProTRY:GUS Plants.
Supplemental Figure 4. Trichome Production on Inflorescences of gis, gis Pro35S:MIR156f, and gis Pro35S:MIM156 Plants.
Supplemental Table 1. Trichome Numbers of the Main Stem of gis and gis Pro35S:MIM156 Plants.
Supplemental Table 2. Oligonucleotide Primers.
Acknowledgments
We thank the ABRC for Arabidopsis T-DNA insertion lines, Jia-Wei Wang, Detlef Weigel, and Yinbo Gan for sharing materials and for their helpful discussions. This research was supported by grants from the National Natural Science Foundation of China (30630008 and 30721061), the State Key Basic Research Program of China (2010CB126004), and the Chinese Academy of Sciences (KSCX2-YW-N-057).
References
- Baurle I., Dean C. (2006). The timing of developmental transitions in plants. Cell 125: 655–664 [DOI] [PubMed] [Google Scholar]
- Benlloch R., Berbel A., Serrano-Mislata A., Madueno F. (2007). Floral initiation and inflorescence architecture: A comparative view. Ann. Bot. (Lond.) 100: 659–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl R.P., Jach G., Saedler H., Huijser P. (2005). Functional dissection of the plant-specific SBP-domain: Overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352: 585–596 [DOI] [PubMed] [Google Scholar]
- Bouyer D., Geier F., Kragler F., Schnittger A., Pesch M., Wester K., Balkunde R., Timmer J., Fleck C., Hülskamp M. (2008). Two-dimensional patterning by a trapping/depletion mechanism: The role of TTG1 and GL3 in Arabidopsis trichome formation. PLoS Biol. 6: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D., Ratcliffe O., Vincent C., Carpenter R., Coen E. (1997). Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83 [DOI] [PubMed] [Google Scholar]
- Broun P. (2005). Transcriptional control of flavonoid biosynthesis: A complex network of conserved regulators involved in multiple aspects of differentiation in Arabidopsis. Curr. Opin. Plant Biol. 8: 272–279 [DOI] [PubMed] [Google Scholar]
- Cardon G.H., Hohmann S., Nettesheim K., Saedler H., Huijser P. (1997). Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: A novel gene involved in the floral transition. Plant J. 12: 367–377 [DOI] [PubMed] [Google Scholar]
- Chien J.C., Sussex I.M. (1996). Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 111: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Digiuni S., Schellmann S., Geier F., Greese B., Pesch M., Wester K., Dartan B., Mach V., Srinivas B.P., Timmer J., Fleck C., Hulskamp M. (2008). A competitive complex formation mechanism underlies trichome patterning on Arabidopsis leaves. Mol. Syst. Biol. 4: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch J.J., Chen M., Sanders M., Hillestad M., Ndkium S., Idelkope B., Neizer J., Marks M.D. (2003). A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130: 5885–5894 [DOI] [PubMed] [Google Scholar]
- Esch J.J., Chen M.A., Hillestad M., Marks M.D. (2004). Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J. 40: 860–869 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J.M., Valli A., Todesco M., Mateos I., Puga M.I., Rubio-Somoza I., Leyva A., Weigel D., Garcia J.A., Paz-Ares J. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Galway M.E., Masucci J.D., Lloyd A.M., Walbot V., Davis R.W., Schiefelbein J.W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166: 740–754 [DOI] [PubMed] [Google Scholar]
- Gan Y., Kumimoto R., Liu C., Ratcliffe O., Yu H., Broun P. (2006). GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell 18: 1383–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y., Liu C., Yu H., Broun P. (2007). Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134: 2073–2081 [DOI] [PubMed] [Google Scholar]
- Gandikota M., Birkenbihl R.P., Hohmann S., Cardon G.H., Saedler H., Huijser P. (2007). The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49: 683–693 [DOI] [PubMed] [Google Scholar]
- Gianfagna T.J., Carter C.D., Sacalis J.N. (1992). Temperature and photoperiod influence trichome density and sesquiterpene content of Lycopersicon hirsutum f. hirsutum. Plant Physiol. 100: 1403–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A.Y., Zhu Q.H., Gu X., Ge S., Yang J., Luo J. (2008). Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 418: 1–8 [DOI] [PubMed] [Google Scholar]
- He Y., Amasino R.M. (2005). Role of chromatin modification in flowering-time control. Trends Plant Sci. 10: 30–35 [DOI] [PubMed] [Google Scholar]
- Hülskamp M., Schnittger A., Folkers U. (1999). Pattern formation and cell differentiation: Trichomes in Arabidopsis as a genetic model system. Int. Rev. Cytol. 186: 147–178 [DOI] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Ito T., Wellmer F., Yu H., Das P., Ito N., Alves-Ferreira M., Riechmann J.L., Meyerowitz E.M. (2004). The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430: 356–360 [DOI] [PubMed] [Google Scholar]
- Johnson H.B. (1975). Plant pubescence - Ecological perspective. Bot. Rev. 41: 233–258 [Google Scholar]
- Kardailsky I., Shukla V.K., Ahn J.H., Dagenais N., Christensen S.K., Nguyen J.T., Chory J., Harrison M.J., Weigel D. (1999). Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Kirik V., Simon M., Huelskamp M., Schiefelbein J. (2004a). The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268: 506–513 [DOI] [PubMed] [Google Scholar]
- Kirik V., Simon M., Wester K., Schiefelbein J., Hülskamp M. (2004b). ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 55: 389–398 [DOI] [PubMed] [Google Scholar]
- Klein J., Saedler H., Huijser P. (1996). A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 250: 7–16 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Kaya H., Goto K., Iwabuchi M., Araki T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286: 1960–1962 [DOI] [PubMed] [Google Scholar]
- Komeda Y. (2004). Genetic regulation of time to flower in Arabidopsis thaliana. Annu. Rev. Plant Biol. 55: 521–535 [DOI] [PubMed] [Google Scholar]
- Kropat J., Tottey S., Birkenbihl R.P., Depege N., Huijser P., Merchant S. (2005). A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl. Acad. Sci. USA 102: 18730–18735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J.C., Brown M.L., Schiefelbein J. (2003). How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annu. Rev. Plant Biol. 54: 403–430 [DOI] [PubMed] [Google Scholar]
- Liang X., Nazarenus T.J., Stone J.M. (2008). Identification of a consensus DNA-binding site for the Arabidopsis thaliana SBP domain transcription factor, AtSPL14, and binding kinetics by surface plasmon resonance. Biochemistry 47: 3645–3653 [DOI] [PubMed] [Google Scholar]
- Litt A., Irish V.F. (2003). Duplication and diversification in the APETALA1/FRUITFULL floral homeotic gene lineage: Implications for the evolution of floral development. Genetics 165: 821–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks M.D. (1997). Molecular genetic analysis of trichome development in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 137–163 [DOI] [PubMed] [Google Scholar]
- Martin C., Glover B.J. (2007). Functional aspects of cell patterning in aerial epidermis. Curr. Opin. Plant Biol. 10: 70–82 [DOI] [PubMed] [Google Scholar]
- Masucci J.D., Rerie W.G., Foreman D.R., Zhang M., Galway M.E., Marks M.D., Schiefelbein J.W. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Mauricio R., Rausher M.D. (1997). Experimental manipulation of putative selective agents provides evidence for the role of natural enemies in the evolution of plant defense. Evolution 51: 1435–1444 [DOI] [PubMed] [Google Scholar]
- Morohashi K., Zhao M., Yang M., Read B., Lloyd A., Lamb R., Grotewold E. (2007). Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol. 145: 736–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima S., Murata M., Sakamoto W., Ogura Y., Motoyoshi F. (1997). Cloning and molecular analysis of the Arabidopsis gene Terminal Flower 1. Mol. Gen. Genet. 254: 186–194 [DOI] [PubMed] [Google Scholar]
- Oppenheimer D.G., Herman P.L., Sivakumaran S., Esch J., Marks M.D. (1991). A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67: 483–493 [DOI] [PubMed] [Google Scholar]
- Payne C.T., Zhang F., Lloyd A.M. (2000). GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156: 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D., Vachon G., Herzog M. (1998). Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant Physiol. 117: 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Estrada L.B., Cano-Santana Z., Oyama K. (2000). Variation in leaf trichomes of Wigandia urens: Environmental factors and physiological consequences. Tree Physiol. 20: 629–632 [DOI] [PubMed] [Google Scholar]
- Pesch M., Hülskamp M. (2004). Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr. Opin. Genet. Dev. 14: 422–427 [DOI] [PubMed] [Google Scholar]
- Ramsay N.A., Glover B.J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10: 63–70 [DOI] [PubMed] [Google Scholar]
- Rerie W.G., Feldmann K.A., Marks M.D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8: 1388–1399 [DOI] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart B.J., Lim L.P., Burge C.B., Bartel B., Bartel D.P. (2002). Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Roslan H.A., Salter M.G., Wood C.D., White M.R.H., Croft K.P., Robson F., Coupland G., Doonan J., Laufs P., Tomsett A.B., Caddick M.X. (2001). Characterization of the ethanol-inducible alc gene-expression system in Arabidopsis thaliana. Plant J. 28: 225–235 [DOI] [PubMed] [Google Scholar]
- Schellmann S., Schnittger A., Kirik V., Wada T., Okada K., Beermann A., Thumfahrt J., Jurgens G., Hulskamp M. (2002). TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 21: 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J. (2003). Cell-fate specification in the epidermis: A common patterning mechanism in the root and shoot. Curr. Opin. Plant Biol. 6: 74–78 [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schnittger A., Folkers U., Schwab B., Jurgens G., Hulskamp M. (1999). Generation of a spacing pattern: The role of triptychon in trichome patterning in Arabidopsis. Plant Cell 11: 1105–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnittger A., Jurgens G., Hulskamp M. (1998). Tissue layer and organ specificity of trichome formation are regulated by GLABRA1 and TRIPTYCHON in Arabidopsis. Development 125: 2283–2289 [DOI] [PubMed] [Google Scholar]
- Shimizu H., Ichimura K. (2002). Effects of the ease of self-pollination on the vase life of cut eustoma flowers. J. Japan Soc. Hort. Sci. 71: 449–451 [Google Scholar]
- Shikata M., Koyama T., Mitsuda N., Ohme-Takagi M. (2009). Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 50: 2133–2145 [DOI] [PubMed] [Google Scholar]
- Simon M., Lee M.M., Lin Y., Gish L., Schiefelbein J. (2007). Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev. Biol. 311: 566–578 [DOI] [PubMed] [Google Scholar]
- Szymanski D.B., Lloyd A.M., Marks M.D. (2000). Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci. 5: 214–219 [DOI] [PubMed] [Google Scholar]
- Telfer A., Bollman K.M., Poethig R.S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654 [DOI] [PubMed] [Google Scholar]
- Wada T., Tachibana T., Shimura Y., Okada K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Walker A.R., Davison P.A., Bolognesi-Winfield A.C., James C.M., Srinivasan N., Blundell T.L., Esch J.J., Marks M.D., Gray J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11: 1337–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Tang W., Zhu C., Perry S.E. (2002). A chromatin immunoprecipitation (ChIP) approach to isolate genes regulated by AGL15, a MADS domain protein that preferentially accumulates in embryos. Plant J. 32: 831–843 [DOI] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wang J.W., Schwab R., Czech B., Mica E., Weigel D. (2008a). Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20: 1231–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Hubbard L., Chang Y., Guo J., Schiefelbein J., Chen J.G. (2008b). Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 8: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Kwak S.H., Zeng Q., Ellis B.E., Chen X.Y., Schiefelbein J., Chen J.G. (2007). TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134: 3873–3882 [DOI] [PubMed] [Google Scholar]
- Wang S., Wang J.W., Yu N., Li C.H., Luo B., Gou J.Y., Wang L.J., Chen X.Y. (2004). Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 16: 2323–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellso S.G., Hoxie R.P. (1982). The influence of environment on the expression of trichomes in wheat. Crop Sci. 22: 879–886 [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A., Wu M.F., Yang L., Wu G., Poethig R.S., Wagner D. (2009). The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 17: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., Sano R., Wada T., Takabayashi J., Okada K. (2009). Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 136: 1039–1048 [DOI] [PubMed] [Google Scholar]
- Zhang F., Gonzalez A., Zhao M., Payne C.T., Lloyd A. (2003). A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhu H.F., Fitzsimmons K., Khandelwal A., Kranz R.G. (2009). CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol. Plant 2: 790–802 [DOI] [PubMed] [Google Scholar]