Iron deficiency induces a range of physiological responses that are controlled by transcriptional alterations concentrated in the root pericycle. The transcriptional regulator POPEYE regulates many of these responses possibly through interaction with iron deficiency response protein ILR3 and the putative E3 ligase protein BRUTUS.
Abstract
Global population increases and climate change underscore the need for better comprehension of how plants acquire and process nutrients such as iron. Using cell type–specific transcriptional profiling, we identified a pericycle-specific iron deficiency response and a bHLH transcription factor, POPEYE (PYE), that may play an important role in this response. Functional analysis of PYE suggests that it positively regulates growth and development under iron-deficient conditions. Chromatin immunoprecipitation-on-chip analysis and transcriptional profiling reveal that PYE helps maintain iron homeostasis by regulating the expression of known iron homeostasis genes and other genes involved in transcription, development, and stress response. PYE interacts with PYE homologs, including IAA–Leu Resistant3 (ILR3), another bHLH transcription factor that is involved in metal ion homeostasis. Moreover, ILR3 interacts with a third protein, BRUTUS (BTS), a putative E3 ligase protein, with metal ion binding and DNA binding domains, which negatively regulates the response to iron deficiency. PYE and BTS expression is also tightly coregulated. We propose that interactions among PYE, PYE homologs, and BTS are important for maintaining iron homeostasis under low iron conditions.
INTRODUCTION
Iron is an essential nutrient for both plants and animals. In most human populations, nutritional iron is obtained predominantly from plants. Consequently, understanding how plants absorb and use iron is important for meeting increasing global nutritional demands. Even though iron is one of the most abundant elements on earth, in most soil environments, it forms highly stable insoluble oxyhydroxides. As a result, plants have evolved different mechanisms to solubilize and absorb iron from soil (Schmidt, 1999; Walker and Connolly, 2008; Palmer and Guerinot, 2009).
Nongraminaceous plants such as Arabidopsis thaliana respond to iron deficiency with increased expression of the bHLH transcription factor FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) (Bauer et al., 2007) whose protein forms heterodimers with bHLH protein bHLH38 or bHLH39 (Yuan et al., 2008). FIT positively regulates expression of FRO2, which encodes an epidermal iron reductase that reduces ferric iron in response to iron deficiency (Robinson et al., 1999). FIT also positively regulates the accumulation of IRT1 protein, which transports reduced Fe2+ into the symplast (Eide et al., 1996; Vert et al., 2002; Colangelo and Guerinot, 2004). Moreover, FIT regulates the expression of >40% of all iron deficiency inducible genes and is essential for normal growth and development under iron sufficient conditions (Colangelo and Guerinot, 2004). An additional bHLH protein, ILR3, has also been shown to play a role in metal ion–mediated auxin sensing within Arabidopsis roots. ILR3 gain-of-function mutants not only display altered iron deficiency–induced metal ion uptake, but also a decreased expression of the three genes homologous to yeast Ccc1p (Rampey et al., 2006). Yeast Ccc1p and its Arabidopsis homolog, VIT1, are both involved in vacuolar iron influx (Lapinskas et al., 1996; Li et al., 2001; Kim et al., 2006). Thus, ILR3 might control metal homeostasis by regulating the expression of genes implicated in intracellular iron transport (Rampey et al., 2006).
In addition to VIT1, other proteins involved in intracellular metal transport are also important for maintaining iron homeostasis in plants. For example, disruption of the vacuolar iron transporters NRAMP3/4, similar to VIT1, causes increased sensitivity to iron deficiency (Lanquar et al., 2005; Kim et al., 2006). Furthermore, IRT1 nonspecifically takes up other metals, such as Zn, Mn, Co, Cd, and Ni, in addition to Fe, under iron-deficient conditions (Vert et al., 2002). Therefore, genes involved in vacuolar sequestration and detoxification of Zn, such as ZIF1, MTPa2 (Arrivault et al., 2006; Haydon and Cobbett, 2007), or Ni, Co, and excess Fe, such as FPN2 (Schaaf et al., 2006; Morrissey et al., 2009), are also primary components of the iron deficiency response (Buckhout et al., 2009). Differential expression and localization of iron reductases FRO2, FRO3, and FRO7 to the root epidermal plasma membrane, root vasculature mitochondria, and shoot plasma membrane, respectively (Jeong and Connolly, 2009), also suggest that there are organ and organelle-specific requirements for reduced iron.
The intercellular distribution of iron between the root and shoot is also essential for tolerating fluctuations in iron availability. Studies show that after iron is loaded into the root xylem from the pericycle, FRD3, a multidrug and toxin efflux protein, facilitates Fe chelation to citrate and subsequent transport of Fe-citrate from the root to the shoot (Green and Rogers, 2004; Grotz and Guerinot, 2006; Durrett et al., 2007). Within developing seeds and young leaf tissues, iron is then thought to be transported from the xylem to the phloem in the form of Fe (II)-nicotianamine complexes via Yellow Stripe-Like1 (YSL1), an Fe transporter localized in the xylem parenchyma (Curie et al., 2009; Klatte et al., 2009). Disruption of FRD3, nicotianamine synthases (NAS1, NAS2, NAS3, and NAS4), or YSL genes leads to chlorosis, reduced growth, and increased sensitivity to iron deficiency (Ling et al., 1999; Rogers and Guerinot, 2002; Waters et al., 2006; Klatte et al., 2009). Thus, proper iron distribution within each plant organ is essential for normal growth and development.
Previously, a systems biology approach, which included high-resolution transcriptional profiling, was used to characterize root transcriptional regulatory networks involved in maintaining iron homeostasis under iron-deficient conditions (Dinneny et al., 2008). In this study, our objective was to identify key regulators of the initialization of iron deficiency response that act in a spatiotemporally specific manner. Through cell type–specific transcriptional profiling, we found that the pericycle may act as a regulatory center for sensing and responding to root iron content. Furthermore, whole root time-course microarray analyses showed that numerous transcription factors are temporally coexpressed when plants are exposed to iron-deficient growth conditions. We screened mutants of these transcription factors looking for alterations in response to iron deficiency. We found two genes that are induced by iron deficiency, one of which encodes a bHLH protein, which we named POPEYE (PYE), and a second gene tightly coregulated with PYE that we called BRUTUS (BTS). Both PYE and BTS appear to play important roles in responding to iron-deficient conditions and interact with the same PYE homologs, including ILR3. Our findings indicate that PYE, possibly in combination with BTS and other regulatory proteins, controls iron homeostasis by regulating the expression of genes involved in iron homeostasis and other biological processes.
RESULTS
A Transcription Factor Involved in Responding to Iron Deficiency Is Highly Expressed in the Pericycle
Rapid activation of iron reductase activity and IRT1 expression are common markers of the iron deficiency response in Arabidopsis roots (Eide et al., 1996; Robinson et al., 1999). In our study, we found that when plants are germinated and grown on iron-sufficient conditions for 5 d and then shifted to iron-deficient conditions, their roots displayed increased iron reductase activity and IRT1 expression within 24 h (see Supplemental Figure 1A online). In addition, plants exposed to iron deprivation displayed increased lateral root emergence (see Supplemental Figure 1B online). These are typical responses to iron deprivation (Römheld and Marschner, 1981; Moog et al., 1995). We also discovered that roots exposed to iron-deficient growth media (Figure 2B) became wavier under our growth conditions than those maintained on iron-sufficient media (see Supplemental Figure 3A online).
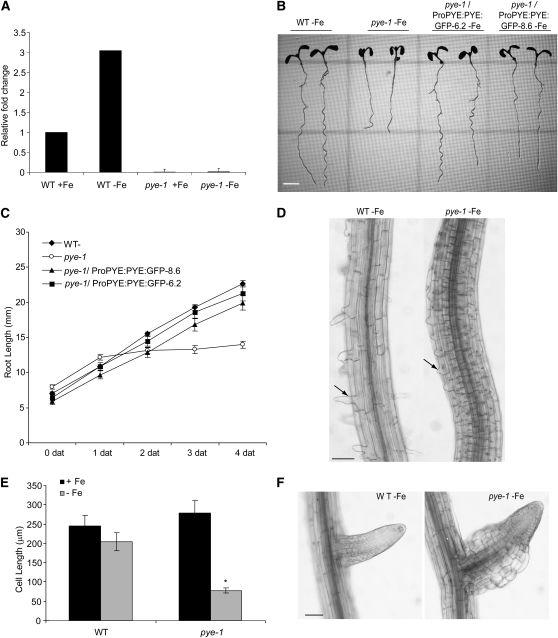
Figure 2.
Effects of Loss of PYE Function on Root Development.
(A) qRT-PCR analysis of PYE expression in wild-type (WT) and pye-1 plants grown on iron-sufficient media (+Fe) for 5 d and then transferred to +Fe or –Fe media for 24 h (±se).
(B) Wild-type, pye-1, and pye-1/ProPYE:PYE:GFP complementation lines germinated and grown on iron-sufficient media for 4 d and then shifted to –Fe media for 4 d.
(C) Time course quantification of root length of plants 0 to 4 d after transfer to –Fe media (±se).
(D) Light microscopy images of root maturation zone of plants germinated on +Fe media for 4 d and then shifted to –Fe media for 4 d. Arrows indicate root hairs.
(E) Quantification of cortex cell length within early maturation zone of plants germinated on +Fe media for 4 d and then shifted to –Fe media for 4 d (±se). Significant differences from the wild type are indicated by *P < 0.0001.
(F) Lateral roots of plants from (D).
Bars = 3 mm in (B) and 50 μm in (D) and (F).
To detect novel regulatory genes that control these physiological and developmental alterations, we queried our transcriptional profile data from roots exposed to iron-deficient media for 0, 3, 6, 12, 24, 48, and 72 h (Dinneny et al., 2008) for genes that encode proteins with DNA binding domains. We selected and screened 38 T-DNA insertion mutants of putative transcription factors whose expression is affected during the iron deficiency time course. We found that a T-DNA insertion in At3g47640, which we named PYE, causes alterations in response to iron deprivation that we describe later in the study.
PYE is over 3-fold upregulated after 72 h of growth on iron-deficient media (Figures 1A and 2A). To assess which biological activities might be coordinated along with PYE, we selected genes from the Dinneny et al. (2008) data set that were at least 1.5-fold differentially regulated by iron deficiency with a Q-value < 10−4. We then used affinity propagation (Frey and Dueck, 2007) with Pearson correlation to group genes with temporal expression patterns similar to PYE. We found that PYE is coexpressed with a group of genes that is enriched in transcription (P value, 10−4.3) and transcriptional repressor (P value, 10−3.2) activity (see Supplemental Figure 1C and Supplemental Data Sets 1A and 1B online).
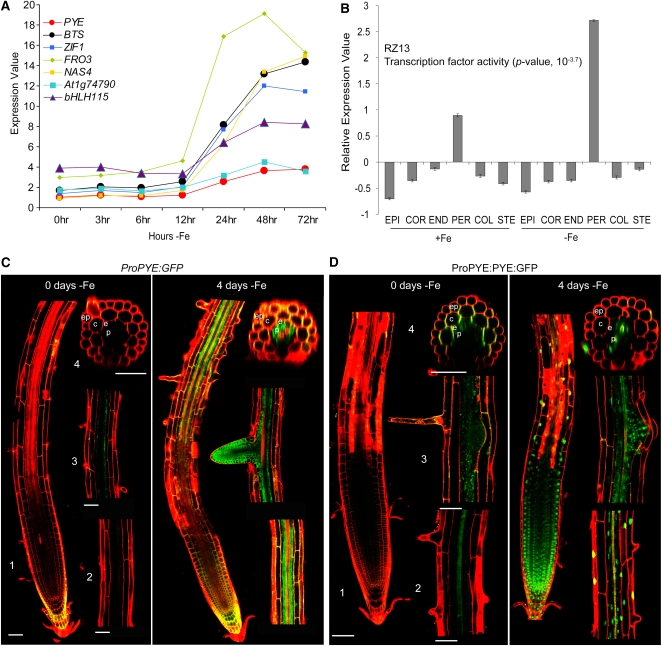
Figure 1.
PYE Expression Pattern and Protein Localization.
(A) Temporal expression pattern of PYE and direct PYE targets, bHLH115 and BTS, in response to iron deficiency (–Fe) derived from Dinneny et al. (2008).
(B) Expression pattern of RZ13, coregulated genes whose expression is induced in the pericycle after 24 h –Fe. Graph represents average normalized value of all genes in the cluster (±se). The enriched GO category is shown, and the associated P value is indicated in parentheses.
(C) ProPYE:GFP induction in pericycle cells after 4 d –Fe.
(D) PYE:GFP localization in all root cell types in the root tip after 4 d –Fe. (1) Root tip including meristematic, elongation, and maturation zones. (2) Early maturation zone. (3) Late maturation zone. (4) Transverse section of early maturation zone. ep, epidermis; c, cortex; e, endodermis; p, pericycle. Bars = 50 μm.
To validate this expression pattern in vivo, we transformed wild-type plants with a construct that has the PYE promoter upstream of green fluorescent protein (ProPYE:GFP). Trace levels of ProPYE:GFP expression were visible in the root vasculature, columella root cap, and lateral root cap under iron sufficiency. However, ProPYE:GFP expression was elevated throughout the root after exposure to iron deficiency with the highest expression occurring in the pericycle in the maturation zone (Figure 1C; see Supplemental Figure 2A online). These findings led us to ask if PYE protein localization is concentrated in this same region. We generated translational fusions with the full-length PYE cDNA fused to the GFP open reading frame downstream of the PYE promoter (ProPYE:PYE:GFP). Notably, we detected trace levels of PYE:GFP in multiple cell types of the root under iron sufficiency and rapid accumulation of PYE:GFP in the nuclei of all cells throughout the root upon exposure to iron deprivation (Figure 1D; see Supplemental Figure 2B online). This finding suggests that although PYE expression is strongest in the pericycle, PYE protein may function in multiple cell types.
Iron Deficiency Induces Pericycle-Specific Enrichment of Transcription Factor Activity
The strong induction of PYE expression in the pericycle led us to investigate what role this cell type plays in responding to iron deficiency. Previous work has shown that FRD3, which encodes an iron deficiency–inducible citrate transporter required for long-distance transport of iron from the root to the shoot, is expressed in and localized to the pericycle (Rogers and Guerinot, 2002; Green and Rogers, 2004). The pericycle surrounds the xylem and phloem cells in which iron is transported throughout the plant. Moreover, the pericycle is the outermost layer of the stele, which is the most transcriptionally active group of cells in the root in response to iron deprivation (Dinneny et al., 2008). Therefore, it is possible that the pericycle plays an important role in mediating the presence or absence of iron between the outer layers of the root and the vasculature.
To determine how the pericycle might be involved in the iron deficiency response, we profiled gene expression in this cell type after 24 h of exposure to iron-deficient media (see Supplemental Data Set 2A online). We then compared the pericycle expression data to that of the epidermis, columella, cortex, endodermis, and stele exposed to 24 h of iron deficiency derived from the Dinneny et al. (2008) data set. Our query identified >4300 genes at least 1.5-fold upregulated or downregulated after 24 h exposure to iron deficiency in any of the six cell types (see Supplemental Data Set 1C online). We again used affinity propagation (Frey and Dueck, 2007) with Pearson correlation to identify groups of genes with similar spatial expression patterns. We found 83 clusters of genes, which we termed radial zone (RZ) clusters (see Supplemental Data Sets 1C and 1D online). Three of these clusters, RZ13, RZ10, and RZ48, contain genes that are induced in the pericycle by iron deprivation (see Supplemental Data Set 1C online). In fact, 159 genes, the largest set of coexpressed genes in the entire radial zone data set, are found in RZ13 alone (see Supplemental Data Set 1C online). Gene ontology (GO) term enrichment indicates that, in contrast with all other groups of coregulated genes, all three of these clusters are enriched in transcription factor activity (Figure 1B; see Supplemental Data Set 1D online). PYE may be one of these transcription factors as it is most highly induced in the pericycle after 24 h of exposure to iron deficiency (Figure 1C; see Supplemental Figure 1D online). Based on this enrichment of transcription factors that respond to iron deficiency, we propose that the pericycle acts as a transcriptional regulatory center for sensing and responding to iron levels.
Loss of PYE Function Alters Response to Iron Deficiency and Disrupts Iron Accumulation
In wild-type plants, expression of PYE is elevated in response to iron deprivation (Figures 1A and 2A). A T-DNA insertion in the third exon of this gene results in a significant decrease in PYE expression (Figure 2A), resulting in inhibition of root growth under iron-deficient conditions (Figures 2B and 2C; see Supplemental Figures 3A and 3B online). pye-1 root growth inhibition is due to decreased elongation of cells compared with the wild type (Figures 2D and 2E; see Supplemental Figure 3C online). In addition, although exposure to iron deficiency causes root hairs to be distorted and misshapen (Muller and Schmidt, 2004; Dinneny et al., 2008), the pye-1 mutation appears to inhibit emergence of root hairs (Figure 2D; see Supplemental Figure 3C online) and causes swelling of cells within the elongation zone of lateral roots, particularly the epidermal and cortex cells only under iron deficiency (Figure 2F; see Supplemental Figure 3D online). Based upon this enlargement of certain cells, in addition to the altered response to iron deprivation, we named the gene associated with this mutant PYE.
We also observed alterations in shoot responses to iron-deficient conditions. The leaves of pye-1 mutants germinated on iron deficiency media became severely chlorotic after 7 d (Figure 3A). The shoots of pye-1 plants germinated and grown on iron-sufficient media were initially green, similar to the wild type (see Supplemental Figure 4A online). When shifted to iron-deficient media, the shoots of both wild-type and pye-1 lines became chlorotic within days. However, this phenotype was more intense in pye-1 mutants (Figure 3B), and pye-1 mutants contained lower chlorophyll content compared with wild-type plants (see Supplemental Figure 4B online).
Figure 3.
Effects of Loss of PYE Function on the Shoot.
(A) Seven-day-old wild-type (WT) and pye-1 plants germinated directly on –Fe media.
(B) Wild-type, pye-1, and pye-1/ProPYE:PYE:GFP complementation lines germinated and grown on iron-sufficient media for 4 d and then shifted to –Fe for 4 d.
(C) Fourteen-day-old plants germinated on pH 5 to 6 soil (left) and pH 7 to 8 soil (right).
Bars = 1 mm in (A) and (B) and 5 mm in (C). Red circles indicate pye-1 seedlings.
To validate that these phenotypes are due to loss of PYE expression, we transformed the PYE translational fusion (ProPYE:PYE:GFP) into pye-1 mutants. ProPYE:PYE:GFP expression led to higher PYE transcript levels compared with the wild type (see Supplemental Figure 4C online), possibly due to positional affects upon the transgene or the absence of certain repressor elements within the promoter fragment we used. Despite this increase in transcript levels, both shoot and root phenotypes were rescued by expression of ProPYE:PYE:GFP in pye-1 mutants exposed to iron-deficient media (Figures 2B, 2C, and 3B; see Supplemental Figure 4A online).
To characterize the pye-1 phenotype on iron-deficient soil, we germinated plants on alkaline soil (pH 7 to 8), in which iron is insoluble, and compared them to growth on soil with standard pH (pH 5 to 6). Although pye-1 mutants were able to germinate on high pH soil, their cotyledons became chlorotic after 6 to 7 d, and seedlings arrested growth before producing true leaves (Figure 3C). Therefore, PYE appears to regulate tolerance to multiple growth conditions in which iron availability is reduced.
To further determine how PYE controls responses to iron deprivation, we analyzed the typical indicators of iron deficiency, including iron reductase activity, using the ferrozine assay (Yi and Guerinot, 1996). Although iron reductase activity was elevated in both wild-type and pye-1 plants in response to iron deprivation, pye-1 mutants showed decreased iron reductase activity compared with the wild type (Figure 4A). Plant roots also respond to iron deprivation by extruding protons into the apoplast (Yi et al., 1994). This response lowers the rhizosphere pH and solubilizes iron, thus increasing iron availability. To determine whether loss of PYE expression alters iron deficiency–induced rhizosphere acidification, we grew plants on iron-sufficient media for 5 d, transferred them to iron-deficient media for 3 d, and then placed them on agar plates containing the pH indicator Bromocresol Purple for 24 h. Wild-type plants exhibit typical acidification of the rhizosphere, which causes the media around the roots to become yellow. By contrast, pye-1 mutants exhibit a visible decrease in rhizosphere acidification as indicated by the lack of discoloration around the root (Figure 4B). These findings suggest that disruption of PYE expression leads to decreased activation of typical markers of the iron deficiency response.
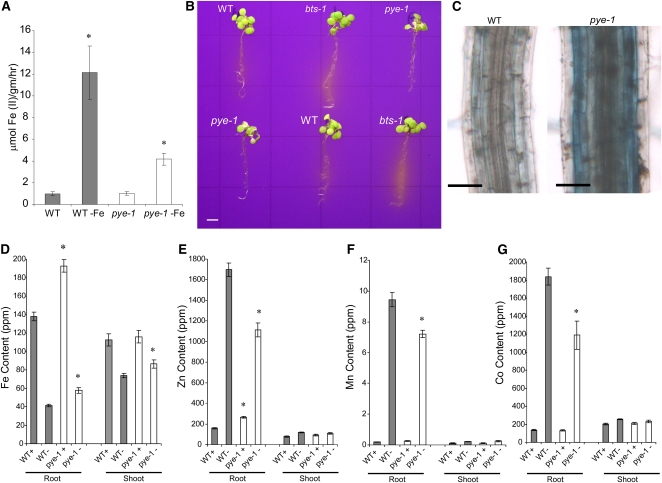
Figure 4.
Iron Deficiency Response of pye-1 Mutants.
(A) Iron reductase activity of wild-type (WT) and pye-1 plants germinated and grown on +Fe media for 5 d and then shifted to –Fe media for 3 d (±se). The ferrozine assay was performed, in quadruplicate, on 10 pooled plant roots. Significant differences from the wild type are indicated by *P < 0.05.
(B) Rhizosphere acidification of 7-d-old wild-type, pye-1, and bts-1 plants exposed to iron-deficient media for 3 d. Plants were then transferred to agar plates containing Bromocresol Purple for 1 d. Acidification is indicated by yellow color around the roots.
(C) Perl-stained maturation zone of 2-week-old plants germinated and grown on iron-sufficient media.
(D) to (G) Fe (D), Zn (E), Mn (F), and Co (G) content of wild-type and pye-1 roots and shoots germinated and grown on +Fe (+) media for 5 d and then shifted to –Fe (–) media and grown for 3 d (±se). Gray indicates the wild type, and white indicates pye-1. Significant differences from the wild type are indicated by *P < 0.05.
Bars = 3 mm in (B) and 50 μm in (C).
We also measured ion content of plants exposed to iron-sufficient and iron-deficient media. Plants commonly contain elevated amounts of Zn, Mn, and Co under iron-deficient conditions (Vert et al., 2002) because IRT1 nonspecifically takes up these metals. Interestingly, roots of pye-1 plants displayed decreased root Zn, Mn, and Co levels under iron deficiency compared with the wild type, whereas no differences were observed in shoots (Figures 4E to 4G). Together, these data further indicate that while pye-1 mutants do exhibit a typical iron deficiency response, this response is muted compared with wild-type plants.
To determine how these alterations affected the iron content of pye-1 mutants, we measured iron content two ways. Perl staining revealed that pye-1 mutants contain visibly elevated amounts of ferric iron in their roots compared with wild-type plants grown on iron-sufficient media (Figure 4C; see Supplemental Figure 5A online) but no differences in visible stainable ferric iron between wild-type and pye-1 leaves (see Supplemental Figure 5B online). Moreover, quantification of root and shoot iron content, using inductively coupled plasma–mass spectrometry, revealed that pye-1 mutants contained significantly elevated concentrations of iron in the root under iron-sufficient and -deficient conditions, as well as in the shoot under iron-deficient conditions (Figure 4D). This suggests that PYE is involved in maintaining iron homeostasis under iron-sufficient conditions. However, it appears to play an even more important role under iron-deficient conditions, since loss of PYE expression results in only partial induction of the typical iron deficiency response in addition to increased accumulation of iron.
Iron Homeostasis Genes and Others Are Regulated by PYE
To further examine the role of PYE, we performed transcriptional profiling of pye-1 mutants and found that loss of PYE function leads to a number of transcriptional changes. The expression level of ~490 genes was affected by iron deprivation in wild-type plants, while only ~130 genes were affected in pye-1 mutants under the same conditions (Q-value < 10−4; >1.2-fold change) (see Supplemental Data Sets 3A and 3B online). Analysis of the expression profiles of all genes affected by iron deficiency in wild-type and pye-1 plants combined with genes differentially expressed between wild-type and pye-1 mutants under both iron-sufficient and -deficient conditions identified 661 genes. Hierarchical clustering with Pearson correlation revealed that these genes fall into eight groups of coexpressed genes (Figure 5A; see Supplemental Data Set 3D online). Notably, genes involved in root hair differentiation, such as MRH2 and MRH6, are repressed in pye-1 mutants under iron-sufficient and -deficient conditions (Figure 5A; see Supplemental Data Set 3D online). Loss of MRH2 or MRH6 function causes alterations in the transition to tip growth and root hair initiation, respectively (Mark et al., 2006), which may explain the alterations in root hair development seen in pye-1 mutants under iron deficiency.
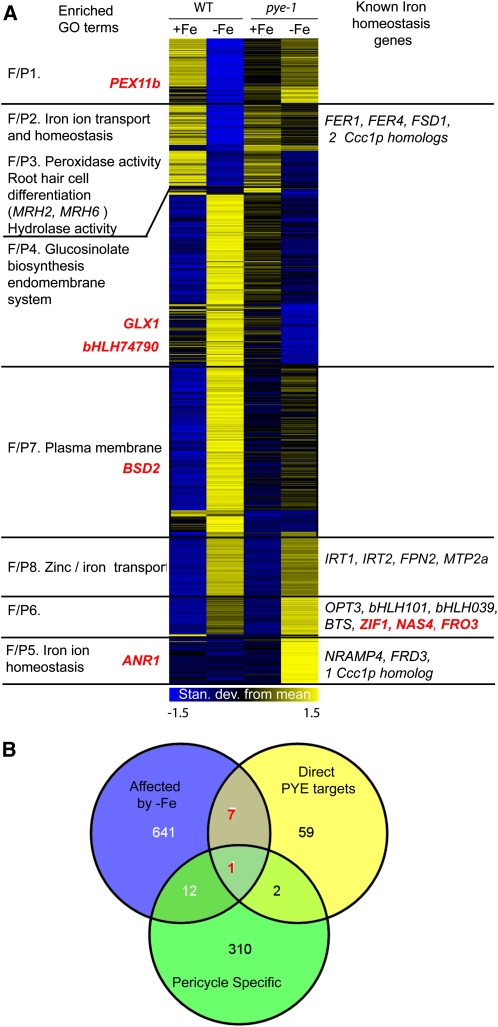
Figure 5.
PYE Transcriptional Regulation of Iron Homeostasis Genes.
(A) Heat map of all 661 genes affected by –Fe in wild-type (WT) roots and pye-1 roots or between wild-type and pye-1 plants under +Fe or –Fe conditions. Genes coregulated by Fe and/or pye-1 (F/P clusters) have similar expression patterns. Left, enriched GO categories associated with each F/P gene cluster. Right, individual genes known to play a role in responding to iron deficiency or involved in iron homeostasis. Direct PYE targets are indicated in red.
(B) Overlap of PYE direct targets based on ChIP-on-chip target analysis, 661 genes affected by –Fe and/or pye-1, and pericycle-specific transcripts. The number of PYE targets that are affected by –Fe are indicated in red.
We also noted that many genes involved in iron homeostasis were differentially expressed in pye-1 mutants (Figure 5A; see Supplemental Data Set 3E online). For example, the iron storage proteins (FER1 and FER4) and oxidative stress proteins, such as FSD1 (superoxide dismutase), were repressed in both wild-type and pye-1 mutants under iron deficiency. However, they were more highly expressed under both iron-sufficient and -deficient conditions in pye-1 than in wild-type plants (Figure 5A; see Supplemental Data Set 3D online), indicating that pye-1 roots experience increased iron storage and oxidative stress under both conditions. By contrast, genes encoding known and putative metal ion transporters (NRAMP4, a Ccc1p-like gene) and metal ion transport facilitators (OPT3 and FRD3) are induced by iron deficiency in wild-type plants and are expressed even more highly in pye-1 mutants under both sufficient and deficient conditions (Figure 5A; see Supplemental Data Set 3D online). These alterations suggest that there is an increased intercellular and intracellular iron movement within pye-1 mutants under both conditions and that PYE acts directly or indirectly to repress this activity in wild-type plants. Interestingly, IRT1, which encodes an epidermal iron transporter that takes up iron from the rhizosphere (Vert et al., 2002), is upregulated by iron deficiency in both wild-type and pye-1 plants. However, IRT1 is not as elevated in pye-1 plants as in wild-type plants by iron deficiency (Figure 5A; see Supplemental Data Set 3D online). Thus, although there is increased iron movement within pye-1 mutants, there is decreased iron uptake from the rhizosphere.
PYE Binds to the Promoters of Genes Involved in Iron Homeostasis
To determine if and how PYE directly regulates iron homeostasis genes, we set out to identify PYE direct targets by performing chromatin immunoprecipitation (ChIP) on roots of pye-1 mutants expressing ProPYE:PYE:GFP grown on iron-deficient media. As a control, we conducted the same experiment on wild-type plants. These samples were hybridized to custom long oligonucleotide (~60-mer) Arabidopsis promoter microarrays (Sozzani et al., 2010). We reasoned that genes that are differentially expressed under iron-deficient conditions in pye-1 should be overrepresented among the direct targets. With this information in hand, we systematically explored the parameter space to optimize our detection settings for enriched regions in the ChIP-on-chip experiments (see Supplemental Figure 8A online). Using highly stringent conditions in which transcriptional targets were mostly enriched, we identified 44 chromosomal regions that could be attributed to >70 PYE direct target genes (see Supplemental Data Set 3F online). Among these are members of the major facilitator superfamily (MFS-1) general substrate transporters, amino acid transporters, hydrolases, and transcription factors.
According to our microarray analysis, eight of the 70 PYE direct targets were also affected in either wild-type plants or pye-1 mutants by 24 h iron deficiency (Figures 5A, red, and 5B, red; see Supplemental Data Set 3F online, bold). However, only 2% of genes affected by iron deficiency in the wild type or pye-1 mutants are expressed only in the pericycle, and only one of the direct PYE targets is pericycle specific (Figure 5B). Thus, PYE downstream targets appear to be expressed throughout the root, which supports the idea that PYE acts in multiple cell types.
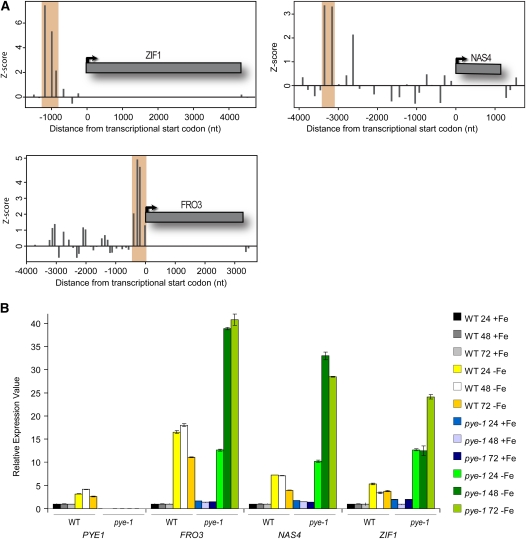
Two of the direct PYE targets (PEX11b and GLX1) encode peroxisomal proteins: one encodes a transcription factor involved in nutrition stress response (ANR1), and one encodes a protein homologous to maize (Zea mays) BSD2, which is involved in posttranslational ribulose-1,5-bis-phosphate carboxylase/oxygenase accumulation (Zhang and Forde, 1998; Brutnell et al., 1999; Desai and Hu, 2008; Reumann et al., 2009). Three of the PYE direct genes, FRO3, NAS4, and ZIF1 (Figure 6A), are known to encode proteins involved in metal ion homeostasis (Haydon and Cobbett, 2007; Jeong and Connolly, 2009; Klatte et al., 2009). To verify that PYE targets are misregulated in pye-1 mutants and determine whether loss of PYE expression affects target gene expression more strongly after longer exposure to iron deficiency, we used quantitative RT-PCR (qRT-PCR) to compare the expression of three PYE direct targets after 24, 48, and 72 h of growth on iron-sufficient and -deficient media. Loss of PYE function causes significantly increased and prolonged expression of ZIF1, FRO3, and NAS4 after exposure to iron deficiency (Figure 6B). Furthermore, the expression of ZIF1 and FRO3, in addition to a sixth PYE direct target with catalytic function (At1g74790), is elevated, then subsequently decreases within 72 h of exposure to iron deficiency in wild-type plants (Figure 1A). Thus, PYE appears to directly regulate a number of genes that may be important for properly responding to iron deprivation.
Figure 6.
PYE Target Binding Sites and PYE-Mediated Transcriptional Regulation.
(A) PYE binding signatures as shown by averaged Z-scores of probe enrichments from ChIP-on-chip experiments (y axis). Genomic positions (x axis) are given relative to the annotated transcription start of the gene. Shaded areas indicate genomic regions that were detected as enriched.
(B) qRT-PCR analysis of PYE and PYE direct targets after 24, 48, and 72 h of exposure of the wild type (WT) and pye-1 to +Fe or –Fe (±sd).
PYE Is Tightly Coregulated with a Second Iron Deficiency Response Regulator, BTS
To learn more about the PYE gene regulatory network, we analyzed PYE using the ATTED-II Network Drawer, which predicts regulatory networks based upon coexpressed genes determined from microarray analyses and predicted cis-elements (Obayashi et al., 2009). This investigation revealed that PYE is tightly coregulated with its direct targets FRO3 and ZIF1 and other well-known iron deficiency response genes, such as OPT3, NRAMP4, bHLH101, and bHLH039, across 58 publicly available experimental microarray data sets (Figure 7A). One of the genes most tightly coregulated with PYE encodes a plant-specific protein containing a putative DNA binding domain (CHY-type zinc finger), a putative E3 ubiquitin-protein ligase domain (RING/FYVE/PHD-type Zinc finger), and six Haemerythrin/HHE cation binding motifs (see Supplemental Figure 6A online), which are known to bind iron and provide a hydrophobic pocket for dioxygen (Stenkamp et al., 1985; Sheriff et al., 1987; Uchida et al., 1990).
Figure 7.
BTS Expression, Coregulation, and Mutant Analysis
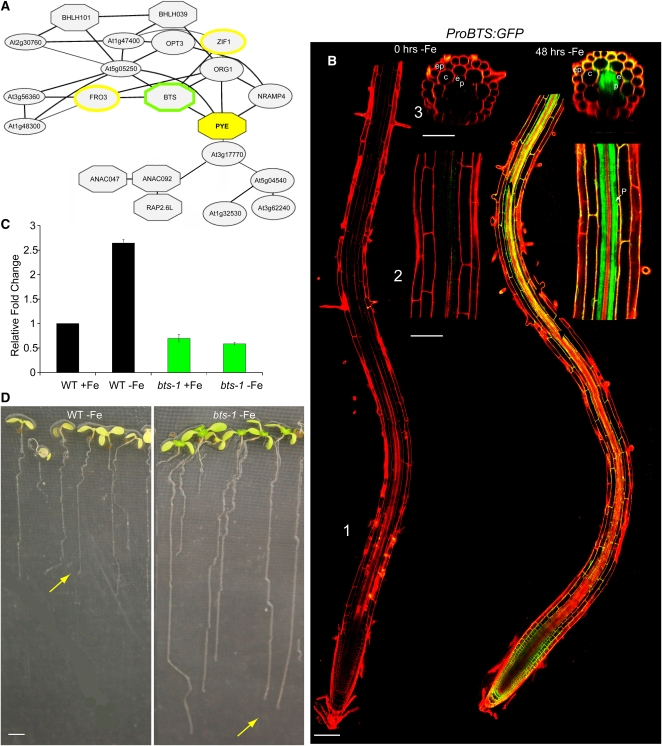
(A) Coexpression analysis of PYE direct target genes. Octagonal blocks are DNA binding domain containing proteins, and the yellow outline indicates PYE direct targets. Green outline indicates BTS.
(B) ProBTS:GFP induction in roots, especially the pericycle, after 48 h of exposure to –Fe. (1) Root tip including meristematic, elongation, and maturation zones. (2) Late maturation zone. (3) Transverse section of early maturation zone. ep, epidermis; c, cortex; e, endodermis; p, pericycle.
(C) qRT-PCR analysis of BTS expression in plants grown on +Fe media for 5 d and then shifted to +Fe or –Fe media for 24 h (±se). WT, wild type.
(D) Eleven-day-old wild-type and bts-1 plants germinated on iron-deficient media. Yellow arrows indicate root tip.
Bars = 100 μm in (B1), 50 μm in (B2) and (B3), and 2 mm in (D).
This protein, which we have named BTS was previously identified as EMB2454 in a screen for embryonic lethal genes (McElver et al., 2001), indicating that it is essential for survival even under standard growth conditions. We characterized its expression pattern using microarray analysis and visual inspection of lines containing a transcriptional GFP fusion construct. We found that, similar to PYE, BTS is upregulated by iron deficiency specifically in the pericycle (Figures 1A and 7B). We also analyzed the growth and development of a partial loss-of-function allele in which BTS induction by iron deficiency is disrupted (Figure 7C). In contrast with pye-1 mutants, bts-1 plants germinated on iron-sufficient media then shifted to iron-deficient media developed longer roots (Figure 7D) that acidified the rhizosphere more than the wild type (Figure 4B), and their shoots were greener and larger than wild-type plants after extended exposure to iron deficiency (Figure 7D). These findings suggest that bts-1 plants have increased tolerance to iron deprivation. Hence, the name BTS, which reflects how this protein appears to play a role in responding to iron deficiency opposite that of PYE.
Based on its putative protein structure, it is possible that BTS may have both transcriptional regulatory capabilities as well as the ability to act as an E3 ligase that interacts with target proteins, causing protein degradation through ubiquitin-mediated conjugation. Suspecting that BTS may interact with PYE or PYE targets, we performed yeast two-hybrid screening with the hybrid BTS protein as bait and a root-specific cDNA library as prey. We found nine interacting proteins with this approach (see Supplemental Figures 7A and 7B online). Two of these proteins, bHLH115 (At1g51070) and bHLH104 (At4g14410), are members of the Group IVc bHLH family, which also includes the iron homeostasis protein ILR3. Moreover, bHLH family IVc is a small subgroup of the bHLH family IV, which also includes PYE (Heim et al., 2003) (Figure 8A; see Supplemental Data Set 4A online).
Figure 8.
PYE and BTS Protein Interactors.
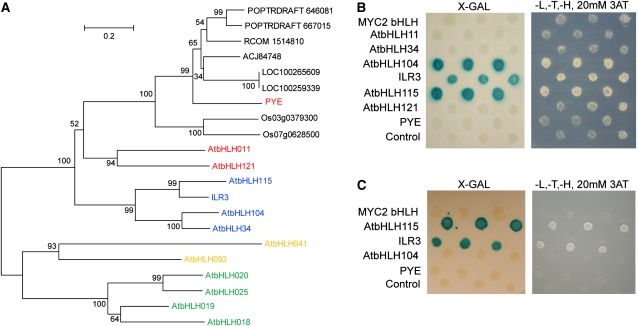
(A) Phylogenetic tree of PYE Arabidopsis homologs and orthologs from Vitis vinifera, Populus trichocarpa, Oryza sativa, Ricinus communis, and Lotus japonicus. Arabidopsis bHLH group IV subgroups a (green), b (red), c (blue), and d (orange) are indicated.
(B) BTS interaction with three of six PYE putative Arabidopsis homologs from bHLH subgroup IVc.
(C) PYE interaction with two PYE homologs from bHLH subgroup IVc. For (B) and (C), interacting colonies were identified by colorimetric detection of β-galactosidase (left) and growth after 5 to 10 d on -Leu (L), -Trp (T), -His (H), and 20 mM 3-AT media (right).
To validate our original yeast two-hybrid screen and further determine whether BTS might interact with additional members of the bHLH IV family, such as PYE, we cloned the open reading frames of the six remaining members of the bHLHIVb and bHLHIVc gene families (At1g51070 [bHLH115], At4g14410 [bHLH104], At4g36060 [bHLH11], At3g19860 [bHLH121], At5g54680 [ILR3], and At3g23210 [bHLH34]) and performed directed yeast two-hybrid analysis. We used BTS as the bait and PYE and the six putative PYE homolog proteins, as well as the bHLH domain of MYC2 as a bHLH domain control, as prey. We found that BTS interacts with three members of subgroup IVc, AtbHLH104, ILR3, and AtbHLH115, but does not interact with PYE (Figure 8B).
Although BTS does not interact with PYE or PYE targets in yeast, we were intrigued by the finding that BTS interacts with proteins so closely related to PYE. Since bHLH proteins commonly form heterodimers and homodimers that interact with downstream targets, we theorized that PYE might form heterodimers with the PYE homologs that interact with BTS. We performed directed yeast two-hybrid analysis using PYE as prey and bHLH104, ILR3, and bHLH115 as baits. We found that PYE interacts specifically with ILR3 and bHLH115 (Figure 8C). bHLH115 is also ~2-fold upregulated within 72 h of iron deficiency in whole roots (Figure 1A). Moreover, ILR3 and AtbHLH115 are induced specifically in the pericycle (P value < 0.0011) and stele (P value < 0.00013), respectively, by iron deficiency (see Supplemental Data Set 2A online; Dinneny et al., 2008). These findings suggest that PYE and ILR3 or bHLH115 could form heterodimers that may interact with BTS.
DISCUSSION
Plants have evolved remarkable abilities to withstand a range of environmental changes. The capacity to take up and store excess iron under sufficient conditions and use it when plants are exposed to low iron conditions is important for tolerating fluctuations in local iron availability. Recent studies have highlighted transcriptional responses to iron deprivation (Dinneny et al., 2008; Buckhout et al., 2009; Yang et al., 2010). In this study, we describe a transcriptional regulator, PYE, which appears to regulate these responses.
A Pericycle-Specific Iron Deficiency Response Is Important for Growth and Development
Previously, using cell type–specific transcriptional profiling, we found that the stele, which includes the pericycle, is the most responsive cell type within the root (Dinneny et al., 2008). In this study, we observed that the largest group of genes coordinately expressed in response to iron deficiency is found in the pericycle. In addition, the iron deficiency response in the pericycle appears to be enriched in GO terms associated with transcription factors. Therefore, we propose that the pericycle serves as a transcriptional regulatory center for the iron deficiency response, which may explain why the transcription factor, PYE, is expressed most highly in the pericycle in response to iron deprivation.
Microarray data and in vivo analysis of ProPYE:GFP expression indicate that PYE is most highly expressed in the pericycle within the maturation zone. However, PYE protein is localized in the nuclei of all cells within the root when plants are exposed to iron deficiency, and the majority of PYE downstream targets are expressed outside of the pericycle in response to iron deficiency (Figure 5B). Furthermore, Perl staining indicates that excess iron is localized to multiple cell types. Thus, after induction by iron deficiency within the pericycle, PYE might then move throughout the root, where it localizes to the nuclei, binds to DNA, and regulates the transcription of its target genes. This would suggest that the sensing of iron deficiency may occur in a dedicated tissue, the pericycle, and that the response is orchestrated over all cell types by transcription factor movement. Alternatively, PYE could be expressed to some extent throughout the root in response to iron deprivation. The accumulation of PYE protein within all cells of the root could be due to differential stability within different cell types and developmental zones.
PYE-Mediated Gene Regulation Is Important for Maintaining Fe Homeostasis
Transcriptional profiling of wild-type plants (Dinneny et al., 2008; Buckhout et al., 2009; this study) reveals that the iron deprivation response causes widespread transcriptional alterations. It appears that many genes must be coordinately regulated before and after exposure to iron deficiency to trigger a sufficient iron deficiency response. Our data indicate that PYE is involved in this coordination. For example, ChIP-on-chip analysis indicates that the FRO3 and NAS4 promoters are directly bound by PYE. In addition, transcriptional analysis indicates that FRO3 and NAS4 are upregulated in wild-type plants by iron deficiency, yet they are upregulated even more in pye-1 mutants by iron deficiency. Thus, PYE appears to directly repress the induction of FRO3 and NAS4 under iron deficiency. FRO3 encodes an iron reductase, while NAS4 is an enzyme that synthesizes nicotianamine, an iron chelator important for intercellular Fe transport (Klatte et al., 2009). Although FRO3 and NAS4 single mutants do not have a phenotype under iron sufficiency or iron deficiency, FRO3 is induced in the vasculature of roots and in the root tip by iron deficiency and localized to the mitochondrial membrane (Jeong and Connolly, 2009). Moreover, quadruple nas mutants (nas4x-1) are sterile with constitutive expression of iron uptake responses and intercostal chlorosis (Klatte et al., 2009). Notably, nas4x-1 and the related chloronerva mutant of tomato (Solanum lycopersicum) accumulate iron that is biologically unavailable and, consequently, exhibit an increase in the iron deprivation response (King, 1991; Klatte et al., 2009).
By contrast, pye-1 mutants, which also exhibit elevated iron content compared with the wild type, display decreased tolerance to iron deprivation and exhibit a weaker iron deficiency response. We propose that this difference could be due to differences in the function of PYE compared with NAS4. PYE is a regulatory protein that regulates the expression of iron deficiency–responsive genes, while NAS is an enzymatic protein involved in the synthesis of NA, an iron chelator important for intercellular iron transport. While lower NAS may disrupt the bioavailability of iron within specific plant organs, triggering a typical iron deprivation response, loss of PYE appears to first affect the global transcriptional response to iron deprivation. These altered transcriptional responses likely result in lower induction of the iron deficiency response, compared with the wild type, and decreased use of iron stores. Consequently, pye-1 mutants exhibit increased iron accumulation in addition to decreased iron deficiency response.
Elevated iron content can lead to the formation of harmful reactive oxygen species and could explain why iron storage proteins FER1, FER4, and superoxide dismutase (FSD1) are constitutively upregulated in pye-1 mutants. This may also explain why pye-1 mutants exhibit decreased expression of IRT1, which uptakes iron from the rhizosphere under iron deficiency and why genes involved in intra- and intercellular iron transport, such as NRAMP4, OPT3, and FRD3, are misregulated in pye-1. IRT1 takes up Zn, Mn, and Co from the rhizosphere under iron deficiency. Thus, decreased Fe uptake by IRT1 likely leads to the decreased Mn and Zn content and decreased expression of Zn, Ni, Mn, and Co detoxification genes we observed in pye-1 roots relative to the wild type. ZIF1, however, is an exception to this observation. The gene that encodes ZIF1, a Zn vacuolar efflux protein involved in Zn detoxification (Haydon and Cobbett, 2007), is upregulated by iron deficiency in pye-1 compared with the wild type. This difference could occur because ZIF1 is a direct PYE target. Therefore, PYE-mediated repression of ZIF1 may be important for maintaining Zn homeostasis under iron deficiency while other Zn detoxification genes (MTPa2) and Ni, Co, and Mn detoxification genes (FPN2 and Ccc1p-like) are misregulated indirectly by PYE.
Since iron is a major component of the photosynthetic and mitochondrial electron transport chains and a component of many enzymatic cofactors, alterations in iron content may also disrupt metabolism, leading to the generation of harmful cytotoxins, in addition to reactive oxygen species. This may explain why genes encoding proteins involved in critical metabolic processes are direct PYE targets. For example, PEX11b, a peroxin involved in light-induced peroxisome proliferation (Desai and Hu, 2008), is downregulated by iron deprivation in wild-type plants, yet unaffected by iron deficiency in pye-1, suggesting that PYE may repress PEX11b. By contrast, a BSD2-like gene is downregulated in pye-1 mutants under both iron-sufficient and -deficient conditions compared with wild-type plants. BSD2 positively regulates the accumulation of ribulose-1,5-bisphosphate carboxylase/oxygenase protein in maize (Brutnell et al., 1999), suggesting that PYE activates key photosynthetic processes. In addition, GLX1, a glyoxalase I homolog (Reumann et al., 2009), is activated by iron deficiency in the wild type and repressed in pye-1 by iron deficiency. Glyoxalase I enzymes catalyze a reaction that causes the detoxification of methylglyoxal, a cytotoxic byproduct of glycolysis. GLX1 has been shown to play a role in salt stress and to be induced by salt, osmotic stress, and phytohormonal stimuli, presumably as a result of elevated glycolysis caused by salt stress (Espartero et al., 1995). Thus, it is possible that PYE both represses and activates specific metabolic processes in response to iron deprivation.
Loss of PYE function leads to several additional physiological and developmental alterations in response to iron deficiency. In addition to becoming chlorotic, pye-1 mutants develop swollen emerging lateral roots, develop shorter primary and lateral roots due to inhibition of epidermal and cortical cell elongation, and arrest growth shortly after exposure to iron-deficient conditions. Although chlorosis and growth arrest are commonly associated with iron deficiency, we were surprised by the developmental alterations pye-1 mutants also displayed. Transcriptional profiling of pye-1 roots helped us identify several genes involved in development that may also be part of the response to iron deficiency. Peroxidases and hydrolases associated with cell wall remolding are upregulated by iron deficiency in wild-type plants. However, these genes appear to be repressed in pye-1 mutants under iron-sufficient and -deficient conditions. This transcriptional alteration may explain why the lateral roots of pye-1 plants are swollen and why the epidermal and cortex cells of the primary root are not as elongated as wild-type plants. In addition, the transcriptional regulator ANR1 is a direct PYE target that is misregulated in pye-1 mutants exposed to iron deprivation. ANR1 is a MADS box transcription factor that positively regulates lateral root development in response to nitrate deficiency (Zhang and Forde, 1998). Therefore, misregulation of ANR1 expression may be associated with disruption of lateral root development exhibited by pye-1 mutants.
Iron deprivation also causes decreased expression of root hair morphogenesis genes, MRH2 and MRH6, in both the wild type and pye-1 mutants. In addition, loss of PYE function results in lower expression of MRH2 and MRH6 under iron-sufficient conditions. Loss of MRH2 and MRH6 function in wild-type plants under normal growth conditions causes developmental alterations in root hair development similar to those seen in pye-1 mutants grown under iron deficiency (Mark et al., 2006). Therefore, lowered expression of these genes in pye-1 mutants under iron-sufficient and -deficient conditions may explain the exacerbated root hair phenotype observed in pye-1 mutants.
While PYE directly regulates ANR1, PYE also appears to indirectly regulate other transcription factors, such as bHLH039 and bHLH101. bHLH039 and bHLH101 are two bHLH subgroup Ib members that are strongly induced by iron deficiency (Yuan et al., 2008). The upregulation of bHLH039 in pye-1 mutants under both normal and iron deficiency conditions is of particular interest because bHLH039 is an FIT binding partner. FIT is a master regulator of the iron deficiency response. Therefore, PYE function may mediate widespread transcriptional changes through interaction with FIT or other factors that are direct PYE targets.
We have shown that genes encoding proteins with different biological functions are affected by loss of PYE function under iron-sufficient conditions (Figure 5A). Furthermore, iron content is elevated in pye-1 roots under iron sufficiency. Expression analysis and ProPYE:GFP analysis using confocal microscopy suggest that PYE is present in trace amounts under iron sufficiency conditions. Thus, PYE appears to play some role in iron homeostasis under iron-sufficient conditions. However, PYE expression is induced by iron deficiency and, in contrast with FIT, another bHLH transcription factor that plays a key role in iron homeostasis, the physiological effects of loss of PYE function are seen primarily under low iron conditions. Taken together, our data indicate that PYE is a unique transcriptional regulator that, when elevated under conditions of low iron availability, regulates the expression of genes involved in a range of biological activities important for responding to iron-deficient conditions.
PYE Interacts with Other Proteins Involved in Iron Homeostasis
bHLH proteins, such as PYE, commonly form heterodimers and homodimers that interact with downstream targets. Therefore, it is possible that PYE may regulate downstream targets with a binding partner, specifically ILR3 or bHLH115. ILR3 plays a role in metal ion–mediated auxin sensing, and microarray analysis of gain of function ilr3-1 mutants reveals decreased expression of three Ccc1p homologs possibly involved in facilitating metal ion transport (Rampey et al., 2006). Notably, Ccc1p homologs are also elevated in pye-1 under both iron-sufficient and -deficient conditions. Therefore, both PYE and ILR3 appear to repress expression of these genes. While we were not able to detect binding of PYE to the promoters of these homologs under our conditions, it is possible that PYE and ILR3 could interact to regulate the expression of Ccc1p homologs and other downstream targets involved in metal iron homeostasis.
Surprisingly, in addition to PYE, ILR3 and bHLH115 also interact with BTS, which is involved in iron homeostasis and tightly coregulated with PYE. Thus, it is tempting to speculate that interactions among PYE, ILR3, bHLH115, and BTS are important for responding to iron deficiency. But the question remains: Why are PYE and BTS both induced in response to –Fe when they appear to play opposite roles in the iron deficiency response? Maintaining iron homeostasis under fluctuating growth conditions requires a balance of many processes, including inhibition and activation of iron uptake from the rhizosphere, intercellular and intracellular iron transport, and alterations in photosynthesis and metabolism. We propose that under conditions of low iron availability, PYE expression is induced so that PYE can help maintain this balance by transcriptionally regulating the expression of genes involved in many of these processes. However, too much or too little of this regulation could disrupt metal ion homeostasis rather than maintain it. Hence, the inhibitory effects of BTS, which we hypothesize acts as a check on PYE activity since it appears to negatively regulate the response to iron deprivation, are induced by this condition in a similar manner to PYE. BTS could be elevated in response to iron deficiency to disrupt PYE function by interacting with PYE binding partners, such as ILR3 and bHLH115. Based on the increased expression of BTS in a pye-1 background under iron deficiency (Figure 5A), it is also possible that BTS is a negatively regulated downstream target of PYE. In that case, elevation of PYE under iron deficiency would inhibit the repressive activity of BTS. Alternatively, BTS and PYE could be regulated independently by separate activators and repressors involved in maintaining iron homeostasis. Future work will explore the relevance of the potential interaction between BTS and PYE.
The discovery of two novel coregulated proteins, one of which is a bHLH transcription factor that plays an important role in iron homeostasis, increases our understanding of the regulatory networks involved in plant responses to the environment and enables us to further explore how spatiotemporal regulation of regulatory factors and their interactors controls the nutritional status of plants.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana ecotype Columbia (Col-0) was used for all experiments. A T-DNA insertion line for pye-1 (SALK_021217) was confirmed using PCR with T-DNA left border primer (LBb1) and gene-specific primers (5′- TTCAAGACCTCATTCACTGGC-3′ and 5′-GGGGATTGATTATGTTTGGTG-3′). bts-1 (SALK_016526) was confirmed with 5′-CCAAATGCGTTCGTAGGTAAG-3′ and 5′-TCAGATTTACACAAATTTGCAGC-3′. After surface sterilization using 70% ethanol for 2 min followed by 30% bleach and 0.02% Triton X-100 for 15 min, seeds were rinsed three times with sterile water and stratified at 4°C for 2 to 4 d before being planted on media. Iron-sufficient (+Fe) media is standard Murashige and Skoog media with 0.05% MES, 1% sucrose, 1% agar, and 0.1 mM FeEDTA substituted for iron sulfate. Iron deficient (−Fe) media is the same, with 300 μM ferrozine, an iron chelator, substituted for iron sulfate. For microarray analysis and all other experiments in which seedlings were shifted from iron-sufficient to either sufficient or deficient media, Nylon mesh (Nitex Cat 03-100/44; Sefar) was placed on top of the solidified media to facilitate transfer. Seeds were evenly placed on the mesh in a single row at a density of ~20 seeds/cm in two rows. For characterization of phenotypes, 8 to 10 seeds were sown 3 to 4 mm apart. Seeds were sown and allowed to germinate on vertically positioned media plates in a Percival incubator with 16 h of daily illumination at 22°C. High pH soil was generated by adding CaO (7.8 gm CaO/kg; Sigma-Aldrich) to soil as in Kim et al. (2006).
Plasmid Construction and Plant Transformation
Standard molecular biology techniques and the Gateway system (Invitrogen) were used for the cloning procedures. Genomic DNA from Arabidopsis ecotype Col-0 was used as the template for amplification of the upstream regulatory promoter sequence for ProBTS:GFP (3000 bp) using 5′-ATGAGATGAAATGTCTTATCTTTAT-3′ and 5′-TTCCCCCAAAGCTTATCTCCGTTTT-3′ and ProPYE:GFP (1120bp) using 5′- ACCGCAAAACTATATATAGTATTT-3′ and 5′-CTTTGCTTTTATTACAGAACAAGA-3′. Each promoter region was cloned into pDONRP4P1R (Invitrogen). For the translational fusions, 5′-ATGGCGACGCCGTTACCAGA-3′ and 5′-GGATGAGGTTGAGCAGTCCGGGGG-3′ were used to clone the BTS open reading frame. 5′-ATGGTATCGAAAACTCCTTC-3′ and 5′-TTCACTGGCTTTCAGCCGCTCTCC-3′ were used to clone the PYE open reading frame. Each open reading frame was cloned into pENTR/D-TOPO (Invitrogen). Multisite cloning (Invitrogen) was used to place each promoter and open reading frame into a pGREEN (Hellens et al., 2000) binary vector derivative containing a NOS terminator with a C-terminal GFP fusion and spectinomycin and BASTA resistance genes (Lee et al., 2006). Each plasmid was transformed into the wild type by the floral dip method. Single gene transformants were detected by T2 3:1 growth segregation on growth media containing BASTA. To determine whether PYE complements the pye-1 mutation, ProPYE:PYE:GFP was transformed into pye-1 homozygous lines. For all transgenic plant lines constructed, five to six different T3 homozygous lines were analyzed. Lines shown in Figures 1C, 1D, 7B, and Supplemental Figure 2 online are representative of typical expression patterns.
Microscopy and Phenotypic Analyses
Root growth rates were examined by marking the position of the root tips daily after shifting to iron-deficient media. Plates were scanned when plants were 8 d old, and ImageJ software was used for root length measurements. Lateral roots and lateral root primordia of 8-d-old plants were measured using light microscopy. Laser scanning confocal microscopy (Zeiss LSM 510) was used for observation of roots of plants stained with 10 μM propidium iodide.
Ferric chelate reductase assays were performed as previously described (Yi and Guerinot, 1996). Chlorophyll content was measured in 2-week-old plants germinated on pH 7 to 8 soil. All leaves were collected and ground to powder in liquid nitrogen. The powder was resuspended in 80% acetone on ice and centrifuged at 10,000g at 4°C for 5 min. Chlorophyll concentrations were calculated from spectroscopy absorbance measurements at 663.2, 646.8, and 470 nm (Lichtenthaler, 1987).
To localize Fe3+, 7- and 14-d-old seedlings grown on iron-sufficient media were vacuum infiltrated with Perl’s stain solution (equal volumes of 4% [v/v] HCl and 4% [w/v] K-ferrocyanide) for 30 min. Seedlings were then incubated for another 30 min in PERL staining solution, washed three times with water, observed, and photographed with a Leica DM500B microscope.
Rhizosphere acidification was performed as previously described (Yi and Guerinot, 1996). Briefly, seeds were germinated on iron-sufficient media for 4 d and then transferred to iron-deficient media for 3 d. The seeds were finally transferred to a 1% agar plate containing 0.006% Bromocresol Purple and 0.2 mM CaSO4 (pH adjusted to 6.5 with NaOH) for 24 h.
Tissue Elemental Analysis
Seeds were evenly placed on the mesh in a single row at a density of ~20 seeds/cm in two rows, germinated, and grown on mesh on iron-sufficient media for 4 d, and then shifted to iron-sufficient or -deficient plates for 3 d. Three plates of seedlings per replicate were combined to generate at least 150 mg of fresh weight. Five replicates of root and shoot samples were collected separately, rinsed in 18 MV water, and analyzed by inductively coupled plasma–mass spectrometry for Li, B, Na, Mg, P, S,K, Ca, Mn, Fe, Co, Ni, Cu, Zn, As, Se, Mo, and Cd as described by Morrissey et al. (2009).
Gene Expression Analysis and Identification of Differentially Expressed Genes
For qRT-PCR analyses, whole roots were sliced off and collected in liquid nitrogen. cDNA was synthesized with Superscript III cDNA synthesis kit (Invitrogen) from RNA extracted with the RNeasy plant mini kit (Qiagen). The primers used for qRT-PCR analysis and their associated gene accession numbers are listed in Supplemental Table 1A online.
For transcriptional profiling of pericycle cells, wild-type seeds from the J2661 enhancer trap line (Levesque et al., 2006) were germinated on iron-sufficient media and then shifted to either iron-sufficient or iron-deficient media for 24 h. Whole roots were sliced off ~0.5 cm from the apex. GFP-marked cells were then collected by fluorescence-activated cell sorting after protoplasting whole roots (Birnbaum et al., 2003). Three replicates of the pericycle samples were collected and analyzed. For global analysis of gene expression of wild-type and pye-1 mutants, whole roots were sliced off ~0.5 cm from the apex and collected in liquid nitrogen after 24 h of exposure to iron-deficient or iron-sufficient media. Two replicates of the whole roots analysis were performed. Total RNA was isolated from the frozen material using the RNeasy plant mini kit (Qiagen). Fragmented cRNA probes were prepared using the two-cycle (pericycle) or one-cycle (whole root) amplification protocol by Affymetrix. Samples were submitted to Expression Analysis, Inc. for hybridization to Arabidopsis whole-genome ATH1 Affymetrix GeneChips.
Normalization of probe sets on all arrays and identification of differentially expressed was performed as described (Levesque et al., 2006; Brady et al., 2007). To detect reproducibility, the Pearson correlation was calculated between replicates, and a cutoff value to 0.92 was used for all experiments except for the radial zone data sets where 0.89 was used. Replicates that did not meet this criterion were not used in the analysis. Raw values for the whole root time course and all radial zones datasets except the pericycle were indicated by Dinneny et al. (2008). Raw values for the pericycle radial zone data set are indicated in Supplemental Data Set 2A online. Raw values of all genes affected in wild-type and pye-1 mutants are indicated in Supplemental Data Sets 3A to 3D online. Affymetrix probe sets that are predicted to hybridize to transcripts from multiple loci were eliminated based on the lookup table published by Affymetrix on May 29, 2008. Probe sets that only detect expression for nuclear-encoded genes were used for further analysis. Genes with a Q-value < 10−4 were declared differentially expressed. For the interpretation of the results, we focused on those with a Q-value < 10−4 and a >1.2- or >1.5-fold change between treatments. To eliminate the effects of protoplasting, genes whose expression was affected by protoplasting in a manner opposite from that seen by the treatment in question were removed from the cell type–specific microarray data sets.
The National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) SuperSeries accession number for both microarray data sets is GSE21582.
Phylogenetic Analysis
Sequences of putative PYE paralogs were aligned using ClustalW (Thompson et al., 1994) (see Supplemental Data Set 4A online). Unrooted phylogenic trees were generated using the neighbor-joining method (Saitou and Nei, 1987). The optimal trees are shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein, 1985). The trees are drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the data set (complete deletion option). Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007).
Yeast Two-Hybrid Analysis
Yeast two-hybrid analysis was performed as previously described (Walhout and Vidal, 2001) using a root-specific cDNA library kindly provided by Ben Scheres. BP cloning (Invitrogen) was used to fuse BTS and PYE, which had both been cloned into pENTR/D TOPO, into pDEST-BD. This reaction generated a 5′ fusion of the yeast Gal4p binding domain (BD) to BTS and PYE (BD:BTS and BD:PYE). These constructs were transformed into yeast strain MaV103 and mated with MaV203, which had been transformed with 300 ng of a root-specific cDNA library fused to the Gal4p activation domain (AD). Interacting colonies were identified by growth after 5 to 10 d on -Leu, -Trp, -His, 20 mM 3-AT media, -Leu, -Trp, -Ura media, and colorimetric detection of β-galactosidase. For directed yeast two-hybrid analysis between BTS, PYE, and PYE homologs, primers listed in Supplemental Table 1B online were used to clone the PYE homologs from Col-0 root-specific cDNA library. Each open reading frame was cloned into pENTR/D-TOPO; the resulting constructs were used along with pDEST-AD in LR cloning (Invitrogen) reactions to fuse each open reading frame to the Gal4p activation domain.
ChIP-on-Chip Assay
pye-1/ProPYE:PYE:GFP and wild-type lines were germinated and grown on iron-deficient media for 5 d. Whole root tissue was fixed and ChIP was performed using a GFP antibody (Abcam) as described previously (Leibfried et al., 2005) with some modifications, including chromatin shearing using a Bioruptor UCD-200 (Diagenode). DNA from the pye-1/ProPYE:PYE:GFP and wild-type ChIPs were blunted using T4 DNA polymerase, cleaned up with the Qiagen reaction cleanup kit, amplified with 22 PCR cycles, and subsequently cleaned up according to the Agilent Mammalian ChIP-on-chip kit protocol. Two micrograms of DNA were fragmented and labeled with Cyanine 3-dUTP and Cyanine 5-dUTP according to the Agilent Mammalian ChIP-on-chip kit protocol. This labeled wild-type and pye-1/ProPYE:PYE:GFP DNA was hybridized to a custom long oligonucleotide (~60-mer) Arabidopsis promoter microarray (NCBI GEO Platform record GPL10326). The probes on this array cover the intergenic region, the first intron, and the 5′ and 3′ untranslated region sequences of nearly all of the genes of the Arabidopsis genome. A total of 244,000 probes are evenly distributed with densities around 125 bp per probe for genes encoding transcription factors and 250 bp per probe for other genes. Hybridization was performed according to the Agilent ChIP-on-chip protocol, and images were obtained using an Agilent microarray scanner (model G2565BA) at a resolution of 5 μm (Sozzani et al., 2010). The NCBI GEO accession number associated with the ChIP-on-chip data is GSE21338.
Signal extraction and initial data processing were done using the Agilent feature extraction software. Two biological replicates were conducted, and two technical replicates performed by dye swapping. Each of the four replicates was treated individually in the subsequent steps. A processed signal ratio for each probe was calculated by dividing the processed signal value of the PYE-GFP sample by the processed signal value of the wild-type sample. To assess the statistical significance for the enrichment, empirical P values were calculated by random sampling of processed signal ratios on a single probe basis. For this, 100,000 randomly sampled processed signal ratios were ranked according their value. From this distribution we derived empirical P values for processed signal ratios with P = r/n, where P is the empirical P value, r is the number of simulated replicates that yield in a signal ratio that is higher than the actual data, and n is the number of replicate samples that have been simulated (in our case 100,000). Whereas the previous steps were performed independently for each replicate, we combined the information from each replicate by calculating the final probe P values by multiplying probe P values over all experiments. Those probe P values were fed into a seed extension algorithm called SeedXrich (Busch et al., 2010). This algorithm systematically combines different parameters to call enriched regions. Those parameters were as follows: (1) length of region in base pairs covered by probes below a defined P value threshold, (2) a local P value minimum (seed), and (3) the number of nucleotides allowed as gaps within called regions. For each combination of parameters, the detected regions were registered. A gene was assigned to an enriched region if that region was present within 4000 bp upstream or 300 bp downstream of the transcription start site, in an intron, or 300 bp downstream of the gene model. Each parameter combination produced a list of called regions and, thus, of assigned genes. The proportion of PYE response genes (genes that are upregulated or downregulated between wild-type and pye-1 mutants under iron deficiency; see Supplemental Data Set 3C online) to all of the assigned genes, which were represented on the ATH1 array, was recorded for each list. To select optimal parameters, we first excluded enriched regions that spanned less than the expected hybridization length of 200 bp. We also had to exclude the upstream regulatory region of PYE because there is an additional copy present in the transgenic line we used for ChIP. We then analyzed the distribution of the lists created by SeedXrich by parameter sweeping for the highest number of PYE responses genes (see Supplemental Figure 8 online). The list was obtained with the following parameters: probe P value seed, P < 5 × 10−9; probe P value, P < 0.006; minimum length of hybridization, 210 bp; maximum gap, 200 bp. The SeedXrich program is available upon request as a python script. To further evaluate our results, we compared them to the results that we would have obtained using the ChIPmix algorithm (Martin-Magniette et al., 2008) in a regular fashion. Compared with SeedXrich, none of the different false discovery rate thresholds that we tested using ChIPmix produced a higher overlap with PYE response genes (see Supplemental Figure 8 online).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At3g47640 (PYE), AT5G13740 (ZIF1), AT1G23020 (FRO3), AT1G56430 (NAS4), AT1G20010 (βtub), At3g18290 (BTS), At4g36060 (bHLH11), At3g23210 (bHLH34), At4g14410 (bHLH104), At5g54680 (bHLH105), At1g51070 (bHLH115), and At3g19860 (bHLH121). Additional accession numbers can be found in Supplemental Table 1 online.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Early Responses to Iron Deficiency.
Supplemental Figure 2. PYE Expression Pattern and Localization.
Supplemental Figure 3. Effects of Loss of PYE Function on Root Development under Iron-Sufficient Conditions.
Supplemental Figure 4. Effects of Loss of PYE Function on Shoot Development under Iron-Sufficient Conditions and PYE Complementation.
Supplemental Figure 5. Perl Stain of pye-1 Roots and Leaves.
Supplemental Figure 6. BTS Protein Alignment.
Supplemental Figure 7. PYE and BTS Yeast Two-Hybrid Analysis.
Supplemental Figure 8. Detection of PYE Direct Targets by Intersecting Genes from the Set of PYE Response Genes (Supplemental Table 3C) to the ChIP-on-Chip Gene Lists.
Supplemental Table 1. Primers and Accession Numbers.
Supplemental Data Set 1. Genes Affected by Iron Deficiency in Wild-Type Roots.
Supplemental Data Set 2. Pericycle-Specific Transcriptional Profile.
Supplemental Data Set 3. Microarray Analysis of Wild-Type and pye-1 Mutants after 24 h of Exposure to +Fe or –Fe Media.
Supplemental Data Set 4. Alignment of PYE and Putative PYE Homologs and Orthologs.
Acknowledgments
We thank Joel Burrill, Krisstonia Spruiell, and Kimberly Lewis for technical assistance, David Orlando for writing the R-script used for running the affinity propagation clustering algorithm, Hongchang Cui for designing and implementing the ChIP-on-chip technology, and Mary Lou Guerinot, Erin Connolly, José Dinneny, Siobhan Brady, and members of the Benfey lab and Guerinot lab for helpful discussion and reviewing of the manuscript. T.A.L. was supported by a National Science Foundation Minority Postdoctoral Research Fellowship. This work was funded by a grant from the National Science Foundation AT2010 Program.
References
- Arrivault S., Senger T., Krämer U. (2006). The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 46: 861–879 [DOI] [PubMed] [Google Scholar]
- Bauer P., Ling H.-Q., Guerinot M.L. (2007). FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol. Biochem. 45: 260–261 [DOI] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960 [DOI] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brutnell T.P., Sawers R.J.H., Mant A., Langdale J.A. (1999). BUNDLE SHEATH DEFECTIVE2, a novel protein required for posttranslational regulation of the rbcL gene of maize. Plant Cell 11: 849–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckhout T.J., Yang T.J., Schmidt W. (2009). Early iron-deficiency-induced transcriptional changes in Arabidopsis roots as revealed by microarray analyses. BMC Genomics 10: 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W., et al. (2010). Transcriptional control of a plant stem cell niche. Dev. Cell 18: 849–861 [DOI] [PubMed] [Google Scholar]
- Colangelo E.P., Guerinot M.L. (2004). The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C., Cassin G., Couch D., Divol F., Higuchi K., Le Jean M., Misson J., Schikora A., Czernic P., Mari S. (2009). Metal movement within the plant: Contribution of nicotianamine and yellow stripe 1-like transporters. Ann. Bot. (Lond.) 103: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai M., Hu J. (2008). Light induces peroxisome proliferation in Arabidopsis seedlings through the photoreceptor phytochrome A, the transcription factor HY5 HOMOLOG, and the peroxisomal protein PEROXIN11b. Plant Physiol. 146: 1117–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny J.R., Long T.A., Wang J.Y., Jung J.W., Mace D., Pointer S., Barron C., Brady S.M., Schiefelbein J., Benfey P.N. (2008). Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Durrett T.P., Gassmann W., Rogers E.E. (2007). The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 144: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D., Broderius M., Fett J., Guerinot M.L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espartero J., Sánchez-Aguayo I., Pardo J. (1995). Molecular characterization of glyoxalase-I from a higher plant; upregulation by stress. Plant Mol. Biol. 29: 1223–1233 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Frey B.J., Dueck D. (2007). Clustering by passing messages between data points. Science 315: 972–976 [DOI] [PubMed] [Google Scholar]
- Green L.S., Rogers E.E. (2004). FRD3 controls iron localization in Arabidopsis. Plant Physiol. 136: 2523–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotz N., Guerinot M.L. (2006). Molecular aspects of Cu, Fe and Zn homeostasis in plants. Biochim. Biophys. Acta 1763: 595–608 [DOI] [PubMed] [Google Scholar]
- Haydon M.J., Cobbett C.S. (2007). A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiol. 143: 1705–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim M.A., Jakoby M., Werber M., Martin C., Weisshaar B., Bailey P.C. (2003). The basic helix-loop-helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Edwards E.A., Leyland N.R., Bean S., Mullineaux P.M. (2000). pGreen: A versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Jeong J., Connolly E.L. (2009). Iron uptake mechanisms in plants: Functions of the FRO family of ferric reductases. Plant Sci. 176: 709–714 [Google Scholar]
- Kim S.A., Punshon T., Lanzirotti A., Li L., Alonso J.M., Ecker J.R., Kaplan J., Guerinot M.L. (2006). Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314: 1295–1298 [DOI] [PubMed] [Google Scholar]
- King J. (1991). The Genetic Basis of Plant Physiological Processes. (London: Oxford University Press). [Google Scholar]
- Klatte M., Schuler M., Wirtz M., Fink-Straube C., Hell R., Bauer P. (2009). The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol. 150: 257–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanquar V., Lelievre F., Bolte S., Hames C., Alcon C., Neumann D., Vansuyt G., Curie C., Schroder A., Kramer U., Barbier-Brygoo H., Thomine S. (2005). Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J. 24: 4041–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapinskas P.J., Lin S.J., Culotta V.C. (1996). The role of the Saccharomyces cerevisiae CCC1 gene in the homeostasis of manganese ions. Mol. Microbiol. 21: 519–528 [DOI] [PubMed] [Google Scholar]
- Lee J.Y., Colinas J., Wang J.Y., Mace D., Ohler U., Benfey P.N. (2006). Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 103: 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibfried A., To J.P.C., Busch W., Stehling S., Kehle A., Demar M., Kieber J.J., Lohmann J.U. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Levesque M.P., Vernoux T., Busch W., Cui H., Wang J.Y., Blilou I., Hassan H., Nakajima K., Matsumoto N., Lohmann J.U., Scheres B., Benfey P.N. (2006). Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol. 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1987). Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 148: 350–382 [Google Scholar]
- Li L., Chen O.S., McVey Ward D., Kaplan J. (2001). CCC1 is a transporter that mediates vacuolar iron storage in yeast. J. Biol. Chem. 276: 29515–29519 [DOI] [PubMed] [Google Scholar]
- Ling H.Q., Koch G., Baumlein H., Ganal M.W. (1999). Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc. Natl. Acad. Sci. USA 96: 7098–7103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark A.J., Marjorie J.R., Nicholas S. (2006). Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J. 45: 83–100 [DOI] [PubMed] [Google Scholar]
- Martin-Magniette M.-L., Mary-Huard T., Berard C., Robin S. (2008). ChIPmix: Mixture model of regressions for two-color ChIP-chip analysis. Bioinformatics 24: i181–i186 [DOI] [PubMed] [Google Scholar]
- McElver J., et al. (2001). Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moog P.R., van der Kooij T.A., Bruggemann W., Schiefelbein J.W., Kuiper P.J. (1995). Responses to iron deficiency in Arabidopsis thaliana: The Turbo iron reductase does not depend on the formation of root hairs and transfer cells. Planta 195: 505–513 [DOI] [PubMed] [Google Scholar]
- Morrissey J., Baxter I.R., Lee J., Li L., Lahner B., Grotz N., Kaplan J., Salt D.E., Guerinot M.L. (2009). The ferroportin metal efflux proteins function in iron and cobalt homeostasis in Arabidopsis. Plant Cell 21: 3326–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M., Schmidt W. (2004). Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol. 134: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37: D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C.M., Guerinot M.L. (2009). Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat. Chem. Biol. 5: 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey R.A., Woodward A.W., Hobbs B.N., Tierney M.P., Lahner B., Salt D.E., Bartel B. (2006). An Arabidopsis basic helix-loop-helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics 174: 1841–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S., Quan S., Aung K., Yang P., Manandhar-Shrestha K., Holbrook D., Linka N., Switzenberg R., Wilkerson C.G., Weber A.P.M., Olsen L.J., Hu J. (2009). In-depth proteome analysis of Arabidopsis leaf peroxisomes combined with in vivo subcellular targeting verification indicates novel metabolic and regulatory functions of peroxisomes. Plant Physiol. 150: 125–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N.J., Procter C.M., Connolly E.L., Guerinot M.L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rogers E.E., Guerinot M.L. (2002). FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell 14: 1787–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römheld V., Marschner H. (1981). Iron deficiency stress induced morphological and physiological changes in root tips of sunflower. Physiol. Plant. 53: 354–360 [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Schaaf G., Honsbein A., Meda A.R., Kirchner S., Wipf D., von Wiren N. (2006). AtIREG2 encodes a tonoplast transport protein involved in iron-dependent nickel detoxification in Arabidopsis thaliana roots. J. Biol. Chem. 281: 25532–25540 [DOI] [PubMed] [Google Scholar]
- Schmidt W. (1999). Review: Mechanisms and regulation of reduction-based iron uptake in plants. New Phytol. 141: 1–26 [Google Scholar]
- Sheriff S., Hendrickson W.A., Smith J.L. (1987). Structure of myohemerythrin in the azidomet state at 1.7/1.3 A resolution. J. Mol. Biol. 197: 273–296 [DOI] [PubMed] [Google Scholar]
- Sozzani R., Cui H., Moreno-Risueno M.A., Busch W., Van Norman J.M., Vernoux T., Brady S.M., Dwitte W., Murray J.A., Benfey P.N. (2010). Spatiotemporal regulation of cell-cycle gene by SHORTROOT links patterning and growth. Nature 466: 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp R.E., Sieker L.C., Jensen L.H., McCallum J.D., Sanders-Loehr J. (1985). Active site structures of deoxyhemerythrin and oxyhemerythrin. Proc. Natl. Acad. Sci. USA 82: 713–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 11: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida T., Yano H., Satake K., Kubota I., Tsugita A. (1990). The amino acid sequence of hemerythrin from Siphonosoma cumanense. Protein Seq. Data Anal. 3: 141–147 [PubMed] [Google Scholar]
- Vert G., Grotz N., Dedaldechamp F., Gaymard F., Guerinot M.L., Briat J.F., Curie C. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout A.J.M., Vidal M. (2001). High-throughput yeast two-hybrid assays for large-scale protein interaction mapping. Methods 24: 297–306 [DOI] [PubMed] [Google Scholar]
- Walker E.L., Connolly E.L. (2008). Time to pump iron: Iron deficiency signaling mechanisms of higher plants. Curr. Opin. Plant Biol. 11: 530–535 [DOI] [PubMed] [Google Scholar]
- Waters B.M., Chu H.H., Didonato R.J., Roberts L.A., Eisley R.B., Lahner B., Salt D.E., Walker E.L. (2006). Mutations in Arabidopsis yellow stripe-like1 and yellow stripe-like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol. 141: 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T.J.W., Lin W.-D., Schmidt W. (2010). Transcriptional profiling of the Arabidopsis iron deficiency response reveals conserved transition metal homeostasis networks. Plant Physiol. 152: 2130–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y., Guerinot M.L. (1996). Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 10: 835–844 [DOI] [PubMed] [Google Scholar]
- Yi Y., Saleeba J.A., Guerinot M.L. (1994). Iron uptake in Arabidopsis thaliana In Biochemistry of Metal Micronutrients in the Rhizosphere, J. Manthey, D. Luster, and D.E. Crowley, eds (Chelsea, MI: Lewis Publishers), pp. 295–307. [Google Scholar]
- Yuan Y., Wu H., Wang N., Li J., Zhao W., Du J., Wang D., Ling H.-Q. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Zhang H., Forde B.G. (1998). An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407–409 [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. (1965). Evolving Genes and Proteins. (New York: Academic Press). [Google Scholar]