There is a great demand to increase meiotic crossovers frequency in order to boost genetic diversity in traditional breeding and genetic studies. This work shows that this can be achieved by manipulating karyotype composition (diploid vs. allotriploid vs. allotetraploid) of plant hybrids.
Abstract
Meiotic crossovers are necessary to generate balanced gametes and to increase genetic diversity. Even if crossover number is usually constrained, recent results suggest that manipulating karyotype composition could be a new way to increase crossover frequency in plants. In this study, we explored this hypothesis by analyzing the extent of crossover variation in a set of related diploid AA, allotriploid AAC, and allotetraploid AACC Brassica hybrids. We first used cytogenetic methods to describe the meiotic behavior of the different hybrids. We then combined a cytogenetic estimation of class I crossovers in the entire genome by immunolocalization of a key protein, MutL Homolog1, which forms distinct foci on meiotic chromosomes, with genetic analyses to specifically compare crossover rates between one pair of chromosomes in the different hybrids. Our results showed that the number of crossovers in the allotriploid AAC hybrid was higher than in the diploid AA hybrid. Accordingly, the allotetraploid AACC hybrid showed an intermediate behavior. We demonstrated that this increase was related to hybrid karyotype composition (diploid versus allotriploid versus allotetraploid) and that interference was maintained in the AAC hybrids. These results could provide another efficient way to manipulate recombination in traditional breeding and genetic studies.
INTRODUCTION
Meiosis and recombination are essential to the life cycles of all sexual eukaryotes. Meiosis is a two-round cell division during which chromosome number is halved (from diploid to haploid) and gametes are generated. In most organisms, accurate separation of maternal and paternal chromosomes during the first division requires that they first be connected to one another by crossovers. Crossovers are one of the products of meiotic double-strand break repair, which occurs during prophase I. This connection is established by the exchange of homologous chromatid arms (manifested as cytological features called chiasmata) plus cohesion between sister chromatids, which are both essential for proper attachment of bipolar homologs to the meiosis I spindle (Whitby, 2005). In addition, crossovers produce new combinations of chromosomes/alleles at different genetic loci, thereby boosting genome variability.
The number and distribution of crossovers during meiosis are subject to very stringent controls (Mézard et al., 2007). Typically, every chromosome undergoes at least one obligate crossover to ensure proper segregation at metaphase I (Jones, 1984). The total number and relative position of crossovers on each chromosome are then limited by a phenomenon called interference (i.e., a crossover in one region reduces the probability that a second crossover occurs simultaneously in an adjacent region, so that the distance between crossovers is larger than would be expected if they occurred independently; Muller, 1916; Jones, 1984). Formation of interfering crossovers (class I crossovers) is catalyzed by a set of specific meiotic proteins called ZMM (for Zip1, Zip2, Zip3, Zip4, Mer3, and Msh4/Msh5; Lynn et al., 2007) and depends on MutL Homolog1 (MLH1; Lhuissier et al., 2007). Not all crossovers are affected by interference; a second class of noninterfering crossovers is catalyzed by other proteins, such as Methyl methansulfonate and UV Sensitive 81 (MUS81), that are responsible for 15 to 30% of all crossovers (see Mézard et al., 2007, for review). The net result of crossover control is at least one (obligate) crossover per chromosome and a tendency for multiple crossovers (more than two) to be rare.
The possibility of increasing crossover frequency has attracted considerable interest because of the obvious practical applications in traditional breeding and genetic studies. Boosting crossovers would effectively speed up the combining of valuable traits from different parents in new elite varieties (Wijnker and de Jong, 2008), accelerate the removal of linkage drag during the introgression of valuable genes from exotic germplasms (Able and Langridge, 2006; Martinez-Perez and Moore, 2008), and facilitate the construction of highly recombinogenic lines and thus help improve genetic maps and positional cloning techniques. Recently, Carlton et al. (2006) showed that the presence of a single pair of univalents during meiosis of Caenorhabditis elegans induces a compensatory increase in crossovers on the recombining chromosomes involved in bivalents of the same nuclei. This observation is in agreement with at least two studies in plants (Parker, 1975; Tease and Jones, 1975). It suggests that chromosomes that carry early recombination intermediates but ultimately fail to form a crossover may cause “the nucleus to linger in a recombination active state,” in which a higher number of crossovers can be completed on the chromosomes that were correctly synapsed (Martinez-Perez and Moore, 2008). Thus, an important direction that studies should take is to analyze the effect of univalents on the frequency of meiotic recombination among the remaining chromosomes. Interspecific hybrids, in which the number of chromosomes that fail to form crossovers can be finely tuned, provide a good way to address this question.
In this study, we analyzed the extent of crossover variation in different closely related Brassica hybrids: a diploid hybrid that undergoes regular meiosis without univalents; a newly formed allotetraploid hybrid that, in theory, can form a few univalents and multivalents; and a digenomic allotriploid hybrid in which systematic univalents are expected. We characterized the meiotic behavior of the diploid, allotriploid, and allotetraploid hybrids produced from the same Brassica rapa genotypes by using different cytogenetic methods (classical techniques and BAC-FISH [for fluorescent in situ hybridization] analyses). We then compared crossover rates between pairs of homologous chromosomes in these hybrids using two independent and complementary methods: the number of class I crossovers in the entire genome was counted using a cytogenetic approach, and genetic distances were used to infer the number of crossovers between one pair of A chromosomes (class I and II). We showed that crossover rates were significantly higher in the allotetraploid hybrid than in the diploid hybrid and were highest in the allotriploid hybrid. Our results also indicated that crossovers remained affected by interference at meiosis of the allotriploid hybrids, in spite of the high frequency of multiple crossovers per chromatid observed in this hybrid. These results are important because they provide another way to manipulate (some of) the mechanisms determining the rate of crossovers and/or to use them in plant breeding programs.
RESULTS
Meiotic Behavior in Hybrids
As a first step to analyze the extent of crossover variation in relation to karyotype composition in Brassica hybrids, we investigated meiotic behavior at prophase I of diploid (ArAr′), allotetraploid (ArAr′CoCo), and allotriploid (ArAr′Co) hybrids produced from the same B. rapa genotypes (Figure 1). We examined meiotic stages by DNA staining of pollen mother cells (PMCs) undergoing meiosis, and we also used BAC-FISH to specifically identify chromosomes from the A and C genomes (see Supplemental Figures 1 and 2 online).
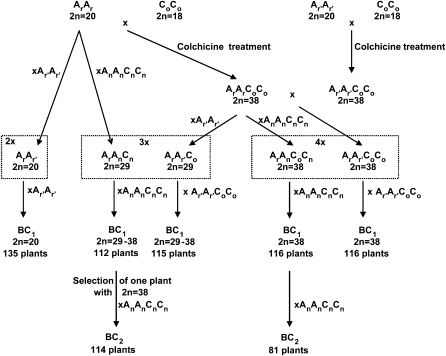
Figure 1.
Schematic Detailing the Production of Hybrids and Segregating Backcross Populations.
ArAr and Ar′Ar′ represent the C1.3 and Z1 B. rapa plants, respectively, CoCo designates the B. oleracea cultivars (RC and HDEM), and AnAnCnCn stands for the B. napus cv Darmor. The genomic structure of F1 hybrids is shown in dotted boxes (2x, 3x, or 4x for diploid, allotriploid, or allotetraploid, respectively).
Meiosis of the diploid hybrid (ArAr′) appeared normal and very regular (see Supplemental Figure 1 online). At diakinesis, 10 bivalent structures were always observed, where chromosomes were maintained together by sister chromatid cohesion and chiasmata. Although bivalent configurations could be classified into several categories, the majority (four to five on average) were ring bivalents, with their two arms bound by chiasmata (see Supplemental Figures 1D and 1E online). Assuming that most other configurations resulted from a single chiasma, we estimated that there were 1.2 to 1.7 crossovers on average on every bivalent.
In the allotetraploid ArAr′CoCo hybrid, the early stages of prophase I could not be distinguished from those in the diploid hybrid. We only observed a few cells with multiple alignments at pachytene (see Supplemental Figure 1G online) and chiasmatic multivalents at diakinesis (see Supplemental Figure 1H online) and metaphase I (Table 1). Approximately 70% of the PMCs displayed 19 bivalents, as expected (Table 1). The BAC-FISH analyses confirmed that bivalents were mostly formed between pairs of A and pairs of C chromosomes (see Supplemental Table 1 and Supplemental Figures 2A and 2B online), in spite of some occasional recombination between homeologous chromosomes (see Supplemental Figures 2C and 2D online). One notable feature of meiosis in the allotetraploid ArAr′CoCo hybrid was the presence of one to three bivalents per PMC that had an atypical compact appearance (see Supplemental Figures 1H and 1I online), as if they were more intimately linked than in the diploid hybrid.
Table 1.
Average Meiotic Behavior at Metaphase I of AAC and AACC Hybrids
| Average No. ofb |
||||||
| Hybridsa | No. of Chromosomes | No. of PMCs | I | II | III | IV |
| ArAr′CoCo | 38 | 41 | 0.19 (0–2) | 18.56 (17–19) | 0.00 | 0.17 (0–1) |
| ArAnCoCn | 38 | 21 | 0.24 (0–2) | 18.33 (17–19) | 0.05 (0–1) | 0.24 (0–1) |
| ArAr′Co | 29 | 62 | 8.79 (5–11) | 10.00 (7–11) | 0.05 (0–1) | 0.02 (0–1) |
| ArAnCn | 29 | 23 | 8.62 (6–11) | 10.05 (9–11) | 0.09 (0–1) | 0.00 |
Values in parentheses indicate the range of variation.
Hybrids produced using the two B. rapa genotypes (ArAr′CoCo and ArAr′Co) are distinguished from those derived using B. napus cv Darmor (ArAnCoCn and ArAnCn).
I stands for univalents, II for bivalents, III for trivalents, and IV for quadrivalents.
Meiosis of the ArAr′Co allotriploid hybrid was obviously disrupted during prophase I. As expected from the uneven number of chromosomes in this hybrid (29), some chromosomes remained as univalents at pachytene (see Supplemental Figure 1J online). At diakinesis, it was sometimes difficult to distinguish bivalents from univalents because the bivalent-like entities displayed a very unusual compact appearance (see Supplemental Figures 1K and 1L online). In particular, we only rarely identified ring bivalents with one terminal chiasma recognizable in each arm at diakinesis. At metaphase I, ~70% of the cells examined in the ArAr′Co hybrid displayed 9 univalents and 10 bivalents (Table 1). The BAC-FISH analyses showed that bivalents were mostly formed by A chromosomes but not exclusively: illegitimate recombination occasionally occurred between A and C chromosomes or even between two C chromosomes (see Supplemental Table 1 and Supplemental Figures 2E and 2F online).
Although it was not possible to reliably estimate chiasma frequencies in the allotetraploid and the allotriploid hybrids, all our observations suggested that bivalents were bound by more crossovers in the allotetraploid and allotriploid hybrids than in the diploid hybrid. Cytogenetic and genetic analyses were thus performed to quantify the difference in crossover rates among the different hybrids.
Estimation of Crossover Rates in the Diploid, Allotriploid, and Allotetraploid Hybrids
Crossover rates were estimated in two independent and complementary ways, by cytologically counting the number of crossovers and by examination of genetic distances. First, we used an anti-Arabidopsis thaliana MLH1 antibody to determine interfering crossover frequencies. MLH1 is one of the eukaryotic homologs of Escherichia coli MutL, which is essential for wild-type levels of crossing over in budding yeast, mammals, and plants (Lhuissier et al., 2007, and refs. therein). In Arabidopsis, immunolocalization of MLH1 antibodies in diakinesis PMCs reveals distinct foci on chromatin, which correspond to positions of interfering (class I) crossovers (Chelysheva et al., 2010). In order to compare the numbers of MLH1 foci in all hybrids, in which not only bivalents but also an additional number of multivalents were observed (Table 1), we chose to relate the number of MLH1 foci to the number of “recombining chromosomes”; this number was estimated in every PMC by dividing the total number of MLH1 foci by the number of chromosomes involved in recombination events (i.e., that showed at least one MLH1 focus).
The diploid hybrid ArAr′ showed on average 16.5 MLH1 foci per nucleus (1.65 crossovers per bivalent), which is in close agreement with our chiasma estimation from 4′,6-diamidino-2-phenylindole (DAPI) observations. As only bivalents were observed at diakinesis of the diploid hybrid, the mean number of MLH1 foci per recombining chromosome was 0.83 (Figures 2A–2C, Table 2). In the allotetraploid hybrid (ArAr′CoCo), we counted an average of 37.6 MLH1 foci per PMC, which corresponded to 1.0 foci per recombining chromosome (Figures 2D–2F, Table 2). This difference was statistically significant from that in the diploid hybrid (one-way analysis of variance; P = 0.0082), representing a 1.2-fold increase in the class I crossover rate. In the allotriploid hybrid, 6 to 10 chromosomes without MLH1 foci (Figures 2I–2L, asterisks) were systematically observed in the PMCs, which corresponded to univalents. An average of 29.3 MLH1 foci per nucleus was scored in the allotriploid hybrid (ArAr′Co), which corresponded to 1.40 foci per recombining chromosome (Figures 2G–2L, Table 2). This was a 1.7-fold increase (one-way analysis of variance; P < 0.0001) in the class I crossover rate compared with the diploid and hybrids and a 1.4-fold increase compared with the allotetraploids. Interestingly, in every nucleus of the allotriploid hybrids, at least five bivalents showed more than two foci, and three to four bivalents repeatedly showed four MLH1 foci (Figures 2I, early diakinesis, and 2L, late diakinesis), whereas in the diploid hybrid, bivalents showed only one or two foci. Together, these findings indicated a genome-wide increase in class I crossover rate in the allotetraploid hybrid and an even bigger increase in the allotriploid hybrid compared with the diploid hybrid.
Figure 2.
MLH1 Immunolocalization in PMCs of the ArAr Diploid ([A]–[C]), ArAr′CoCo Allotetraploid ([D]–[F]), and ArAr′Co Allotriploid ([G]–[L]) Hybrids.
Chromosomes at diakinesis stage were stained with DAPI (red) and the Arabidopsis MLH1 antibody (green). (C), (F), (I), and (L) were generated by merging DAPI and anti-MLH1. Arrows indicate multivalents, and asterisks show univalents. Bars = 10 μm.
Table 2.
Average Number of MLH1 Foci at Diakinesis of the ArAr′, ArAr′CoCo, and ArAr′Co Hybrids
| Hybrids | No. of PMC | Average No. of MLH1 Foci | Average No. of MLH1 Foci per Recombining Chromosomea |
| ArAr′ | 8 | 16.5 (14–19) | 0.83 (0.70–0.95) |
| ArAr′CoCo | 22 | 37.6 (29–46) | 1.00 (0.76–1.21) |
| ArAr′Co | 45 | 29.3 (23–36) | 1.40 (1.09–1.89) |
Values in parentheses indicate the range of variation.
For each PMC, the average number of MLH1 foci per recombining chromosome was estimated by dividing the total number of MLH1 foci by the number of chromosomes showing at least one MLH1 focus.
For the second estimation of crossover rates, we compared genetic map lengths to account for the two classes of crossovers. Comparisons of genetic map lengths were performed using three backcross (BC) progeny that were genotyped with 18 to 19 polymorphic markers positioned along linkage group A7. These comparisons were justified and reliable because we used identical A genotypes and markers to construct the linkage maps.
Map distances were much higher in the progeny of the allotetraploid hybrid (ArAr′CoCo) compared with that of the diploid hybrid (ArAr′). The same molecular markers collectively expanded the total length of linkage group A7 from 52 centimorgan (cM; in the progeny of the diploid hybrid ArAr′) to 96 cM (in the progeny of the allotetraploid hybrid ArAr′CoCo; Figure 3). The proportion of crossovers in ArAr′CoCo was on average 1.9 fold higher than in ArAr′, with significant differences for three of the seven intervals where the χ2 test was feasible, depending on the number of plants in each class. At the scale of the linkage group, considered as a combination of intervals with or without crossovers, the difference between the two hybrids was highly significant (χ2 test, df = 10, P = 5.5e-5).
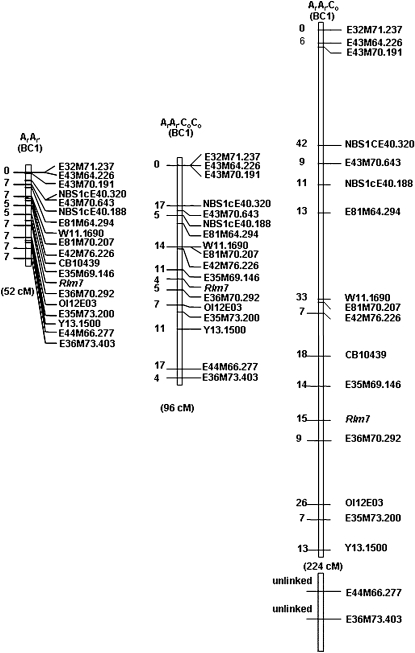
Figure 3.
Maps of the A7 Linkage Group in Progeny of the Diploid (ArAr′), Allotriploid (ArAr′Co), and Allotetraploid (ArAr′CoCo) Hybrids.
Genetic distances, indicated on the left of the linkage group, are expressed in cM and represent the distance between the marker and the annotated marker immediately above.
Map distances were even larger in the progeny of the allotriploid hybrid (ArAr′Co), in which the A7 linkage group was 224 cM long (Figure 3). In addition, two molecular markers (E36M73.403 and E44M66.277), located at the very end of A7, remained unlinked in the ArAr′Co hybrid progeny. The proportion of crossovers in the ArAr′Co hybrid, calculated for every interval between adjacent markers on A7, was on average 6.1-fold higher than in the ArAr′ hybrid (with significant differences observed for the 12 intervals for which a χ2 test was feasible) and 3.1-fold higher than in ArAr′CoCo (with significant differences for 9 of the 13 intervals for which a χ2 test was feasible). At the scale of the linkage group, considered as a combination of intervals with or without crossovers, the differences were highly significant between the ArAr′Co and ArAr hybrids (χ2 test, df = 13, P = 4.6e-58) or between the ArAr′Co and ArAr′CoCo (χ2 test, df = 12, P = 2.6e-27).
It is obvious from our results that genetic map length expansions were higher than chiasma count increases. These discrepancies were not due to experimental error in classifying marker data (Lincoln and Lander, 1992). As misclassifications usually result from markers that appear recombined with both flanking markers, we removed all singletons from the data set, even if some were double checked. The genetic maps obtained for the allotetraploid and allotriploid hybrids with corrected data points still remained inflated compared with chiasma counts. Likewise, genetic map inflation was not related to marker type; removing AFLP markers did not change the extent of the increase between the different maps. Therefore, we are confident that our genetic data are sound.
Our combined cytogenetic and genetic analyses thus demonstrated that crossovers get a boost in the allotetraploid and allotriploid hybrids produced from the B. rapa genotypes (Figure 1). We then examined whether map distance expansions still occurred when the A and C genomes are from different genotypes.
For this, we first reanalyzed some of the data presented by Nicolas et al. (2009) and compared crossover rates between allotriploid (ArAnCn) and allotetraploid (ArAnCoCn) interspecific hybrids produced by crossing either B. rapa C1.3 or synthetic Brassica napus RCC to natural B. napus cv Darmor. At metaphase I, the meiotic configurations of these hybrids were very similar to the allotriploid and allotetraploid hybrids previously described (Table 1). The genetic sizes of the A7 linkage group, based on 12 markers, varied from 54 cM in the BC1 progeny of the allotetraploid hybrid to 239 cM in the BC1 progeny of the corresponding allotriploid hybrid. Although no corresponding diploid control was available, this result confirmed that crossovers get a boost in the allotriploid hybrid compared with the allotetraploid hybrid, irrespective of the origin of the A and C genomes.
Finally, we investigated whether karyotype composition (allotriploid versus allotetraploid) was the major cause of map distance expansion by analyzing genetic distances in two BC2 progeny. These were produced from two BC1 plants with the same genomic structure (2n = 38) but derived from the progeny of either an allotriploid or an allotetraploid hybrid with the B. napus cv Darmor as parent (Figure 1). The genetic sizes of the A7 linkage group were 75.4 cM (using only 10 markers) and 86 cM (using 12 markers) in the two BC2 progeny (see Supplemental Figure 3 online). At the linkage group scale as well as at the interval scale, crossover rates were similar in the two BC2 progeny. At the interval scale, crossover rates in the two BC2 progeny were similar and appeared slightly higher than in the allotetraploid BC1 progeny for two of the three intervals where the χ2 test was feasible (χ2 test, df = 2, P = 0.032 and P = 0.037) and lower than in the allotriploid BC1 progeny for the four intervals for which a test was feasible (Figure 4).
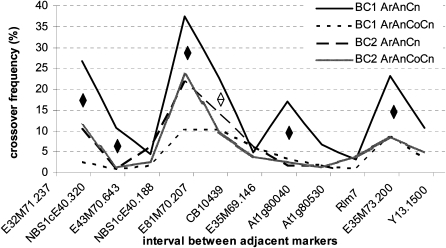
Figure 4.
Crossover Rates in Allotriploid and Allotetraploid Hybrids and BC1 Progeny along the A7 Linkage Group.
Black diamonds indicate that the crossover frequency (as measured by the proportion of plants showing recombination for each interval) estimated in the BC1 progeny of ArAnCn differed from that estimated using any of the other three progeny. White diamonds indicate that crossover frequencies in the BC1 progeny of ArAnCn are significantly different from both BC1 and BC2 progeny of ArAnCnCn but not significantly different from the BC2 progeny of ArAnCn. See Figure 1 for BC1 progeny lineage. A False Discovery Rate correction was applied to account for pairwise multiple comparisons.
These findings confirmed that crossover rates increased as a result of karyotype composition (allotriploid versus allotetraploid). One manifestation of this increase was the occurrence of multiple crossovers per bivalent (Figure 2). Genetic data indicated that the mean number of exchange points per chromatid was 3.0 in the ArAr′Co hybrid compared with 0.5 in the ArAr′ hybrid and 0.9 in the ArAr′CoCo hybrid (see Supplemental Figure 4 online). Likewise, the mean number of exchange points per chromatid in the other genetic background was 1.7 (ranging from 0 to 3) in the ArAnCn hybrid compared with 0.5 (ranging from 0 to 2) in the ArAnCoCn hybrid. This increase in the number of crossovers per chromatid led us to investigate whether these multiple crossovers were still affected by interference.
Analysis of Crossover Interference in the Progeny of ArAr′Co and ArAnCn Hybrids
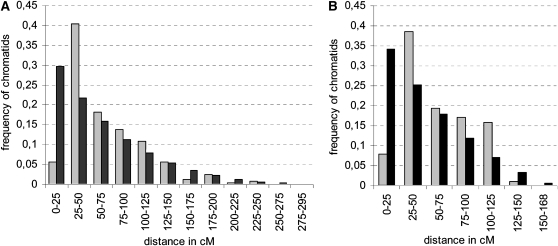
The distance between adjacent crossovers, expressed as the percentage of crossovers observed in this interval over the whole population, was calculated for every chromatid showing at least two crossovers. This analysis could only be performed using the progeny of the allotriploid hybrids, because the number of multiple crossovers in the populations derived from diploid and allotetraploid hybrids was too low for statistical analyses. We calculated 230 and 88 distances in the progeny of ArAr′Co and ArAnCn, respectively. The distributions of these observed genetic distances between adjacent crossovers were significantly different (α = 5%) from the distributions expected if the position of one crossover was independent of the location of the other one(s) (Figure 5). In particular, the shortest distances (from 0 to 25 cM) were observed at frequencies 4.3- to 5.2-fold lower than expected in the progeny of ArAnCn and ArAr′Co, respectively. Likewise, on the 22 intervals of the linkage group that were shorter than 40 cM and carried at least three markers, only 18 and 12 double crossover events were observed in ArArCo and ArAnCn, respectively, whereas 44 and 26 events were expected, respectively, if there was no interference. These two analyses led to the same conclusion: that multiple crossovers were not located independently from one another. Therefore, interference was still affecting crossover distribution even if multiple crossovers occurred very frequently.
Figure 5.
Distribution of the Distances between Adjacent Crossovers Expressed in cM.
The histograms in black represent the theoretical distribution, which assumes that the positions of multiple (more than one) crossovers are independent from one another. The histogram in gray represents the observed distribution in the BC1 progeny of ArAr′Co (A) or ArAnCn (B) hybrids.
DISCUSSION
Variations in the number of crossovers depend on environmental (e.g., temperature; Francis et al., 2007), developmental (Francis et al., 2007), sex-specific (Drouaud et al., 2007), genetic (Robbins et al., 1995; Sanchez-Moran et al., 2002; Esch et al., 2007), or genomic (Dvorak et al., 1998; Barth et al., 2001; Dooner, 2002) cues. It was recently shown that when some chromosomes in a nucleus fail to form crossovers, this may lead to a compensatory increase in crossovers on other recombining chromosomes (Carlton et al., 2006). Based on this result, which echoed older work on Crepis capillaris and in Hypochoeris radicata (Parker, 1975; Tease and Jones, 1975), new ways can be envisaged to manipulate recombination in plants. In this study, we explored this prospect by analyzing meiotic recombination in allotriploid and allotetraploid Brassica hybrids.
Our cytogenetic and genetic data demonstrate in two independent ways that crossover rate increases from the diploid to the allotetraploid hybrids and from the allotetraploid to the allotriploid hybrids. This increase is not sex-specific, as it was detected in both male (cytogenetic survey) and female (genetic survey) meiosis. Considering that genetic map length expansions (Figure 3) were higher than chiasma count increases (Table 2), it could be hypothesized that crossovers get a higher boost during female as compared with male meiosis. This hypothesis remains tentative, and several alternative explanations could be proposed.
First, using MLH1 immunolocalization, we observed only interfering (class I) crossovers, whereas with genetic analyses, we observed the results of both interfering and noninterfering crossovers. It is possible, therefore, that the number of noninterfering crossovers got a boost in the allotetraploid and allotriploid hybrids, which would increase the genetic maps without increasing the number of MLH1 foci. This hypothesis is difficult to test, because it is not possible to specifically label noninterfering crossovers; for example, although antibodies were successfully raised against Arabidopsis MUS81 (upon which noninterfering crossovers depend), the number of MUS81 foci seems to exceed the number of noninterfering crossovers expected in Arabidopsis (Higgins et al., 2008). In addition, the occurrence of residual crossovers in the yeast and Arabidopsis msh4 mus81 double mutants suggests that a third recombination pathway exists (activated in a double mutant background at least), which has not yet been characterized (Whitby, 2005; Higgins et al., 2008). However, if noninterfering crossovers were responsible, we would have observed many bivalents with chiasmata not labeled with MLH1, which we did not. Likewise, our genetic analyses showed that interference was still constraining crossover distribution during meiosis in allotriploid Brassica hybrids, although we do not have sufficient statistical power to determine if the interference level varied between the hybrids.
Second, the ability to form an increased number of crossovers could be different between bivalents. Cytogenetic observations showed that MLH1 foci were not homogeneously distributed between bivalents at meiosis in the allotriploid hybrid. In each nucleus, some bivalents appeared linked by four crossovers, whereas other bivalents showed no more than two crossovers (Figure 2). Thus, the bivalent corresponding to linkage group A7 could be one of the highly recombinant chromosome pairs. In that case, the genetic distances on A7 should be compared with the highest numbers of MLH1 foci observed on a chromosome pair. Unfortunately, it was not possible to test this hypothesis by BAC-FISH labeling A7 specifically at meiosis because of the whole genome triplication that occurred after the Arabidopsis–Brassiceae split (Lysak et al., 2005). It must also be emphasized that the nonhomogeneous distribution of crossovers between bivalents could also be explained, at least in part, by the occurrence of CC and AC bivalents (see Supplemental Table 1 online) that were formed between divergent chromosomes and therefore should display fewer MLH1 foci.
Finally, increased map length expansion could reflect gametic selection, which usually favors balanced gametes. It is usually assumed that recombinant gametes are selected when pairing failure occurs, because this would increase the chance of producing balanced gametes. This would result in an increase in the overall frequency of recombinant progeny relative to the crossover rate (Canady et al., 2006). “Pairing failure” is limited in the allotriploid and allotetraploid hybrids (Table 1), which certainly reduces the role of gametic selection in our material.
Thus, although we cannot conclude which of these (or combination of) factors explained the variation in amplitude, the crossover rate clearly increases in the allotetraploid and allotriploid hybrids (Figure 2, Table 2; see Supplemental Figure 1 online), irrespective of the origin of the A and C genomes. We effectively observed that a similar increase in crossover rates occurred between allotriploid and allotetraploid hybrids produced with B. napus cv Darmor (Nicolas et al., 2009), indicating that the Ar, An, Co, and Cn genomes did not significantly alter this outcome. This does not mean that genotypic variation cannot introduce additional variation in the number of crossovers that can be formed; comparing the genetic length of A7 estimated in the BC1 progeny of the ArAr′CoCo (75.4 cM) and ArAnCoCn (50 cM) allotetraploid hybrids using the same nine markers indicated that there were significantly fewer crossovers when B. napus cv Darmor is used as parent. It rather indicates that, using the same haploid complements from the same two genotypes, varying karyotype composition (allotriploid versus allotetraploid) was a reliable cause of crossover rate increase. This is consistent with other published observations. For example, different allotriploid Lolium hybrids showed more chiasmata per cell than the corresponding diploid hybrids (Gymer and Whittington, 1975; Jenkins, 1985a, 1985b, 1986). Likewise, a comparison of genetic maps between diploid and allotetraploid Gossypium (Brubaker et al., 1999; Desai et al., 2006) and Brassica (Suwabe et al., 2008) species showed larger genetic lengths in the allotetraploid than in their diploid counterparts (however, for Brassica, see Ferreira et al., 1994; Teutonico and Osborn, 1994; Parkin et al., 1995). A few counterexamples were also reported. White and Jenkins (1988) and Jenkins and White (1988) observed that chiasma frequency was higher in Scilla autumnalis allotetraploid hybrids than in the corresponding allotriploid hybrid. Thus, variations in crossover rate according to karyotype structure may be lineage-dependent.
As yet, we do not know the genetic/genomic basis for the increase in crossovers in allotetraploid and allotriploid hybrids; however, some potential causes seem improbable. It is very unlikely that recombination between Brassica homeologous chromosomes would have resulted in a net increase in recombinant chromosomes, as proposed in allotriploid interspecific Festuca × Lolium hybrids (Thomas et al., 1988; Zwierzykowski et al., 1999). Although the frequent trivalents observed at meiosis in the Festuca × Lolium hybrids effectively led to chromosomes with additional interstitial break points in the progeny (Zwierzykowski et al., 1999), the situation is clearly different in our study. Homeologous recombination rarely occurred between A and C chromosomes in the allotetraploid and allotriploid Brassica hybrids we analyzed (Table 1; see Supplemental Table 1 and Supplemental Figure 2 online).
As an alternative, it is tempting to link the higher number of crossovers observed in the allotriploid hybrid to the occurrence of chromosomes that remain as univalents (Table 1; Leflon et al., 2006). This interpretation is consistent with previous reports (Parker, 1975; Tease and Jones, 1975; Carlton et al., 2006) in which chromosome-specific desynaptic “mutants” showed a compensatory increase in crossover frequency among unaffected bivalents. In all these studies, the magnitude of the compensatory effect was generally small and roughly correlated to the relative proportion of chromosomes that remained as univalents. Most commonly, affected meiocytes contained just one pair of univalents at meiosis, and the compensating increase in chiasma frequencies on the remaining two to three bivalents approximated 25 to 30%. In our study, the increase in crossover frequency was greater, which was probably associated with the fact that a larger number of chromosomes remained unpaired during meiosis in the allotriploid hybrids.
The situation is less straightforward for the allotetraploid hybrid, in which the presence of only a few univalents does not satisfactorily explain the observed increase in crossover frequency. In this hybrid, our results suggest that the change in ploidy is associated with an increase in recombination. This hypothesis is consistent with the previous reports mentioned above (Brubaker et al., 1999; Desai et al., 2006; Suwabe et al., 2008), suggesting that this could be a general trend. If this is true, then ploidy-related crossover increases would have profound implications in both the fields of evolutionary science and agricultural research, because polyploidy represents a hallmark in plant evolution (Soltis et al., 2009; Wood et al., 2009).
In C. elegans, Carlton et al. (2006) showed that increased crossover rates were associated with delayed meiotic progression, presumably due to the presence of unrepaired double-strand breaks. These authors observed that this delay resulted in the Rad51 recombinase persisting for longer than usual on chromosomes and proposed that this may allow supernumerary crossovers to form. Likewise, Wang et al. (2010) recently reported that in a rice (Oryza sativa) mutant for the central element protein ZEP1 of the synaptonemal complex, MER3, a component of the interfering crossover pathway, persisted for longer and crossover frequency increased. Therefore, these findings suggest that future studies should be directed toward investigating the control of meiotic progression (Martinez-Perez and Moore, 2008) as well as meiotic protein dynamics. This would especially be of interest in crop plants, where the appearance/disappearance of meiotic proteins could alter the recombination pattern across the genome. We expect that such analyses will soon be possible in diploid, allotetraploid, and allotriploid Brassica hybrids, through the development and use of antibodies against meiotic proteins.
METHODS
Plant Materials
The strategy we used to produce hybrids and segregating backcross populations is detailed in Figure 1.
One single plant, C1.3 (ArAr, 2n = 20), was selected within the Brassica rapa Chicon variety (an old nonhomogeneous French forage variety) and crossed as the female to a B. rapa doubled haploid line (Ar′Ar′), called Z1 (kindly provided by Agriculture and Agri-Food Canada). The resulting ArAr′ F1 hybrid was then backcrossed as female to Z1, and a progeny of 135 plants was generated.
The same two B. rapa plants were crossed, as males, with two Brassica oleracea double haploid lines, RC and HDEM (provided by M. Manzanares-Dauleux, Institut National de la Recherche Agronomique Le Rheu). The resulting F1 interspecific ArCo and Ar′Co hybrids were colchicine doubled to produce two resynthesized Brassica napus plants named RCC (RC × C1.3; ArArCoCo) and EMZ (HDEM × Z1; Ar′Ar′CoCo), respectively. The RCC-resynthesized B. napus was then crossed as female to Z1 to produce the ArAr′Co hybrid; this allotriploid hybrid, which carried exactly the same pairs of A chromosomes as the former ArAr′ diploid hybrid, was crossed as female to EMZ, and a backcross progeny of 115 plants was produced. The RCC-resynthesized B. napus (ArArCoCo) was crossed as female to EMZ (Ar′Ar′CoCo) in order to analyze crossover rates in allotetraploid hybrids: one single F1 hybrid (ArAr′CoCo) was backcrossed as female to EMZ to produce a progeny containing 116 plants.
Two other progeny of 112 and 116 plants, deriving from crosses between Darmor, a winter B. napus cultivar (AnAnCnCn), and C1.3 (the hybrid was named ArAnCn in this paper) and between RCC and Darmor (the hybrid was named ArAnCoCn in this paper), respectively, were also used in this study. This plant material is presented in detail by Nicolas et al. (2009).
Two second-generation backcross progeny were produced by crossing as female one plant from the progeny of the ArAnCn hybrid and one plant from the progeny of the ArAnCoCn hybrid to Darmor (Figure 1). These two plants were selected from the first-generation backcross progeny, because they contained 38 chromosomes and all the alleles from Ar on one of the analyzed linkage groups (A7).
Meiotic Observation of Backcross Parents
Antibodies
The anti-Arabidopsis thaliana MLH1 polyclonal antibody was described by Chelysheva et al. (2010), and the purified serum was used at a dilution of 1:15.
DAPI Observations and MLH1 Immunolocalization
Anthers containing PMCs at prophase I were fixed in 1:3 (v/v) acetic acid:ethanol and stored at −20°C. Then anthers were incubated in 1% acetocarmine for 5 min, and those that were at the correct meiotic stage were squashed in 45% acetic acid. The slides were mounted in DAPI (2 μg/mL) in Vectashield antifade mounting medium. Fluorescence immunolocalization of MLH1 was performed on squashes according to the method described by Chelysheva et al. (2010). All observations were made using a Leica (http://www.leica.com) DMRXA2 microscope; photographs were taken using a CoolSNAP HQ (Roper; http://www.roperscientific.com) camera driven by Open-LAB 4.0.4 software. All images were further processed with OpenLAB 4.0.4 or Adobe Photoshop 7.0 (http://www.adobe.com).
Meiotic Behavior
For metaphase I analyses, samples of young floral buds were fixed in Carnoy’s solution (alcohol:chloroform:acetic acid, 6:3:1 [v/v]) for 24 h at room temperature and stored until use in 50% ethanol at 4°C. Anthers were then squashed and stained with 1% acetocarmine to analyze the meiotic behavior or squashed in 50% acetic acid for BAC-FISH experiments.
BAC-FISH
Two BAC clones, B. oleracea BAC 14O06 (Howell et al., 2002) and B. napus BAC 54B2 (provided by B. Chalhoub, Unité de Recherches en Génomique Végétale), were labeled by random priming with Alexa 488-5-dUTP and biotin-14-dUTP (Invitrogen, Life Technologies), respectively. The BAC clone 54B2 hybridizes to three A-genome chromosomes in B. napus, and the BAC clone 14O06 allows all C-genome chromosomes to be specifically distinguished in B. napus (Leflon et al., 2006; Nicolas et al., 2007).
Chromosomal preparations were incubated in RNase A (100 ng/μL) and pepsin (0.05%) in 10 mmol of HCl, fixed with paraformaldehyde (1%), dehydrated in an ethanol series (70, 90, and 100%), and air-dried. The hybridization mixture consisted of 50% deionized formamide, 10% dextran sulfate, 2× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), 1% SDS, and labeled probes (100 ng per slide). Chromosome preparations and predenatured (92°C for 6 min) probes were denatured at 82°C for 30 s. In situ hybridization was performed overnight in a moist chamber at 37°C. After hybridization, slides were washed for 5 min in 50% formamide in 2× SSC at 42°C, followed by several washes in 4× SSC–Tween. For indirect detection of BAC 54B2 DNA with biotin, we visualized the probe using avidin–Texas red (Vector Laboratories). The chromosomes were mounted and counterstained in Vectashield (Vector Laboratories) containing 2.5 μg/mL DAPI. Fluorescence images were captured using a CoolSnap HQ camera (Photometrics) on an Axioplan 2 microscope (Zeiss) and analyzed using MetaVue (Universal Imaging).
Molecular Analyses
Genomic DNA was extracted from young leaves according to the method of Doyle and Doyle (1990). The DNA concentration was adjusted to 10, 50, and 1 ng/μL for random amplification of polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), and single sequence repeat assays or specific markers, respectively. Molecular markers were selected from published maps (Foisset et al., 1996; Piquemal et al., 2005; Leflon et al., 2007) for their position on linkage group A7. Two RAPD markers (W11 and Y13; Foisset et al., 1996), two single sequence repeat markers (Ol12E03 and CB10439; Piquemal et al., 2005), two specific markers developed from Arabidopsis sequences (At1g80040 and At1g80530; Leflon et al., 2007), one SSAP (for sequence-specific amplification polymorphism) marker with two genotyped fragments (NBS1cE40; Leflon et al., 2007), and 11 AFLP markers (prefixed with E; Leflon et al., 2007) were used in the analysis. In figures, RAPD, SSAP, and AFLP markers are suffixed by the size of the amplified fragment. PCR and electrophoresis were performed using the same protocols as described by Leflon et al. (2007).
Construction of Genetic Linkage Maps and Statistical Analyses
Segregation of molecular markers in the BC1 and BC2 progeny was analyzed using χ2 tests. Segregating markers were scored for each plant, and linkage analysis was performed with MAPMAKER/EXP version 3.0 (Lincoln et al., 1992). Linkage groups were established with a threshold Logarism of Odds score of 4.0 and a maximum recombination frequency of 0.4. The Kosambi function was used to evaluate the genetic distances in cM between linked markers.
The heterogeneity of crossover rates among populations was assessed for every interval between adjacent markers using χ2 tests.
To analyze crossover interference, the theoretical distribution of genetic lengths between adjacent crossovers on a chromatid was determined by assuming that the positions of crossovers on the linkage group are independently, uniformly distributed random variables, as proposed by Drouaud et al. (2006). The distribution probability function of distances between two adjacent crossovers depends on the number of crossovers on the chromatid:
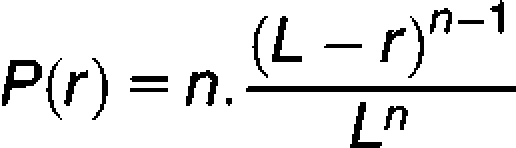 |
where r is the distance between two adjacent crossovers, evaluated in percentage of recombination, L is the genetic length of the chromatid, and n is the number of crossovers. For example, when two crossovers are formed, the probability of finding a distance of 0 to 1/4 L between them is
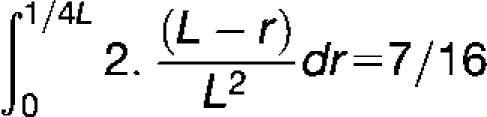 |
The theoretical distribution of genetic lengths between adjacent crossovers was calculated according to the observed frequency of chromatids with two, three, or more exchange points. The theoretical distribution of genetic lengths between adjacent crossovers, if there was no interference, was compared with the observed distribution using χ2 tests.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 2. Representative Examples of Meiosis I in PMCs of the Diploid, Allotetraploid, and Allotriploid Hybrids.
Supplemental Figure 2. BAC-FISH Analyses of PMCs in Metaphase I in the ArAr′CoCo and ArAr′Co Hybrids.
Supplemental Figure 3. Maps of the A7 Linkage Group in BC2 Progeny of the Allotriploid (ArAnCn) and Allotetraploid (ArAnCoCn) Hybrids.
Supplemental Figure 4. Number of Exchange Points per Chromatid Transmitted from the Hybrids to Their Progeny.
Supplemental Table 1. Comparison of Meiotic Configurations Observed at Metaphase I between the Allotriploid and Allotetraploid Hybrids.
Supplementary Material
Acknowledgments
We thank J.C. Letanneur (Institut National de la Recherche Agronomique [INRA] Rennes) for plant material production, S. Delaunay (INRA Rennes) for molecular analyses, Biogenouest for technical assistance, and C. Mezard, M. Grelon, R. Mercier, and J. Drouaud (INRA Versailles) for their help in data interpretation and critical review of the manuscript. This work was carried out with the financial support of the Agence Nationale de la Recherche, the French National Research Agency, under the Programme Biodiversité project ANR-05-BDIV-015, Effet de la Polyploïdie sur la Biodiversité et l’Évolution du Génome des Plantes, and under the program BLANC07-3_188863, project Unraveling Crossover Pathways with Arabidopsis thaliana and Crop Relatives.
References
- Able J.A., Langridge P. (2006). Wild sex in the grasses. Trends Plant Sci. 11: 261–263 [DOI] [PubMed] [Google Scholar]
- Barth S., Melchinger A.E., Devezi-Savula B., Lubberstedt T. (2001). Influence of genetic background and heterozygosity on meiotic recombination in Arabidopsis thaliana. Genome 44: 971–978 [DOI] [PubMed] [Google Scholar]
- Brubaker C.L., Paterson A.H., Wendel J.F. (1999). Comparative genetic mapping of allotetraploid cotton and its diploid progenitors. Genome 42: 184–203 [Google Scholar]
- Canady M.A., Ji Y., Chetelat R.T. (2006). Homeologous recombination in Solanum lycopersicoides introgression lines of cultivated tomato. Genetics 174: 1775–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton P.M., Farruggio A.P., Dernburg A.F. (2006). A link between meiotic prophase progression and crossover control. PLoS Genet. 2: 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L., Grandont L., Vrielynck N., Le Guin S., Mercier R., Grelon M. (2010). An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: Immunodetection of cohesins, histones and MLH1. Cytogenet. Genome Res. (in press). [DOI] [PubMed] [Google Scholar]
- Desai A., Chee P.W., Rong J., May O., Paterson A.H. (2006). Chromosome structural changes in diploid and tetraploid A genomes of Gossypium. Genome 49: 336–345 [DOI] [PubMed] [Google Scholar]
- Dooner H.K. (2002). Extensive interallelic polymorphisms drive meiotic recombination into a crossover pathway. Plant Cell 14: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J.J., Doyle J.L. (1990). Isolation of plant DNA from fresh tissue. Focus 12: 13–15 [Google Scholar]
- Drouaud J., Camilleri C., Bourguignon P.Y., Canaguier A., Berard A., Vezon D., Giancola S., Brunel D., Colot V., Prum B., Quesneville H., Mezard C. (2006). Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots.” Genome Res. 16: 106–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouaud J., Mercier R., Chelysheva L., Bérard A., Falque M., Martin O., Ziani V., Brunel D., Mezard C. (2007). Sex-specific crossover distributions and variations in interference level along Arabidopsis thaliana chromosome 4. PLoS Genet. 3: 1096–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak J., Luo M.C., Yang Z.L. (1998). Restriction fragment length polymorphism and divergence in the genomic regions of high and low recombination in self-fertilizing and cross-fertilizing Aegilops species. Genetics 148: 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch E., Szymaniak J.S., Yates H., Pawlowski W.P., Buckler E.S. (2007). Using crossover breakpoints in recombinant inbred lines to identify quantitative trait loci controlling the global recombination frequency. Genetics 177: 1851–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M.E., Williams P.H., Osborn T.C. (1994). RFLP mapping of Brassica napus using doubled haploid lines. Theor. Appl. Genet. 89: 615–621 [DOI] [PubMed] [Google Scholar]
- Foisset N., Delourme R., Barret P., Hubert N., Landry B.S., Renard M. (1996). Molecular-mapping analysis in Brassica napus using isozyme, RAPD and RFLP markers on a doubled haploid progeny. Theor. Appl. Genet. 93: 1017–1025 [DOI] [PubMed] [Google Scholar]
- Francis K.E., Lam S.Y., Harrison B.D., Bey A.L., Berchowitz L.E., Copenhaver G.P. (2007). Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 3913–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymer P.T., Whittington W.J. (1975). Hybrids between Lolium perenne and Festuca pratensis. III. Meiosis and fertility. New Phytol. 74: 295–306 [Google Scholar]
- Higgins J.D., Buckling E.F., Franklin C.F.H., Jones G.H. (2008). Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54: 152–162 [DOI] [PubMed] [Google Scholar]
- Howell E.C., Barker G.C., Jones G.H., Kearsey M.J., King G.J., Kop E.P., Ryder C.D., Teakle G.R., Vicente J.G., Armstrong S.J. (2002). Integration of the cytogenetic and genetic linkage maps of Brassica oleracea. Genetics 161: 1225–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins G. (1985a). Synaptonemal complex formation in hybrids of Lolium temulentum × Lolium perenne (L.). I. High chiasma frequency diploid. Chromosoma 92: 81–88 [Google Scholar]
- Jenkins G. (1985b). Synaptonemal complex formation in hybrids of Lolium temulentum × Lolium perenne (L.). II. Triploid. Chromosoma 92: 387–390 [Google Scholar]
- Jenkins G. (1986). Synaptonemal complex formation in hybrids of Lolium temulentum × Lolium perenne (L.). III. Tetraploid. Chromosoma 93: 413–419 [Google Scholar]
- Jenkins G., White J. (1988). Elimination of multivalents during meiotic prophase in Scilla autumnalis. II. Tetraploid. Genome 30: 940–946 [Google Scholar]
- Jones G.H. (1984). The control of chiasma distribution. Symp. Soc. Exp. Biol. 38: 293–320 [PubMed] [Google Scholar]
- Leflon M., Brun H., Eber F., Delourme R., Lucas M., Vallée P., Ermel M., Balesdent M., Chèvre A.M. (2007). Detection, introgression and localization of genes conferring specific resistance to Leptosphaeria maculans from Brassica rapa into B. napus. Theor. Appl. Genet. 115: 897–906 [DOI] [PubMed] [Google Scholar]
- Leflon M., Eber F., Letanneur J.C., Chelysheva L., Coriton O., Huteau V., Ryder C.D., Barker G., Jenczewski E., Chèvre A.M. (2006). Pairing and recombination at meiosis of Brassica rapa (AA) × Brassica napus (AACC) hybrids. Theor. Appl. Genet. 113: 1467–1480 [DOI] [PubMed] [Google Scholar]
- Lhuissier F.G.P., Offenberg H.H., Wittich P.E., Vischer N.O.E., Heyting C. (2007). The mismatch repair protein MLH1 marks a subset of strongly interfering crossovers in tomato. Plant Cell 19: 862–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln S.E., Daly M.J., Lander E.S. (1992). Constructing Genetic Linkage Maps with MAPMAKER/EXP 3.0: A Tutorial and Reference Manual. Technical Report 3rd ed. (Cambridge, MA: Whitehead Institute; ). [Google Scholar]
- Lincoln S.E., Lander E.S. (1992). Systematic detection of errors in genetic linkage data. Genomics 14: 604–610 [DOI] [PubMed] [Google Scholar]
- Lynn A., Soucek R., Börner G.V. (2007). ZMM proteins during meiosis: Crossover artists at work. Chromosome Res 15: 591–605 [DOI] [PubMed] [Google Scholar]
- Lysak M.A., Koch M.A., Pecinka A., Schubert I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15: 516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., Moore G. (2008). To check or not to check? The application of meiotic studies to plant breeding. Curr. Opin. Plant Biol. 11: 222–227 [DOI] [PubMed] [Google Scholar]
- Mézard C., Vignard J., Drouaud J., Mercier R. (2007). The road to crossovers: Plants have their say. Trends Genet. 23: 91–99 [DOI] [PubMed] [Google Scholar]
- Muller H.J. (1916). The mechanisms of crossing over. Am. Nat. 50: 193–221 [Google Scholar]
- Nicolas S.D., Leflon M., Monod H., Eber F., Coriton O., Huteau V., Chèvre A.M., Jenczewski E. (2009). Genetic regulation of meiotic crossovers between related genomes in Brassica napus haploids and hybrids. Plant Cell 21: 373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas S.D., Le Mignon G., Eber F., Coriton O., Monod H., Clouet V., Huteau V., Lostanlen A., Delourme R., Chalhoub B., Ryder C.D., Chèvre A.M., et al. (2007). Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics 175: 487–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker J.S. (1975). Chromosome-specific control of chiasma formation. Chromosoma 49: 391–406 [Google Scholar]
- Parkin I.A.P., Sharpe A.G., Keith D.J., Lydiate D.J. (1995). Identification of the A and C genomes of amphidiploid Brassica napus (oilseed rape). Genome 38: 1122–1131 [DOI] [PubMed] [Google Scholar]
- Piquemal J., Cinquin E., Couton F., Rondeau C., Seignoret E., Doucet I., Perret D., Villeger M.J., Vincourt P., Blanchard P. (2005). Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theor. Appl. Genet. 111: 1514–1523 [DOI] [PubMed] [Google Scholar]
- Robbins T.P., Gerats A.G.M., Fiske H., Jorgensen R.A. (1995). Suppression of recombination in wide hybrids of petunia hybrids as revealed by genetic mapping of marker transgenes. Theor. Appl. Genet. 90: 957–968 [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E., Armstrong S.J., Santos J.L., Franklin F.C.H., Jones G.H. (2002). Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162: 1415–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis D.E., Albert V.A., Leebens-Mack J., Bell C.D., Paterson A.H., Zheng C., Sankoff D., dePamphilis C.W., Wall P.K., Soltis P.S. (2009). Polyploidy and angiosperm diversification. Am. J. Bot. 96: 336–348 [DOI] [PubMed] [Google Scholar]
- Suwabe K., Morgan C., Bancroft I. (2008). Integration of Brassica A genome genetic linkage map between Brassica napus and B. rapa Genome 51: 169–176 [DOI] [PubMed] [Google Scholar]
- Tease C., Jones G.H. (1975). Chromosome-specific control of chiasma formation in Crepis capillaris. Chromosoma 57: 33–49 [Google Scholar]
- Teutonico R.A., Osborn T.C. (1994). Mapping of RFLP and qualitative trait loci in Brassica rapa and comparison to the linkage maps of Brassica napus, Brassica oleracea, and Arabidopsis thaliana. Theor. Appl. Genet. 89: 885–894 [DOI] [PubMed] [Google Scholar]
- Thomas H., Morgan W.G., Humphreys M.W. (1988). The use of triploid hybrids for introgression in Lolium species. Theor. Appl. Genet. 76: 299–304 [DOI] [PubMed] [Google Scholar]
- Wang M., Wang K., Tang D., Wei C., Li M., Shen Y., Chi Z., Gu M., Cheng Z. (2010). The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22: 417–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby M.C. (2005). Making crossovers during meiosis. Biochem. Soc. Trans. 33: 1451–1455 [DOI] [PubMed] [Google Scholar]
- White J., Jenkins G. (1988). Elimination of multivalents during meiotic prophase in Scilla autumnalis. I. Diploid and triploid. Genome 30: 930–939 [Google Scholar]
- Wijnker E., de Jong H. (2008). Managing meiotic recombination in plant breeding. Trends Plant Sci. 13: 640–646 [DOI] [PubMed] [Google Scholar]
- Wood T.E., Takebayashi N., Barker M.S., Mayrose I., Greenspoon P.B., Rieseberg L.H. (2009). The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA 106: 13875–13879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwierzykowski Z., Lukaszewski A.J., Naganowska B., Lesniewska A. (1999). The pattern of homoeologous recombination in triploid hybrids of Lolium multiflorum with Festuca pratensis. Genome 42: 720–726 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.