This work examines light wavelength–specific electrophysiological signaling and cellular light memory in Arabidopsis. Animals have their network of neurons, synapses, electrophysiological circuits and memory, but plants have their network of chloroplasts (connected by stromules), photoelectrophysiological signals transduced by bundle sheath cells, and cellular light memory.
Abstract
Although light is essential for photosynthesis, excess light can damage the photosynthetic apparatus and deregulate other cellular processes. Thus, protective integrated regulatory responses that can dissipate excess of absorbed light energy and simultaneously optimize photosynthesis and other cellular processes under variable light conditions can prove highly adaptive. Here, we show that the local and systemic responses to an excess light episode are associated with photoelectrophysiological signaling (PEPS) as well as with changes in nonphotochemical quenching and reactive oxygen species levels. During an excess light incident, PEPS is induced by quantum redox changes in photosystem II and in its proximity and/or by changes in glutathione metabolism in chloroplasts. PEPS is transduced, at least in part, by bundle sheath cells and is light wavelength specific. PEPS systemic propagation speed and action potential are dependent on ASCORBATE PEROXIDASE2 function. Excess light episodes are physiologically memorized in leaves, and the cellular light memory effect is specific for an excess of blue (450 nm) and red (650 nm) light of similar energy. It is concluded that plants possess a complex and dynamic light training and memory system that involves quantum redox, reactive oxygen species, hormonal, and PEPS signaling and is used to optimize light acclimation and immune defenses.
INTRODUCTION
When plant leaves are exposed to full sunlight, only a portion of the absorbed light energy is used for CO2 fixation (Asada, 1999; Karpinski et al., 1999). Environmental stresses, like a sudden increase in light intensity, changes in temperature, or limitation in water accessibility, depress efficiency of CO2 assimilation due to reduction of stomatal conductance, but do not depress foliar absorption of light energy (e.g., Niyogi, 2000; Mullineaux and Karpinski, 2002; Holt et al., 2004; Baker, 2008). This results in an increase of the excitation energy in excess (EEE) over that required for optimal photosynthetic metabolism. In the classical view, the failure to dissipate EEE can be highly damaging to plants and often manifests as chlorosis, bleaching, or bronzing of leaves due to imbalanced metabolism of reactive oxygen species (ROS) (e.g., Karpinski et al., 1999; Niyogi, 2000; Apel and Hirt, 2004; Laloi et al., 2007; Mühlenbock et al., 2008; Van Breusegem et al., 2008). However, it was recently revealed that EEE-induced permanent photodamage could be due to induction of programmed cell death, regulated by a very similar or the same genetic system as for the hypersensitive response in disease resistance and for systemic acquired resistance (SAR) (Jabs et al., 1996; Dangl and Jones, 2001; Kwak et al., 2003; Mateo et al., 2004; Mühlenbock et al., 2007, 2008).
Plants have developed a highly responsive and flexible physiology, which allows them to function under short- and long-term fluctuations and rapid changes in their natural environment. Plants also are able to integrate and simultaneously process multiple stimuli and prioritize their responses (Mullineaux and Karpinski, 2002; Mittler, 2002; Mühlenbock et al., 2007, 2008; Pfannschmidt et al., 2009). Light acclimation processes in plants act to dissipate EEE and optimize photosynthesis under variable light conditions. Therefore, plants have nonphotochemical quenching (NPQ) and photochemical quenching (qp) mechanisms that control EEE dissipation (Niyogi, 2000; Holt et al., 2004; Ruban et al., 2007; Baker, 2008) and ROS metabolism (Willekens et al., 1997; Asada, 1999; Karpinska et al., 2000; Mateo et al., 2004; Laloi et al., 2007; Mühlenbock et al., 2008). When photosystem II (PSII) is exposed to excess light, acidification of the lumen increases; this triggers a change in PSII light harvesting antenna function from light absorption to EEE dissipation by NPQ (Pascal et al., 2005; Ruban et al., 2007; Baker, 2008; Johnson et al., 2008). During that time, various components of the photosynthetic electron transport chain become transiently more reduced or oxidized and thus deregulate ROS production. This, in turn, changes the chloroplast stromal redox state (e.g., Asada, 1999; Karpinska et al., 2000; Kruk and Karpinski, 2006). Such redox changes are regulated by NPQ and qp mechanisms that are prerequisites for induction of various light acclimatory and defense responses, such as systemic acquired acclimation (SAA) and SAR (Karpinski et al., 1999; Mateo et al., 2004; Mühlenbock et al., 2007, 2008), shifting of transcription programs in the chloroplasts and in the nucleus (Karpinski et al., 1999; Pfannschmidt et al., 1999, 2009; Mullineaux et al., 2000; Apel and Hirt, 2004; Fey et al., 2004; Rossel et al., 2007; Mühlenbock et al., 2008), and inducing state transitions and other light acclimatory mechanisms (Larkin et al., 2003; Bellafiore et al., 2005; Pascal et al., 2005; Barneche et al., 2006; Laloi et al., 2007).
Recently, we have shown that stomatal aperture is regulated, at least in part, by the quantum redox events in the proximity of the PSII, for example, by changes in NPQ and the redox status of the glutathione and plastoquinone (PQ) pools that influence ROS/hormonal homeostasis (Karpinski et al., 1999; Mateo et al., 2004; Mühlenbock et al., 2008). We further established that cellular hormonal/ROS homeostasis is changed not only in leaves experiencing conditions that promote EEE, but also in naive leaves undergoing SAA (not previously exposed to excess light). Using photosystem I and PSII-specific light wavelengths, photosynthetic electron transport inhibitors, and null mutants of LESIONS SIMULATING DISEASE1, ENHANCED DISEASE SUSCEPTIBILITY1, and PHYTOALEXIN DEFICIENT4 (lsd1, eds1, and pad4) as a test system for deregulated light acclimatory and defense responses, we found that programmed cell death propagation signals and SAR depend on stomatal conductance, photorespiration, and hormonal/ROS cellular homeostasis that is regulated, at least in part, by quantum redox signals from PSII and its proximity (Karpinski et al., 1997, 1999; Karpinska et al., 2000; Ball et al., 2004; Mateo et al., 2004; Chang et al., 2004; Mühlenbock et al., 2007, 2008).
Laboratory-based stress research has traditionally focused on only one well-defined response to a single environmental stress. By contrast, plants are subjected to many simultaneous challenges in their natural environment, bringing about the simultaneous activation of many signaling pathways that are combined in a regulatory network (Mittler, 2002; Mühlenbock et al., 2007, 2008; Koussevitzky et al., 2008). Moreover, mechanisms that control stomatal aperture play a pivotal role in the regulation of SAA, SAR, innate immunity, and programmed cell death (e.g., Kwak et al., 2003; Mateo et al., 2004; Melotto et al., 2006; Rossel et al., 2007; Mühlenbock et al., 2008). It is also known that plant leaves solve their problem of optimal gas exchange, transpiration, and photosynthesis in a way similar to that defined by the algorithm of the cellular automation (Peak et al., 2004). Cellular automation proposed by John von Neumann and Oskar Morgenstern in 1944 is a mathematical model for a dynamic system that is discrete in time and in space. It depends on local interactions, but global phenomena emerge on these interactions (Gramss et al., 1998). Basic elements of the cellular automation are regular lattice of cells in which at each time step each cell is in one defined “s” state. It has a defined neighborhood of cells behaving in such way that the state of each cell at time step “t+1” is a function of some of its surrounding (neighbors) cells at time step “t.” Therefore, findings of Peak et al. (2004) and our results (Mühlenbock et al., 2008) imply that there must be a molecular mechanism (automation) that coordinates functioning of signaling networks that govern light acclimation, immune defenses, photosynthesis, transpiration, and subsequent developmental processes in plants. Despite the importance of this information, surprisingly little is known about these signaling interactions and their coordination.
Changes in electrical potential of the cellular plasma membrane are well-documented regulators of different signaling cascades in animals and plants (Burdon-Sanderson, 1873; Darwin, 1875; Bose, 1926; Higinbotham, 1973; Pickard, 1973; Bowles, 1990; Wildon et al., 1992; Trewavas, 2003; Davies, 2004; Lautner et al., 2005). Local changes in the plasma membrane electrical potential in response to light, hereafter called photoelectrophysiological signaling (PEPS), were described before and are the most rapid physiological reactions known in plants (in the order of μs to ms) (Wheeler and Brownlee, 2008). In this study, we have found that PEPS is a new component of signaling cascades that potentially regulates light acclimatory and defense responses, such as SAA and SAR. We also demonstrated that leaves are able to physiologically memorize different excess light episodes and use this stored information, for example, for improving their acclimation and immune defenses, thereby improving the survival chances of the whole plant under prolonged periods of low light. This finding has broad implications in the study of plant light acclimatory responses, plant–microbe interactions, bacterial pathogenesis, and diseases, but also in the molecular ecophysiology of a plant behavior.
RESULTS
Local and Systemic Responses to Excess Light Are Characterized by Changes in NPQ and ROS
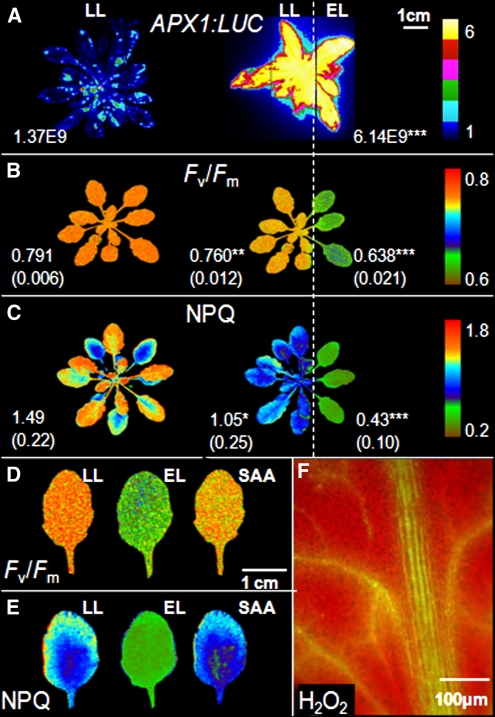
In transgenic Arabidopsis thaliana rosettes partially exposed to excess light, induction of APX1:LUC in both exposed and unexposed leaves shows that excess light produces a systemic effect leading to induction of SAA (Figure 1A) (Karpinski et al., 1999). At the same time, reduction in the maximal photochemical efficiency (Fv/Fm) and NPQ was observed and indicates photooxidative stress and higher dissipation of EEE as heat, in both directly exposed leaves and in leaves undergoing SAA (Figures 1B and 1C). However, the changes in Fv/Fm and NPQ have a different pattern. Changes in Fv/Fm in directly exposed leaves and leaves undergoing SAA have patchy character, whereas changes in NPQ varied, with the lowest NPQ values in the central vein region and the highest values toward the edge of the leaf (Figures 1D and 1E; see Supplemental Figure 5 online). Furthermore, we detected sectors of different NPQ values in leaves undergoing SAA, but in leaves experiencing conditions that promote EEE, NPQ was strongly reduced and its foliar gradient was flattened (Figures 1C and 1E; see Supplemental Figure 5B online). Such a pattern of NPQ in leaves undergoing SAA is similar to that observed in control plants, but it is characterized by a higher gradient differences and lower average NPQ. The lowest NPQ observed in the central vein region of systemic leaves (Figure 1E; see Supplemental Figure 5B online) is associated with the highest foliar H2O2 levels (Figures 1F and 2E). The highest foliar H2O2 levels, induction of APX2:LUC, and programmed cell death were observed previously proximal to the central vein in leaves undergoing SAA (Karpinski et al., 1999; Mullineaux et al., 2006; Mühlenbock et al., 2008).
Figure 1.
Induction of SAA Is Associated with Systemic Patchy Changes in Maximal Photochemical Efficiency (Fv/Fm) and Systemic Gradient-Like Changes of Foliar NPQ.
Arabidopsis transgenic APX1:LUC and Col-0 rosettes were grown at low-light conditions (LL; 100 μmol photons m−2 s−1) and were partially exposed to excess light (EL; 2000 μmol photons m−2 s−1). Left, controls that were LL grown with no excess light exposure; right, partially exposed rosettes, with the leaves to the right of the dotted line exposed to excess light. Fv/Fm and NPQ values ([A] to [C]) are expressed as means from three independent experiments. Significant differences in comparison to plants from LL conditions are indicated accordingly to Student’s test (*P ≤ 0.05, **P ≤ 0.005, and ***P ≤ 0.001).
(A) CCD image of luciferase activity in relative light units (LUs) in LL-grown APX1:LUC rosette and rosette that was partially exposed to EL for 60 min. Dark blue is equal to 1 relative light unit (RLU); yellow is equal to 6 LU.
(B) Image of maximal quantum efficiency of PSII (Fv/Fm) in leaves of LL-grown rosette and rosette that was partially exposed to EL for 60 min.
(C) Image of the NPQ in leaves of LL-grown rosette and rosette that was partially exposed to EL for 60 min.
(D) Patchy pattern of Fv/Fm in leaf of LL-grown rosette, leaf exposed to EL for 60 min, and leaf undergoing SAA.
(E) Gradient-like pattern of NPQ in leaf of LL-grown rosette, leaf exposed to EL for 60 min, and leaf undergoing SAA.
(F) Fluorescence image of H2O2 production. H2O2 was labeled with 2’,7’-dichlorofluorescin diacetate in veins of leaf undergoing SAA for 60 min.
Figure 2.
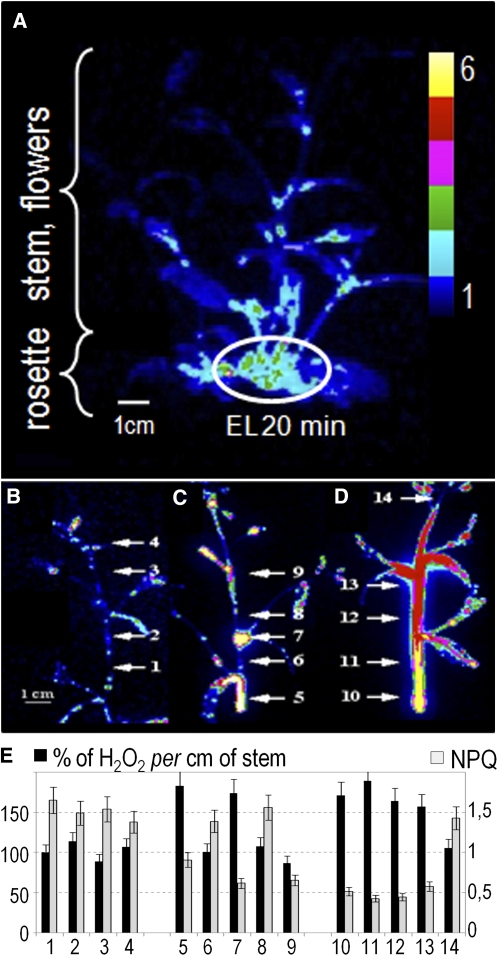
Induction of SAA in the Inflorescence Stem Is Characterized by Wave-Like Changes of APX1:LUC Expression, NPQ, and H2O2 Levels.
Arabidopsis transgenic APX1:LUC flowering rosettes grown in low-light conditions (LL; 100 μmol photons m−2 s−1) were partially exposed to excess light (EL; 2000 μmol photons m−2 s−1).
(A) CCD image of luciferase activity in RLUs in LL-grown APX1:LUC rosette and rosette that was partially exposed to EL for 20 min. Dark blue is equal to 1 RLU; yellow is equal to 6 LU.
(B) LU image of inflorescence stem of LL-grown plants.
(C) LU image of LL-exposed inflorescence stem undergoing SAA for 60 min.
(D) LU image of LL-exposed inflorescence stem undergoing SAA for 80 min.
(E) H2O2 levels and NPQ were measured in 1-cm sectors of inflorescence stems (indicated by arrows and numbered from 1 to 14 in [B] to [D]). H2O2 levels are shown as relative values in comparison to sector 1 = 100%. The results show representative data of three independent experiments expressed as mean, with bars indicating sd.
To investigate further the nature of SAA, we wondered how systemic signals spread to other plant organs. To test this, we examined NPQ, APX1:LUC induction, and H2O2 levels in the inflorescence stems of plants where part of the rosette had been exposed to excess light. Indeed, a slightly different pattern of NPQ from that observed in leaves was observed in the inflorescence stem undergoing SAA (Figures 2A to 2D). However, like in leaves that were undergoing SAA, NPQ in inflorescence stems undergoing SAA was lower in regions where APX1:LUC induction and H2O2 levels were higher (Figures 2C and 2E). These results assured us that the local and systemic induction of APX1:LUC, changes of NPQ, Fv/Fm, foliar levels of ROS (Figures 1 and 2), hormones, and programmed cell death reported before (Mühlenbock et al., 2008) appear in a specific pattern. They reflect changes of NPQ (Figures 1C, 1E, and 2E) such that zones of higher APX1:LUC expression and H2O2 levels have lower NPQ.
The inverse relationship between induction of APX1 and APX2 and the reduction of NPQ was confirmed by measurements of foliar APX1 and APX2 transcript levels in npq4 null mutants. The NPQ4 gene encodes PsbS protein, a member of the chlorophyll a/b binding, light-harvesting complex family of proteins that binds zeaxanthin. PsbS function is essential for EEE- and zeaxanthin-dependent conformational changes in the thylakoid membrane that are necessary for ΔpH-dependent regulation of NPQ (Li et al., 2000; Peterson and Havir, 2001). Foliar APX2 transcript levels, in low light–acclimated npq4 leaves, were significantly higher than those observed in control plants (see Supplemental Figure 1B online) and remained unchanged during excess light episodes. Foliar APX1 and APX2 transcript levels in cry1/cry2, phyA/phyB, and phot1/phot2 null mutants were also preinduced in low light, but their further induction after exposure to excess light was clear (see Supplemental Figures 1A and 1B online). All these double recessive mutants except phyA/phyB also have reduced NPQ in ambient light conditions (see Supplemental Figure 1C online). Double phyA/phyB recessive mutants have significantly higher NPQ in comparison to control wild-type plants. These results strongly suggest that these mutants are susceptible to low-light conditions that normally do not evoke photooxidative stress. Moreover, APX1 and APX2 expression in all these mutants was not suppressed, as in experiments that combined excess light and DCMU (Figure 4) where NPQ was strongly reduced (almost to zero).
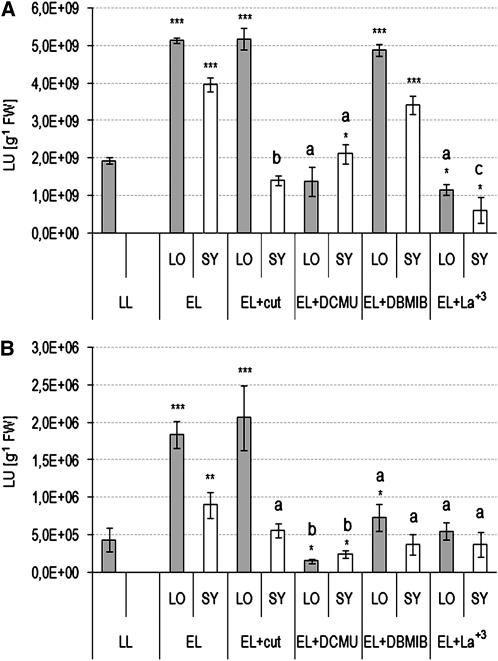
Figure 4.
Quantum Redox Changes in PSII and Its Proximity and Calcium Signaling Regulate Local and Systemic Induction of APX1:LUC and APX2:LUC.
Arabidopsis Col-0, transgenic APX1:LUC, and APX2:LUC rosettes grown at low-light (LL; 100 μmol photons m−2 s−1) conditions were exposed to excess white light (EL; 1500 μmol photons m−2 s−1 applied to LO, local leaves) and treated with 8 μM DCMU, 14 μM DBMIB, or 10 μM LaCl3 or had physically interrupted leaf petiole vasculature (cut) in the local leaves 15 min before EL exposure. Measurements were performed in LO and systemic leaves that were not directly treated (SY).
(A) and (B) Expression of APX1:LUC and APX2:LUC, respectively, in control LO exposed to 1.5 h EL, in LO with physically interrupted vascular tissues in the petiole, and exposed to 1.5 h EL in LO treated with DCMU, with DBMIB or LaCl3 and exposed to 1.5 h EL, and in leaves undergoing SAA under LL conditions (SY). The results show representative data from 30 independent experiments expressed as mean, with bars indicating sd. Significant differences in comparison to plants from LL conditions are indicated (*P ≤ 0.05, **P ≤ 0.005, and ***P ≤ 0.001). Letters indicate significant differences in LO and SY in comparison to control LO or SY, respectively (a, P ≤ 0.05; b, P ≤ 0.005).
Others and we have suggested that a propagated ROS/hormonal signal was the main trigger of light acclimatory responses (Karpinski et al., 1999; Fryer et al., 2003; Rossel et al., 2007; Mühlenbock et al., 2008, Galvez-Valdivieso et al., 2009). However, the pattern of systemic changes observed in inflorescence stems undergoing SAA, including APX1:LUC induction and changes of NPQ and ROS levels, would be linear, not wavy (Figure 2). Therefore, we asked ourselves if SAA has another yet unknown signaling mechanism, different from that of linear hormone transport, activation and diffusion of ROS, or gaseous hormones.
Leaves Exposed to Excess Light-Induced Systemic and Light Wavelength–Specific PEPS
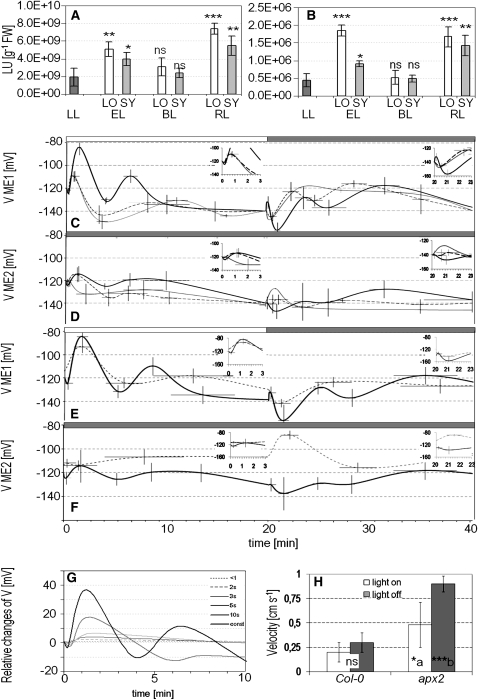
It has been demonstrated that electrical signals regulate plant stress responses (Burdon-Sanderson, 1873; Darwin, 1875; Bose, 1926; Pickard, 1973; Bowles, 1990; Wildon et al., 1992; Lautner et al., 2005). In Figures 3A and 3B, we show that excess of white and red (650 ± 20 nm wavelength) but not blue (450 ± 20 nm wavelength) light, of similar energy of the red light, is able to induce APX1:LUC and APX2:LUC in directly exposed leaves and in leaves undergoing SAA. Taking into consideration that these two transgenic lines are robust molecular markers of SAA (Karpinski et al., 1997, 1999; Mullineaux et al., 2000; Fryer et al., 2003; Ball et al., 2004; Chang et al., 2004), we concluded that red light, but not blue light, is the main inducer of SAA. Simultaneous measurements of the plasma membrane electrical potential in cells of the bundle sheath layer of the central leaf vein in exposed leaves and in leaves undergoing SAA indicate that white light induces PEPS with an action potential of ~40 to 50 mV in directly exposed leaves (Figures 3C and 3G) and 25 to 30 mV in leaves undergoing SAA in the twilight zone (Figure 3D). During the first seconds of light exposure, we observed hyperpolarization followed by depolarization and repolarization (Figures 3C and 3G). Red and blue light exposure also induced PEPS with a slightly different pattern of changes (weaker depolarization and deeper repolarization), and with lower action potential values than observed for white light (Figure 3C). The characteristic feature, differentiating the action of these three different lights, are specific changes of plasma membrane potential in directly exposed leaves and in leaves undergoing SAA (Figures 3C and 3D). In the first seconds after switching off the white light, in the directly exposed leaf, we observed depolarization followed by hyperpolarization and repolarization in several minutes with an amplitude of 25 to 30 mV (Figure 3C), whereas in leaves undergoing SAA, switching off the light is signalized with an inverse order of events, hyperpolarization followed by depolarization and repolarization with the amplitude of ~15 mV (Figure 3D). For red and blue light, we observed an opposite order of events in comparison with that observed in white light (Figures 3C and 3D).
Figure 3.
SAA Is Dependent on Spectral Composition of Light and Is Associated with Wavelength-Specific Changes in PEPS That Is Regulated by APX2 Expression.
Arabidopsis Col-0, transgenic APX1:LUC, APX2:LUC, and apx2-1 null mutant rosettes grown at low-light (LL; 100 μmol photons m−2 s−1) conditions were exposed to excess white light (EL; 1500 μmol photons m−2 s−1), excess blue light (BL; 80 μmol of photons m−2 s−1 of 450 ± 20 nm wavelength; [A] to [F]), or excess red light (RL; 120 μmol photons m−2 s−1 of 650 ± 20 nm wavelength; [A] to [F]).
(A) and (B) Expression of APX1:LUC and APX2:LUC measured as luciferase activity in RLUs in LL-grown transgenic rosettes as well as in rosettes exposed to 1.5 h of EL, 4 h BL, and 4 h RL. The results show representative data from pooled leaf samples of three independent experiments expressed as mean, with bars indicating sd. Significant differences in relation to plants grown at LL conditions are indicated (*P ≤ 0.05, **P ≤ 0.005, and ***P ≤ 0.001).
(C) and (D) Changes in the plasma membrane electrical potential of bundle sheath cells of foliar central vein (later called PEPS) were recorded during 20-min exposure to white EL (white bar above panel, light switch on) followed by 20-min twilight zone (<1 μmol photons m−2 s−1) period (gray bar, light switch off). Leaves were exposed to white EL (black line), BL (dashed line), and RL (gray line). PEPS was measured in exposed leaves ([C]; electrode 1) and in leaves undergoing SAA in the twilight zone ([D]; electrode 2). Inset shows changes in PEPS during the first minutes directly after switching light on or off.
(E) and (F) Changes in PEPS in Col-0 (black line) and null apx2-1 (dashed line). PEPS was recorded in central veins during 20-min exposure to white EL (white bars, light switch on) followed by 20-min twilight zone (<1 μmol photons m−2 s−1) period (gray bar, light switch off). Plasma membrane electrical potential was measured in exposed leaves ([E]; electrode 1) and in leaves undergoing SAA in the twilight zone ([F]; electrode 2). Inset shows changes in PEPS during the first minutes after light switching.
(G) Changes of PEPS pattern in leaves exposed to a single flash of white EL for the period of <1 s and for periods of 2, 3, 5, and 10 s and for continuous EL.
(H) The propagation velocity of PEPS measured between two single bundle sheath cells of two different leaves (local and systemic) in Arabidopsis Col-0 and apx2-1. The velocity was determined after measurements of physical distance between electrodes and delay in PEPS changes in systemic leaves after light switch on or off in the locally exposed leaf. Significant differences in relation to wild-type plants are indicated (*P ≤ 0.05 and ***P ≤ 0.001). The letters indicate significant differences between light switch on and off treatment of the same type of plants. The results showed representative data from 30 independent experiments and at least 20 cells and are expressed as mean, with bars indicating sd of PEPS amplitude and time.
Changes in PEPS in response to excess white light in the apx2-1 recessive mutant (Figures 3E and 3F) were different from that observed in the wild type but similar to that for red light (Figures 3C and 3D). This similarity was observed in both directly exposed leaves and in leaves undergoing SAA during switching on and switching off the light. Another characteristic feature was higher values of action potential in apx2-1 mutant in leaves undergoing SAA than those observed for Columbia-0 (Col-0).
Several seconds of excess light illumination is sufficient to induce PEPS with maximal action potential (Figure 3G). We also determined that the propagation speed of PEPS between two different leaves is ~0.3 cm s−1 for switching off light and 0.2 cm s−1 for switching on light (Figure 3H). The speed of the PEPS between different leaves depends on APX2 gene expression. APX2 is exclusively expressed in the bundle sheath cell layer (Fryer et al., 2003), and the systemic propagation of PEPS between different leaves is more than 2 times faster in the null apx2-1 mutant than that in wild-type plants (Figure 3H).
The above results allow us to conclude that PEPS is induced in response to conditions promoting EEE and its amplitude depends on the duration of the excess light episode and the light spectral composition. PEPS pattern and its systemic propagation speed in bundle sheath cells depend on specific expression of APX2 in these cells. Therefore, we concluded that PEPS is a signaling component of light acclimatory responses, such as SAA.
Changes in NPQ, PQ, and Ca2+ Signaling Are Required for Induction of SAA Molecular Markers
Connecting quantum redox changes of different PSII components with the physiological signaling is very difficult, since the absolute time required for such changes is several orders of magnitude shorter than that required for physiological signal transduction (Karpinski et al., 1997; Knight, 2000). An effective experimental way that connects quantum redox changes, such as changes in NPQ and redox status of PQ pool, with physiological signaling is the use of selected light wavelengths and photosynthetic electron transport inhibitors (Pfannschmidt et al., 1999; Mateo et al., 2004; Kruk and Karpinski, 2006; Mühlenbock et al., 2008; Pfannschmidt et al., 2009). Treatment of leaves with DCMU blocks reduction of the secondary electron acceptor, QB in PSII, and in consequence, slows down the oxidation of QA and reduction of the PQ pool. Moreover, DCMU treatment has been shown to strongly reduce NPQ (Doron et al., 2008; Goss et al., 2008) since DCMU treatment equalizes Fm and F’m values (NPQ = Fm/F’m – 1). DCMU also inhibit excess light-mediated induction of stomata closure and acclimatory and immune defense responses (Mühlenbock et al., 2008). DCMU treatment generates some similar physiological and molecular effects as treatment of plants with light enriched in 700-nm wavelength (Mühlenbock et al., 2008) and is highly specific. In our experiments, we measured NPQ with separate dark adaptation periods (40 min before and 40 min after appropriate excess light treatment) to determine the new Fm value. Measuring NPQ in such a way mirrors changes induced by DCMU. Therefore, the reduced NPQ value observed after partial excess light exposure is mostly due to increase of F0 and reduction of Fm values in directly exposed leaves and in leaves undergoing SAA.
In contrast with DCMU, treatment of leaves with 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB) slows down the rate of oxidation of the PQ pool by inhibiting the function of the cytochrome b6/f complex. DBMIB treatment generates some similar effects as exposure of leaves to light enriched in 650-nm wavelength and is able to induce stomata closure and light acclimatory and immune defense responses under low-light conditions (Karpinski et al., 1997, 1999; Mühlenbock et al., 2008). DBMIB has been shown to promote induction of programmed cell death and reduction of stomata aperture under low light, whereas DCMU produces the opposite effect under excess light (Mühlenbock et al., 2008).
DCMU treatments of a single leaf and its petiole blocked the induction of APX1:LUC and APX2:LUC in a leaf exposed to excess light and reduced or blocked the induction of these transgenes in leaves undergoing SAA (Figures 4A and 4B). In the case of DBMIB treatment, we observed different effects in comparison with the DCMU treatment (Figures 4A and 4B). After DBMIB treatment, APX1:LUC was induced in local and systemic leaves to similar levels as in control plants. However, APX2:LUC was induced to a lower extent in directly exposed and DBMIB treated leaves, and APX2:LUC was not induced in leaves undergoing SAA. In the systemic leaves that were not treated with DCMU or DBMIB, we could not detect (up to 1 h after treatment) any significant changes in chlorophyll a fluorescence parameters (e.g., Fv/Fm; see Supplemental Figure 2 online). Therefore, significant translocation of DCMU and DBMIB from locally treated leaf to the systemic leaves during 1 h of the experiment is unlikely. In systemic leaves, where xylem, phloem, and bundle sheath tissues were mechanically interrupted in the leaf petiole exposed to excess white light (see Supplemental Figures 3B and 3C online), we observed a lack of systemic induction of APX1:LUC and APX2:LUC similarly to those in DCMU and LaCl3 experiments for both analyzed transgenes and in the DBMIB experiment for APX2:LUC (Figures 4A and 4B). However, in a leaves exposed to excess light, APX1:LUC and APX2:LUC induction was not significantly suppressed like it was in the DCMU-treated leaf. To obtain more information about other possible signaling components regulating APX1, APX2 gene expression, and general SAA, we used the Ca2+ channel blocker LaCl3. This blocker prevented APX1:LUC and APX2:LUC induction in directly exposed leaves and in leaves undergoing SAA. It is also important to note that induction of APX1 and APX2 expression in response to excess white light was not blocked in cry1/cry2, phyA/phyB, and phot1/phot2 recessive double mutants, and NPQ in these mutants in low light was significantly deregulated (see Supplemental Figures 1A and 1B online).
We demonstrated before that the second step of glutathione synthesis in the chloroplasts is important for regulation of APX2 expression and light acclimatory and immune defense responses (Ball et al., 2004). Karpinska et al. (2000) suggested previously that redox status of the glutathione and PQ pools are dependent on each other during conditions generating EEE. Therefore, we measured redox status of the PQ pool (Kruk and Karpinski, 2006) in recessive cad2 and rax1 null mutants (Ball et al., 2004). In these mutants, under low-light conditions, the photoactive PQ pool was reduced to significantly lower levels than that of wild-type plants (Table 1). NPQ was also reduced in these mutants, however insignificantly (see Supplemental Figure 1C online).
Table 1.
Deficiency in Glutathione Synthesis in Arabidopsis Chloroplasts Deregulates Redox Status of the PQ Pool
| Arabidopsis (Genotype)a | PQ Pool (% Total)b | Redox State of PQ Pool in the Dark (% Reduced) | Redox State of PQ Pool in the Light(% Reduced) | Redox State of PQNP (% Reduced)c |
| Col-0 | 32.5 ± 3.3 | 15.4 ± 5.7 | 40.6 ± 6.2 | 60.4 ± 5.2 |
| cad2 | 34.6 ± 2.8 | 24.8 ± 7.4 | 17.9 ± 6.1** | 71.0 ± 5.4 |
| rax1-1 | 29.7 ± 5.3 | 23.5 ± 12.0 | 9.6 ± 8.9** | 67.8 ± 8.6 |
The presented data are representative for three independent experiments of pooled leaves samples. Significant differences in comparison to Col-0 are indicated (n = 3, **P ≤ 0.005).
Arabidopsis Col-0 and null mutants cad2 and rax1-1 were grown under low-light conditions (100 μmol photons m−2 s−1). Five-week-old plants were used for the measurements.
Photoactive PQ pool was measured as described by Kruk and Karpinski (2006). Redox state of the photoactive PQ pool in the dark was measured 1 to 1.5 h before the end of the dark period and 2.5 to 3 h after the onset of the photoperiod.
Nonphotoactive PQ pool was determined as described by Kruk and Karpinski (2006).
The above experiments demonstrate that quantum redox changes in PSII and in its proximity (e.g., changes in NPQ and in redox status of the glutathione and PQ pools regulate APX1 and APX2 expression, but not cryptochromes, phototropins, or phytochromes) influenced the regulation of APX1 and APX2 expression. Moreover, we showed that Ca2+ signaling could be also implicated in the regulation of APX1 and APX2 expression and that intact vascular tissues are required for systemic induction of APX1:LUC and APX2:LUC.
Changes in NPQ, Redox Status of the Glutathione and PQ Pools, and Ca2+ Signaling Deregulated PEPS
The above results lead us to the following question: Do changes in NPQ, redox status of the glutathione and PQ pools, and Ca2+ signaling deregulate PEPS? In the case of mechanical damage made to a leaf petiole, we observed reduced amplitude of PEPS in the leaf with damaged veinal tissues and exposure excess light. The damaged leaf was not able to induce PEPS changes in systemic leaves in which the amplitude of PEPS changes was nearly zero (flat line) (Figures 5C and 5D). It is important to know that in this experiment, vascular tissues of a leaf petiole were interrupted, while other petiole mesophyll cells were intact (see Supplemental Figure 3C online). Therefore, it is unlikely that PEPS could bypass through other types of cells. Similar effects were observed in the case of LaCl3. In systemic leaves (for local LaCl3 treatment), PEPS was reduced to almost flat line (Figure 5J). In the local LaCl3-treated leaf, we observed deregulation of PEPS in excess light exposed leaf during switching on the light but not during switching off the light (Figure 5I) in comparison to control leaves.
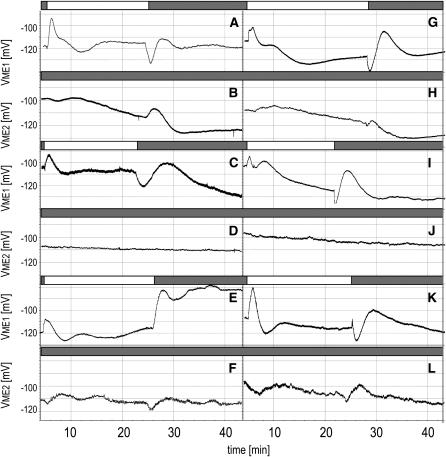
Figure 5.
Quantum Redox Changes in PSII and Its Proximity and Calcium Signaling Regulate Local and Systemic PEPS.
Arabidopsis Col-0 and cad2 null mutants (deficient in glutathione synthesis in chloroplasts) grown at low-light conditions (LL; 100 μmol photons m−2 s−1) were exposed to excess white light (EL; 1500 μmol photons m−2 s−1 applied on local leaves) and treated with 8 μM DCMU, 14 μM DBMIB, or 10 μM LaCl3 or with physically interrupted leaf petiole vasculature in local leaf petiole 15 min before EL incident. Measurements were made in local and systemic (not directly treated) leaves.
(A) PEPS changes measured in EL exposed local leaves (ME1, electrode 1).
(B) PEPS changes measured in systemic leaves undergoing SAA in the twilight zone (ME2, electrode 2).
(C) PEPS changes measured in EL exposed local leaves with physically interrupted vasculature of the leaf petiole (ME1, electrode 1).
(D) PEPS changes measured in systemic leaves undergoing SAA in the twilight zone of rosette where petiole vasculature of EL-exposed local leaves was mechanically interrupted (ME2, electrode 2).
(E) PEPS changes measured in local leaves partially treated with drops of DCMU (ME1, electrode 1).
(F) PEPS changes measured in systemic leaves undergoing SAA in the twilight zone of rosette where EL-exposed local leaf and leaf petiole were treated with DCMU (ME2, electrode 2).
(G) PEPS changes measured in local leaves partially treated with drops of DBMIB (ME1, electrode 1).
(H) PEPS changes measured in systemic leaves undergoing SAA in the twilight zone of rosette where EL-exposed leaf and leaf petiole were treated with DBMIB (ME2, electrode 2).
(I) PEPS changes measured in local leaves partially treated with drops of 10 μmol LaCl3 (ME1, electrode 1).
(J) PEPS changes measured in systemic leaves undergoing SAA in the twilight zone of rosette where EL-exposed leaf and leaf petiole were treated with LaCl3 (ME2, electrode 2).
(K) PEPS changes measured in EL exposed local leaves of cad2 mutant (ME1, electrode 1).
(L) PEPS changes measured in systemic leaves of cad2 mutant undergoing SAA in the twilight zone (ME2, electrode 2).
In DCMU experiments, we observed deregulation of PEPS in leaves directly treated with DCMU and in systemic leaves (Figures 5E and 5F). After switching on the excess light, for the DCMU-treated leaf, we observed reduced amplitude of PEPS (Figure 5E) in comparison with the control leaves (~20 mV; Figure 5A). After switching off the light, for the DCMU-treated leaf, we observed only large depolarization (~35 mV), and a new, much higher action potential was established (Figure 5E). In systemic leaves, which were not directly treated with DCMU and were kept in twilight zone, the amplitude of PEPS was reduced to ~12 to 10 mV or lower (Figure 5F), whereas in control systemic leaves, the corresponding amplitude was ~25 mV (Figure 5B). We used a concentration of DCMU (8 μM) that only inhibits electron transport to 30 to 50% (see Supplemental Figure 2A online), and systemic PEPS was reduced by ~50%. In contrast with DCMU treatment, in leaves treated with DBMIB (Figures 5G and 5H), we observed higher amplitude of PEPS (40 mV) during switching off the light in exposed leaves (Figure 5G). In systemic leaves for local DBMIB treatment, we observed also higher amplitude of PEPS during switching off the light. In the DBMIB (14 μM) experiment, APX1:LUC was significantly induced, but APX2:LUC was not, nor significantly suppressed like in the case of DCMU (Figure 4). This could be due to fact that we used a new experimental system (see Supplemental Figures 3 and 4 online) where plants are submerged in a solution that allows measurements of PEPS. Moreover, for the first time we used a combination of excess light and DBMIB (before we used DBMIB only in low light). Such a combination can generate slightly different effects than that reported before (e.g., Karpinski et al., 1997, 1999; Mühlenbock et al., 2008).
We also wondered if PEPS in the recessive cad2 mutant, which is impaired in glutathione synthesis (Figures 5K and 5L), is deregulated. After switching on the light, for the cad2 leaf, amplitude of PEPS was higher than that observed in the case of control plants (~45 mV; Figure 5K). After switching off the light, in the cad2 leaf, we observed a similar profile of PEPS changes, but the amplitude was also higher, like in DBMIB local leaves (Figures 5I and 5K). In systemic leaves, the amplitude of PEPS in cad2 was reduced to a maximum of ~15 mV (Figure 5L), whereas in control systemic leaves, the corresponding amplitude was ~25 mV (Figures 3D and 5B).
These experiments demonstrate that quantum redox changes in PSII and in its proximity (e.g., changes in NPQ and in redox status of the glutathione and PQ pools) are involved in regulation/deregulation of PEPS. Moreover, we showed that Ca2+ signaling and intact foliar vascular tissues are required for systemic induction of PEPS, and APX1:LUC and APX2:LUC expression. The above experiments indicate that PEPS and APX1 and APX2 gene expression are regulated/deregulated by the same mechanisms.
Cellular Light Memory
According to our knowledge, mechanisms of training and memorizing in plants are not well known, and it is widely accepted that plants do not have such capacities (e.g., Trewavas, 2003). However, in fact, SAA is a training process of naive cells, chloroplasts, and PSII (cells and PSII that never experienced excess light or conditions that promote EEE), shown by the cells and PSII that are actually experiencing such conditions. The above results (Figures 1 to 5), work done by Peak et al. (2004) and Mott and Peak (2007), and our own work (Mühlenbock et al., 2008) suggest that SAA could be a process based on collective dynamics and emergent, distributed computation in plants, performed by their leaves. This could be concluded based on the fact that stomatal conductance and quantum redox events in PSII, reflected by chlorophyll fluorescence, play an essential role in the regulation of SAA and SAR (Karpinski et al., 1999; Mateo et al., 2004; Mühlenbock et al., 2008). If leaves are able to perform emergent, complex, and distributed computation, as suggested before (Peak et al., 2004; Mott and Peak, 2007), we should ask ourselves if a plant cell is able to differentially memorize excess light incidents and spectral composition of light and effectively use this memorized information for improving their survival chances. Therefore, we performed the following experiments.
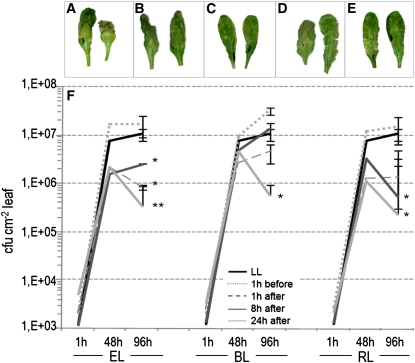
Leaves were infected with the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 1 h before and 1, 8, and 24 h after excess light incidents (induced by excess of white, blue, and red light separately) (Figure 6). In control plants, under low-light conditions, disease symptoms developed and infection progressed (Figures 6A and 6F). In the case of plants infected 1 h before excess light incidents, for all wavelengths used, disease progressed even faster than that observed in control plants (Figures 6B and 6F). However, in plants infected 1, 8, and 24 h after EEE incidents, we observed different results. In these cases, development of disease symptoms was effectively inhibited at all times of infection after excess white light incidents (Figures 6C and 6F). In the case of blue light, development of disease symptoms was only effectively stopped in the case of infection 24 h after a blue light incident (Figures 6D and 6F), and in the case of red light, 8 and 24 h after the light incident (Figures 6E and 6F). It is important to note that the provided energy of red and blue light was similar in all experiments, and it was similar to that present in the white light. Therefore, we conducted an additional statistical analysis of the total light memory effect. After summarizing results from all infections made after specific excess light incidents, we observed significant difference for white (P = 7.41E-06) and red light (P = 1.16E-04), but not for blue light (P = 0.327) in comparison with the control light conditions. When we summarized all results from infections made before and after specific excess light incidents, we did not observe any significant difference in comparison to control values in low-light conditions.
Figure 6.
Different Cellular Light Memory Effects on Immune Defenses after Episodes of Excess White, Blue, (450-nm Wavelength), and Red (650-nm Wavelength) Light.
Arabidopsis leaves of rosette grown at low-light conditions (LL; 100 μmol photons m−2 s−1) were inoculated with virulent P. syringae pv tomato DC3000 strains 1 h before and 1, 8, and 24 h after exposure to excess white light (EL; 1500 μmol photons m−2 s−1, 1 h), excess blue light (BL; 80 μmol photons m−2 s−1; 4 h), and excess red light (RL; 120 μmol photons m−2 s−1; 4 h). Infections were performed under LL, and infected plants were grown up to 96 h under LL. Results from four independent experiments with nine repetitions (n = 36) for each light condition are expressed as mean, with bars indicating sd. Significant differences in comparison with LL laboratory conditions are indicated according to Student's t test (*P ≤ 0.05 and **P ≤ 0.005).
(A) Representative examples of foliar disease symptoms developed 96 h after infection preformed in LL conditions.
(B) Representative examples of foliar disease symptoms developed 96 h after infection performed 1 h before EL incident.
(C) Representative examples of foliar disease symptoms developed 96 h after infection performed 24 h after EL incident.
(D) Representative examples of foliar disease symptoms developed 96 h after infection performed 24 h after BL incident.
(E) Representative examples of foliar disease symptoms developed 96 h after infection performed 24 h after RL incident.
(F) Bacterial growth was measured 1, 48, and 96 h after infections performed 1 h before and 1, 8, and 24 h after EL, BL, and RL incidents. Bacterial growth is expressed as a colony-forming unit per leaf area (cfu/cm−2).
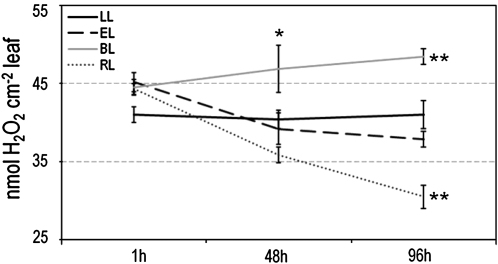
Cellular light memory is also characterized by the changes in other parameters developing in time, such as foliar H2O2 levels (Figure 7) and chlorophyll a fluorescence (see Supplemental Figure 4 online). H2O2 levels were initially higher after all excess light incidents. However, 48 and 96 h after excess light incidents, foliar H2O2 levels were different for all light treatments. Four days after incident of excess white light, foliar H2O2 level were similar to that observed in control plants from low-light conditions (Figure 7). In the case of blue light, foliar H2O2 levels 96 h after light incident were significantly higher, while for the red light were significantly lower (Figure 7). These results are in agreement with induction of APX1:LUC and APX2:LUC by red but not by blue light (Figures 3A and 3B).
Figure 7.
Cellular Excess Light Memory of White, Blue (450-nm Wavelength), and Red (650-nm Wavelength) Light Episodes Expressed by Changes of Foliar H2O2 Levels.
Arabidopsis Col-0 rosettes grown under low-light conditions (LL; μmol photons m−2 s−1) were exposed to excess white light (EL; 1500 μmol photons m−2 s−1; 1 h), excess blue light (BL; 80 μmol photons m−2 s−1; 4 h), and excess red light (RL; 120 μmol photons m−2 s−1; 4 h). Foliar H2O2 levels were determined 1, 48, and 96 h after excess light incidents. The results show data from three independent experiments and five repetitions (n = 15) expressed as mean, with bars indicating sd. Significant differences in comparison with LL conditions are indicated according to Student's t test (*P ≤ 0.05, **P ≤ 0.005, and ***P ≤ 0.001).
Taken together, these results indicate that different excess light incidents can be physiologically memorized and that the different spectral composition of light is memorized differently. Moreover, the order of events (infections before or after appropriate excess light incidents) is important for the cellular light memory effect and immune defenses. The above results suggest that, when plants are infected prior to an excess light incident, virulent bacteria could initiate a process, which can no longer be counteracted by the plant, even if excess light is provided later. It thus seems that virulent bacteria can erase the light memory mechanism if they succeed in infecting plants before the excess light incident and SAA induction.
DISCUSSION
Light energy absorbed in excess regulates light acclimatory and immune defenses in plants; however, this signaling is complex and involves many simultaneous, networked pathways (Karpinski et al., 1999; Mateo et al., 2004; Rossel et al., 2007; Mühlenbock et al., 2008; Galvez-Valdivieso et al., 2009). This complexity is not well characterized and understood. Particularly, it is not demonstrated how the excess light energy absorbed by PSII antennae is exchanged into redox hormonal signaling. Moreover, it is not clear how signals are transduced from chloroplast to the nucleus and from chloroplasts to plasma membrane and to other cells and organelles. Previously, we linked reduced PSII antenna size (reduction in absorption of light energy) and higher NPQ in the cao mutant (Mateo et al., 2004; Klenell et al., 2005) with light-2 dependent (650-nm wavelength) suppression of the runaway cell death phenotype of lsd1. We concluded that light absorption, NPQ, and qp mechanisms are essential for regulation of intra- and extracellular ROS/hormonal homeostasis, stomatal conductance, photosynthesis, and photorespiration that regulate light acclimation, programmed cell death, and immune defenses (Mateo et al., 2004; Klenell et al., 2005; Mühlenbock et al., 2007, 2008; Galvez-Valdivieso et al., 2009). In fact, light acclimatory responses, such as SAA, are a complex and dynamic training system for naive cells, chloroplasts, and PSII that did not previously experience excess light episodes, by cells, chloroplasts, and PSII that are in the process of experiencing such conditions (Karpinski et al., 1999). Here, we demonstrate that SAA spreads to other plant organs, for example, the inflorescence stem, is associated with contrariwise changes in NPQ and ROS, and that PEPS could be an important component of the signaling network for SAA (Figures 1 to 5, Table 1). We have also found that light acclimation responses are associated with a wavelength-specific cellular light memory effect that lasts for several days or longer (Figures 6 and 7).
Quantum-Redox Regulation of Light Acclimatory Responses
The patchy character of changes in Fv/Fm in leaves undergoing SAA in comparison with the gradient pattern of NPQ changes suggests a specific character and the role of NPQ in SAA (Figures 1C to 1E, 2C, and 2E). Systemic changes in NPQ are intriguing, since the available data in the literature indicate that NPQ changes only appear in leaves directly exposed to excess light (Niyogi, 2000; Holt et al., 2004; Baker, 2008). Therefore, we ask ourselves: What is the meaning and mechanism of systemic changes in NPQ? The others’ (Niyogi, 2000; Kulheim et al., 2002) and our (Mateo et al., 2004; Klenell et al., 2005) data indicate that changes in NPQ are prerequisites for light acclimation, immune defenses, and survival in the natural fluctuating light environment. Systemic reduction of NPQ correlates with an increase in H2O2 (Figures 1E, 1F, and 2B to 2E) and with reduction of stomatal conductance (Mateo et al., 2004; Mühlenbock et al., 2008); therefore, systemic changes in NPQ and other chlorophyll fluorescence parameters must be an active and physiologically regulated process. Therefore, it is highly intriguing to determine what mechanisms and algorithms are used by plant leaves to calculate patchy changes in Fv/Fm and gradient wave-like NPQ changes in directly exposed leaves and leaves undergoing SAA. Peak et al. (2004) suggested that dynamic and emergent chlorophyll fluorescence and stomatal aperture change patterns are calculated in leaves by mathematical algorithms similar to the cellular automation (von Neumann and Morgenstern, 1944). However, Peak et al. (2004) did not identify the biological hardware that regulates these dynamic, emergent, and antagonistic changes of chlorophyll fluorescence and stomatal aperture. Our results (Mühlenbock et al., 2008) indicated that ROS, induction of marker genes, programmed cell death, immune defenses, and changes in stomatal conductance in response to excess light are inversely regulated by light enriched in 650 and 700 nm or by DCMU and DBMIB treatment. Here, we identified parts of the biological hardware that regulates changes described by Peak et al. (2004).
It is known that chloroplasts are connected with each other as well as with the nucleus and plasma membrane by stromules that form a cellular network of extended chloroplast envelope membranes (Senn, 1908; Kwok and Hanson, 2003a, 2003b; Hanson and Sattarzadeh, 2008). Moreover, chloroplasts connected by stromules can communicate with each other (Köhler et al., 1997). Our results indicate that communication between chloroplasts of distant cells in different organs is possible at the whole-plant level with, for example, PEPS transduced from chloroplasts to other chloroplasts or nucleus or plasma membrane by a organized global membrane network. The electrophysiological nature of PEPS obtained from studies in animals, plants, and our results strongly suggests involvement of plasma membrane ion channels in signal propagation and in induction of SAA markers (e.g., Figures 4A, 4B, 5I, and 5J).
Programmed cell death in leaves undergoing SAA was mainly induced near vascular foliar tissues (Mühlenbock et al., 2008). Systemic induction of APX2 expression was also observed in bundle sheath cells that surround vascular tissues (Karpinski et al., 1999; Fryer et al., 2003), and recently ABA signaling between vascular and surrounding bundle sheath cells during light acclimatory responses was demonstrated (Galvez-Valdivieso et al., 2009). Here, we observed the lowest NPQ in the proximity of a leaf central vein in leaves undergoing SAA (Figure 1E; see Supplemental Figure 5 online). We also detected higher expression of APX1:LUC and higher H2O2 levels in inflorescence stem in regions with reduced NPQ (Figure 2E). Such systemic changes in NPQ must be induced by systemic changes in the chloroplast lumen acidification and subsequent activation of the xanthophyll cycle, and as a consequence this must induce ROS/hormonal response loops (Karpinski et al., 1999; Demming-Adams and Adams, 2000; Fryer et al., 2003; Chang et al., 2004; Havaux et al., 2004; Pascal et al., 2005; Mullineaux et al., 2006; Ruban et al., 2007; Johnson et al., 2008; Mühlenbock et al., 2008; Galvez-Valdivieso et al., 2009). This, in turn, leads, for example, to reduction of stomatal conductance and a photorespiratory burst of ethylene and ROS in some sectors of a leaf and in a consequence to induction of programmed cell death in adjacent cells (Mühlenbock et al., 2008).
In previous work, we reasoned that induction of molecular markers of light acclimatory responses (APX1 and APX2) is regulated, at least in part, by redox changes of the glutathione and PQ pools (Karpinski et al., 1997, 1999; Karpinska et al., 2000; Fryer et al., 2003; Ball et al., 2004; Chang et al., 2004; Mühlenbock et al., 2008). This was mainly concluded based on the fact that DCMU blocked or diminished electron transfer from QB to the PQ pool, reduced nearly completely NPQ, strongly diminished chloroplastic H2O2 production (Ślesak et al., 2003), and blocked or diminished excess light induction of APX1 and APX2. Recently, we demonstrated that DCMU/DBMIB effect could be induced by light-1 and light-2, respectively (Mühlenbock et al., 2008) (light-1 enriched in 700-nm wavelength and light-2 enriched in 650-nm wavelength). The results presented in Figure 4 confirm this observation, and additionally, we demonstrated here that deficiency in glutathione synthesis in chloroplasts that deregulate APX2 expression in normal low-light conditions (Ball et al., 2004) also deregulates redox status of the PQ pool (Table 1). Such possible redox interactions of glutathione and PQ pools during excess light responses that regulate pH gradient across the thylakoid membrane and APX1 and APX2 gene expression was suggested before (Karpinska et al., 2000). Therefore, the results presented here (Table 1) prove that the redox changes of glutathione and PQ pools and regulation of, for example, APX2 gene expression are indeed physiologically and genetically linked to each other. Similar links for glutathione and salicylic acid synthesis was also demonstrated before (Mateo et al., 2006).
In this work, we also demonstrated that red but not blue excess light mainly induces APX1 and APX2 expression and that functional APX2 is required for induction of normal PEPS in response to excess light incident (Figure 3). Therefore, we tested if other light sensing mechanisms, such as cryptochromes, phytochromes, and phototropins, could be involved in the regulation of APX1 and APX2 expression. Deficiency in functional CRY1 and CRY2, PHYA and PHYB, and PHOT1 and PHOT2 proteins did not block or diminished excess light induction of APX1 and APX2, like it is observed in combined excess light and DCMU experiments and in the case of APX2 also in excess light and DBMIB experiments (Figure 4; see Supplemental Figure 1 online). On the contrary, APX1 and APX2 preinduction in low light and further induction in response to excess white light was observed in all these mutants except for APX2 in npq4 (see Supplemental Figures 1A and 1B online). These results indicate that the presence of active CRY1 and CRY2, PHYA and PHYB, and PHOT1 and PHOT2 influenced APX1 and APX2 expression and light acclimatory responses, for example, due to the lack of normal chloroplast avoidance response or phenolic compound synthesis. The absence of these responses makes mutant plants sensitive to low-light intensities and indicates a secondary regulatory effect.
In npq4 null mutants (with inactive S-protein in PSII [PsbS]), which show constitutively strongly diminished NPQ (see Supplemental Figure 1C online) and are not able to acclimate to natural variable light conditions (Kulheim et al., 2002), relative foliar APX2 transcript levels were always high (10-fold), regardless of low or excess white light treatments (see Supplemental Figure 1B online). However, this was not observed for APX1 (see Supplemental Figure 1A online). These results confirm our previous interpretations that specific quantum redox events in PSII and its proximity are indeed important for the regulation of APX2 expression during light acclimatory responses and confirm that APX2 is indeed a robust molecular marker of light acclimatory responses, such as SAA, and retrograde chloroplast to nucleus signaling.
Quantum Redox Regulation of PEPS
Peak et al. (2004) demonstrated that reduction in stomatal conductance was inversely correlated with increase of chlorophyll fluorescence (Peak et al., 2004; Mott and Peak, 2007). The data from their studies demonstrate that chlorophyll fluorescence changes are an accurate surrogate for stomatal aperture. The evidence presented in the literature indicates that the function of stomata cells and the mechanism regulating their aperture play essential roles in light acclimation and immune defenses (Kwak et al., 2003; Mateo et al., 2004; Melotto et al., 2006). We demonstrated before that spectral composition of light and quantum redox events in PSII and its proximity are important for the regulation of stomatal aperture and, as a consequence, for SAA and SAR (Mateo et al., 2004; Mühlenbock et al., 2008; Chang et al., 2009). However we, as well as Melotto et al. (2006), did not take into consideration the fact that stomata aperture in leaves changes in cycles with constant rate (Peak et al., 2004; Mott and Peak, 2007). Peak et al. (2004) demonstrated that stomatal aperture and chlorophyll fluorescence values are actually computed by leaves with the use of a mathematical algorithm similar to the cellular automation that optimizes transpiration and photosynthesis.
If plant leaves indeed perform emergent and dynamic computing as suggested by Peak et al. (2004), do they have other attributes of electronic computation, such as electrical signaling and memory? In animals, electric signals transduced by neuron cells organized in a network are essential for life (Trewavas, 2003; Spencer et al., 2004; Baluska et al., 2005). These signals regulate coordination of movements, vision, hearing, sense of smell, taste, and temperature, and many other inputs from the environment. The neuron network in animals is responsible for learning and memory processes (Nagai et al., 1996; Firestein, 2001). In plants, systemic electrical signaling transduced by a specialized network of cells is not well described, and the electrical signaling network was considered as primitive and simple, in comparison with that in animals (Bowles, 1990; Trewavas, 2003; Davies, 2004). Moreover, plants are widely considered to be passive organisms that are not in need of rapid electrical long-distance signaling and excitability (Baluska et al., 2005). Electrical signaling in plants is used for signaling injury (Fromm and Eschrich, 1989; Wildon et al., 1992), potential dangers of injury (e.g., touch), in mimosa (Koziolek et al., 2004), of drugs (Mwesigwa et al., 2000; Volkov et al., 2001) and others, or in other specialized plants it is used for hunting of insects (Higinbotham, 1973). Our results demonstrate a different and more complicated picture of the PEPS network and its potential role in light acclimation and immune defense in plants (Lautner et al., 2005).
It was suggested earlier that generation of electrophysiological signaling in plants is made in keeping with the all-or-none law, according to which the maximum spike is obtained once a threshold stimulus is given, and increasing the intensity of the stimulus does not increase the response (Higinbotham, 1973; Davies, 2004). Our results indicate that PEPS requires relatively high light energy input (Figure 3G), but different action potentials depend on the applied spectrum and the provided light energy (Figures 3 and 5). We demonstrated that specific PEPS are induced by different light wavelengths, are transduced, at least in part, by bundle sheath cell layer, and that PEPS is deregulated by quantum redox events in PSII and its proximity (e.g., changes in NPQ and in redox status of the glutathione and PQ pools). Such changes regulate, at least in part, APX1 and APX2 gene expression (Figures 1 to 4), stomatal conductance, programmed cell death, and in consequence SAA and SAR (Karpinski et al., 1999; Mühlenbock et al., 2008). Moreover, PEPS propagation speed and action potential depend on functional APX2 (Figures 3E, 3F, and 3H), which is exclusively expressed in the bundle sheath cell layer (Fryer et al., 2003). It is probably transducing information to the naive cells, which never experienced excess light episodes. PEPS propagation from cell to cell in the bundle sheath layer may generate changes in cellular redox status of naive adjacent vascular or mesophyll cells and, thus, in naive chloroplasts and PSII; therefore, it may induce changes in ROS/hormonal homeostasis that regulate light acclimation and immune defenses (Karpinski et al., 1999; Mullineaux et al., 2000, 2006; Mühlenbock et al., 2008; Galvez-Valdivieso et al., 2009). This reasoning is supported by several facts.
First, PEPS is deregulated in local and systemic leaves of cad2 mutant plants (Figures 5K and 5L) that have altered redox status of the PQ pool (Table 1), have deregulated APX2 expression, and are not able to acclimate to high light intensities and induce normal immune defenses (Ball et al., 2004).
Second, DCMU and DBMIB differentially deregulated PEPS in leaves directly exposed to excess light and in leaves undergoing SAA (Figures 5E to 5H). This strongly correlates with differential regulation of APX1:LUC and to a weaker extent with APX2:LUC.
Third, blue and red excess light of similar energies differentially regulate PEPS, APX1 and APX2 gene expression, and immune defenses in directly exposed leaves and in leaves undergoing SAA (Figures 3, 6, and 7). It is well known that PSII will be more excited than PSI by excess of red light of 650-nm wavelength, so-called light-2 that induces state-2 transition regulated by overreduction of the PQ pool (Bellafiore et al., 2005; Pascal et al., 2005; Barneche et al., 2006). Excess blue light (450-nm wavelength) will equally excite both PSII and PSI and does not cause overreduction of the PQ pool, and it is not able to induce state-2 transition. This is confirmed by the fact that red, but not blue excess light, induced APX1:LUC and APX2:LUC and immune defenses (Figures 3, 6, and 7).
Fourth, we observed a strong similarity between patterns of PEPS in apx2 mutants in local and systemic leaves in response to switched on and off white excess light incidents to that induced by excess red light in wild-type plants. This result suggests a specific role of APX2 in the regulation of excess red light signals transduced by bundle sheath cells (Figures 1 to 3). This is also confirmed by the fact that APX2:LUC was induced by red but not by blue light of the same energy and that the known blue and red light photoreceptors do not specifically regulate excess light induction of APX2 (see Supplemental Figure 1B online).
Fifth, the propagation speed of PEPS is deregulated in apx2-1 null mutants (Figure 3H) in such a way that lack of functional APX2 maximizes the propagation speed of PEPS.
Finally, DCMU and LaCl3 treatment of light-exposed local leaves and mechanical breaking of petiole vasculature lead to restricted (strongly reduced) systemic PEPS changes and to absence of induction of SAA molecular markers (APX1:LUC and APX2:LUC; Figures 4 and 5).
Cellular Light Memory
A definition of memory and intelligence for plants was proposed by Trewavas (2003): adaptively variable growth and development during the lifetime of the individual. In animals, memory is connected with intelligence in such a way; the more intelligent the organism is, the greater the degree of individual adaptively variable behavior. Because this definition was used to describe intelligence in other organisms than humans, we used this definition in our experimental system. Do plants exhibit memory and behavior that result from memorized previous events? Our data (Figures 6 and 7; see Supplemental Figure 4 online) indicate that plants possess memory of previous light incidents, hereafter called cellular light memory, which is used for optimization of future light acclimatory and immune defense responses. In other words, plants can store and use information from the spectral composition of light to anticipate changes that might appear in the near future in the environment, for example, for anticipation of pathogen attack (Figures 6 and 7). Therefore, plants have to possess a mechanism for processing of the memorized information.
Here, we have to ask, why do plants evolve mechanisms in which excess light and its spectral composition regulate immune defenses? A straightforward answer to this question is difficult, and several answers could be given. One possible answer is that in dense canopy, light intensities are strongly reduced; therefore, the majority of leaves are in shade (low or very low light) and thus are prone to, for example, pathogen attack (Figure 6) (Mühlenbock et al., 2008). SAA is in fact a mechanism in which plants use the disadvantages of being partly exposed to excess light (condition that generates EEE) to strengthen, for example, immune defenses in the dense canopy zone. This explains why plants possess a natural capacity to absorb more light energy than that required for photosynthesis. They need this excess energy for optimization of acclimatory and immune defense responses.
CONCLUSIONS
From the work of Peak et al. (2004), Mott and Peak (2007), Melotto et al. (2006), from our own work (Karpinski et al., 1999; Fryer et al., 2003; Chang et al., 2004, 2009; Mateo et al., 2004; Klenell et al., 2005; Mühlenbock et al., 2007, 2008), and the experiments presented here, we conclude that plants solve their optimal light acclimation (SAA), immune defense (SAR and innate immunity), photosynthesis, and transpiration by a computational algorithm (cellular automation) in which input, output, and processing of the data are all accomplished using the same hardware. Our experiments identified some parts of this hardware, which includes quantum redox changes in PSII and in its proximity (e.g., in NPQ and redox status of the glutathione and PQ pools), PEPS, ROS/hormonal circuits, and finally the cellular light memory. Probably, it is the most elegant system that evolved in complex photosynthetic organisms, since it uses absorbed photons energy in excess by some leaves to improve survival chances of a whole plant. Animals have their network of neuron synapses, electrophysiological circuits, and memory, but plants have their network of chloroplasts (connected by stromules), PEPS circuits transduced by bundle sheath cells, and cellular light (quantum) memory. We are aware that these results suggest that plants have the capacity to be trained and memorize; indeed, leaves in the dark are able to not only see the light (Foyer and Noctor, 1999; Karpinski et al., 1999), but also are able to differently remember its spectral composition and use this memorized information to increase their survival chances.
METHODS
Plant Material
Arabidopsis thaliana ecotype Col-0 and transgenic lines with hybrid reporter gene construct, for example, ASCORBATE PEROXIDASE1 (APX1) and APX2 promoter fused in frame with firefly LUCIFERASE (LUC) gene APX1:LUC and APX2:LUC (Karpinski et al., 1999), were used. The following mutants were also analyzed: recessive npq4 mutant, kindly provided by K. Niyogi Laboratory, University of California, Berkeley, CA; phot1/phot2 by M. Wada Laboratory, Kyushu University, Fukuoka Japan; phyA-211 and phyB-9 from E. Schäfer Laboratory, Freiburg University, Germany; and cad2 and rax1-1 null mutants deficient in γ-GSH1 synthetase (Ball et al., 2004). Recessive null mutant apx2-1 was isolated from activation-tagged primary lines of Arabidopsis ecotype Col-7 (M2; first and second donation from SALK). The mutant was backcrossed to Col-0 (and reciprocally) to assert Mendelian segregation of the Basta resistance and the lack of APX2 expression traits in the F2 by t test. Plants were cultivated under short photoperiod (9 h), temperature 22/18°C (day/night, respectively), relative humidity of 70 to 80%, and illumination from a mixture of fluorescence tubes L30W/77-fluora and 30W41-827 lumilux (OSRAM), with the intensity of 100 ± 20 μmol photons m−2 s−1. For experiments, 6- and 10-week-old plants were used.
Light, Bacterial, and Pharmacological Treatment
Arabidopsis rosettes were fully or partly exposed to excess white light (2000 or 1500 μmol photons m−2 s−1; 1 h), blue (80 μmol photons m−2 s−1; 4 h, 450 ± 20 nm) and red light (120 μmol photons m−2 s−1; 4 h, 650 ± 20 nm) supplied by light-emitting diode panels (Photon System Inst.). The above light conditions provided similar energy at the indicated spectral regions. Heat emission from the light source was insignificant. Otherwise, low light (100 ± 20 μmol photons m−2 s−1) conditions were used. During electrophysiological experiments, leaves were exposed to excess light of 1500 μmol photons m−2 s−1 supplied by a halogen lamp. Light was directed on a single leaf by optical fiber and selective filters (Jenaer Glaswerk Schott and Gen.) of 650- and 450-nm wavelength were used when required.
In experiments with mechanically damaged petioles and photosynthetic electron transport inhibitors, 15 min before the excess light, leaves were treated as follows: main veins in petioles of exposed leaves were mechanically interrupted with a wood splinter (see Supplemental Figure 3 online) or leaves and their petiole were treated with 14 μM DBMIB, 8 μM DCMU, or 10 μM lanthanide chloride (LaCl3) as described earlier (Karpinski et al., 1999; Mühlenbock et al., 2008). Systemic leaves were kept at low light (100 ± 20 μmol photons m−2 s−1) conditions (Figures 1, 2, and 4) or in the twilight zone (<0.1 μmol photons m−2 s−1) for experiments presented in Figures 4 and 5. In experiments with DCMU and DBMIB partially treated rosettes (Figures 3 and 5), the action of inhibitors was monitored by measurement of chlorophyll a fluorescence parameters (see Supplemental Figure 2 online). These measurements ensured us that the effects of DCMU and DBMIB were not significantly spreading to systemic leaves.
In the pathogen proliferation tests, leaves of Col-0 plants were inoculated with Pseudomonas syringae pv tomato DC3000 strains by subepidermal injections 1 h before and 1, 8, and 24 h after exposure to excess light episodes and bacterial growth was measured at 1, 48, and 96 h after infection (for details, see figure legends) as described before (Rusterucci et al., 2001; Mühlenbock et al., 2008).
Imaging and Measurements of Transgene Induction and Expression
Expression of APX1:LUC was imaged in leaves after removal of plants from the given treatment and spraying them with 1 mM luciferin (Promega). Leaves were kept in the dark for ~15 min and imaged with a Peltier-cooled CCD camera (Wright Instruments) for 4 min. APX1:LUC induction was quantified in three leaves of each rosette. Collected samples (~5 mg) were also grinded in 0.5 mL lysis buffer (Promega kit). About 50 μL of the homogenate was placed under a luminometer tube (Berthold), and 50 μL of luciferin assay was added 10 s before the measurement. LUs or RLUs were expressed per gram fresh weight of leaves.
Chlorophyll Fluorescence Parameters
Chlorophyll fluorescence parameters were determined with a portable fluorescence monitoring system (FMS1) and the manufacturer's software (Hansatech). Images of the NPQ and Fv/Fm were generated as described by Barbagallo et al. (2003) using a FluorImager and the associated software (Technologica and Photon System Inst.). Chlorophyll fluorescence terminology is explained in detail elsewhere (Maxwell and Johnson, 2000; Baker, 2008).
Plasma Membrane Electrical Potential Measurements
Electrical potential difference across the plasma membrane was measured by impaling a cell with a microelectrode filled with 1 M KCl connected to a microelectrode preamplifier. Impalements were made using a three-dimensional micromanipulator, the tip of one microelectrode was inserted into veins of the locally treated leaves, and the tip of a second electrode was placed in one of the systemic leaves. Implementation of the microelectrode was made under ×80 magnification, and the number of cell layers through which the microelectrode had penetrated was detected by specific changes of the displayed electrical potential of the instrument. All experiments were performed in a chamber (see Supplemental Figure 3B online) filled with a solid transparent silicon layer (3 mm) and with buffered solution containing 5 mM MES and 0.1 mM CaCl2, pH 6, at room temperature (22°C to 24°C). The chamber and fiber optics were arranged in such a way that systemic part of Arabidopsis rosettes was isolated from direct light provided by optic fibers and was in the twilight zone (<0.1 μmol photons m−2 s−1). The reference electrode (Ag/AgCl) was immersed in this solution. The light was turned on and off during the experiment, and the action potential was measured simultaneously for 30 to 40 min in two separated single bundle sheath cells (in two different leaves) by a coupled system of Axoclamp-2B (Axon Instruments), and the simultaneous dual signal was recorded by the Axoscope 10.1 program.
The Analysis of the Redox State of PQ
HPLC was used to estimate the concentration of oxidized PQ and reduced PQ (PQH2) in leaves according to the protocol described in detail by Kruk and Karpinski (2006). PQ and PQH2 concentrations were determined based on the area of the peaks of standard PQ and PQH2 solutions. The concentration of oxidized and reduced PQ standards was determined using ε255 = 17.94 mM−1 cm−1 for PQ and ε290 = 3.39 mM−1 cm−1 for PQH2, both in absolute ethanol.
Hydrogen Peroxide Measurements
Total H2O2 content was measured in fresh plant material by a fluorometric assay with homovanillic acid according to Ishikawa et al. (1993). Samples were homogenized in 0.6 mL of ice-cold 5% TCA and centrifuged. The reaction mixture contained 1% supernatant, 1.25 mM homovanillic acid, l unit of horseradish peroxidase (Sigma-Aldrich), and 25 mM potassium phosphate buffer, pH 7.5. The fluorescence was measured at the excitation wavelength of 315 nm and emission of 425 nm.
For fluorescence microscopy analysis of hydrogen peroxide, leaves were sprayed with 2’,7’-dichlorofluorescein diacetate just before the experiment.
Statistical Analysis
All experiments were performed in at least three independent experimental repetitions, and number of replicates for each experiment is given in the figure legends. Results are expressed as means ± sd. Student's t test (for equality of means) was used for comparisons among light treatments, and a difference at P < 0.05 was considered weakly significant, P < 0.005 significant, and P < 0.001 strongly significant.
RNA Isolation, cDNA Preparation, Primer Design, and PCR
A standard protocol was used for total RNA extraction and first-strand cDNA synthesis. Primer pairs for quantification of APX1 mRNA (forward 5′-CGTCTGATGCTGACTACGCT-3′ and reversed 5′-ACTACGCTTGCATCGTCGCT-3′) and of APX2 mRNA (forward 5′-CGGTTGGTAGTTGAAGAAGTC-3′ and reverse 5′-AAGAAAGCTGTTCAGAGATGC-3′) transcripts were used for PCR reaction. Harvested leaf tissue or other specified parts of the plant were frozen and ground to a fine powder in liquid nitrogen. RNA extraction was performed using a Qiagen Rneasy plant mini kit (Qiagen) followed by DNA-free kit (Ambion). The first cDNA strand was synthesized with a RETROscript kit (Ambion). RT-PCRs were performed using specific primers custom made from Invitrogen and combined with 18S RNA as an internal standard (QuantumRNA 18S; Ambion). Quantitative RT-PCR was performed using a 2:8 molar ratio of 18S primers/competimers (this ratio was experimentally determined before; Mühlenbock et al., 2008). Quantitative analysis was performed with ethidium bromide staining after 5, 10, 15, and 20 RT-PCR cycles in comparison to the internal standard. Such DNA concentrations gave linear responses of the sample and the internal 18S RNA standard and were calculated in such a way that relative APX1 and APX2 gene induction values were in the range of the relative induction values obtained by a luciferase assay in APX1:LUC and APX2:LUC transgenenic plants exposed to excess light. This RT-PCR method was successfully used before in our laboratory (Mühlenbock et al., 2008; Chang et al., 2009). The relative expression was determined by the ratio of sample band intensity/internal standard band intensity using a densitometer system (GS-800; Bio-Rad Laboratories). A pool of samples from three plants for each line was collected, and two independent experiments were performed with several technical repetitions per experiment.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: AT1G07890 (APX1), AT3G09640 (APX2), AT2G21730 (CAD2), AT4G08920 (CRY1), AT1G04400 (CRY2), AT2G925080 (GSH1), AT1G44575 (NPQ4), AT1G09570 (PHYA), AT2G18790 (PHYB), AT3G45780 (PHOT1), and AT5G58140 (PHOT2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. APXs Transcript Levels and NPQ Are Deregulated in Photoreceptors and npq4 Null Mutants.
Supplemental Figure 2. The Effect of DCMU and DBMIB on Time Course of Maximum Photochemical Efficiency (Fv/Fm) Measured in Local (LO) and Systemic (SY) Leaves.
Supplemental Figure 3. Experimental System for Measurement of Plasma Membrane Electrical Potential.
Supplemental Figure 4. Cellular Light Memory Expressed by Changes of Maximum Photochemical Efficiency (Fv/Fm) and Efficiency of Photosystem II (ΦPSII).
Supplemental Figure 5. Induction of the Systemic Acquired Acclimation Is Associated with Systemic Patchy Changes in Maximal Photochemical Efficiency (Fv/Fm) and Systemic Gradient-Like Changes of Foliar Nonphotochemical Quenching.
Acknowledgments
This article is in memory of Markus Klenell. We are grateful to our colleagues, Christine Foyer and Philip Mullineaux, for valuable comments made on our manuscript and to Dietmar Funk for comments and advise concerning PEPS measurements. This work was financed by the Welcome 2008/1 project operated within the Foundation for Polish Science Welcome Program cofinanced by the European Regional Development Fund and from Knut and Alice Wallenberg Foundation (Sweden) (to S.K.). M.S.-H. is grateful for the financial support from European Union Committee, Maria Skłodowska-Curie fellowship MEIF-CF-2005-024914, and MERG-CT-2007-207350.
References
- Apel K., Hirt H. (2004). Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Asada K. (1999). The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 601–639 [DOI] [PubMed] [Google Scholar]
- Baker N.R. (2008). Chlorophyll fluorescence: A probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59: 89–113 [DOI] [PubMed] [Google Scholar]
- Ball L., Accotto G.-P., Bechtold U., Creissen G., Funck D., Jimenez A., Kular B., Leyland N., Mejia-Carranza J., Reynolds H., Karpinski S., Mullineaux P.M. (2004). An Arabidopsis mutant with raised ASCORBATE PEROXIDASE2 expression reveals glutathione as a direct modulator of stress responsive gene expression. Plant Cell 1: 2448–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baluska F., Volkmann D., Menzel D. (2005). Plant synapses: Actin-based domains for cell-to-cell communication. Trends Plant Sci. 10: 106–111 [DOI] [PubMed] [Google Scholar]
- Barbagallo R.P., Oxborough K., Pallett K.E., Baker N.R. (2003). Rapid noninvasive screening for perturbations of metabolism and plant growth using chlorophyll fluorescence imaging. Plant Physiol. 132: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barneche F., Winter V., Crèvecur M., Rochaix J.-D. (2006). ATAB2 is a novel factor in the signalling pathway of light-controlled synthesis of photosystem proteins. EMBO J. 25: 5907–5918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellafiore S., Bameche F., Peltier G., Rochaix J.D. (2005). State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature 433: 892–895 [DOI] [PubMed] [Google Scholar]
- Bose J.C. (1926). The Nervous Mechanism of Plants. (London: Langmans, Green and Company; ). [Google Scholar]
- Bowles D.J. (1990). Defence-related proteins in higher plants. Annu. Rev. Biochem. 59: 873–907 [DOI] [PubMed] [Google Scholar]
- Burdon-Sanderson J. (1873). Note on the electrical phenomena which accompany irritation of the leaf of Dionea muscipula in the excited and unexcited states. Proc. R. Soc. Lond. 21: 491–496 [Google Scholar]
- Chang C.C.C., Ball L., Fryer M., Baker N.R., Karpinski S., Mullineaux P. (2004). Induction of ASCORBATE PEROXIDASE 2 expression in wounded Arabidopsis leaves does not involve known wound-signalling pathways but is associated with changes in photosynthesis. Plant J. 38: 499–511 [DOI] [PubMed] [Google Scholar]
- Chang C.C.C., Ślesak I., Jorda L., Sotnikov A., Melzer M., Miszalski Z., Mullineaux P.M., Parker J.E., Karpinska B., Karpinski S. (2009). Arabidopsis chloroplastic glutathione peroxidases play a role in cross talk between photooxidative stress and immune responses. Plant Physiol. 150: 670–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl J.L., Jones J.D.G. (2001). Plant pathogens and integrated defense responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Darwin C. (1875). Insectivorous Plants. (London: John Murray; ). [Google Scholar]
- Davies E. (2004). New functions for electrical signals in plants. New Phytol. 161: 607–612 [DOI] [PubMed] [Google Scholar]
- Demming-Adams B., Adams W.W. (2000). Harvesting sunlight safely. Nature 403: 371–374 [DOI] [PubMed] [Google Scholar]
- Doron E., Itzhak O., Nir K., Aaron K. (2008). Changes in the photosynthetic reaction centre II in the diatom Phaeodactylum tricornutum result in non-photochemical fluorescence quenching. Environ. Microbiol. 10: 1997–2007 [DOI] [PubMed] [Google Scholar]
- Fey V., Wagner R., Brautigam K., Wirtz M., Hell R., Dietzmann A., Leister D., Oelmuller R., Pfannschmidt T. (2004). Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J. Biol. Chem. 280: 5318–5328 [DOI] [PubMed] [Google Scholar]
- Firestein S. (2001). How the olfactory system makes sense of scents. Nature 413: 211–218 [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G. (1999). Leaves in the dark see the light. Science 284: 599–601 [DOI] [PubMed] [Google Scholar]
- Fromm J., Eschrich W. (1989). Correlation of ionic movements with phloem unloading and loading in barley leaves. Plant Physiol. Biochem. 27: 577–585 [Google Scholar]
- Fryer M.J., Ball L., Oxborough K., Karpinski S., Mullineaux P.M., Baker N.R. (2003). Control of Ascorbate Peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J. 33: 691–705 [DOI] [PubMed] [Google Scholar]
- Galvez-Valdivieso G., Fryer M.J., Lawson T., Slattery K., Truman W., Smirnoff N., Asami T., Davies W.J., Jones A.M.J., Baker N.R., Mullineaux P.M. (2009). The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 21: 2143–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss R., Opitz C., Lepetit B., Wilhelm C. (2008). The synthesis of NPQ-effective zeaxanthin depends on the presence of a transmembrane proton gradient and a slightly basic stromal side of the thylakoid membrane. Planta 228: 999–1009 [DOI] [PubMed] [Google Scholar]
- Gramss T., Bornholdt S., Gross M., Mitchell M., Pellazzari T. (1998). Non-standard Computation: Molecular Computing, Cellular Automata, Evolutionary Algorithms, Quantum Computers. (New York: Wiley; ). [Google Scholar]
- Hanson M.R., Sattarzadeh A. (2008). Dynamic morphology of plastids and stromules in angiosperm plants. Plant Cell Environ. 31: 646–657 [DOI] [PubMed] [Google Scholar]
- Havaux M., Dall’Osto L., Cuine S., Giuliano G., Bassi R. (2004). The effect of zeaxanthin as the only xanthophyll on the structure and function of the photosynthetic apparatus in Arabidopsis thaliana. J. Biol. Chem. 279: 13878–13888 [DOI] [PubMed] [Google Scholar]
- Higinbotham N. (1973). Electropotentials of plant cells. Annu. Rev. Plant Physiol. 24: 25–46 [Google Scholar]
- Holt N.E., Fleming G.R., Niyogi K.K. (2004). Toward an understanding of the mechanism of nonphotochemical quenching in green plants. Biochemistry 43: 8281–8289 [DOI] [PubMed] [Google Scholar]
- Ishikawa T., Takeda T., Shigeoka S., Hirayama O., Mitsunaga T. (1993). Hydrogen peroxide generation in organelles of Euglena gracilis. Phytochemistry 33: 1297–1299 [Google Scholar]
- Jabs T., Dietrich R.A., Dangl J.L. (1996). Initiation of runaway cell death in an Arabidopsis mutant by extracellular superoxide. Science 273: 1853–1856 [DOI] [PubMed] [Google Scholar]
- Johnson M.P., Davison P.A., Ruban A.V., Horton P. (2008). The xanthophyll cycle pool size controls the kinetics of non-photochemical quenching in Arabidopsis thaliana. FEBS Lett. 582: 262–266 [DOI] [PubMed] [Google Scholar]
- Karpinska B., Wingsle G., Karpinski S. (2000). Antagonistic effects of hydrogen peroxide and glutathione on acclimation to excess excitation energy in Arabidopsis. IUBMB Life 50: 21–26 [DOI] [PubMed] [Google Scholar]
- Karpinski S., Escobar C., Karpinska B., Creissen G., Mullineaux P. (1997). Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. Plant Cell 9: 627–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski S., Reynolds H., Karpinska B., Wingsle G., Creissen G., Mullineaux P. (1999). Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 284: 654–657 [DOI] [PubMed] [Google Scholar]
- Klenell M., Morita S., Tiemblo-Olmo M., Mühlenbock P., Karpinski S., Karpinska B. (2005). Photosystem II light harvesting antenna organization controlled by cpSRP43 plays an essential role in regulation of Arabidopsis thaliana photooxidative stress tolerance. Plant Cell Physiol. 46: 118–126 [DOI] [PubMed] [Google Scholar]
- Knight H. (2000). Calcium signaling during abiotic stress in plants. Int. Rev. Cytol. 195: 269–324 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S., Suzuki N., Huntington S., Armijo L., Sha W., Cortes D., Shulaev V., Mittler R. (2008). Ascorbate Peroxidase 1 plays a key role in the response of Arabidopsis thaliana to stress combination. J. Biol. Chem. 283: 34197–34203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziolek C., Grams T.E.E., Schreiber U., Matyssek R., Fromm J. (2004). Transient knockout of photosynthesis mediated by electrical signals. New Phytol. 161: 715–722 [DOI] [PubMed] [Google Scholar]
- Kruk J., Karpinski S. (2006). An HPLC-based method of estimation of the total redox state of plastoquinone in chloroplasts, the size of the photochemically active plastoquinone-pool and its redox state in thylakoids of Arabidopsis. Biochim. Biophys. Acta 1757: 1669–1675 [DOI] [PubMed] [Google Scholar]
- Köhler R.H., Cao J., Zipfel W.R., Webb W.W., Hanson M.R. (1997). Exchange of protein molecules through connections between higher plant plastids. Science 276: 2039–2042 [DOI] [PubMed] [Google Scholar]
- Kulheim C., Agren J., Jansson S. (2002). Rapid regulation of light harvesting and plant fitness in the field. Science 297: 91–93 [DOI] [PubMed] [Google Scholar]
- Kwak J.M., Mori I.C., Pei Z.M., Leonhardt N., Torres M.A., Dangl J.L., Bloom R.E., Bodde S., Jones J.D., Schroeder J.I. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok E.Y., Hanson M.R. (2003a). Microfilaments and microtubules control the morphology and movement of non-green plastids and stromules in Nicotiana tabacum. Plant J. 35: 16–26 [DOI] [PubMed] [Google Scholar]
- Kwok E.Y., Hanson M.R. (2003b). Stromules and the dynamic nature of plastid morphology. J. Microsc. 214: 124–137 [DOI] [PubMed] [Google Scholar]
- Laloi C., Stachowiak M., Pers-Kamczyc E., Warzych E., Murgia I., Apel K. (2007). Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana Proc. Natl. Acad. Sci. USA 104: 672–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin R.M., Alonso J.M., Ecker J.R., Chory J. (2003). GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299: 902–906 [DOI] [PubMed] [Google Scholar]
- Lautner S., Grams T.E.E., Matyssek R., Fromm J. (2005). Characteristics of electrical signals in poplar and responses in photosynthesis. Plant Physiol. 138: 2200–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.P., Björkmann O., Shih C., Grossman A.R., Rosenquist M., Jansson S., Niyogi K.K. (2000). A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature 403: 391–395 [DOI] [PubMed] [Google Scholar]
- Mateo A., Funck D., Mühlenbock P., Kular B., Mullineaux P.M., Karpinski S. (2006). Controlled levels of salicylic acid are required for optimal photosynthesis and redox homeostasis. J. Exp. Bot. 57: 1795–1807 [DOI] [PubMed] [Google Scholar]
- Mateo A., Mühlenbock P., Rustérucci C., Chi-Chen C., Miszalski Z., Karpinska B., Parker J.E., Mullineaux P.M., Karpinski S. (2004). The LESION SIMULATING DISEASE (LSD1) gene is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 136: 2818–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K., Johnson G.N. (2000). Chlorophyll fluorescencea practical guide. J. Exp. Bot. 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Mittler R. (2002). Oxidative stress, antioxidants, and stress tolerance. Trends Plant Sci. 7: 405–410 [DOI] [PubMed] [Google Scholar]
- Mott K.A., Peak D. (2007). Stomatal patchiness and task-performing networks. Ann. Bot. (Lond.) 99: 219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenbock P., Plaszczyca M., Mellerowicz E., Karpinski S. (2007). Lysigenous aerenchyma formation in Arabidopsis is controlled by LESION SIMULATING DISEASE1. Plant Cell 19: 3819–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenbock P., Szechyńska-Hebda M., Plaszczyca M., Baudo M., Mullineaux P.M., Parker J.E., Karpińska B., Karpiński S. (2008). Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20: 2339–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux P., Ball L., Escobar C., Karpinska B., Creissen G., Karpinski S. (2000). Are diverse signalling pathways integrated in the regulation of Arabidopsis antioxidant defence gene expression in response to excess excitation energy? Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 1531–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullineaux P., Karpinski S. (2002). Signal transduction in response to excess light: Getting out of the chloroplast. Curr. Opin. Plant Biol. 5: 43–48 [DOI] [PubMed] [Google Scholar]
- Mullineaux P.M., Karpinski S., Baker N.R. (2006). Spatial dependence for hydrogen peroxide-directed signaling in light-stressed plants. Plant Physiol. 141: 346–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwesigwa J., Collins D.J., Volkov A.G. (2000). Electrochemical signaling in green plants: effects of 2,4-dinitrophenol on variation and action potentials in soybean. Bioelectrochemistry 51: 201–205 [DOI] [PubMed] [Google Scholar]
- Nagai T., Kim D.J., Delay R.J., Roper S.D. (1996). Neuromodulation of transduction and signal processing in the end organs of taste. Chem. Senses 21: 353–365 [DOI] [PubMed] [Google Scholar]
- Niyogi K.K. (2000). Safety valves for photosynthesis. Curr. Opin. Plant Biol. 3: 455–460 [DOI] [PubMed] [Google Scholar]
- Pascal A.A., Liu Z., Broess K., van Oort B., van Amerongen H., Wang C., Horton P., Robert B., Chang W., Ruban A. (2005). Molecular basis of photoprotection and control of photosynthetic light-harvesting. Nature 436: 134–137 [DOI] [PubMed] [Google Scholar]
- Peak D., West J.D., Messinger S.M., Mott K.A. (2004). Evidence for complex, collective dynamics and emergent, distributed computation in plants. Proc. Natl. Acad. Sci. USA 101: 918–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R.B., Havir E.A. (2001). Photosynthetic properties of an Arabidopsis thaliana mutant possessing a defective PsbS gene. Planta 214: 142–152 [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T., Braeutigam K., Wagner R., Dietzel L., Schroeter Y., Steiner S., Nykytenko A. (2009). Potential regulation of gene expression in photosynthetic cells by redox and energy state: Approaches towards better understanding. Ann. Bot. (Lond.) 103: 599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T., Nilsson A., Allen J. (1999). Photosynthetic control of chloroplast gene expression. Nature 397: 625–628 [Google Scholar]
- Pickard B.G. (1973). Action potentials in higher plants. Bot. Rev. 39: 172–201 [Google Scholar]
- Rossel J.B., Wilson P.B., Hussain D., Woo N.S., Gordon M.J., Mewett O.P., Howell K.A., Whelan J., Kazan K., Pogson B.J. (2007). Systemic and intracellular response to photooxidative stress in Arabidopsis. Plant Cell 19: 4091–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban A.V., Berera R., Ilioaia C., van Stokkum I.H.V., Kennis J.T.M., Pascal A.A., van Amerongen H., Robert B., Horton P., van Grondelle R. (2007). Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450: 575–578 [DOI] [PubMed] [Google Scholar]
- Rusterucci C., Aviv D.H., Holt B.F., III, Dangl J.L., Parker J.E. (2001). The disease resistance signaling components EDS1 and PAD4 are essential regulators of the cell death pathway controlled by LSD1 in Arabidopsis. Plant Cell 13: 2211–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn G. (1908). Die Gestalts- und Lageveränderung der Pflanzen-Chromatophoren. (Leipzig, Germany: Engelmann; ). [Google Scholar]
- Ślesak I., Karpinska B., Surówka E., Miszalski Z., Karpinski S. (2003). Redox changes in the chloroplast and hydrogen peroxide are essential for regulation of C3-CAM transition and photooxidative stress responses in the facultative CAM plant Mesembryanthemum crystallinum L. Plant Cell Physiol. 44: 573–581 [DOI] [PubMed] [Google Scholar]
- Spencer G.J., Hitchcocka I.S., Genever P.G. (2004). Emerging neuroskeletal signalling pathways: A review. FEBS Lett. 559: 6–12 [DOI] [PubMed] [Google Scholar]
- Trewavas A. (2003). Aspects of plant intelligence. Ann. Bot. (Lond.) 92: 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Breusegem F., Bailey-Serres J., Mittler R. (2008). Unravelling the tapestry of networks involving reactive oxygen species in plants. Plant Physiol. 147: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov A.G., Collins D.J., Mwesigwa J. (2001). Plant electrophysiology: Pentachlorophenol induces fast action potentials in soybean. Plant Sci. 153: 185–190 [DOI] [PubMed] [Google Scholar]
- von Neumann J., Morgenstern O. (1944). Theory of Games and Economic Behavior. (Princeton, NJ: Princeton University Press; ). [Google Scholar]
- Wheeler G.L., Brownlee C. (2008). Ca2+ signalling in plants and green algae: Changing channels. Trends Plant Sci. 13: 506–514 [DOI] [PubMed] [Google Scholar]
- Wildon D.C., Thain J.F., Minchin P.E.H., Gubb I.R., Reilly A.J., Skipper Y.D., Doherty H.M., Odonnell P.J., Bowles D.J. (1992). Electrical signaling and systemic proteinase-inhibitor induction in the wounded plant. Nature 360: 62–65 [Google Scholar]
- Willekens H., Chamnongpol S., Davey M., Schraudner M., Langebartels C., Van Montagu M., Inzé D., Van Camp W. (1997). Catalase is a sink for H2O2 and is indispensable for stress defence in C3 plants. EMBO J. 16: 4806–4816 [DOI] [PMC free article] [PubMed] [Google Scholar]