This study examines how the transcription factor APETALA2 suppresses flowering by mapping direct targets of AP2 on a genome-wide scale and comparing the map to changes in gene expression. The results indicate an unexpected level of complexity in the interactions of transcription factors with one another and their targets.
Abstract
The Arabidopsis thaliana transcription factor APETALA2 (AP2) has numerous functions, including roles in seed development, stem cell maintenance, and specification of floral organ identity. To understand the relationship between these different roles, we mapped direct targets of AP2 on a genome-wide scale in two tissue types. We find that AP2 binds to thousands of loci in the developing flower, many of which exhibit AP2-dependent transcription. Opposing, logical effects are evident in AP2 binding to two microRNA genes that influence AP2 expression, with AP2 positively regulating miR156 and negatively regulating miR172, forming a complex direct feedback loop, which also included all but one of the AP2-like miR172 target clade members. We compare the genome-wide direct target repertoire of AP2 with that of SCHLAFMÜTZE, a closely related transcription factor that also represses the transition to flowering. We detect clear similarities and important differences in the direct target repertoires that are also tissue specific. Finally, using an inducible expression system, we demonstrate that AP2 has dual molecular roles. It functions as both a transcriptional activator and repressor, directly inducing the expression of the floral repressor AGAMOUS-LIKE15 and directly repressing the transcription of floral activators like SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1.
INTRODUCTION
For both plants and animals, the timing of the transition from immature to reproductive growth must be tightly regulated to maintain its adaptive value. In flowering plants, for example, a diminished competence for tracking environmental cues and responding appropriately is correlated with declining species abundance or even extinction (Willis et al., 2008). It is therefore no surprise that a finely nuanced gene regulatory network evolved to translate environmental and endogenous information, integrate the diverse inputs, and commit resources to initiate flowering when the time is most favorable. The emerging view of the regulatory network governing the reproductive transition in Arabidopsis thaliana is that of a tuned balance of repressors and activators, both classes operating throughout the plant tissues and at every level in the flowering time network topology (Kobayashi and Weigel, 2007; Giakountis and Coupland, 2008; Yant et al., 2009). Both positive and negative inputs are conveyed along initially distinct, but later crosstalking pathways to the shoot apex, where the transition is finally realized in morphological changes.
Extensive genetic and molecular studies have described several cardinal flowering time pathways: the photoperiod, vernalization, gibberellic acid, and autonomous pathways, each of which exerts either a floral promoting, neutral, or inhibiting state at any given time in response to different cues. The photoperiod pathway, for example, measures light quantity and quality in the leaves and promotes flowering as soon as daylength, or photoperiod, meets a particular threshold. When this threshold photoperiod is sensed, FLOWERING LOCUS T (FT) expression is upregulated in the leaves. The FT protein is then thought to travel to the shoot apex (Jaeger et al., 2006; Lifschitz et al., 2006; Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007; Tamaki et al., 2007; Komiya et al., 2009), where it interacts with the bZip transcription factor FD to orchestrate the transition to flowering, including the expression of the MADS domain protein APETALA1 (AP1) (Abe et al., 2005; Wigge et al., 2005).
Transcriptional repressors play a major role in flowering. The MADS domain transcription factor FLOWERING LOCUS C (FLC) represses FT expression and represents an important intersection of the photoperiod, vernalization, and autonomous pathways. FLC and another MADS domain transcription factor, SHORT VEGETATIVE PHASE, cooperate to repress directly the expression of the flowering pathway integrators FT, FD, and the MADS domain protein SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) by binding to their genomic loci (Lee et al., 2007; Li et al., 2008). Eventually this repressor, FLC, is itself repressed when plants experience extended periods of cold, or vernalization, during which an antisense RNA at the FLC locus (Swiezewski et al., 2009) initiates localized histone demethylase activity (Liu et al., 2010). In the absence of vernalization, inhibition of FLC expression is finally achieved later in development by members of the autonomous flowering pathway, FCA and FY (reviewed in Simpson et al., 2004), ensuring eventual flowering. In addition to this balancing act of FT promotion and transcriptional repression, another level of repression occurs when FT reaches the shoot apex. TERMINAL FLOWER1 (TFL1), itself a floral repressor with a high degree of homology to FT is thought to compete directly with FT for heterodimerization with FD at the shoot apex (Abe et al., 2005; Hanzawa et al., 2005; Wigge et al., 2005; Ahn et al., 2006; Giakountis and Coupland, 2008).
Aside from MADS domain repressors, another major group of transcriptional repressors plays a role in this complex network. It was recently discovered that the daylength-dependent upregulation of FT is directly repressed by AP2 domain–containing proteins from two related families. First, the TEMPRANILLO1 (TEM1) and TEM2 proteins were shown to repress flowering redundantly by repressing FT transcription (Castillejo and Pelaz, 2008). TEM1 belongs to a family of RAV-like AP2-DNA domain-containing loci that encode proteins with an AP2 domain and a B3-type DNA binding domain. It is most strongly expressed in leaves, where it represses flowering by directly binding to the 5′ untranslated region (UTR) of the FT genomic locus (Castillejo and Pelaz, 2008). A second, larger family of AP2-DNA domain-containing proteins is divided to two lineages: the ANT and euAP2 lineage (Kim et al., 2006). The euAP2 lineage contains six members that are predicted to be targeted by a major developmental microRNA and floral promoter, microRNA172 (miR172). This clade consists of the three TARGET OF EAT (TOE) genes (TOE1 to TOE3), SCHLAFMÜTZE (SMZ), its paralog SCHNARCHZAPFEN (SNZ), and AP2 (Park et al., 2002; Aukerman and Sakai, 2003; Schmid et al., 2003).
Although pleiotropic functions have been obvious in the analysis of ap2 mutants, the extensive redundancy among the AP2-like miR172 target clade members has long obscured their role in flowering time. Recent work has shown that overexpression of AP2, SMZ, SNZ, TOE1, or TOE2 caused late flowering (Aukerman and Sakai, 2003; Schmid et al., 2003; Chen, 2004; Jung et al., 2007) and that a quadruple smz snz toe1 toe2 mutant flowered earlier than any single or double mutant (Jung et al., 2007; Mathieu et al., 2009). When expressed in the leaves, SMZ is capable of repressing flowering by directly binding to the FT genomic locus, although misexpression of SMZ at the shoot apex has a negligible effect on flowering time (Mathieu et al., 2009). Therefore, given that AP2 is well known to be expressed and have functions in the shoot apex, we hypothesized that the partial redundancy observed among members of the clade may be due in part to tissue-specific expression of particular members, but also to differences in gene regulatory interactions with downstream targets.
AP2 is involved in a wide variety of developmental processes at the shoot apex, including the regulation of the stem cell niche (Wurschum et al., 2006), floral organ determination (Bowman et al., 1989), and the control of seed mass (Ohto et al., 2005). This extensive pleiotropy suggests a highly connected regulatory role (Carrera et al., 2009), the elucidation of which should be informative for understanding how AP2 influences such a variety of developmental processes.
Here, we describe repression of flowering by AP2 and its partially redundant family members, all targets of miR172. We reveal its mechanistic basis by high-resolution mapping of AP2 binding sites across the Arabidopsis genome and inducible gene expression analysis. AP2 represses flowering and flower development by binding directly to, and repressing the transcription of, the key flowering loci SOC1 and AGAMOUS and by binding to the miR172 genomic locus and repressing its transcription. Surprisingly, AP2 also functions as a bifunctional transcription factor, directly activating other floral repressors, the MADS domain protein AGAMOUS-LIKE15 (AGL15) (Adamczyk et al., 2007) and miR156 (Wang et al., 2009; Wu et al., 2009) by binding directly to their genomic loci.
RESULTS
AP2 Represses Flowering in a Daylength-Independent Manner
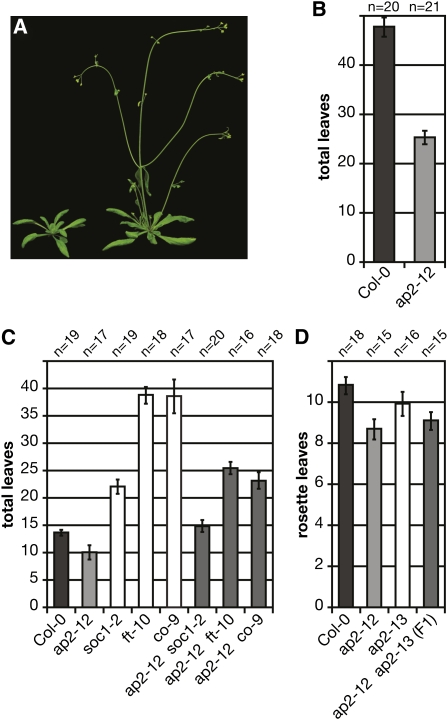
In addition to its better-known role in floral organ identity, a previous study indicated that AP2 was necessary for the late flowering induced by sucrose application (Ohto et al., 2001). When subjected to continuous light, an ap2 mutant flowered earlier than did controls (Ohto et al., 2005). To investigate the effect of AP2 mutations on flowering time under more ecologically relevant daylengths, we obtained a strong AP2 T-DNA insertion allele (Alonso et al., 2003), SALK_071140 (hereafter called ap2-12). We found that the transition to flowering was accelerated in this ap2 mutant under both noninductive short-day (SD; 8-h photoperiod) and inductive long-day (LD; 16-h photoperiod) conditions (Figure 1; see Supplemental Table 1 online). This effect was most marked under SD conditions, where the homozygous mutant produced only 25.3 ± 1.4 (2xSEM) leaves before flowering compared with wild-type Columbia-0 (Col-0) controls, which produced 47.8 ± 2.1 leaves (P < 0.0001, unpaired t test; Figure 1B). Under LD conditions, where the transition to flowering is greatly accelerated in Arabidopsis, ap2-12 mutants still flowered early, producing 10.0 ± 0.5 leaves before flowering, compared with the wild type, which produced 13.5 ± 0.5 leaves (P < 0.0001, unpaired t test; Figure 1C). Direct sequencing of the ap2-12 insertion site showed that the disruption was in the fifth intron, 18 nucleotides 5′ from the exon boundary, and AP2 mRNA levels in the mutant were >16-fold lower than wild-type controls, as measured by quantitative RT-PCR (qRT-PCR) (see Supplemental Figure 1 online). Flowers from ap2-12 plants clearly exhibited the characteristic ap2 loss-of-function phenotype.
Figure 1.
Influence of AP2 on Flowering Time and Genetic Interactions of AP2 with Other Flowering Time Mutants.
(A) Col-0 (left) and ap2-12 (right) plants grown in LDs.
(B) ap2-12 flowers early in SD conditions.
(C) ap2-12 flowers early under LD conditions and attenuates all late flowering mutants tested.
(D) ap2-12 and ap2-13 exhibit a dosage-dependent effect and are allelic. F1 plants flower with an intermediate number of leaves.
In (A), (C), and (D), plants were grown in LD conditions. Flowering time is given as total leaf number (rosette leaves plus cauline leaves) in (B) and (C) and rosette leaf number in (D). Error bars indicate 2xSEM.
To confirm further that the cause of early flowering was directly attributable to a lesion in AP2 and not a second site mutation, we examined the flowering time of an independently derived ap2 mutant allele. This line, ap2-13, was also early flowering, producing 11.8 ± 0.6 total leaves compared with the wild type (13.2 ± 0.4; total leaves in Supplemental Table 1 online, P = 0.0013, unpaired t test; rosette leaves in Figure 1D). F1 progeny of a cross of ap2-12 and ap2-13 all flowered early with an intermediate number of total leaves between the two parents (11.0 ± 0.4; see Supplemental Table 1 online). Thus, independent lesions in AP2 caused an early flowering phenotype, strongly implicating the loss of AP2 itself as the cause of the early flowering. Furthermore, the intermediate flowering of the F1 progeny suggests a quantitative effect.
Because the closely related euAP2 clade member SMZ interacts with the photoperiod pathway to repress FT transcription directly (Mathieu et al., 2009), we investigated whether there might also be an interaction of ap2 mutations with the photoperiod pathway. We therefore crossed ap2-12 with late flowering mutants soc1-2, ft-10, and co-9, hypothesizing that the effect of losing a repressor will be more evident if the floral promoting stimulus is attenuated, similar to what we had observed in SD conditions. In all three late flowering backgrounds the effect of ap2 loss of function was clear (Figure 1C; see Supplemental Table 1 online). In fact, soc1-2 ap2-12 flowered only one leaf later than wild-type Col-0 plants, whereas soc1-2 flowered 8.3 leaves later. When crossed to ap2-12, the strong mutants ft-10 and co-9 flowered 13.2 and 15.3 leaves earlier than the single mutant, respectively. Thus, the relative effect of soc1, ft, and co mutations was similar in the ap2 mutant background, but the introduction of the ap2 mutation produced additive early flowering in each line commensurate with the extent of late flowering in each single mutant. We conclude from these results that AP2 acts as a bona fide floral repressor that delays flowering in Arabidopsis under both inductive and noninductive photoperiods.
Extreme Early Transition of the Hexuple AP2-Like miR172 Target Mutant
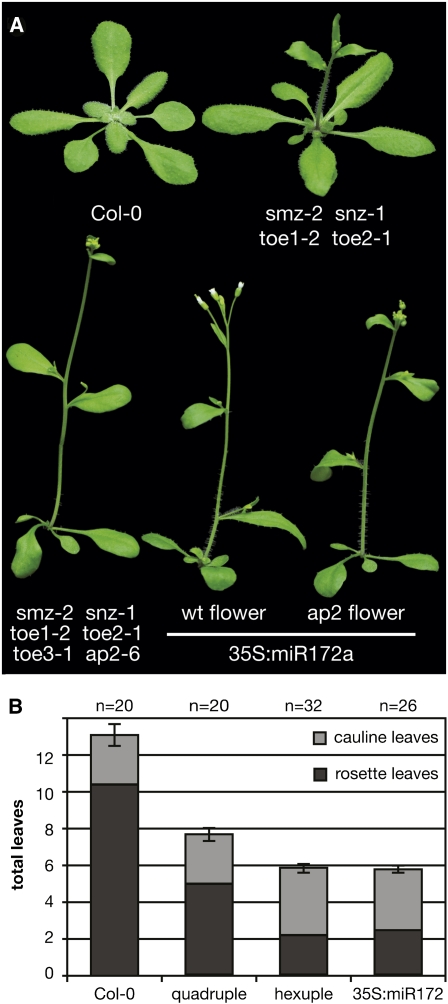
Including AP2, a clade of six AP2 domain-containing transcription factors (TOE1, TOE2, TOE3, SMZ, and SNZ) comprises the predicted target set of miR172. We previously observed that the quadruple mutant, toe1 toe2 smz snz, flowered significantly earlier than did Col-0, toe1, or toe1 toe2 double mutants (Mathieu et al., 2009). This quadruple mutant, however, did not flower as early as 35S:miR172 (Aukerman and Sakai, 2003; Chen, 2004), suggesting that there might exist further functional redundancy among the miR172 targets. Furthermore, as AP2 has a clear flowering time phenotype and TOE3 is a top direct regulatory target of the AP2-clade member SMZ (Mathieu et al., 2009), we were interested in observing the effect of a full clade, hexuple mutant loss of function.
To ensure that this suite of six genes constitutes the entire functional module of miR172 effectors, we combined a quadruple toe1 toe2 smz snz mutant with toe3-1 and ap2-12 to create a hexuple mutant miR172 target line that carries lesions in all six loci. Flowering with only 2.1 ± 0.1 rosette leaves, this hexuple mutant was significantly earlier than either Col-0 (10.4 ± 0.4 rosette leaves) or the quadruple mutant (4.9 ± 0.3 rosette leaves; Figure 2, Table 1) and phenocopied the 35S:miR172 plants very closely, which flowered after producing 2.4 ± 0.2 rosette leaves. For the quadruple, hexuple, and the miR172 overexpressor, total leaf numbers were much lower than in Col-0, but the number of leaves produced during the reproductive transition, or cauline leaves, actually increased to similar levels in both the hexuple mutant and miR172 overexpressor relative to the wild type. This points to a disconnect in this clade’s function before and after the transition from vegetative growth to reproductive growth and flowering.
Figure 2.
The Hexuple AP2-Like miR172 Mutant Recapitulates the miR172 Overexpressor.
(A) miR172 target clade quadruple (smz-2 snz-1 toe1-2 toe2-1) mutant, hexuple (smz-2 snz-1 toe1-2 toe2-1 toe3-1 ap2-6) mutant, and 35S:miR172a plants flower earlier than Col-0 control plants.
(B) Cauline (gray) and rosette (black) leaves to flower in mutants pictured in (A). All hexuple mutant plants and >75% of 35S:miR172 plants exhibited the AP2 floral homeotic phenotype (i.e., lack petals). Plants were grown in long days. Error bars indicate 2xSEM of the total leaf number.
Table 1.
Flowering Time of Hexuple and Quadruple AP2-Like miR172 Target Mutants and miR172 Overexpressor Plants
| (23°C, LD) | Rosette Leaves | Cauline Leaves | Total Leaves | 2xSEM | Range | n |
| Col-0 | 10.4 | 2.7 | 13.1 | 0.6 | 11 - 16 | 20 |
| smz-2 snz-1 toe1-2 toe2-1 | 4.9 | 2.7 | 7.6 | 0.2 | 7 - 9 | 20 |
| smz-2 snz-1 toe1-2 toe2-1 toe3-1 ap2-6 | 2.1 | 3.7 | 5.8 | 0.2 | 5 - 7 | 32 |
| 35S:miR172, T1 | 2.4 | 3.3 | 5.7 | 0.3 | 5 - 7 | 26 |
For each genotype, the mean of the total leaf number, the deviation from the mean (2× the standard error of the mean), the range of values found for each genotype, and the number of plants examined are given.
An earlier developmental phase occurring just before the transition to flowering, the juvenile to adult phase change, was also perturbed in the hexuple mutant. A hallmark of the transition from vegetative to reproductive phase in Arabidopsis is the appearance of trichomes on the abaxial (lower) leaf surface. In the hexuple mutants, this transition in trichome distribution was precocious. The hexuple mutants also displayed floral homeotic transformations typical of ap2 mutants. Finally, stigmatic papillae were occasionally evident on the tips of rosette leaves, similar to what has been observed in cauline leaves in 35S:miR172 overexpressors (see Supplemental Figure 2 online; Chen, 2004), suggesting derepression of reproductive transition genes in the leaves. Consistent with this, we observed strong induction of AG mRNA and that of its paralogs SHATTERPROOF1 (AGL1; SHP1), SHATTERPROOF2 (AGL5; SHP2), and SEEDSTICK (AGL11; STK) in the leaves of the hexuple mutant by qRT-PCR (see Supplemental Figure 2 online).
Genome-Wide Identification of AP2 Binding Sites
To understand better the mechanism of AP2-mediated repression of the reproductive transition, we mapped the genome-wide binding profile of AP2 by performing chromatin immunoprecipitation coupled to ultra-high-throughput deep sequencing (ChIP-seq) and also performed inducible gene expression studies.
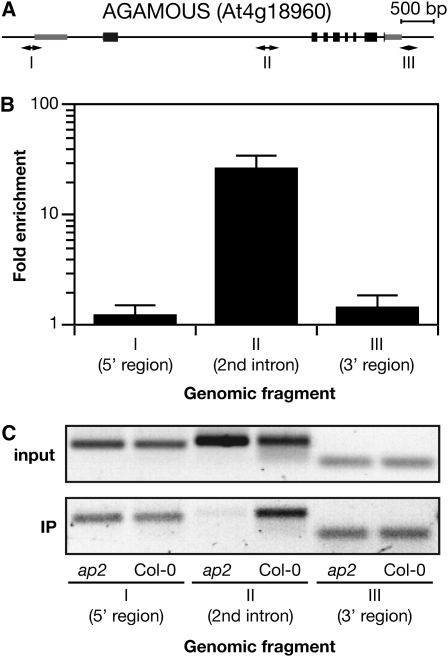
Based on genetic studies (Bowman et al., 1991; Drews et al., 1991), AP2 has been shown to repress AG expression most likely through multiple elements in the second intron (Bomblies et al., 1999; Deyholos and Sieburth, 2000). Hypothesizing that the mechanism underlying this repression might be direct chromatin binding, we tested raw technical and biological replicate ChIP samples by qPCR for enrichment of AG intron binding. All technical and biological replicates tested exhibited greatly enriched levels of binding at the AG intron, but not for two negative control regions flanking the AG locus (Figure 3). Independent confirmation ChIP experiments of a different mutant line, ap2-2, were performed with the same antibody with similar results (T. Dinh and X. Chen, unpublished data). As all pull downs exhibited enriched binding to AG, we processed two biological replicates by standard Illumina ChIP-seq library generation protocols to create single read libraries and sequenced them on an Illumina Genome Analyzer GAII.
Figure 3.
AP2 Binds Directly to the Second Intron of AG.
(A) ChIP-qPCR. AG locus pictured with ChIP amplicons I, II, and III.
(B) qPCR analysis of triplicate biological replicate samples for binding enrichment of AP2 in the AG second intron. Replicates from independent experiments were measured to produce mean and 2xSEM for regions mapped in (A).
(C) Abundance of one biological replicate of the PCR products used to create (B). PCR product for input (top) and immunprecipitated (bottom) DNA is shown for Col-0 and ap2-12 for the three amplicons (I, II, and III) tiling the AG locus.
After filtering for read quality and nonrepetitive mapability, between 5.2 and 14.2 million sequencing reads were uniquely mapped to the TAIR9 genome per sample (see Supplemental Table 2 online). We used SHORE (Ossowski et al., 2008) for mapping to the genome and performed enrichment analyses with an algorithm we developed specifically for Arabidopsis ChIP-seq peak calling. High confidence binding sites were identified by determining the overlap between highly significantly (false discovery rate [FDR] < 10−10) enriched bound regions in both biological replicates that also were enriched by a per base excess enrichment cutoff (see Methods for details). A total of 2275 regions in the nuclear genome were found to be significantly enriched in both independent biological replicates by these stringent criteria (see Supplemental Data Set 1 online). To ensure that our algorithm performed at least as well as published peak detection methods, we analyzed our AP2 data set using CisGenome (Ji et al., 2008) and compared the results to those obtained using our algorithm. Whereas CisGenome identified most of the targets that our algorithm did (2158 of 2275 at FDR < 0.01 in CisGenome), our script allowed us more transparency over the peak calling, so we continued analysis with this algorithm. The average width of these 2275 regions was 248 bp (Table 2).
Table 2.
Summary Characteristics of AP2 Peak Regions as Determined by ChIP-Seq
| Description | Value |
| Average peak region width (bases) | 248 |
| Number of bound regions at FDR < 10−10 + per base excess >0.25 | 2275 |
| Peaks <4 kb upstream from TSS or <2 kb downstream from TES | 2247 |
TSS, transcription start site; TES, transcription end site.
Genome-Wide Physical Distribution and Functional Classification of Bound Loci
Of the bound regions flanking genes, more were associated with regions <1 kb upstream of transcription start sites or 5′ UTRs than with the regions <1 kb downstream of transcription end sites or 3′ UTRs (see Supplemental Figure 3 online), similar to what has been observed for other Arabidopsis transcription factors, such as SEP3 (Kaufmann et al., 2009) or AGL15 (Zheng et al., 2009). Of those top 200 bound regions, 66 were located <1 kb upstream of a transcription start site. Several enrichments were evident for particular gene ontology (GO) categories among the 1780 bound loci for which GO assignments exist. Genes associated with organ development (GO:0048513) and transcription regulator activity (GO:0030528) were significantly overrepresented at a FDR P < 0.0005 among the list of AP2-bound loci, indicating functional specificity of target binding. Among the other biological processes found to be significantly (P < 0.01) overrepresented were shoot development (GO:0048367), shoot morphogenesis (GO:0010016), flower development (GO:0009908), transcription (GO:0006350), and DNA binding (GO:0003677).
AP2 Directly Binds Key Flowering Time Loci
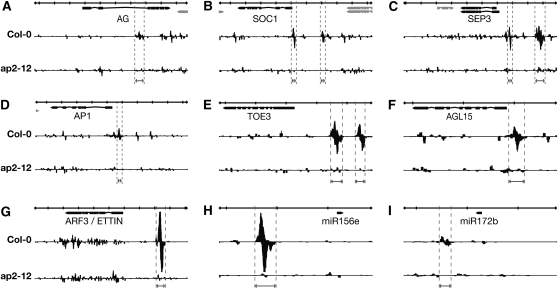
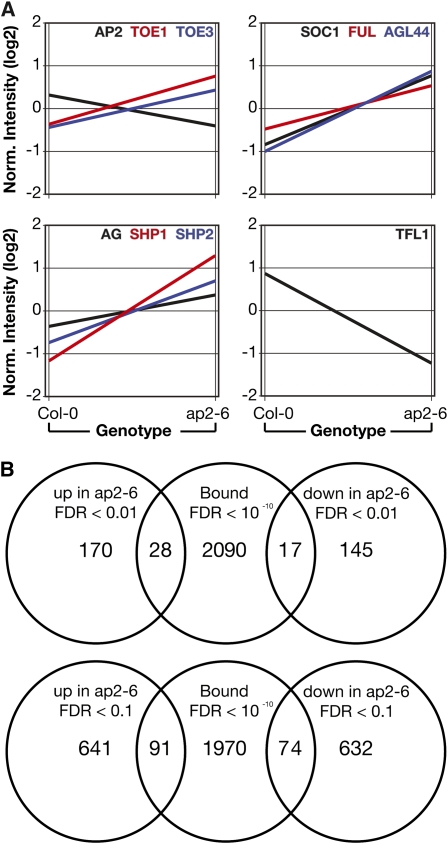
GO category enrichment indicated that AP2 occupies many loci involved in shoot development and morphogenesis in addition to floral development, consistent with the pleiotropic phenotypes of ap2 mutants. We hypothesized, however, that its role in the control of flowering time might involve loci just upstream of these well-described downstream floral organ development processes. Loci marking the transition to flowering, AP1 and SOC1, were bound with high confidence (FDR < 10−10) in both biological replicates, along with the floral organ identity genes SEPALLATA3 (SEP3) and AG (Figures 4A to 4D). The AP2-bound regions were all positioned in the 5′ sequences <1.6 kb upstream of the transcription start sites for SOC1, AP1, and SEP3; the region in AG, however, was in the 3′ end of the second intron (Figure 4A). The specific region bound in the AG intron corresponded closely with our ChIP qPCR (Figure 3), confirming those results (see Supplemental Figure 4 online).
Figure 4.
ChIP-Seq Reveals Direct AP2 Targets.
GBrowse traces of mapped ChIP-seq reads. In each panel, the top row indicates scale, with bold vertical lines indicating 1-kb increments. Gene models are shown under the scale bar, and single biological samples are pictured below. Forward reads are mapped above each line and reverse reads below. Regions adjacent to or inside introns of AG, SOC1, SEP3, AP1, TOE3, AGL15, ARF3, miR156, and miR172 are bound with high confidence (FDR < 10−10) in both biological replicates versus control samples in the ap2 mutant line. Vertical dotted lines delimit the position of the peak regions.
Interestingly, AP2 also directly bound to its own genomic locus and to those of clade members TOE3, SMZ, TOE1, and SNZ, for a total of five of the six AP2-like miR172 targets, consistent with the previously observed feedback regulation of AP2 (Schwab et al., 2005). In fact, the TOE3, AP2, SOC1, and SEP3 loci each contained multiple AP2-bound regions (Figures 4B, 4C, and 4E). The SMZ locus also had two high ranking regions <1 kb upstream of its transcription start site. Interestingly, another floral repressor, the MADS domain protein AGL18 (Adamczyk et al., 2007) was the nearest locus to multiple AP2-bound sites.
Gene Expression Changes in Response to AP2 Loss of Function
To understand the global effects of AP2 on the shoot transcriptome and to identify direct targets for which AP2 DNA binding may have an effect on transcript levels, we analyzed expression data from the inflorescences of ap2-6 mutants and Col-0 from the AtGenExpress developmental gene expression atlas (Schmid et al., 2005) (see Supplemental Data Set 2 online). As expected, AP2 RNA levels were significantly (RankProducts, percentage false positives [pfp] < 0.05) downregulated in ap2 mutant plants (Figure 5A). Consistent with its highly connected gene regulatory role and repressor function, 198 nuclear-encoded genes were upregulated and 162 were downregulated in ap2 mutant inflorescences (pfp < 0.01) relative to the wild type.
Figure 5.
Overlap of Genome-Wide Binding and Expression.
(A) Gene expression changes in ap2-6 influorescences as determined by Affymetrix microarray analysis.
(B) Overlap of loci bound by AP2 (determined by ChIP-seq) with transcripts differentially expressed in ap2 mutants (from [A]). Top Venn diagram indicates transcripts changed at RankProducts pfp < 0.01, and bottom Venn diagram indicates transcripts changed at RankProducts pfp < 0.1.
Elevated levels of several transcripts influencing flowering time were detected in ap2 mutants. While levels of CONSTANS, FT, and TWIN SISTER OF FT transcripts were not significantly different from the wild type, downstream of these genes, SOC1, FRUITFULL (FUL), and AG levels were significantly (pfp < 0.01) elevated in the ap2 mutant (Figure 5A), consistent with AP2 repressing these genes.
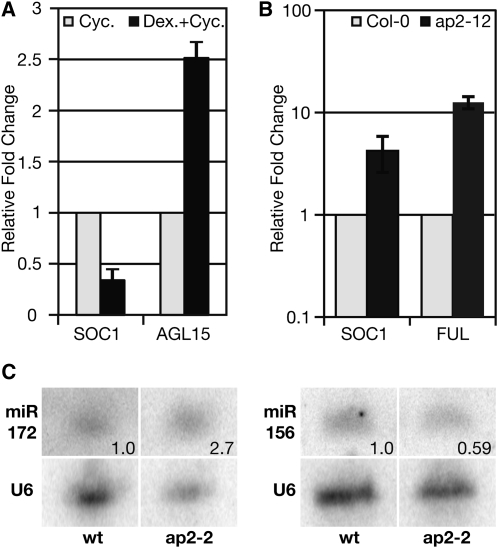
To confirm the effect of ap2 loss of function in the native context at the time the commitment to flowering is made but before reproductive structures are morphologically evident, we measured the mRNA abundance of two key redundant floral activators, SOC1 and FUL, by qRT-PCR on the aerial parts of 8-d-old LD-grown ap2-12 seedlings. Similar to the array data from bolting ap2-6 inflorescences (Figure 5A), both SOC1 and FUL transcript levels were upregulated in this early transition ap2 mutant seedling tissue (Figure 6B).
Figure 6.
AP2 Activates and Directly Represses Flowering Time Genes.
(A) Direct activation of AGL15 or repression of SOC1 was assayed by qRT-PCR in 35S:AP2m3-GR seedlings grown for eight LDs and treated with 10 μM CYC in the presence (black) or absence (gray) of 10 μM DEX. Data shown are representative of one biological replicate and three technical replicates from which sd was calculated. These data are representative of quadruplicate biological replicates, all of which showed similar results.
(B) SOC1 and FUL transcripts are upregulated in ap2 mutants undergoing the transition to flowering. Error bars indicate sd of triplicate biological replicates.
(C) miR172 and miR156 participate in a complex feedback loop with AP2. Small RNA gel blotting to detect miR172 and miR156 levels in wild-type versus ap2-2 dissected inflorescence tissue. The number on the gel images indicates normalized relative abundance of the small RNAs normalized to the loading control (U6). This assay was repeated twice with similar results.
Extensive feedback regulation has been observed in the clade of AP2-like miR172 targets, and we accordingly observed significantly upregulated TOE1 and TOE3 transcript levels in the ap2 mutants (Figure 5A). Of the transcripts downregulated in the ap2 mutant, only one stood out in regard to flowering: another floral repressor, TFL1, was significantly (pfp < 0.01) downregulated in the ap2 mutant (Figure 5A). Finally, as lipid transfer proteins have been implicated in TFL1 function (Sohn et al., 2007) and in pollen tube growth (Chae et al., 2009), we note that the four most upregulated transcripts in ap2 plants encoded lipid transfer proteins. Two of these four lipid transfer protein loci are bound by AP2, but the TFL1 locus is not. It has recently been reported, however, that AP1, which we identify as a direct transcriptional target of AP2, itself directly binds the TFL1 locus (Kaufmann et al., 2010).
Regulators of the Floral Transition and Floral Development Are Direct AP2 Targets
Having identified genome-wide AP2 binding sites and surveyed the transcriptome for genes that were differentially expressed in ap2 mutant plants, we sought to determine whether there was overlap between these data sets, which would indicate direct targets for AP2 (Figure 5B). In all, 91 loci were directly bound by AP2 and also upregulated in the ap2 mutant (binding FDR 10−10; expression FDR < 0.1; see Supplemental Data Set 3 online), suggesting that AP2 directly represses these genes. Seventy-four loci were directly bound by AP2 but downregulated in the ap2 mutant, suggesting the possibility that AP2 might also function as a positive regulator of transcription.
In surveying the set of these directly bound and regulated loci, we observed a synergistic effect. Particular GO categories, which were somewhat enriched in the list of bound genes and in the list of misregulated genes, were much more enriched in the overlap list of genes that were both bound and regulated by AP2 (see Supplemental Figure 5 online). Biological processes implicated in gynoecium, flower, carpel, floral whorl, and floral organ development all demonstrated this trend, as did the molecular functions transcription regulator activity, DNA binding, and regulation of transcription.
Several genes that play central roles in flowering and floral organ identity were among these AP2 direct targets, indicating that the influence of AP2 on flowering time is quite direct. These include the MADS domain genes SOC1, AG, and its paralogs SHP1 and SHP2, all of which were upregulated in the ap2 mutant and bound by AP2 (binding FDR 10−10; expression FDR < 0.05). Another MADS domain gene that was bound by AP2 and upregulated in the ap2 mutant is AGL44. This gene is strongly upregulated over the course of the floral transition but until now has not been closely associated with flowering. In addition, the AP2-like miR172 targets TOE1 and TOE3 and the locus encoding the DELLA protein RGA-like1 were directly bound and upregulated by AP2. Of the 74 directly bound loci whose transcript levels were downregulated (pfp < 0.1) in the ap2 mutant, only one, AGL15, stood out in relation to flowering time (binding FDR 10−10; expression FDR = 0.07). Interestingly, this gene was recently shown to act redundantly with AGL18 as a repressor of flowering in a daylength-independent manner, like AP2 (Adamczyk et al., 2007).
The combined analysis of genome-wide direct binding and global gene expression changes indicates that AP2 regulates flowering by modulating the expression of a range of known flowering time and organ identity genes. Furthermore, the expression of other floral repressors is dependent on AP2, with one, AGL15, both directly bound by AP2 (Figure 4F) and downregulated in the ap2 mutant. This raises the possibility that AP2 may act not only as a direct repressor but also as a bifunctional transcriptional regulator that either directly activates or directly represses the transcription of particular target genes.
Inducible AP2 Confirms Direct Targets and a Bifunctional Molecular Role
To explore whether these AP2 regulatory interactions were direct or indirect, we employed an inducible expression system. We expressed a translational fusion of AP2m3 (a miR172-resistant version of AP2) (Chen, 2004) to the hormone binding domain of the rat glucocorticoid (GR) receptor under the control of a constitutive promoter, the cauliflower mosaic virus 35S. Treatment with the GR ligand dexamethasone (DEX) allows AP2m3-GR entry to the nucleus, while administration of cycloheximide (CYC) inhibits protein synthesis, allowing the detection of direct AP2 transcriptional targets by quantitative PCR. The functionality of AP2m3-GR was revealed by the typical AP2m3 floral phenotype and the late flowering induced by DEX treatment (see Supplemental Figure 6 online; Chen, 2004). Using this system, we observed significant downregulation of SOC1 transcript levels in response to DEX treatment in the absence of protein synthesis, indicating that AP2 directly represses the transcription of SOC1 in 8-d-old seedlings when the decision to flower is being made (Figure 6A). By contrast, we observed significant upregulation of the directly bound floral repressor AGL15 in response to DEX treatment, confirming that AP2 can act also as a direct transcriptional activator (Figures 6A and 4F).
AP2 Participates in a Feedback Loop with Its Regulators miR172 and miR156
We observed that several of the highest confidence AP2 binding sites from the ChIP-seq experiment were associated with microRNAs that regulate developmental processes, such as flowering and phase transition. The most striking of these was the #2 genome-wide peak (as rank sum of biological replicate rankings), which was centered immediately upstream of miR156e (Figure 4H). This microRNA affects the juvenile-to-adult phase transition (Wu et al., 2009) and an endogenous flowering pathway (Wang et al., 2009) by negatively regulating the expression of miR172b (Wu et al., 2009). This latter microRNA, miR172b, was another high-confidence target of AP2 (Figure 4I), making these two microRNAs the highest-ranking microRNAs in the AP2 target list. These interwoven connections between AP2 and the two microRNAs that regulate it suggest a highly buffered regulatory network controlling the closely related developmental processes underlying phase transition and flowering time.
We sought to determine whether the expression of these microRNAs is changed as a result of changes in AP2 expression, which would indicate that this binding has a functional consequence. We therefore performed small RNA gel blots on inflorescence tissue from ap2-2 mutants and compared the abundance of these microRNA families to that in wild-type plants. Levels of miR172 in ap2 mutants were 270% of those in the wild type, while levels of miR156 in ap2 mutants were 59% of those in the wild type (Figure 6C), indicating that AP2 not only binds to but also negatively regulates miR172 and positively regulates miR156. Thus, multiple lines of evidence (ChIP-seq and gene expression changes in response to AP2) point to this selective and direct positive and negative regulation of developmental microRNAs by AP2.
AP2 Target Binding during AP2-Mediated Floral Repression in Leaves
Based on the well-known ap2 floral homeotic phenotype, we initially expected to observe a genetic role for AP2 primarily in the regulation of loci involved in early floral whorl development. Surprisingly, however, AP2 bound to numerous loci influencing the transition from vegetative growth to flowering, even in the developing inflorescence. We therefore followed these data to investigate whether active AP2-mediated floral repression resulted in binding to the same loci. Because several of the members of the AP2-like miR172 target clade, including AP2 itself, are expressed in vegetative apices and in leaves (Okamuro et al., 1997; Schwab et al., 2005; see Supplemental Figure 7 online), we reasoned that AP2 may repress flowering by mechanisms similar to the closely related SMZ (Mathieu et al., 2009). We therefore employed a strategy that would provide direct comparisons to the previous SMZ ChIP-chip experiments (Mathieu et al., 2009), while also assaying AP2 binding during active repression of flowering by AP2 with an independent antibody to corroborate our native inflorescence ChIP-seq.
We prepared a translational fusion of an miR172 targeting–resistant version of AP2 (rAP2) (Schwab et al., 2005) to green fluorescent protein (GFP) driven by the 35S promoter. The rAP2 transcript carries silent mutations within the miR172 target site. An important advantage of this strategy is that it allows the demonstration of AP2 binding to target loci during the strong floral repression of 35S:rAP2, which results in a very clear floral repression phenotype directly attributable to rAP2 function (Chen, 2004). For a genome-wide view, we performed ChIP followed by hybridization to Affymetrix genome tiling arrays (ChIP-chip). This AP2-GFP fusion protein retains activity, as evidenced by extremely late flowering and crinkly leaves, which recapitulates the 35S:AP2m3 phenotype (Chen, 2004). As a negative control for 35S:rAP2-GFP plants, we used a nuclear-localized GFP expressed from the same promoter. Eight tiling arrays were hybridized with material from two biological replicates of each genotype and two technical ChIP replicates of each biologically independent sample. This also serves as an independent corroboration of our ChIP-seq experiment employing an anti-AP2 antibody and native expression.
By ChIP-chip of the leaves with an independent GFP antibody, we detected high confidence binding to many of the same target loci that we found by native ChIP-seq of inflorescence tissue, especially those among the top targets. The same loci encoding ARF3/ETTIN, SOC1, SEP3, AP1, TOE1, TOE3, SMZ, SNZ, RGA1, AGL15, and AGL44 were all bound with an FDR < 0.05 (CisGenome default analysis as in Mathieu et al., 2009; see Supplemental Figure 8 and Supplemental Data Set 4 online). However, we could not detect any trace of binding to the AG intron in the leaf tissues (see Supplemental Figure 9 online), despite hybridizing multiple independent samples. This may suggest that the native ChIP-seq experiment can detect AG binding because of the presence of particular cofactors at the apex that were not available in the tissues assayed by ChIP-chip. For the other loci listed above (and others), however, strong binding was detected in both tissues (see Supplemental Figures 9C and 9D online). Furthermore, expression analysis in the inducible AP2m3-GR line during the time the commitment to flowering is made indicates that key floral genes SOC1 and AGL15 (Figure 6A) are directly regulated by AP2. qPCR expression analyses on the ap2-12 loss-of-function line (Figure 6B) show that the functionally redundant SOC1 and FUL are negatively regulated by AP2 when the decision to flower is being made. Taken together, these data indicate that AP2 can influence the expression of these targets during early vegetative growth, in inflorescences, and also when actively repressing flowering in an overexpression context.
Whereas nearly all of the major flowering-related genes were similarly enriched in leaf ChIP-chip as in inflorescence ChIP-seq experiments, surprisingly, we did not detect binding to miR172b or miR156e in the leaf tissue by ChIP-chip. This is not likely due to a difference in sensitivity between these methods, as both loci are well bound in inflorescence tissue, with miR156e a genome-wide top target (Figure 4H; see Supplemental Data Set 1 online). Because AP2-dependent expression of these transcripts was confirmed by expression analysis in inflorescence tissue (Figure 6C), we conclude instead that these different tissues may have contrasting chromatin accessibility profiles, possibly accounting for this striking difference. Findings such as these suggest the need for tissue-specific chromatin accessibility surveys to interpret better the data generated in genome-wide ChIP experiments. This would also better distinguish whether different levels of binding for a particular locus occur in different tissues as a result of the differential presence of particular transcriptional cofactors or contrasting epigenetic regulation.
DISCUSSION
Considered generally a transcriptional repressor, AP2 is a versatile protein, with effects not only on flowering time, but also on ovule and seed coat development, floral organ morphogenesis, and the maintenance of the stem cell niche. The degree of pleiotropy exhibited by ap2 mutants is unusual. This is perhaps more surprising given that AP2 is a member of a group of closely related targets of a major developmental microRNA, miR172. While this pleiotropy fits the general theme of floral repressor phenotypes (Pouteau et al., 2004; Del Olmo et al., 2009), why is it so clearly associated with AP2 and not the other AP2-like miR172 targets, which are partially redundant with AP2 and one another, at least in the governance of flowering time? How the phenotypic output and function of these targets is coordinated is currently not well understood, but elucidating their direct transcriptional target repertoires promises to clarify better this functional divergence.
We set out to address the following question: How does AP2 suppress flowering? Clearly, AP2 and the other members of the miR172 target clade share some direct network connections to achieve this end, but some findings are specific to each protein. SMZ expression, for example, is relatively concentrated in the young seedling as evidenced by β-glucuronidase staining (Mathieu et al., 2009), where it directly represses FT transcription. AP2, on the other hand, has been previously shown to act at the shoot apical meristem (Wurschum et al., 2006). Similarly, we found AP2 to repress directly the reproductive genes AG and SOC1 at the shoot apex, while it did not repress FT in leaves, as assayed in both tissues by genome-wide, direct chromatin binding approaches. This indicates that AP2 primarily acts as a flowering time integrator at the shoot apex. AP2, however, also directly bound the SOC1 and FUL loci in our ChIP-chip experiment, and consistent with this, SOC1 and FUL transcript levels were upregulated in the aerial tissues of ap2 mutants. We also observed this upregulation in ap2 mutant inflorescence arrays. The functional redundancy of SOC1 and FUL has been shown to affect dramatically both flowering time and meristem determinacy (Melzer et al., 2008). Another of the best bound AP2 direct targets was ARF3/ETTIN (Figure 4G). ARF3/ETTIN is involved in floral organ identity and the promotion of adult leaf traits (Sessions et al., 1997; Fahlgren et al., 2006; Hunter et al., 2006) and is therefore a potential mediator of the precocious phase change observed in our AP2 clade hexuple mutant. Overall, the complex pleiotropy of AP2, and the interplay of a gene traditionally considered a floral organ specificity factor with flowering time, fits with an emerging picture of shared function between floral patterning and flowering time genes more generally (Liu et al., 2009).
Perhaps the most well-known function for AP2 is as an A-class floral homeotic gene, promoting sepal and petal identity and opposing C class function (Bowman et al., 1991; Drews et al., 1991; Bomblies et al., 1999). Here, we observe that AP2 directly targets not only the C locus exemplum AG but also its ancient paralogs the PLE genes (Kramer et al., 2004; Causier et al., 2005), represented in Arabidopsis by SHP1 and SHP2. Interestingly, targeting is present, although these lineages have undergone subfunctionalization since divergence: while AG is involved in floral meristem determinacy, carpel and stamen identity, and ovule development (Bowman et al., 1989), SHP1 and SHP2 are more functionally constrained, contributing to ovule identity and influencing particular tissue types in fruit development (Liljegren et al., 2000; Pinyopich et al., 2003). Whether AP2 orthologs in other species might exhibit targeting of the AG and PLE lineages would be an interesting question to address. We hope that this data set will facilitate further analysis of this question and, more broadly, precisely how AP2 orchestrates both flowering time and gynoecium development.
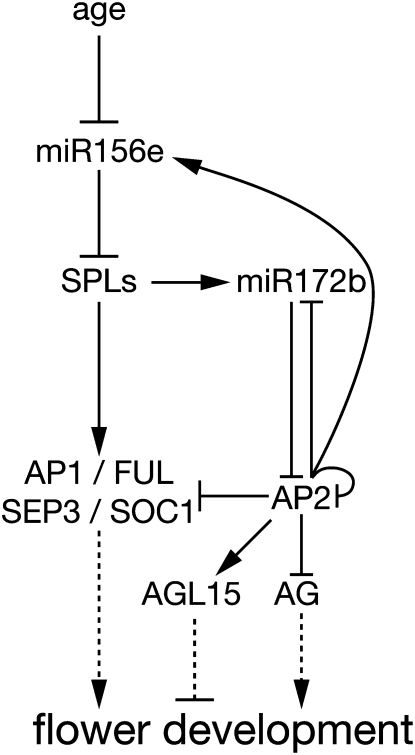
One of the most unexpected findings of this study was that AP2 can function not only as a direct repressor but also as a transcriptional activator of divergent gene classes that share a common function: one, a transcription factor that represses flowering, and the other, a microRNA that also represses flowering. Direct activation of the floral repressor AGL15 is evidenced by inducible gene expression assays in the absence of protein synthesis and direct chromatin binding by ChIP-seq. Functionally, this relationship makes sense given the floral repressor roles of all three genes. The second instance we demonstrate of direct bifunctional activation and repression by AP2 is its direct regulation of its own regulators, miR156 and miR172. Again, in this case, the direction of regulation is logical: AP2 directly promotes the expression of the floral repressor miR156, which, in turn represses the repressor of AP2, miR172, a floral activator (Figure 7). That this repression is even evident in microRNA abundance is all the more significant, as there are multiple family members of each of the two microRNAs (miR156a-h and miR172a-e), but only one family member of each microRNA is bound by AP2, miR172b, and miR156e. Thus, while evident, the expression difference is by necessity only moderately changed in the ap2 mutant due to the redundant activity of other microRNA family members.
Figure 7.
Salient AP2 Direct Network Connections to Flowering Time and Organ Development Genes.
AP2 participates in multiple feedback loops by directly binding flowering time loci and modulating their expression in logical, reinforcing circuits. AP2 directly represses the expression of flowering promoting transcripts AP1, FUL, SEP3, and SOC1 by binding to their genomic loci. At the same time, AP2 directly promotes the expression of miR156e, which represses the floral transition by repressing the SPLs. Additional, dual feedback loops via the repressor of AP2, miR172b, are evident, with AP2 directly repressing miR172b and also reinforcing this direct action on miR172b through increased miR156 expression. Finally, AP2 directly promotes the expression of the unrelated MADS domain floral repressor, AGL15, while directly repressing the expression of AG, a key MADS domain floral identity gene.
While the observation that AP2 influences flowering time redundantly with its fellow miR172 targets was no great surprise, we did not anticipate that lesions in AP2 alone would have such substantial effects, with the mutant flowering 23 leaves earlier than controls in short days. This incomplete redundancy among the AP2-like miR172 targets suggests a diversity of mechanisms by which family members fine-tune flowering time, making this miR172 target node an interesting model for discrete gene regulatory modules in complex organisms. The eventual recapitulation of the 35S:miR172 phenotype in the hexuple mutant demonstrates at the phenotypic level that these six genes, AP2, SMZ, SNZ, TOE1, TOE2, and TOE3, constitute the entire complement of effectors of this microRNA, which is consistent with the absence of additional predicted miR172 targets involved in the floral transition.
The recent advent of whole-genome approaches to the question of direct transcriptional interactions can facilitate new insights into how transcription factors coordinate their actions across the genome. We recently mapped the genome-wide binding profile of SMZ (Mathieu et al., 2009). Thus, we now have an opportunity to compare the direct target repertoires of these family members. Indeed, many high confidence flowering loci were targeted by both the AP2 and SMZ proteins: TOE3 was bound with the highest confidence values, and the loci SMZ, SNZ, AP2, AP1, SOC1, and SEP3 were also bound by both SMZ and AP2. Other loci encoding transcription factors not obviously involved in flowering, such as TOPLESS RELATED1 (TPR1), TPR2, and AGL44 (ANR1) were bound by both transcription factors, in addition to 92 other protein coding loci (see Supplemental Table 3 online). The significance of the regulation of these genes by AP2 and SMZ is currently unknown but indicates additional functions that may have been obscured by the extreme redundancy in this clade. On a genome-wide scale, AP2 did bind to many of the same loci as SMZ did: Of the 372 protein coding gene loci most closely associated with an SMZ-bound region, over 27% (102) also are the nearest annotated locus to a high confidence AP2-bound region, pointing to both shared function and divergence.
Because the AP2 ChIP-seq experiment reported here was more sensitive than the SMZ ChIP-chip experiment, we can compare results only unidirectionally; an absence from the SMZ data set and presence in the AP2 data set (as in the case of AG, for example) cannot necessarily be interpreted that AP2 targets a locus that SMZ definitely does not. We can, however, make inferences in the reverse direction. Loci that we observed in the less sensitive SMZ ChIP-chip experiment that we did not observe in the more sensitive AP2 ChIP-seq experiment are almost certainly meaningful differences. Indeed, a divergence in target repertoire is clearly evident: the flowering time loci FT and TEM1 were among the top 434 genomic regions bound by SMZ (FDR < 0.05) but not the top 2275 genomic regions bound by AP2 in inflorescences (FDR < 10−10). However, because the 35S:rAP2-GFP ChIP-chip experiment was performed on leaf tissue from the same promoter and assay as the SMZ experiment, comparisons are valid. In contrast with the native AP2 ChIP-seq experiments on inflorescence tissue, we failed to detect binding to AG in leaf tissue but did detect high confidence binding of AP2 to the TEM1 locus in leaf tissue, consistent with the expression and functional domains of these direct targets. Thus, while AP2 and SMZ are partially redundant, sharing some direct transcriptional targets, each has a distinct repertoire that may differ from tissue to tissue.
Understanding the level of complexity inherent in developmental gene regulatory networks has only just begun, as the ability to analyze genome-wide binding profiles of key transcription factors is in its infancy. A handful of studies have reported binding profiles for the transcription factors SEP3, AGL15, AP1, SMZ, and now, AP2. Results of these studies indicate widespread interaction between AP2-like and MADS domain transcription factors, a pattern that we will surely see extended to many transcription factor classes. More generally, they indicate an unexpected level of complexity in the interactions of these transcription factors with one another and their targets. Furthermore, it may be expected that such studies will likely illuminate new aspects of transcriptional regulation: for example, transcription factors generally considered to be solely activators or repressors may be discovered to have more subtle and particular direct effects on the transcription of a diversity of target genes. The mechanisms underlying these diverse functions may only be speculated upon here but may indicate the activity of cofactors that may help control the transcriptional activating or suppressing roles, as has been indicated for other transcription factors.
METHODS
Sequences of oligonucleotide primers used in this work are given in Supplemental Table 4 online.
Plasmid Construction
The cDNA of a miR172-resistant version of AP2, AP2m3 (Chen, 2004), was amplified by PCR using oligos AP2ProKPNF and AP2cDNANOTR using pAP2:AP2m3 (Zhao et al., 2007) as a template, digested by KpnI-NotI, and cloned into the pENTR1A vector (Invitrogen). A Gateway destination vector (pBI-GR-GW) containing the 35S promoter and the GR was subsequently used to generate the 35S:AP2m3-GR plasmid. To generate the 35S:rAP2-3xYFP transgenic construct, 3xYFP-NLS (Heisler et al., 2005) was cloned into a modified Gateway entry plasmid (pJL-Blue) in between the attL1 and attL2 recombination sites to create pHW100. The AP2 cDNA lacking the stop codon was released from plasmid pRS276 as an EcoRI (blunted)/SpeI fragment and cloned in front of the 3xYFP-NLS in the pJL-Blue vector to generate an entry plasmid containing the AP2-3xYFP-NLS open reading frame. Mutations rendering AP2 resistant to miR172 (Schwab et al., 2005) were introduced by site-directed mutagenesis using oligos G-1895 and G-1896 to create pHW154. For plant transformation, the entry plasmid containing the rAP2-3xYFP-NLS open reading frame was recombined into a pGREEN-IIS based binary plasmid carrying the cauliflower mosaic virus 35S promoter in front of the Gateway recombination cassette (Mathieu et al., 2007) to create pHW159.
Plant Transformation and Growth
The AP2m3-GR construct was transformed into Agrobacterium tumefaciens strain GV3101 and introduced into a population segregating for ap2 via the floral dip method. Positive transformants were selected for kanamycin resistance. AP2m3-GR transgenic lines with a single-locus T-DNA insertion were subsequently identified based on the 3:1 segregation ratio between kanamycin resistance to sensitivity. The 35S:rAP2-3xYFP construct was transformed into A. tumefaciens strain ASE, and T1 transformants were selected on kanamycin.
Plant Material
Wild-type plants were of the Col-0 accession in all experiments except in comparisons to ap2-2, where we used the Landsberg erecta accessions. All T-DNA insertion mutants used in this work are in Col-0 accession except ap2-2, which was in Landsberg erecta. ap2-2, toe1-2, toe2-1, soc1-2, ft-10, co-9, and 35S:miR172a have been described before (see Supplemental Table 5 online). Mutant plants were confirmed by PCR-based genotyping
Growth Conditions
All plants, except those from the GR experiments, were grown in growth chambers in a controlled environment (23°C, 65% relative humidity). Plants were raised on soil under a mixture of Cool White and Gro-Lux Wide Spectrum fluorescent lights, with a fluence rate of 125 to 175 μmol m−2 s−1. All light bulbs were of the same age. LD is defined as 16 h light and 8 h dark and SDs as 8 h light and 16 h dark. For flowering time measurements, plants were randomized with the respective controls, and the flowering time phenotype was determined without prior knowledge of the genotype.
Total RNA Extraction and Quantitative Real-Time PCR
Total RNA was extracted from plant tissue using the Plant RNeasy kit (Qiagen) according to the manufacturer’s instructions. Two micrograms of total RNA was DNase I-treated and single-stranded cDNA was synthesized using oligo(dT) and the RevertAid first strand cDNA synthesis kit (Fermentas). Quantitative real-time PCR was performed on an Opticon Continuous fluorescence detection system (MJR) using the Platinum SYBR Green qPCR Supermix-UDG (Invitrogen). Gene expression was calculated relative to β-tubulin using the ΔΔCT method. For qRT-PCR, all results are reported for triplicate reactions. For the GR experiments, cDNA was synthesized from total RNA isolated from chemically treated seedlings. qRT-PCR to detect SOC1 and AGL15 transcript levels was performed using the Bio-Rad Real-Time PCR SYBR Green Mix. Three biological replicates were performed. Oligonucleotide primers used for qRT-PCR are listed in Supplemental Table 4 online.
Microarray Expression Analysis
Analysis of inflorescence transcriptomes in Arabidopsis thaliana ap2-6 and Col-0 plants was performed on data from the AtGenExpress data set (Schmid et al., 2005). Lists of statistically significantly expressed genes were calculated for pairwise comparisons between the two genotypes using RankProducts (version 2.6.0) implemented in R (version 2.4.0; GUI 1.17) on gcRMA (version 2.6.0) normalized expression estimates (Breitling et al., 2004; Wu et al., 2004).
Cross-Linking, Chromatin Isolation, and ChIP-Seq
The entire experiment from harvest through deep sequencing was repeated twice to produce independent biological replicates. ChIP-seq was performed with an antibody raised against the C terminus of AP2 amino acids 289 to 432 (right after the second AP2 DNA binding domain until the stop codon). Col-0 plants were used to determine binding sites in the fully native context, and ap2-12 plants were used as negative controls for any nonspecific background antibody binding and pull down. To maximize AP2 levels, we harvested clusters of young influorescences at the beginning of bolting, but before flowers opened, when AP2 expression is high. Briefly, clusters of just-bolting inflorescences were harvested and fixed as described previously (Gomez-Mena et al., 2005). Frozen tissue was ground, filtered three times through Miracloth (Calibrochem), and washed as described previously thorough buffers M1, M2, and M3 (Gomez-Mena et al., 2005). Nuclear pellets were resuspended in sonic buffer as described (1 mM PEFA BLOC SC [Roche Diagnostics] was substituted for PMSF), split into technical duplicate samples, and sonicated with a Branson sonifier at continuous pulse (output level 3) for eight rounds of 2 × 6 s and allowed to cool on ice between rounds. Immunoprecipitation reactions were performed by incubating chromatin with 2.5 μL anti-rabbit AP2 antiserum overnight at 4°C as described (Gomez-Mena et al., 2005). The immunoprotein-chromatin complexes were captured by incubating with protein A-agarose beads (Santa Cruz Biotechnology), followed by consecutive washes in immunoprecipitation buffer and then elution as described (Gomez-Mena et al., 2005). Immunoprotein-DNA was then incubated consecutively in RNase A/T1 mix (Fermentas) and Proteinase K (Roche Diagnostics) as described, after which DNA was purified using Minelute columns (Qiagen) (Gomez-Mena et al., 2005). ChIP samples we tested for enrichment by qPCR and then deep sequencing libraries were produced by standard Illumina protocols.
ChIP-Seq Analysis
Standard Illumina base calling software was used to base call the 42 nucleotide sequence reads. We used SHORE (Ossowski et al., 2008) for read mapping and coverage analysis. The raw reads were pruned of low quality bases at their 3′ end exactly as described (Ossowski et al., 2008) before mapping them to the unmasked TAIR9 genome using GenomeMapper (Schneeberger et al., 2009), allowing for up to four mismatching nucleotides and no gaps.
A two-step procedure was applied to the mapped reads to identify regions significantly enriched in the positive sample when compared with the control sample. First, a sliding window approach was used to identify potentially enriched candidate regions in the ChIP sample. Second, these genomic regions were quantified and used as an input to a statistical test to assess the significance of the enrichment in ChIP versus the control sample.
Briefly, using uniquely mapping reads only, we first calculated the fragment coverage graph for the genome (i.e., each position in the genome was annotated with the number of overlapping DNA fragments to obtain the coverage depth for each base). To achieve this, each read was extended in the 3′ direction to 130 bp, corresponding to half the experimentally observed average fragment size of the immunoprecipitated DNA of ~200 to 300 bp. For detection of the described candidate peak regions, a 2-kb wide sliding window was applied to the fragment coverage graph. The sliding window was moved over the genome in single base steps, and in each step the potential enrichment of the central base was evaluated. Then, a one-sided Poisson test was applied, with the test parameter λ set to the local average coverage. This local average was calculated as the accumulated coverage values of all bases inside the 2-kb window, divided by the number of bases in the same range having a coverage value above zero. Positions with zero fragment coverage were considered inaccessible by the ChIP experiment and thus were not included in the calculation. Any position whose coverage had a P value below 0.05 assigned by the Poisson test was considered a part of a potentially enriched region. For further analysis, we considered only consecutive stretches of positions with a P value below 0.05 that were longer than the approximate DNA fragment size of 130 bp. To reduce further the number of potentially enriched regions, we checked for unwarranted high average coverage in the control sample in each of these candidate regions. A candidate was discarded if the coverage mean in the control sample in the region corresponding to a potential peak in the positive sample was larger than the median average control coverage plus a tolerance of three standard deviations in the peak regions.
To assess the significance of the remaining enrichment candidates, a one-sided binomial test was applied to the read count data for each region. For any potential peak region, the test parameter N was set to the total number of reads mapping to the considered region in both the ChIP and the control assay. Using this test, a P value was assigned to the number of reads in the ChIP sample mapping to the region. To calculate the probability of success “r” required as a parameter for the binomial test, we first computed a scaling factor “s” for the control sample and the chromosome containing the considered region. The complete chromosome sequence was subdivided into 400-bp bins, and for each bin, the numbers of mapped reads for the positive sample as well as the control sample were recorded. Then, s was chosen such that the median ChIP sample read count for all bins equaled the median control sample read count multiplied by s. Using this, the binomial test parameter r was calculated as r = s/(s + 1).
Finally, the resulting binomial test P values were transformed into FDRs using the Benjamini-Hochberg correction method. To provide further quality measures regarding the fidelity of peak regions, we also calculated the per base excess, which is defined as the excess of coverage averaged per base for the positive versus the control experiment ([chip reads – (s * control reads)]/ peak width). In the end, only peak regions with FDR < 10−10 and a per base excess > 0.25 in both replicates were retained and are reported in Supplemental Data Set 1 online. We selected a particularly stringent FDR of 10−10 to limit our list of enriched regions to the most significant. This was employed as protection that our method might potentially underestimate the FDR for less significant regions. It is conceivable that using a binomial distribution to calculate P values could have the effect of producing low P values if it does not perfectly model the true tag distribution between experiment and control in every case, for example, if the variance of the true distribution is larger. Alternatively, it may be that AP2 binds an unusually large number of regions in the genome at a lower affinity. In any case, we chose to err on the side of higher peak calling stringency. ChIP-seq traces were visualized using GBrowse (Stein et al., 2002). All scripts and source code are available upon request.
EasyGO (Zhou and Su, 2007) was used to perform GO analysis.
Inducible AP2-GR Experiments
Since ap2-2 plants do not grow well on selection media, ap2-2 plants were genotyped for the transgene prior to chemical treatments. 35S:AP2m3-GR ap2-2 seedlings were treated with 10 μM CYC with or without 10 μM DEX and 0.015% Silwet L-77 for 6 h. Four biological replicates were performed.
RNA Isolation and Blot Analysis
Dissected inflorescence tissue was obtained and total RNA isolation was performed using TRI reagent (Molecular Research Center). RNA gel blotting to detect microRNAs was done as previously described (Gy et al., 2007) with 6 μg of total RNA. Hybridization was performed at 50°C in buffer containing 5× SSC, 20 mM Na2HPO4, pH 7.2, 7% SDS, and 2× Denhardt’s solution. The 5′-end labeled (32P) antisense oligonucleotides (see Supplemental Table 4 online) were used to detect miR156, miR172, and U6 (internal control). The membrane was washed with a solution containing 1× SSC and 1% SDS. Radioactive signals were then obtained and quantified using the Typhoon PhosphorImager system.
ChIP-Chip
Leaf tissue from 5-week-old plants was processed for ChIP as above, except we used an anti-GFP antibody (Abcam ab290). Raw ChIP was recovered and amplified using the Sigma-Aldrich WGA GenomePlex kit after we performed a comparison to other systems, which showed this protocol gives improved amplification consistency and minimal amplification bias, in accordance with a previous study (O'Geen et al., 2006). One microgram of DNA was fragmented, labeled, and hybridized to Affymetrix Arabidopsis tiling 1.0F arrays (Affymetrix). Chromatin size distribution and fragmentation performance was confirmed on an Agilent Bioanalyzer prior to array hybridization (Agilent Technologies).
Primary Tiling Array Analysis
Tiling array data were processed using the CisGenome suite (Ji et al., 2008). Briefly, raw .CEL files were quantile normalized, and peaks were called using TileMapv2. Analysis was performed in MA mode with window size 5, and only peaks detected with a FDR of better than 0.05 were analyzed. EasyGO was used to do gene ontology-based enrichment analysis (Zhou and Su, 2007). Genome-wide visualization was performed with Affymetrix Integrated Genome Browser after normalization with Affymetrix Tiling Array Software (Affymetrix).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AP2 (At4g36920), SMZ (At3g54990), SNZ (At2g39250), TOE1 (At2g28550), TOE2 (At5g60120), TOE3 (AT5G67180), AGL15 (AT5G13790), ARF3 (AT2G33860), CO (AT5G15840), FT (AT1G65480), TFL1 (AT5G03840), SOC1 (AT2G45660), FUL (AT5G60910), SEP3 (AT1G24260), AG (AT4G18960), SHP1 (AT3G58780), SHP2 (AT2G42830), STK (AT4G09960), AP1 (AT1G69120), miR156e (AT5G11977), and miR172b (AT5G04275). AP2 ChIP-seq data are available from the Gene Expression Omnibus database (GSE21301). AP2 tiling array data are freely available from the ArrayExpress database (E-MEXP-2653). AtGenExpress data on Arabidopsis development (Schmid et al., 2005) are available from The Arabidopsis Information Resource database.
Author Contributions
L.Y., M.S., J.M., F.O., T.D., and X.C. conceived of and designed the experiments. L.Y., T.D., J.M., M.S., and H.W. performed the experiments. L.Y., F.O., M.S., and T.D. analyzed the data. L.Y. and M.S. wrote the article.
Supplemental Data
The following materials are available in the online version of this article:
Supplemental Figure 1. AP2 mRNA Levels Are Decreased in ap2-12.
Supplemental Figure 2. Hexuple AP2-Like miR172 Target Mutant Exhibits Ectopic Activation of Flower Organ Identity Programs.
Supplemental Figure 3. Genome-Wide Physical Distribution of AP2-Bound Regions.
Supplemental Figure 4. Overlay of AG Amplicons Tested by qPCR with ChIP-Seq Data from Native AP2 Antibody ChIP-Seq.
Supplemental Figure 5. Gene Ontology of Target Sets Bound, Repressed, or Both Bound and Repressed by AP2.
Supplemental Figure 6. Induction of AP2-GR Promotes Late Flowering.
Supplemental Figure 7. AP2 mRNA Expression throughout Arabidopsis thaliana Development.
Supplemental Figure 8. ChIP-Chip Reveals AP2 Targets in Leaf Tissue during Functional Floral Repression.
Supplemental Figure 9. Comparison of AP2 Direct Targets in Different Tissues.
Supplemental Table 1. Flowering Time of ap2, miR172 Target Clade, and Photoperiod Pathway Mutants.
Supplemental Table 2. ChIP Sequencing and Mapping Summary.
Supplemental Table 3. Genomic Loci Containing Both AP2- and SMZ-Bound Regions.
Supplemental Table 4. Oligonucleotides Used in This Work.
Supplemental Table 5. Mutant Lines Used throughout This Work.
Supplemental Data Set 1. Excel Spreadsheet Containing Information on Chromosomal Regions That Were Significantly Enriched in the AP2 ChIP-Seq Experiment.
Supplemental Data Set 2. Excel Spreadsheet Containing Results from RankProducts Pairwise Comparisons (pfp < 0.01 or pfp < 0.1; 100 Permutations) between Genotypes.
Supplemental Data Set 3. AP2_Excel Spreadsheet Containing Information of the Overlap of Direct AP2 Targets Generated from Supplemental Data Sets 1 and 2.
Supplemental Data Set 4. Excel Spreadsheet Containing Information on Chromosomal Regions That Were Significantly Enriched in the AP2-GFP ChIP-Chip Experiment.
Acknowledgments
We thank the Nottingham Arabidopsis Seed Centre for seeds. We also thank Schallum Werner for 35S:miR172 T1 seeds, Rebecca Schwab for the gift of a plasmid containing the AP2 open reading frame without stop codon, Patricia Springer for the pBI-GR-GW gateway destination vector, Jürgen Berger for assistance with scanning electron microscopy, members of the Schmid lab for discussion, and Detlef Weigel and Kirsten Bomblies-Yant for comments on the manuscript. F.O. was supported through BMBF-GABI Trilateral Grant TRANSNET to Detlef Weigel, H.W. was supported by a doctoral fellowship from Boehringer Ingelheim Fonds, and T.T.D. was supported by a National Science Foundation IGERT training grant (DGE0504249). This work was supported by a grant from the National Institutes of Health (GM61146) to X.C. and Deutsche Forschungsgemeinshaft grants (SCHM1560/3-1 and SCHM1560/5-1) to M.S. and the Max Planck Society.
References
- Abe M., Kobayashi Y., Yamamoto S., Daimon Y., Yamaguchi A., Ikeda Y., Ichinoki H., Notaguchi M., Goto K., Araki T. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309: 1052–1056 [DOI] [PubMed] [Google Scholar]
- Adamczyk B.J., Lehti-Shiu M.D., Fernandez D.E. (2007). The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 50: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Ahn J.H., Miller D., Winter V.J., Banfield M.J., Lee J.H., Yoo S.Y., Henz S.R., Brady R.L., Weigel D. (2006). A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 25: 605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aukerman M.J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K., Dagenais N., Weigel D. (1999). Redundant enhancers mediate transcriptional repression of AGAMOUS by APETALA2. Dev. Biol. 216: 260–264 [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112: 1–20 [DOI] [PubMed] [Google Scholar]
- Breitling R., Armengaud P., Amtmann A., Herzyk P. (2004). Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Carrera J., Rodrigo G., Jaramillo A., Elena S.F. (2009). Reverse-engineering the Arabidopsis thaliana transcriptional network under changing environmental conditions. Genome Biol. 10: R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C., Pelaz S. (2008). The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18: 1338–1343 [DOI] [PubMed] [Google Scholar]
- Causier B., Castillo R., Zhou J., Ingram R., Xue Y., Schwarz-Sommer Z., Davies B. (2005). Evolution in action: Following function in duplicated floral homeotic genes. Curr. Biol. 15: 1508–1512 [DOI] [PubMed] [Google Scholar]
- Chae K., Kieslich C.A., Morikis D., Kim S.C., Lord E.M. (2009). A gain-of-function mutation of Arabidopsis lipid transfer protein 5 disturbs pollen tube tip growth and fertilization. Plant Cell 21: 3902–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303: 2022–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier L., Vincent C., Jang S., Fornara F., Fan Q., Searle I., Giakountis A., Farrona S., Gissot L., Turnbull C., Coupland G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Del Olmo I., Lopez-Gonzalez L., Martin-Trillo M.M., Martinez-Zapater J.M., Pineiro M., Jarillo J.A. (2009). EARLY IN SHORT DAYS 7 (ESD7) encodes the catalytic subunit of DNA polymerase epsilon and is required for flowering repression through a mechanism involving epigenetic gene silencing. Plant J. 61: 623–636 [DOI] [PubMed] [Google Scholar]
- Deyholos M.K., Sieburth L.E. (2000). Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell 12: 1799–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews G.N., Bowman J.L., Meyerowitz E.M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65: 991–1002 [DOI] [PubMed] [Google Scholar]
- Fahlgren N., Montgomery T.A., Howell M.D., Allen E., Dvorak S.K., Alexander A.L., Carrington J.C. (2006). Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr. Biol. 16: 939–944 [DOI] [PubMed] [Google Scholar]
- Giakountis A., Coupland G. (2008). Phloem transport of flowering signals. Curr. Opin. Plant Biol. 11: 687–694 [DOI] [PubMed] [Google Scholar]
- Gomez-Mena C., de Folter S., Costa M.M., Angenent G.C., Sablowski R. (2005). Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132: 429–438 [DOI] [PubMed] [Google Scholar]
- Gy I., Gasciolli V., Lauressergues D., Morel J.B., Gombert J., Proux F., Proux C., Vaucheret H., Mallory A.C. (2007). Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19: 3451–3461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa Y., Money T., Bradley D. (2005). A single amino acid converts a repressor to an activator of flowering. Proc. Natl. Acad. Sci. USA 102: 7748–7753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hunter C., Willmann M.R., Wu G., Yoshikawa M., de la Luz Gutierrez-Nava M., Poethig S.R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133: 2973–2981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger K.E., Graf A., Wigge P.A. (2006). The control of flowering in time and space. J. Exp. Bot. 57: 3415–3418 [DOI] [PubMed] [Google Scholar]
- Jaeger K.E., Wigge P.A. (2007). FT protein acts as a long-range signal in Arabidopsis. Curr. Biol. 17: 1050–1054 [DOI] [PubMed] [Google Scholar]
- Ji H., Jiang H., Ma W., Johnson D.S., Myers R.M., Wong W.H. (2008). An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 26: 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J.H., Seo Y.H., Seo P.J., Reyes J.L., Yun J., Chua N.H., Park C.M. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19: 2736–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Muino J.M., Jauregui R., Airoldi C.A., Smaczniak C., Krajewski P., Angenent G.C. (2009). Target genes of the MADS transcription factor SEPALLATA3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7: e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Wellmer F., Muiño J.M., Ferrier T., Wuest S.E., Kumar V., Serrano-Mislata A., Madueño F., Krajewski P., Meyerowitz E.M., Angenent G.C., Riechmann J.L. (2010). Orchestration of floral initiation by APETALA1. Science 328: 85–89 [DOI] [PubMed] [Google Scholar]
- Kim S., Soltis P.S., Wall K., Soltis D.E. (2006). Phylogeny and domain evolution in the APETALA2-like gene family. Mol. Biol. Evol. 23: 107–120 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Weigel D. (2007). Move on up, it's time for change–Mobile signals controlling photoperiod-dependent flowering. Genes Dev. 21: 2371–2384 [DOI] [PubMed] [Google Scholar]
- Komiya R., Yokoi S., Shimamoto K. (2009). A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 136: 3443–3450 [DOI] [PubMed] [Google Scholar]
- Kramer E.M., Jaramillo M.A., Di Stilio V.S. (2004). Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Yoo S.J., Park S.H., Hwang I., Lee J.S., Ahn J.H. (2007). Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 21: 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C., Shen L., Wu Y., Chen H., Robertson M., Helliwell C.A., Ito T., Meyerowitz E.M., Yu H. (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15: 110–120 [DOI] [PubMed] [Google Scholar]
- Lifschitz E., Eviatar T., Rozman A., Shalit A., Goldshmidt A., Amsellem Z., Alvarez J.P., Eshed Y. (2006). The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc. Natl. Acad. Sci. USA 103: 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren S.J., Ditta G.S., Eshed Y., Savidge B., Bowman J.L., Yanofsky M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Liu C., Xi W., Shen L., Tan C., Yu H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16: 711–722 [DOI] [PubMed] [Google Scholar]
- Liu F., Marquardt S., Lister C., Swiezewski S., Dean C. (2010). Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327: 94–97 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Warthmann N., Kuttner F., Schmid M. (2007). Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 17: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Mathieu J., Yant L.J., Murdter F., Kuttner F., Schmid M. (2009). Repression of flowering by the miR172 target SMZ. PLoS Biol. 7: e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer S., Lens F., Gennen J., Vanneste S., Rohde A., Beeckman T. (2008). Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat. Genet. 40: 1489–1492 [DOI] [PubMed] [Google Scholar]
- O'Geen H., Nicolet C.M., Blahnik K., Green R., Farnham P.J. (2006). Comparison of sample preparation methods for ChIP-chip assays. Biotechniques 41: 577–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M., Onai K., Furukawa Y., Aoki E., Araki T., Nakamura K. (2001). Effects of sugar on vegetative development and floral transition in Arabidopsis. Plant Physiol. 127: 252–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M.A., Fischer R.L., Goldberg R.B., Nakamura K., Harada J.J. (2005). Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. USA 102: 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro J.K., Caster B., Villarroel R., Van Montagu M., Jofuku K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94: 7076–7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Clark R.M., Lanz C., Warthmann N., Weigel D. (2008). Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 18: 2024–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G.S., Savidge B., Liljegren S.J., Baumann E., Wisman E., Yanofsky M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Pouteau S., Ferret V., Gaudin V., Lefebvre D., Sabar M., Zhao G., Prunus F. (2004). Extensive phenotypic variation in early flowering mutants of Arabidopsis. Plant Physiol. 135: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schmid M., Uhlenhaut N.H., Godard F., Demar M., Bressan R., Weigel D., Lohmann J.U. (2003). Dissection of floral induction pathways using global expression analysis. Development 130: 6001–6012 [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Hagmann J., Ossowski S., Warthmann N., Gesing S., Kohlbacher O., Weigel D. (2009). Simultaneous alignment of short reads against multiple genomes. Genome Biol. 10: R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Palatnik J.F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8: 517–527 [DOI] [PubMed] [Google Scholar]
- Sessions A., Nemhauser J.L., McColl A., Roe J.L., Feldmann K.A., Zambryski P.C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124: 4481–4491 [DOI] [PubMed] [Google Scholar]
- Simpson G.G., Quesada V., Henderson I.R., Dijkwel P.P., Macknight R., Dean C. (2004). RNA processing and Arabidopsis flowering time control. Biochem. Soc. Trans. 32: 565–566 [DOI] [PubMed] [Google Scholar]
- Sohn E.J., Rojas-Pierce M., Pan S., Carter C., Serrano-Mislata A., Madueno F., Rojo E., Surpin M., Raikhel N.V. (2007). The shoot meristem identity gene TFL1 is involved in flower development and trafficking to the protein storage vacuole. Proc. Natl. Acad. Sci. USA 104: 18801–18806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein L.D., Mungall C., Shu S., Caudy M., Mangone M., Day A., Nickerson E., Stajich J.E., Harris T.W., Arva A., Lewis S. (2002). The generic genome browser: A building block for a model organism system database. Genome Res. 12: 1599–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S., Liu F., Magusin A., Dean C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Tamaki S., Matsuo S., Wong H.L., Yokoi S., Shimamoto K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316: 1033–1036 [DOI] [PubMed] [Google Scholar]
- Wang J.W., Czech B., Weigel D. (2009). miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Kim M.C., Jaeger K.E., Busch W., Schmid M., Lohmann J.U., Weigel D. (2005). Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309: 1056–1059 [DOI] [PubMed] [Google Scholar]
- Willis C.G., Ruhfel B., Primack R.B., Miller-Rushing A.J., Davis C.C. (2008). Phylogenetic patterns of species loss in Thoreau's woods are driven by climate change. Proc. Natl. Acad. Sci. USA 105: 17029–17033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Park M.Y., Conway S.R., Wang J.W., Weigel D., Poethig R.S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Irizarry R.A., Gentleman R., Martinez-Murillo F., Spencer F. (2004). A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99: 909–917 [Google Scholar]
- Wurschum T., Gross-Hardt R., Laux T. (2006). APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell 18: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Schmid M. (2009). Just say no: Floral repressors help Arabidopsis bide the time. Curr. Opin. Plant Biol. 12: 580–586 [DOI] [PubMed] [Google Scholar]
- Zhao L., Kim Y., Dinh T.T., Chen X. (2007). miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J. 51: 840–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Ren N., Wang H., Stromberg A.J., Perry S.E. (2009). Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21: 2563–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Su Z. (2007). EasyGO: Gene Ontology-based annotation and functional enrichment analysis tool for agronomical species. BMC Genomics 8: 246. [DOI] [PMC free article] [PubMed] [Google Scholar]