This study analyzed a transducin β-like gene (TIG1) from the fungal rice pathogen Magnaporthe oryzae. TIG1 was found to interact with several conserved core proteins to form a histone deacetylase complex, which is critical for invasive growth and conidiogenesis in the rice blast fungus.
Abstract
Magnaporthe oryzae is the most damaging fungal pathogen of rice (Oryza sativa). In this study, we characterized the TIG1 transducin β-like gene required for infectious growth and its interacting genes that are required for plant infection in this model phytopathogenic fungus. Tig1 homologs in yeast and mammalian cells are part of a conserved histone deacetylase (HDAC) transcriptional corepressor complex. The tig1 deletion mutant was nonpathogenic and defective in conidiogenesis. It had an increased sensitivity to oxidative stress and failed to develop invasive hyphae in plant cells. Using affinity purification and coimmunoprecipitation assays, we identified several Tig1-associated proteins, including two HDACs that are homologous to components of the yeast Set3 complex. Functional analyses revealed that TIG1, SET3, SNT1, and HOS2 were core components of the Tig1 complex in M. oryzae. The set3, snt1, and hos2 deletion mutants displayed similar defects as those observed in the tig1 mutant, but deletion of HST1 or HOS4 had no detectable phenotypes. Deletion of any of these core components of the Tig1 complex resulted in a significant reduction in HDAC activities. Our results showed that TIG1, like its putative yeast and mammalian orthologs, is one component of a conserved HDAC complex that is required for infectious growth and conidiogenesis in M. oryzae and highlighted that chromatin modification is an essential regulatory mechanism during plant infection.
INTRODUCTION
The ascomycetous fungus Magnaporthe oryzae is the causal agent of rice blast, which is one of the most destructive fungal diseases of rice (Oryza sativa) throughout the world (Dean et al., 2005; Wilson and Talbot, 2009). It produces three-celled pyriform conidia for dispersal. The infection process is initiated with the attachment and germination of conidia on the plant surface. An appressorium, a highly specialized infection structure, forms at the tip of the germ tube and penetrates the plant cuticle and cell wall (Tucker and Talbot, 2001). After penetration, the fungus forms unbranched primary invasive hyphae, which in turn differentiate into bulbous invasive hyphae. M. oryzae is a hemibiotrophic fungus that does not kill infected plant cells during the early stages of infection. Invasive hyphae are enclosed within the host cell membrane (Kankanala et al., 2007). Although it is not clear when necrotrophic growth begins, plant cells eventually die due to infectious growth of M. oryzae. Abundant conidia are produced on lesions that develop on rice plants during the late stages of infection to reinitiate the infection cycle. Under suitable conditions, infected seedlings can be killed by infection with M. oryzae, and panicle blast can cause severe yield losses.
The M. oryzae–rice pathosystem is a model system for studying fungal–plant interactions. In the past decade, there have been extensive studies on the molecular mechanisms that regulate appressorium morphogenesis and penetration in M. oryzae (Zhao et al., 2007; Wilson and Talbot, 2009). In addition to ALB1, BUF1, and RSY1, which are required for melanin synthesis, many genes that are important for appressorium formation and penetration have been characterized (Ebbole, 2007; Xu et al., 2007), including PTH12, MMT1, TPS1, CYP1, PLS1, MIG1, and MHP1 (Clergeot et al., 2001; Tucker et al., 2004; Wilson et al., 2007; Mehrabi et al., 2008; Kim et al., 2009). Among these genes found to be important for early plant infection processes are several components of cAMP signaling and two mitogen-activated protein (MAP) kinase pathways. In M. oryzae, the cAMP-PKA pathway regulates the recognition of hydrophobic surfaces and initiation of appressorium formation (Mitchell and Dean, 1995; Fang and Dean, 2000). The Pmk1 MAP kinase pathway is required for appressorium formation and maturation (Zhao et al., 2007). It is also essential for root infection (Sesma and Osbourn, 2004) and the proper regulation of the mobilization of storage carbohydrate and lipid reserves from conidia to appressoria (Thines et al., 2000). The other MAP kinase pathway essential for pathogenesis in M. oryzae is the MPS1 cascade, which is dispensable for appressorium formation but required for appressorial penetration (Xu et al., 1998; Jeon et al., 2008; Mehrabi et al., 2008). These two pathways also have been shown to be important for plant infection in other phytopathogenic fungi, including some species that do not form appressoria (Rispail et al., 2009).
Compared with our knowledge of appressorium formation and penetration, our knowledge of the molecular mechanisms involved in the differentiation and growth of invasive hyphae in infected plant cells is limited. Although several genes are known to be important for infectious growth in planta (for reviews, see Ebbole, 2007; Xu et al., 2007; Wilson and Talbot, 2009), most of them, such as the PMK1 MAP kinase and PDE1 P-type ATPase genes, also are involved in other developmental and infection processes. The corresponding mutants normally have pleiotropic defects. Only a few mutants, including the abc1, des1, and pth8 mutants, have no obvious defects in growth and appressorium-mediated penetration but are defective in plant infection. These genes must have cellular functions that are specific for invasive hyphae, such as the ABC1 transporter gene for avoiding toxic plant defense compounds (Urban et al., 1999) and DES1 for suppressing the plant defense response (Chi et al., 2009). Recently, microarray analysis has been used to identify genes specifically or highly expressed in invasive hyphae (Mosquera et al., 2009). Many of these genes expressed in planta have never been detected in vitro and encode biotrophy-associated secreted (BAS) proteins. Some, but not all, BAS proteins localize to biotrophic interfacial complexes (Mosquera et al., 2009). Because none of the BAS genes that have been functionally characterized are essential for pathogenicity (Mosquera et al., 2009), their functions in plant colonization and infectious growth are not clear.
In the wheat scab fungus Fusarium graminearum, the TBL1-like gene FTL1 was identified as a novel fungal pathogenicity factor by random insertional mutagenesis (Ding et al., 2009). FTL1 encodes a protein that is putatively orthologous to yeast SIF2 and mammalian TBL1. The ftl1 mutant was nonpathogenic. However, the molecular mechanism underlying its defects in plant infection is not clear. Because its infection processes, particularly fungal–plant interactions after plant penetration, are not well understood, F. graminearum is not suited for detailed characterization of this novel pathogenicity factor. In this study, we identified and characterized the TIG1 gene, an FTL1 ortholog, in the model plant pathogenic fungus M. oryzae. The tig1 mutant formed appressoria but was nonpathogenic. It was defective in the differentiation and growth of invasive hyphae in planta. The mutant had increased sensitivities to oxidative stress and other plant defensive compounds. Using affinity purification and mass spectrometry analyses, we identified several Tig1-associated proteins that are homologous to components of the yeast Set3 complex, including two histone deacetylases (HDACs). Coimmunoprecipitation assays were used to confirm the interactions among Tig1, Snt1, Set3, and Hos2. Mutants lacking any one of these genes had similar defects in plant infection and conidiation. HDAC activities and histone acetylation levels were also affected in these mutants disrupted in the Tig1 complex. Our data indicate that Tig1 is a component of a well-conserved HDAC complex and that chromatin modification is an essential regulatory mechanism during plant infection.
RESULTS
Identification of the TIG1 Transducin β-Like Gene in M. oryzae
In the M. oryzae genome, MGG_03198.5 shares >50% amino acid identity with F. graminearum FTL1 and yeast SIF2. It has an N-terminal LisH domain (residues 6 to 38) and six WD40 repeats (residues 536 to 626) toward the C terminus. The cDNA of TIG1 (for TBL1-like gene required for invasive growth) was amplified and sequenced. Four predicted introns in the TIG1 coding region were confirmed by sequence analysis. Although no known protein domain was identified, the amino acid sequence between the LisH domain and WD40 repeats was conserved between TIG1 and its orthologs from Neurospora crassa and F. graminearum (see Supplemental Figure 1 online).
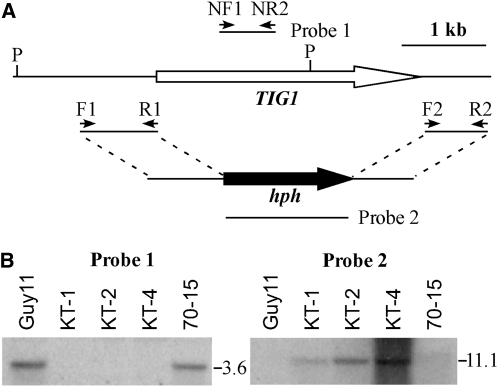
To determine the function of TIG1 in M. oryzae, a gene replacement construct (Figure 1A) was obtained by ligation-PCR and transformed into Guy11 (Chao and Ellingboe, 1991). The resulting hygromycin-resistant transformants were screened by PCR and analyzed by DNA gel blot hybridization. When hybridized with a TIG1 fragment, a 3.6-kb PstI band was observed in the wild type but not in the tig1 deletion mutant strains (Figure 1B). When hybridized with the hygromycin phosphotransferase gene (hph), the tig1 mutant but not the wild type had an 11.1-kb band that was characteristic of the gene replacement event (Figure 1B). The tig1 mutant KT-1 (Table 1) grew slightly slower than the wild type but was significantly reduced in conidiation. In comparison with Guy11, KT-1 produced ~100-fold fewer conidia (Table 2).
Figure 1.
The TIG1 Gene Replacement Construct and Mutants.
(A) Schematic diagram of the genomic region of the TIG1 and hph genes. Primers F1, R1, F2, R2, NF1, and NR1 were used to generate the TIG1 gene replacement constructs and mutant screens. P, PstI.
(B) DNA gel blots of PstI-digested genomic DNA were hybridized with a TIG1 fragment (left panel) and the hph gene (right panel) as the probes. Guy11 and 70-15 are the wild-type strains. KT-1, KT-2, and KT-4 are three independent tig1 null mutants.
Table 1.
Wild-Type and Mutant Strains of M. oryzae Used in This Study
| Strain | Genotype Description | Reference |
| Guy11 | Wild-type (MAT1-2, avr-Pita) | Chao and Ellingboe (1991) |
| 70-15 | Wild-type (MAT1-1, AVR-Pita) | Chao and Ellingboe (1991) |
| Ku80 | MgKu80 deletion mutant of Guy11 | Villalba et al. (2008) |
| KT-1 | tig1 deletion mutant of Guy11 | This study |
| KT-2 | tig1 deletion mutant of 70-15 | This study |
| KT-4 | tig1 deletion mutant of 70-15 | This study |
| ECT23 | Ectopic transformant of Guy11 | This study |
| CT-1 | KT-1 transformant complemented with TIG1 | This study |
| KT1NG1 | KT-1 transformant expressing the TIG1-GFP construct | This study |
| TigRG1 | 70-15 transformant expressing PRP27-TIG1-GFP | This study |
| KT1RG1 | KT-1 transformant expressing PRP27-TIG1-GFP | This study |
| TFg1 | 70-15 transformant expressing TIG1-3xFLAG fusion | This study |
| SetG1 | 70-15 transformant expressing SET3-GFP fusion | This study |
| HosG1 | 70-15 transformant expressing HOS2-GFP fusion | This study |
| TS-29 | TIG1-3xFLAG and SET3-GFP transformant | This study |
| HT-26 | TIG1-3xFLAG and HOS2-GFP transformant | This study |
| HS-9 | SET3-3xFLAG and HOS2-GFP transformant | This study |
| KH2-13 | hos2 deletion mutant of 70-15 | This study |
| CH-1 | hos2/HOS2 complementation strain of KH2-13 | This study |
| KH4-81 | hos4 deletion mutant of 70-15 | This study |
| KH1-23 | hst1 deletion mutant of 70-15 | This study |
| ECT1 | An ectopic transformant of 70-15 | This study |
| KS1-1 | snt1 deletion mutant of Ku80 | This study |
| KS3-1 | set3 deletion mutant of Ku80 | This study |
| CS-1 | set3/SET3 complementation strain of KS3-1 | This study |
| ECT2 | An ectopic transformant of Ku80 | This study |
Table 2.
Vegetative Growth, Conidiation, and Appressorium Formation in the Wild Type and Transformants Generated in This Study
| Strain | Growth Rate on CM (mm/d) | Conidiation (×105 Spores/Plate) | Appressorium Formation (%)a |
| Guy11 (wild type) | 6.3 ± 0.1 | 280.0 ± 6.2 | 99.5 ± 0.1 |
| KT-1 (tig1) | 5.4 ± 0.2 | 3.2 ± 0.7 | 98.7 ± 0.2 |
| ECT23 (ectopic) | 6.1 ± 0.2 | 267.3 ± 5.6 | 97.8 ± 0.2 |
| CT-1 (tig1/TIG1) | 6.1 ± 0.1 | 289.7 ± 4.6 | 95.3 ± 0.1 |
| 70-15 (wild type) | 6.2 ± 0.1 | 77.5 ± 2.5 | 96.7 ± 0.1 |
| KT-4 (tig1) | 5.4 ± 0.1 | 0.9 ± 0.2 | 90.6 ± 2.6 |
| KH2-13 (hos2) | 4.1 ± 0.2 | 0.6 ± 0.1 | 92.1 ± 0.1 |
| KH4-81(hos4) | 5.5 ± 0.1 | 7.5 ± 2.5 | 94.0 ± 0.1 |
| KH1-23(hst1) | 6.2 ± 0.2 | 26.7 ± 2.9 | 93.6 ± 0.1 |
| ECT1 (ectopic) | 6.1 ± 0.1 | 49.2 ± 6.3 | 96.9 ± 0.1 |
| Ku80 | 6.3 ± 0.1 | 170.0 ± 10.0 | No data |
| KS3-1(set3) | 3.4 ± 0.1 | 1.7 ± 0.3 | No data |
| KS1-1(snt1) | 4.1 ± 0.1 | 0.4 ± 0.1 | No data |
| ECT2 (ectopic) | 6.3 ± 0.2 | 150.0 ± 12.6 | No data |
Percentage of germ tubes that formed appressoria.
The tig1 Deletion Mutant Is Defective in Plant Infection
When seedlings of rice cultivar CO-39 were spray inoculated with conidia from Guy11 and an ectopic transformant, abundant lesions formed (Figure 2A). On leaves inoculated with the tig1 mutant KT-1, no typical blast lesions were observed, but small dark brown spots were formed occasionally (Figure 2A). Similar results were obtained in infection assays with seedlings of barley (Hordeum vulgare) cultivar Golden Promise. The tig1 mutant failed to cause blast lesions and green islands on barley leaves. In injection infection assays, only limited necrosis was observed at the wound sites inoculated with KT-1 (Figure 2B). When the dark-brown spots and necrotic areas caused by the tig1 mutant were excised, surface sterilized, and incubated on water agar plates for 72 h, we failed to detect fungal growth or conidiation (Figure 2C). Under the same conditions, abundant fungal growth and conidiation were observed over the necrotic areas or lesions caused by Guy11 (Figure 2C). These results indicate that the tig1 mutant was nonpathogenic and failed to colonize plant tissues through wounds. The rare, small, brown leaf spots caused by the mutant were not true blast lesions and were likely associated with plant defense responses.
Figure 2.
Infection Assays with the tig1 Mutant.
(A) Rice leaves sprayed with conidia from the wild-type strain Guy11, an ectopic transformant ECT23, and tig1 mutant KT-1.
(B) Injection assays with Guy11 and KT-1. Inoculation with 0.25% gelatin was the negative control. The tig1 mutant only caused limited necrosis at the wound site.
(C) Assays for fungal growth on surface-sterilized rice leaves inoculated by spray (top panels) or injection (bottom panels). Abundant hyphae and conidia were observed on leaves inoculated with Guy11. No fungal growth was observed on rare brown spots or limited necrotic regions at the wound sites caused by the tig1 mutant.
[See online article for color version of this figure.]
To determine whether the TIG1 function varies among strains, we also generated the tig1 deletion mutant in strain 70-15 (Chao and Ellingboe, 1991). Mutants KT-2 and KT-4 were identified and confirmed by DNA gel blot analysis to lack TIG1 (Figure 1B). These 70-15 tig1 null mutants displayed the same defects in growth and conidiation as mutant KT-1 (Table 2). On seedlings of rice cultivar Nipponbare, KT-2 and KT-4 failed to cause typical blast lesions in spray or injection infection assays (see Supplemental Figure 2 online).
To confirm that the phenotypes observed in these mutants were directly related to deletion of TIG1, mutant KT-4 (MAT1-1) was crossed with Guy11 (MAT1-2). A total of 19 ascospore progeny were isolated. Eight of these progeny were resistant to hygromycin and had similar defects as the tig1 mutant. The remaining 11 hygromycin-sensitive progeny had the wild-type phenotype, indicating that defects observed in the tig1 mutant cosegregated with the hygromycin resistance marker. In addition, we reintroduced the wild-type TIG1 allele into mutant KT-1. The resulting transformant CT-1 (Table 1) exhibited normal virulence on rice seedlings and produced abundant conidia (Table 2), demonstrating that the inactivation of TIG1 was responsible for defects of the tig1 mutant.
TIG1 Is Required for Infectious Growth after Penetration of Rice Cells
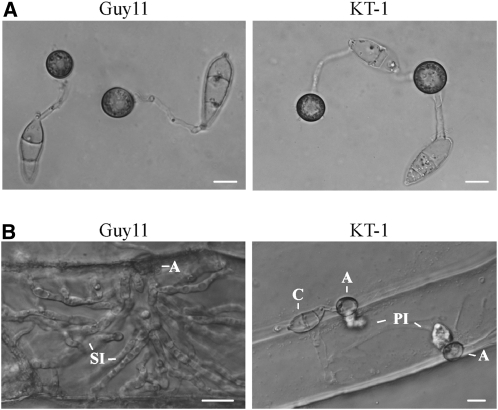
When assayed for appressorium formation on artificial hydrophobic surfaces, the tig1 mutant KT-1 produced abundant melanized appressoria (Figure 3A). No obvious defects in appressorium formation were observed (Table 2). On onion epidermal cells, the mutant exhibited normal levels of appressorium formation. However, it was defective in the penetration of onion epidermal cells and differentiation of invasive hyphae. By 48 h, Guy11 penetrated and formed invasive hyphae inside onion epidermal cells (Figure 3B). Under the same conditions, no invasive hyphae were observed in KT-1. Even up to 72 h, the unbranched primary invasive hyphae formed by KT-1 were blocked in the differentiation and growth of invasive hyphae.
Figure 3.
Appressoria Formation and Penetration of Onion Epidermal Cells.
(A) Melanized appressoria formed by the wild type (Guy11) and tig1 mutant (KT-1) strains on plastic cover slips.
(B) Penetration assays with onion epidermal cells. By 48 h, invasive hyphae were observed in plant cells penetrated by Guy11 but not KT-1. A, appressorium; C, conidium; PI, primary invasive hyphae; SI, secondary invasive hyphae.
Bars = 10 μm.
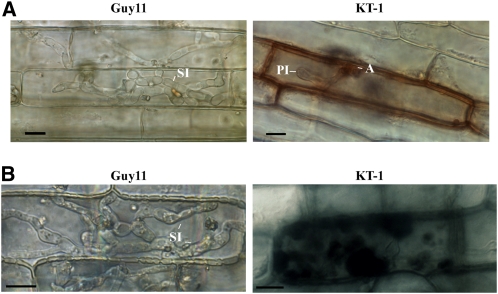
We also assayed appressorium penetration and infectious growth using rice leaf sheath epidermal cells. The tig1 mutant formed appressoria but failed to develop invasive hyphae in plant cells (Figure 4A). By 48 h, invasive hyphae formed by Guy11 began to invade nearby plant cells. Under the same conditions, unbranched primary invasive hyphae formed by the mutant had only limited growth in penetrated plant cells (Figure 4A). Infected plant cells often had discolored cell walls, and fungal hyphae appeared to be restricted and unhealthy in comparison with those formed by Guy11 (Figure 4A). When stained with 3,3′-diaminobenzidine (DAB), reactive oxygen species (ROS) accumulated around invasive hyphae in plant cells penetrated by the mutant (Figure 4B). By contrast, invasive hyphae formed by Guy11 did not trigger significant ROS accumulation in the host (Figure 4B). These results indicate that the tig1 deletion mutant is defective in its ability to overcome plant defense responses and maintain biotrophic growth after penetration. When stained with aniline blue, stronger fluorescence signals were observed in barley leaf epidermal cells penetrated by the tig1 mutant than the wild type (see Supplemental Figure 3 online), suggesting that primary infectious hyphae of the mutant face enhanced callose deposition from the host.
Figure 4.
Penetration Assays with Rice Leaf Sheath Epidermal Cells.
(A) Extensive invasive hyphae were developed by Guy11 inside plant cells by 72 h after inoculation. The tig1 mutant (KT-1) had only limited growth of primary invasive hyphae.
(B) When stained with DAB, rice cells penetrated by the tig1 mutant accumulated ROS around fungal hyphae. No obvious ROS accumulation was observed in plant cells penetrated by Guy11. A, appressorium; PI, primary invasive hyphae; SI, secondary invasive hyphae.
Bars = 10 μm.
The tig1 Mutant Has an Increased Sensitivity to H2O2 and Plant Defense-Related Proteins
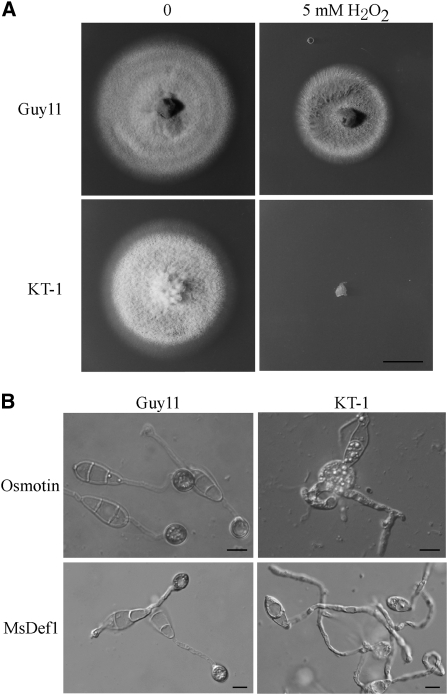
Because plant cells accumulated ROS in the region surrounding primary invasive hyphae of the tig1 mutant, we tested the sensitivity of the mutant to H2O2. When hydrogen peroxide was added to complete medium (CM) plates, fungal growth was decreased with increasing concentrations of H2O2. However, the reduction in the growth of the tig1 mutant was more severe than that in the wild type. Hyphal growth was completely inhibited by 5 mM H2O2 in the tig1 mutant KT-1 but not in Guy11 (Figure 5A). We also assayed the effects of two plant proteins, osmotin and MsDef1, that are toxic to fungal pathogens (Coca et al., 2000; Ramamoorthy et al., 2007). In the presence of 50 μg/mL osmotin, appressorium formation was normal in Guy11 but blocked in KT-1 on hydrophobic surfaces (Figure 5B). Osmotin treatment tended to stimulate the formation of multiple germ tubes and unmelanized apical or intercalary swollen bodies. The tig1 mutant also was more sensitive to MsDef1 than Guy11. In the presence of 20 μM MsDef1, melanized appressoria were formed by Guy11 but not by KT-1 (Figure 5B). Unlike osmotin, MsDef1 treatment did not cause KT-1 germ tubes to swell.
Figure 5.
Increased Sensitivities of the tig1 Mutant to Hydrogen Peroxide, Osmotin, and MsDef1.
(A) CM cultures of the wild-type (Guy11) and tig1 mutant (KT-1) strains in the presence or absence of 5 mM H2O2. Bar = 1 cm.
(B) In the presence of 50 μg/mL osmotin (top panels) or 20 μM MsDef1 (bottom panels), melanized appressoria were formed by Guy11 but not KT-1 by 24 h after inoculation. Osmotin treatment resulted in the formation of multiple germ tubes and apical or intercalary swollen bodies in the tig1 mutant. Bars = 10 μm.
Identification of TIG1-Interacting Proteins by Affinity Purification
TIG1 is putatively orthologous to yeast SIF2, which is a member of the Set3 complex involved in the late stages of ascospore formation (Pijnappel et al., 2001; Cerna and Wilson, 2005). To determine whether TIG1 functions in a similar complex in M. oryzae, we generated a TIG1-3xFLAG construct and transformed it into 70-15. In transformant TFg1 (Table 1), a 72-kD band of the expected size of Tig1-3xFLAG was detected with an anti-FLAG antibody in proteins isolated from vegetative hyphae grown in liquid CM medium (see Supplemental Figure 4 online).
For affinity purification, total proteins were isolated from transformant TFg1 and mixed with anti-FLAG M2 beads. Proteins bound to M2 beads were eluted and digested with trypsin and identified by mass spectrometry analysis (see Methods). Table 3 lists proteins that were copurified with the TIG1-3xFLAG fusion. MGG_09174.5, MGG_01663.5, MGG_02488.5, MGG_10447.5, and MGG_05727.5 (Table 3) are orthologous to yeast SNT1, HOS2, HST1, CPR1, and HOS4, respectively, which are members of the Set3 complex (Pijnappel et al., 2001; Mou et al., 2006). They were consistently identified as TIG1-interacting genes in four independent biological replicates. Other proteins that coimmunoprecipitated with Tig1 included MGG_09602.5 and MGG_01362.5 (Table 3). While MGG_09602.5 has no homologous sequence in Saccharomyces cerevisiae and appears to be unique to filamentous fungi, putative orthologs of other genes exist in yeast. MGG_01362.5 is homologous to yeast Cdc28, which interacts with Hos4 (Ubersax et al., 2003), but does not belong to the Set3 complex.
Table 3.
Putative TIG1-Interacting Genes Identified by Affinity Purification
| Gene ID | Yeast Homolog |
| MGG_01633.5 | HOS2 |
| MGG_09174.5 | SNT1 |
| MGG_02488.5 | HST1 |
| MGG_10447.5 | CPR1 |
| MGG_05727.5 | YIL112w (HOS4) |
| MGG_06453.5 | YCR028C |
| MGG_09602.5 | No homolog |
| MGG_00446.5 | YGL019W (CKB1) |
| MGG_01826.5 | YGL174W (BUD13) |
| MGG_01362.5 | YBR160W (CDC28) |
| MGG_03741.5 | YGR266W |
TIG1 Is a Member of a Conserved HDAC Protein Complex
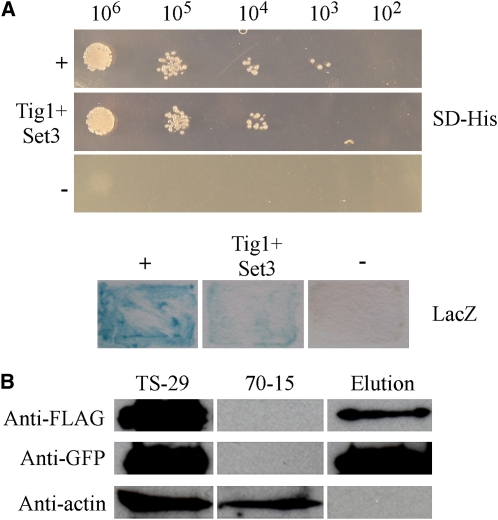
The putative orthologs of all members of the yeast Set3 complex except Set3 itself copurified with Tig1 (Table 3). We amplified the predicted open reading frame (ORF) of MGG_01558 (named SET3 in this study) and cloned it into the pAD-GAL4 prey vector. The TIG1 gene was cloned into the corresponding bait vector. Growth on SD-His plates and LacZ activity were detected in yeast cells expressing the SET3 prey and TIG1 bait constructs (Figure 6A), indicating that SET3 interacted with TIG1. However, their interaction was relatively weak in comparison with the positive control for yeast two-hybrid assays (Figure 6A).
Figure 6.
Yeast Two-Hybrid and Coimmunoprecipitation Assays for the TIG1-SET3 Interaction.
(A) Yeast transformants expressing the TIG1 bait and SET3 prey constructs were assayed for growth on SD-Leu-Trp-His plates (SD-His) and β-galactosidase (LacZ) activities. +, Positive control; −, negative control.
(B) Immunoblot analysis of total proteins (TS-29) and proteins eluted from the anti-FLAG M2 beads (Elution) of transformant TS-29 that expressed the TIG1-3xFLAG and SET3-GFP constructs. Total proteins isolated from the wild-type strain (70-15) were included as the control. Top, middle, and bottom images represent detection with anti-FLAG, anti-GFP, and anti-actin antibodies, respectively.
[See online article for color version of this figure.]
To confirm the TIG1-SET3 interaction, we generated the SET3-green fluorescent protein (GFP) construct and cotransformed it into 70-15 with the TIG1-3xFLAG fusion. The resulting hygromycin-resistant transformant TS-29 (Table 1) was confirmed by PCR to contain both constructs, and successful transformation was further confirmed by immunoblot analysis with anti-FLAG and anti-GFP antibodies. Total proteins were isolated from transformant TS-29 and bound to anti-FLAG M2 beads. Proteins bound to the bead were eluted, separated on 12% SDS-PAGE gels, and transferred onto nitrocellulose membrane. As expected, the Tig1-3xFLAG protein was detected with an anti-FLAG antibody in both the eluted and total proteins (Figure 6B). A protein band of the expected size of the Set3-GFP fusion was also detected with the anti-GFP antibody in both the elution and total proteins (Figure 6B), indicating that Set3 coimmunoprecipitated with Tig1. The anti-actin antibody detected the actin band in total proteins isolated from the transformant, but not in the eluted proteins (Figure 6B).
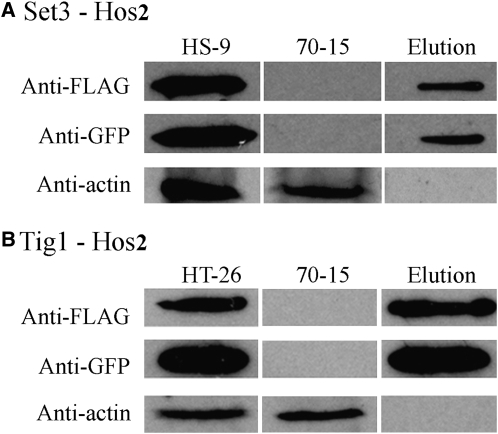
Similarly, coimmunoprecipitation (co-IP) assays were conducted to confirm the interactions between SET3 and HOS2 (Figure 7A) and TIG1 and HOS2 (Figure 7B) in M. oryzae. We generated the SET3-3xFLAG and HOS2-GFP constructs and transformed them into strain 70-15 in pairs or individually with the TIG1 fusion construct. Transformants HT-26 and HS-9 (Table 1), expressing the TIG1-3xFLAG/HOS2-GFP and SET3-3xFLAG/HOS2-GFP fusion constructs, respectively, were identified. The same GFP fusion band was detected in both the total proteins isolated from vegetative hyphae and proteins eluted from anti-FLAG M2 beads in both HT-26 (Figure 7A) and HS-9 (Figure 7B). These data indicate that Hos2 interacts with Set3 and Tig1 in vivo. The results of these co-IP assays suggest that Tig1 belongs to a M. oryzae protein complex that is similar to the yeast Set3 complex.
Figure 7.
Co-IP Assays for Interactions between Components of the Tig1 Complex.
Co-IP assays of transformants HS-9 (A) and HT-26 (B) that expressed the SET3-FLAG/HOS2-GFP and TIG1-3xFLAG/HOS2-GFP constructs, respectively. Immunoblots of total proteins isolated from each transformant and proteins eluted from the anti-FLAG M2 beads (Elution) were detected with the anti-FLAG, anti-GFP, or anti-actin antibody. Total proteins isolated from the wild-type strain 70-15 were included as the control.
SNT1, HOS2, and SET3 Are Required for Plant Infection
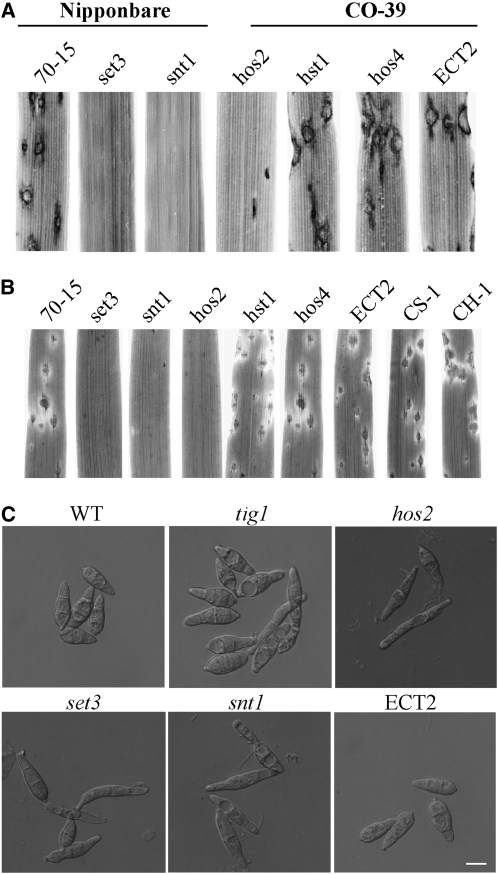
To determine the functions of Tig1-interacting proteins, we constructed deletion mutants of SNT1, HOS2, SET3, HST1, and HOS4 by gene replacement. The putative CPR1 ortholog, CYP1, was not included in this study because it is known to be involved in plant infection in M. oryzae (Viaud et al., 2002). The snt1, set3, hos4, hos2, and hst1 deletion mutants (Table 1) were identified by PCR and confirmed by DNA gel blot analysis (see Supplemental Figure 5 online). Like the tig1 mutant, snt1, set3, and hos2 mutants were nonpathogenic on seedlings of susceptible rice cultivars (Figure 8A) and barley (Figure 8B). These three mutants were also greatly impaired in conidiation (Table 2). By contrast, conidiation was not significantly impaired in the hst1 and hos4 mutants (Table 1), and these mutants were as virulent as the wild type (Figures 8A and 8B), indicating that HST1 and HOS4 are dispensable for plant infection and the function of the Tig1 complex.
Figure 8.
Pathogenicity and Conidium Morphology of Mutants Disrupted in the TIG1 Complex.
(A) Leaves of rice cultivar Nipponbare or CO-39 were inoculated with conidia from 70-15, the set3, snt1, hos2, hst1, and hos4 mutants, and an ectopic transformant of Ku80 (ECT2).
(B) Barley leaves sprayed with the same set of strains described above. The hst1 and hos4 mutant had normal virulence, but the set3, snt1, and hos2 mutants were nonpathogenic. CH-1 and CS-1 were complemented transformants of the hos2 and set3 null mutant, respectively.
(C) Conidia of 70-15, tig1 and set3 mutants, and ectopic transformant ECT2. Bar = 10 μm.
In addition to reduced conidiation, conidia formed by the tig1, snt1, set3, and hos2 mutants varied significantly in size and morphology from those formed by the wild type (Figure 8C). In general, conidia from these mutants were still three celled, but most of them were narrower than wild-type conidia. Some conidia produced by the tig1, snt1, set3, and hos2 mutants no longer had the normal pyriform shape (Figure 8C). A reduction in conidiation and abnormal spore morphology indicate that TIG1, SNT1, SET3, and HOS2 play an important role in conidiogenesis in M. oryzae. We performed a complementation analysis, in which wild-type HOS2 and SET3 genes were reintroduced into the hos2 and set3 mutants, respectively. The resulting complemented transformants (CH-1 and CS-1; Table 1) exhibited normal virulence (Figure 8B), conidiation, and conidium morphology.
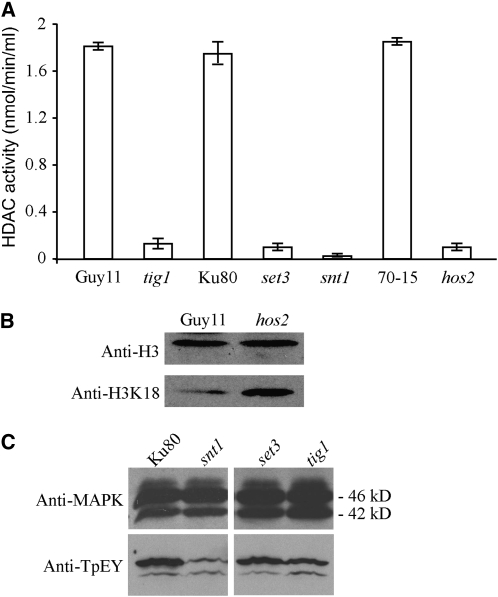
HDAC Activities and Histone Acetylation in the Tig1 Complex Mutants
Because the yeast Set3 complex is involved in histone deacetylation, it is likely that Tig1, Set3, Hos2, and Snt1 are components of a similar HDAC complex in M. oryzae. Therefore, we assayed HDAC activities in protein extracts isolated from protoplasts of the tig1, set3, hos2, and snt1 mutants. In comparison with the wild-type strains, the tig1, set3, snt1, and hos2 mutants were highly reduced in HDAC activities (Figure 9A). The three wild-type strains, Guy11, 70-15, and Ku80, used for generating these mutants had similar HDAC activities in three biological replicates (Figure 9A). These results indicate that Tig1, Set3, Hos2, and Snt1 are required for the activity of the HDAC complex in M. oryzae. Because TIG1, SET3, and SNT1 have no predicted functions in histone modification, their effects on histone acetylation must be related to their participation in an HDAC complex.
Figure 9.
Assays for HDAC Activity and Histone H3 Acetylation in M. oryzae Strains.
(A) HDAC activities were assayed with proteins isolated from the wild-type strains Guy11, 70-15, and Ku80 and from the tig1 (KT-1), set3 (KS3-1), snt1 (KS1-1), and hos2 (KH2-13) mutants. Median and sd were calculated from three biological replicates.
(B) Immunoblots of nuclear proteins isolated from Guy11 (wild type) and the hos2 mutant were detected with the anti-histone 3 antibody (anti-H3; top panel) or the anti-H3K18Ac antibody (anti-H3K18; bottom panel). The level of H3K18 acetylation was higher in the mutant than in Guy11.
(C) Immunoblots of total proteins from Ku80 and the set3, snt1, and tig1 mutants were detected with an anti-MAPK antibody (top panel) and an anti-TpEY antibody (bottom panel). Both the Mps1 (46-kD) and Pmk1 (42-kD) MAP kinases could be detected.
Since reduced HDAC activities may lead to increased histone acetylation, we assayed the acetylation level of histone H3 in the hos2 mutant. In nuclear proteins isolated from Guy11, the expression level of histone 3 was similar to that of the mutant. However, when detected with an anti-H3K18Ac antibody that is specific for K18 acetylation of histone H3 (Figure 9B), Guy11 had weaker signals than the hos2 mutant, indicating that the hos2 mutant had higher levels of H3K18 acetylation than Guy11.
Disruption of the Tig1 Complex Affects the Activation of Mps1 MAP Kinase
Since the Set3 complex functions upstream of the Slt2 MAP kinase during secretory stress responses (Cohen et al., 2008), we assayed the activation of Mps1 in the mutants disrupted in the Tig1 complex. In vegetative hyphae, the expression of Mps1 was not affected by SNT1, TIG1, or SET3 deletion (Figure 9C). However, the phosphorylation level of Mps1 was lower in the snt1, tig1, and set3 mutants than in Ku80 (Figure 9C), indicating a reduction in Mps1 activation in these mutants disrupted in the Tig1 complex. By contrast, phosphorylation of Pmk1 was not affected or was slightly increased in the snt1, tig1, and set3 and mutants (Figure 9C).
Expression and Localization of TIG1-, SET3-, and HOS2-GFP
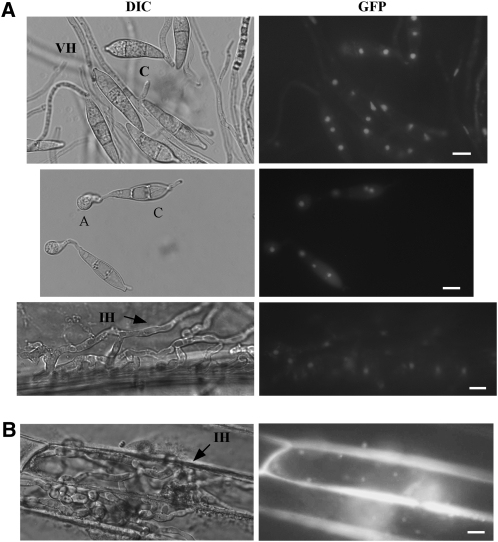
Although Tig1 has no predictable nuclear localization signal (NLS) sequences, it may localize to the nucleus as one component of a HDAC complex. To test this hypothesis, a TIG1-GFP fusion construct was transformed into the tig1 mutant KT-1. Transformant KTlNG1 (Table 1), which expresses the TIG1-GFP fusion, exhibited normal growth, conidiation, and virulence, indicating that the defects in the tig1 mutant were complemented. In KT1NG1, GFP signal was observed in the nucleus in conidia, appressoria, and invasive hyphae (Figure 10A). In vegetative hyphae, only a faint GFP signal was observed in the nuclei of some hyphal compartments (Figure 10A). When the TIG1-GFP fusion was overexpressed with the RP27 promoter in transformant KT1RP1 (Table 1), a stronger GFP signal was observed in the nuclei of vegetative hyphae (see Supplemental Figure 6 online), suggesting that lower expression of TIG1-GFP was responsible for weak GFP signals in vegetative hyphae in KT1NG1.
Figure 10.
Expression and Localization of the GFP Fusion Proteins.
(A) In transformant KT1NG1, which expresses the TIG1-GFP fusion, GFP signals were observed in the nucleus in conidia (top), appressoria (middle), and invasive hyphae (bottom). A, appressoria; C, conidia; VH, vegetative hyphae; IH, invasive hyphae.
(B) Nuclear localization was also observed in the invasive hyphae of transformant CS-1, which expresses the SET3-GFP fusion.
Bars = 10 μm.
We also generated GFP fusion constructs with SET3 and HOS2. In transformants of 70-15 expressing these constructs (Table 1), the GFP signal was also observed in the nucleus in conidia, appressoria, and invasive hyphae (Figure 10B). Similar to the TIG1-GFP transformant, the expression of SET3-GFP was lower in vegetative hyphae than invasive hyphae, but a faint GFP signal could be observed in some nuclei. However, in transformants expressing the HOS2-GFP fusion, GFP signals were weak or not detectable in vegetative hyphae.
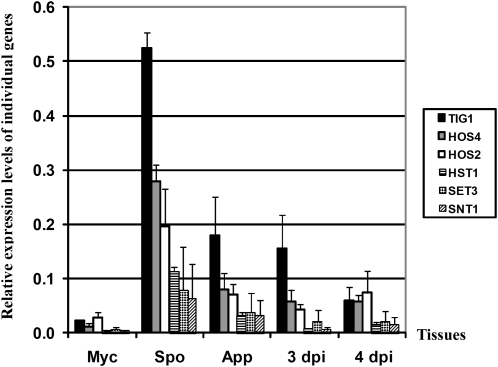
Expression Profiles of Selected Components of the Tig1 Complex
We used quantitative RT-PCR (qRT-PCR) to quantify the transcripts of TIG1, HOS2, HOS4, HST1, SET3, and SNT1 at different fungal developmental and infection stages. All of these genes were expressed at a relatively low level in vegetative hyphae and had their highest expression levels in conidia (Figure 11). In comparison with their expression levels in vegetative hyphae, transcription of TIG1, HOS2, HOS4, HST1, SET3, and SNT1 increased 23-, 6-, 25-, 46-, 16-, and, 27-fold, respectively, in conidia. These results were in agreement with the weaker GFP signal observed in vegetative hyphae than in conidia in transformants expressing the TIG1-GFP or HOS2-GFP fusion.
Figure 11.
Expression Patterns of Genes Associated with TIG1.
Expression levels of TIG1, HOS2, SNT1, HOS4, SET3, and HST1 in vegetative hyphae (Myc), conidia (Spo), 24-h-old appressoria (App), and infected rice leaves harvested at 3 or 4 DAI. qRT-PCR was used to quantify transcripts of these genes relative to that of the constitutive reference gene ILV5 using the 2−ΔΔCT method. Median and sd were calculated from three biological replicates.
The expression of TIG1, HOS2, HOS4, HST1, SET3, and SNT1 was higher in appressoria than in vegetative hyphae (5- to 13-fold; Figure 11), but they all had the highest expression level in conidia (Figure 11). Transcripts of these six genes were not detectable by qRT-PCR during the early stages of infection (1 to 2 d after inoculation [DAI]), likely as a consequence of a combination of their low expression levels (<0.1-fold of the reference gene ILV5 encoding an acetohydroxyacid reductoisomerase) and a low percentage of fungal RNA in these infected rice leaves (<1% of total RNA). Among all of the genes assayed by qRT-PCR, only the expression of CYP1 was detectable during the early infection stages (1 to 2 DAI; see Supplemental Figure 7 online), which is likely related to its expression level being 2- to 4-fold higher than that of the reference gene ILV5. At later infection stages (3 to 4 DAI), transcripts of all of these genes were detectable by qRT-PCR. At 4 DAI, the expression level of TIG1, HOS2, HOS4, HST1, SET3, and SNT1 was 4-, 3-, 5-, 6-, 4-, and 6-fold higher in infected leaves than in vegetative hyphae, respectively (Figure 11). These data indicate that all of the proposed components of the Tig1 complex, except for CYP1, share the same expression pattern at the different fungal developmental and infection stages.
DISCUSSION
In this study, we identified and characterized the TIG1 gene and protein complex in M. oryzae. TIG1 and its orthologs are conserved from yeast to human (Pijnappel et al., 2001; Mou et al., 2006). TIG1 shares higher similarity with mammalian TBL1 than with yeast SIF2. It has an N-terminal LisH domain and six WD domains in the C-terminal region. No other genes in M. oryzae have similar structural features. We also noticed that many of the TIG1 orthologs from filamentous fungi have an intron in the LisH domain and another intron immediately before the stop codon. These two introns at the termini of TIG1 ORF may be evolutionarily conserved for regulating the expression or stability of TIG1 mRNA.
In budding yeast, Sif2 is involved in transcriptional repression of meiosis-specific genes (Pijnappel et al., 2001; Cerna and Wilson, 2005). In M. oryzae, the tig1 mutant was female sterile but male fertile. Among 30 ascospore progeny from a tig1 × TIG1 cross, 18 carried the tig1 null allele, suggesting that TIG1 deletion has no effect on ascospore formation and viability. However, conidiation was significantly reduced in the tig1 and other Tig1 complex-deficient mutants (Table 2). Conidium morphology was also abnormal in these mutants (Figure 8C). Because asexual reproduction plays a critical role in the infection cycle, the Tig1 complex may have been adapted to regulate conidiation in M. oryzae and other plant pathogens. In a number of phytopathogenic fungi, MAP kinases involved in the yeast pheromone response and filamentous growth pathways have evolved to regulate appressorium formation and other plant infection processes (Zhao et al., 2007; Rispail et al., 2009).
Similar to ftl1 mutants in F. graminearum (Ding et al., 2009), the tig1 mutant was nonpathogenic. Close examination indicates that the differentiation of invasive hyphae was blocked in the tig1 mutant. In plant cells penetrated by the tig1 mutant, only limited growth of primary invasive hyphae was observed. DAB staining indicated that tig1 primary invasive hyphae were surrounded by a high concentration of ROS. Plant cells appeared to mount strong defense responses and displayed HR-like cell death. The tig1 mutant also had increased sensitivities to PR protein osmotin and plant defensin MsDef1 (Coca et al., 2000; Ramamoorthy et al., 2007). It is possible that attenuated invasive growth of the tig1 mutant triggered strong defense responses in host cells. Rapid accumulation of high concentrations of ROS and other plant defense compounds at the penetration site may block the proliferation of the tig1 mutant and subsequently result in the death of penetrated plant cells, which may be responsible for the rare, small brown spots formed on susceptible rice seedlings inoculated with the tig1 mutant (Figure 2A).
In comparison with extensive studies on appressorium formation and function, the regulation of postpenetration infection processes is not well characterized in M. oryzae (Ebbole, 2007; Xu et al., 2007; Wilson and Talbot, 2009). Unlike other nonpathogenic mutants that are blocked in either appressorium formation or appressorium-mediated penetration, mutants disrupted in the Tig1 HDAC complex were defective in the differentiation and growth of bulbous secondary invasive hyphae. These results indicate that chromatin modification plays a critical role in regulating fungal invasive growth. Because histone deacetylation is normally associated with a suppression of gene expression (Pijnappel et al., 2001; Brosch et al., 2008), the Tig1 complex may be involved in the repression of genes that are detrimental to the differentiation and biotrophic growth of secondary invasive hyphae. The fact that M. oryzae uses a global regulatory mechanism such as histone modification for invasive growth indicates that a large number of genes may need to be repressed simultaneously in invasive hyphae. Further characterization of the Tig1 complex will be helpful in understanding the regulatory networks that regulate fungal–plant interactions after penetration and development of invasive hyphae.
In mammalian cells, TBL1 and TBLR1 are transducin β-like proteins associated with the N-CoR (nuclear receptor corepressor) and SMRT (silencing mediator for retinoid and thyroid hormone receptors) complex (Guenther et al., 2001; Yoon et al., 2003; Zhang et al., 2006). SMRT proteins have an N-terminal domain that binds to nuclear receptors and a C-terminal region that interacts with HDACs. In yeast, Sif2 is a component of the Set3 HDAC complex that consists of Sif2, Hos2, Set3, Snt1, Hst1, Hos4, and Cpr1 (Pijnappel et al., 2001; Cerna and Wilson, 2005; Mou et al., 2006). Several of these proteins are conserved from yeast to human. The M. oryzae genome contains putative orthologs of every component of the yeast Set3 complex. All of these putative orthologs except SET3 were coimmunoprecipitated with the Tig1-3xFLAG fusion. However, only a weak interaction was detectable between Tig1 and Set3 in yeast two-hybrid assays. The Tig1-Set3 interaction was further confirmed by co-IP assays. We also confirmed the Tig1-Hos2 and Set3-Hos2 interactions by co-IP. SNT1 was not included in the co-IP assays because of its size (~8 kb) and the difficulty of making fusion constructs. Overall, our results suggest that Tig1 functions in a protein complex that is similar to the yeast Set3 complex. The Tig1 complex of M. oryzae likely contains Tig1, Set3, Hos2, Snt1, Hst1, Hos4, and Cyp1.
Similar to the tig1 mutant, the set3, hos2, and snt1 mutants had altered conidium morphology and were defective in plant infection. By contrast, the hst1 and hos4 mutants had no obvious defects in conidiogenesis and pathogenesis. These results indicate that TIG1, SET3, HOS2, and SNT1, but not HST1 and HOS4, are involved in regulating similar biological processes in M. oryzae. In yeast, SIF2, SET3, HOS2, and SNT1 are core components of the Set3 complex. Our results indicate that essential components of the M. oryzae Tig1 complex are Tig1, Set3, Hos2, and Snt1. Deletion of any one of these components may disrupt the integrity and function of this Tig1 complex. The putative Cpr1 ortholog, CYP1, is known to be involved in calcium signaling and to be required for virulence in M. oryzae (Viaud et al., 2002).
Based on the similarities in composition, the Tig1 complex appeared to function as a Set3-like HDAC complex in M. oryzae. The HDAC activities in the tig1, set3, hos2, and snt1 mutants were significantly reduced in comparison with that of the wild type. HDACs catalyze the removal of acetyl groups from Lys residues in histones and nonhistone proteins (Ficner, 2009). The tig1, set3, hos2, and snt1 mutants had increased levels of H3K18 acetylation. Further indirect evidence that these proteins are essential components of a Set3-like HDAC complex is that Tig1-, Set3-, and Hos2-GFP fusion proteins all localized to the nucleus. Tig1 has no predictable NLS sequence. Its association with other components of the complex may be responsible for nuclear localization of the Tig1-GFP fusion proteins. Unlike Tig1, the Set3 and Snt1 proteins have putative NLS sequences.
Recently, the core components of the Set3 complex, including Hos2, Set3, Sif2, and Snt1, have been shown to be important for efficient responses to secretory stress via the Slt2 MAP kinase pathway in yeast (Cohen et al., 2008). In M. oryzae, MPS1 and BCK1, putative orthologs of yeast Slt2 and Bck1, are important for fungal cell wall integrity and pathogenesis (Xu et al., 1998; Jeon et al., 2008). In this study, we showed that phosphorylation of Mps1 was reduced in the tig1, set3, and snt1 mutants (Figure 9). Similar to the mps1 mutant (Xu et al., 1998), conidiation was significantly reduced in the tig1 mutant. It is likely that the Tig1 HDAC complex plays a role in the activation of the Mps1 pathway in response to certain stresses or signals in M. oryzae.
As a key component of the Tig1 complex, HOS2 had increased expression levels in conidia, appressoria, and infected rice leaves at 3 to 4 DAI (Figure 11), and expression was highest in conidia. This expression pattern is shared by all the genes encoding components of the Tig1 HDAC complex, except for CYP1 (see Supplemental Table 5 online). HOS2 is a class II HDAC gene. In yeast, HDA1 and HOS3 are two other class II HDAC genes (Trojer et al., 2003), but they are not part of the Set3 complex. The M. oryzae genome contains HDAC genes that are putatively orthologous to yeast HDA1 and HOS3. While the expression of HDA1 (MGG_01076.5) was higher in appressoria than in conidia, HOS3 (MGG_06043.5) had an expression pattern similar to that of HOS2 in vegetative hyphae, conidia, and appressoria (see Supplemental Figure 7 online). Deletion of HDA1 and HOS3 had no obvious effects on fungal growth, conidiogenesis, and appressorium formation. Unlike the hos2 mutant, hda1 and hos3 null mutants were still pathogenic on rice and barley leaves (see Supplemental Figure 8 online). While the hda1 mutant was delayed in symptom development for about 1 d, the hos3 mutant had slightly reduced virulence. These results suggest that Hos2 is the main class II HDAC in M. oryzae.
TIG1-interacting genes, including SET3, SNT1, and HOS2, are also conserved in the genomes of other filamentous fungi that have been sequenced. However, none of the putative SET3 and SNT1 orthologs had previously been functionally characterized in plant pathogenic fungi. For HOS2, its putative ortholog in Cochliobolus heterostrophus is an important pathogenicity factor, but its role in conidiogenesis is not clear (Baidyaroy et al., 2001). In F. graminearum, we also generated mutants that lack the putative orthologs of yeast SET3, HOS2, and SNT1. Similar to the ftl1 mutant, the Fgset3, Fghos2, and Fgsnt1 mutants were defective in plant infection and conidiogenesis (J.-R. Xu, unpublished data), indicating that the Tig1-HDAC complex plays a similar role in M. oryzae and F. graminearum, and possibly also in other fungal pathogens, by regulating genes important for pathogenesis and conidiogenesis. To date, the molecular mechanisms that regulate the expression of genes that are specifically expressed or highly induced in invasive hyphae, such as BAS or other effector genes (Mosquera et al., 2009), are not clear in M. oryzae. Our results indicate that the Tig1 HDAC complex regulates the differentiation and growth of secondary invasive hyphae after penetration. The identification and characterization of downstream target genes of this well-conserved HDAC complex will enhance our understanding of the regulatory networks that regulate gene expression in invasive hyphae or during the early stages of plant colonization.
METHODS
Culture Conditions and Genetic Manipulations
All the wild-type and mutant strains used in this study (Table 1) were cultured on oatmeal agar or CM plates as described (Talbot et al., 1993; Park et al., 2006; Villalba et al., 2008). Mycelia harvested from 2-d-old 5×YEG (0.5% yeast extract and 1% glucose) cultures shaken at 150 rpm were used for isolation of genomic DNA and protoplasts (Sweigard et al., 1998; Zhao et al., 2005). Hygromycin- or zeocin-resistant transformants were selected on media supplemented with 250 μg/mL hygromycin B or 150 μg/mL zeocin (Invitrogen). Protoplast transformation and genetic crosses were performed as described previously (Talbot et al., 1993; Zhao and Xu, 2007).
Appressorium Formation and Plant Infection Assays
Conidia harvested from 10-d-old oatmeal agar cultures were resuspended to 1 × 104 spores/mL in water for appressorium formation and penetration assays and to 1 × 105 spores/mL in 0.25% gelatin for plant infection assays (Li et al., 2007; Mosquera et al., 2009). Two-week-old seedlings of rice (Oryza sativa) cultivar CO-39, Nipponbare, or Sariceltik and 8-d-old seedlings of barley (Hordeum vulgare) cultivar Golden Promise or Plaisant were used for infection assays (Park et al., 2004). To isolate Magnaporthe oryzae from infected leaves, wound inoculation sites and areas of typical blast lesions or brown spots were excised, surface sterilized, and incubated on 2% water agar as described (Xu and Hamer, 1996). Fungal growth and conidiation were observed after incubation for 2 to 3 d. For staining with DAB (Sigma-Aldrich), rice leaf sheath samples were incubated in 1 mg/mL DAB solution, pH 3.8, for 8 h and destained with ethanol/acetic acid (94/4, v/v) for 1 h before examination. For aniline blue staining (Bhambra et al., 2006), barley leaves were sampled 72 h after inoculation and incubated in 1 M KOH at 70°C for 20 min. After washing three times with water and once with 67 mM K2HPO4, pH 9.0, the samples were stained with 0.05% aniline blue and examined with a Nikon epifluorescence microscope. For RNA isolation, infected leaves of cultivar Sariceltik were sampled at 1, 2, 3, and 4 DAI. Osmotin (Coca et al., 2000) and MsDef1 (Ramamoorthy et al., 2007) were added to conidium suspensions to final concentrations of 20 and 50 μM, respectively for assaying their effects on germination and appressorium formation.
Manual Annotation of TIG1 and SNT1
Sequencing analysis of TIG1 cDNA clones showed that the ORF of MGG_03198 was incorrectly predicted in version 6 of the M. oryzae genome annotation (incorrect start codon and first two introns). MGG_03198.5 (version 5) was similar to the cDNA sequence. The SNT1 gene also differs in version 5 (MGG_09174.5) and version 6 (MGG_14558.6 and MGG_14559.6). Other genes described in this article are similar in versions 5 and 6 of the genome annotation.
Molecular Manipulations
RNA was isolated from mycelia or infected rice leaves (Vergne et al., 2007) with TRIzol reagent (Invitrogen) and purified with the DNA-free kit (Ambion). First-strand cDNA was synthesized with the M-MLV reverse transcriptase (Invitrogen). qRT-PCR was performed with the ABI 7700 sequence detection system (Applied Biosystem) using QuantiTect SYBR-green PCR Master Mix (Qiagen) as described (Flaherty and Dunkle, 2005). Primers used to amplify selected genes in qRT-PCR reactions are listed in Supplemental Table 1 online. ACT1 (Li et al., 2007), TUB1 (MGG_00604.5), or ILV5 (MGG_01808.5) was used as the endogenous constitutive reference gene. The relative quantification of each transcript was calculated by the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Immunoblot Analysis
Total proteins were isolated from vegetative hyphae as described (Bruno et al., 2004). Proteins separated on SDS-PAGE gels were transferred onto a nitrocellulose membrane with a Bio-Rad electroblotting apparatus. The expression and activation of Mps1 and Pmk1 MAP kinases were detected with the PhophoPlus p44/42 MAP kinase antibody kit (Cell Signaling Technology). The horseradish peroxidase–conjugated secondary antibody and SuperSignal West Femto chemiluminescent substrate from Pierce were used for antigen antibody detections. The monoclonal anti-GFP (Roche), anti-FLAG (Sigma-Aldrich), and anti-actin (Sigma-Aldrich) antibodies were used at a 1:1000 to 1:2000 dilution for immunoblot analysis.
Generation of the Gene Replacement Mutants
The ligation-PCR approach (Zhao et al., 2004) was used to generate the TIG1 gene replacement construct. The 0.9-kb upstream and 0.8-kb downstream flanking sequences of TIG1 were amplified with primers F1/R1 and F2/R2 (see Supplemental Table 1 online). The resulting PCR products were digested with FseI and AscI, respectively, and ligated with the hph gene from pCX63. After ligation, a 3.1-kb TIG1 replacement construct was amplified with primers F1/R2 and transformed into protoplasts of Guy11 or 70-15. Hygromycin-resistant transformants were screened by PCR and confirmed by DNA gel blot analysis using the TIG1 fragment, which was amplified with primers NF1/NR2, and the hph gene as probes (Figure 1A). For complementation, a 4.1-kb fragment containing the entire TIG1 gene (with a 1.5-kb promoter region) was cloned into pYK11 (Park et al., 2006) and transformed into the tig1 mutant KT-1.
The same approach was used to generate gene replacement constructs for other components of the TIG1 complex. Primers used to amplify the flanking sequences of HOS2, SNT1, SET3, HST1, and HOS4 are listed in Supplemental Table 1 online. The ligation-PCR products (Zhao et al., 2004) were transformed into protoplasts of Guy11, 70-15, or Ku80 (Villalba et al., 2008) to generate the hos2, snt1, set3, hst1, and hos4 deletion mutants (Table 1). Putative knockout mutants were identified by PCR screens and confirmed by DNA gel blot analysis.
Construction of TIG1-GFP and TIG1-3xFLAG Fusion Constructs
To construct the TIG1-GFP fusion, the TIG1 gene was amplified with primers TNG-F/TGFP-R (see Supplemental Table 1 online) and cotransformed with XhoI-digested pKB04 into Saccharomyces cerevisiae strain XK1-25 as described (Bourett et al., 2002; Bruno et al., 2004). Plasmid pSD11 was rescued from the resulting Trp+ yeast transformants, confirmed by sequence analysis to contain the TIG1-GFP fusion, and transformed into the tig1 mutant KT-1. The resulting zeocin-resistant transformants, which exhibited wild-type colony morphology and growth rate, were analyzed by DNA gel blot hybridization. The expression of the TIG1-GFP fusion construct was observed under a Nikon E-800 epifluorescence microscope.
To generate the TIG1-3xFLAG fusion construct, the TIG1 coding region was amplified with primers RPF and RPR (Supplemental Table 1). Primer RPR contains three copies of the FLAG epitope sequence followed by a termination codon. The resulting PCR products were co-transformed with XhoI-digested pDL2 (Bourett et al., 2002) into XK1-25 (Bruno et al., 2004). The TIG1-3xFLAG fusion vector was recovered from yeast transformants and transformed into 70-15.
Affinity Purification and Mass Spectrometry Analysis
About 150 to 200 mg of freshly harvested mycelia were resuspended in 2 mL of extraction buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 50 mM NaF, 2 mM PMSF, 5 mM EDTA, 1 mM EGTA, 1% Triton X-100, and 10% glycerol) and 10 μL of protease inhibitor cocktail (Sigma-Aldrich). After homogenization with a Biospec mini bead beater (Bruno et al., 2004), the lysate was centrifuged at full speed in a microcentrifuge for 20 min at 4°C. The supernatants were further centrifuged at 45,000 rpm at 4°C for 1 h to remove cell debris. About 50 μL of anti-FLAG M2 beads (Sigma-Aldrich) were added to capture Tig1-interacting proteins, following the instructions of the manufacturer. After incubation at 4°C for 2 h, the beads were washed three times each with 500 μL of lysis buffer, 50 mM TMAB (1 M trimethylammonium bicarbonate; Fluka), and sterile distilled water. Proteins binding to the beads were eluted with 150 μL of 50 mM TMAB containing 0.1% Rapigest and digested with trypsin as described (Tao et al., 2005; Zhou et al., 2007). Tryptic peptides were analyzed by nanoflow liquid chromatography–tandem mass spectrometry on a high-resolution hybrid linear ion trap orbitrap mass spectrometer (LTQ-Orbitrap XL; ThermoFisher) coupled to an Agilent Nanoflow LC system. The tandem mass spectrometry data were queried against the National Center for Biotechnology Information nonredundant M. oryzae protein database using the SEQUEST algorithm (Tabb et al., 2001) on the Sorcerer IDA server (SageN). Putative TIG1-interacting genes were identified by MS analysis, and four biological replicates were performed. Proteins that bound nonspecifically to the anti-FLAG antibody had been identified in our preliminary studies (Supplemental Dataset 1) and were removed from the list of proteins copurified with Tig1-3xFLAG for further analysis (see Supplemental Table 2 online).
Co-IP Assays
The yeast gap repair approach (Bourett et al., 2002; Bruno et al., 2004) was used to generate the SET3-3xFLAG, HOS2-GFP, and SET3-GFP fusion constructs. Primers used to amplify the SET3 and HOS2 genes are listed in Supplemental Table 1 online. The resulting fusion constructs were cotransformed into protoplasts of 70-15 in pairs. Transformants containing the TIG1-3xFLAG/SET3-GFP, TIG1-3xFLAG/HOS2-GFP, and SET3-3xFLAG/HOS2-GFP constructs were identified by PCR and confirmed by DNA gel blot hybridization. The expression of the GFP and 3xFLAG fusion proteins was further confirmed by immunoblot analyses. For co-IP assays, total proteins were isolated and incubated with the anti-FLAG M2 beads as described above. Proteins eluted from M2 beads were analyzed by immunoblot detection with the anti-FLAG (Sigma-Aldrich) and anti-GFP (Roche) antibodies.
Yeast Two-Hybrid Assays
The HybridZap2.1 yeast two-hybrid system (Stratagene) was used to assay protein–protein interactions. The TIG1 ORF was cloned into pBD-GAL4 as the bait vector pSD17. The prey construct of SET3 was generated with pAD-GAL4-2.1. The resulting bait and prey vectors were transformed into yeast strain YRG-2 (Stratagene) with the Alkali-Cation yeast transformation kit (MP Biomedicals). The Leu+ and Trp+ transformants were isolated and assayed for growth on SD-Trp-Leu-His medium and the expression of the LacZ reporter gene as described (Zhao et al., 2005). The positive and negative controls were provided in the HybridZap2.1 XR library construction kit (Stratagene).
HDAC Activity Assays
Protoplasts of Guy11, 70-15, Ku80, and the tig1, hos2, set3, and snt1 mutants were resuspended to 4 × 108 protoplasts/mL in lysis buffer (10 mM Tris-HCl, pH 7.5, 10 mM NaCl, 15 mM MgCl2, 250 mM sucrose, 0.5% Nonidet P-40, 0.1 mM EGTA, and 200 μM PMSF). Nuclei and crude nuclear extracts were prepared from disrupted protoplasts as described (Ding et al., 2009) and assayed for HDAC activities with the HDAC activity assay kit (Cayman Chemical Company) following the instructions provided by the manufacturer. Fluorescent signals in samples of different mutants were detected with a plate reader (Synergy HT; Bio-TEK). The concentration of deacetylated compounds was calculated with the deacetylation standard curve and used to estimate HDAC activity (nmol/min/mL) using the formula provided in the HDAC activity assay kit.
Histone Acetylation Assays
Vegetative hyphae harvested from 100 mL of 2-d-old 5xYEG cultures were resuspended in 400 μL lysis buffer (10 mM Tris-HCl, pH 7.4, 300 mM sorbitol, 600 mM NaCl, 5 mM MgCl2, and 5 mM EDTA) with a mixture of protease and phosphatase inhibitors (1 μg/mL of aprotinin, leupeptin, and pepstatin A, 1 mM PMSF, 1 μM microcystin-LR, and 2 mM p-hloromercuriphenylsulfonic acid). Mycelia were homogenized with acid-washed glass beads in a Biospec minibead beater using three 40-s pulses applied at 1-min intervals on ice. The lysate was separated from the glass beads and centrifuged at 16,000g for 15 min at 4°C. The resulting supernatants containing whole-cell extracts were separated on 15% SDS-PAGE gels, transferred to a nitrocellulose membrane, and assayed for histone acetylation as described (Briggs et al., 2001). Monoclonal anti-H3 (Abcam) and anti-H3K18Ac (Abcam) antibodies were used for detection.
Accession Numbers
Sequence data for the M. oryzae genes from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: TIG1, EDK00306; SET3, EDK04728; SNT1, EDJ98032; HOS2, EDK04649; HST1, EDJ98541; HOS4, EDK06449; CYP1, BAB59119; HDA1, EDK02244; HOS3, EDJ96334; ACT1, EDJ99269; TUB1, EDK02768; and ILV5, EDK04462. Others are as follows: the putative ortholog of TIG1 in Neurospora crassa, XP_963679; FTL1 of Fusarium graminearum, XP_380508.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of Amino Acid Sequences of Tig1 and Its Putative Orthologs from Neurospora crassa, Fusarium graminearum, and Saccharomyces cerevisiae (Sif2).
Supplemental Figure 2. KT-2 and KT-4 Failed to Cause Typical Blast Lesions on Rice Leaves.
Supplemental Figure 3. The Tig1 Mutant Triggered Enhanced Callose Deposition in Plant Cells.
Supplemental Figure 4. Immunoblot Analysis of the Expression and Immunoprecipitation of Tig1-3xFLAG Fusion Proteins with an anti-FLAG Antibody.
Supplemental Figure 5. Targeted Deletion of the SNT1, SET3, HOS4, HOS2, and HST1 Gene.
Supplemental Figure 6. GFP Signals in the PRP27-TIG1-GFP Transformant.
Supplemental Figure 7. Expression Patterns of HOS3, HDA1, and CYP1.
Supplemental Figure 8. Infection Assays with hda1 and hos3 Mutants.
Supplemental Table 1. PCR Primers Used in This Study.
Supplemental Data Set 1. Magnaporthe oryzae Proteins Nonspecifically Copurified with 3xFLAG.
Acknowledgments
We thank Larry Dunkle and Charles Woloshuk at Purdue University for critical reading of this manuscript and Scott Briggs for assistance with histone acetylation assays. We also thank Xinhua Zhao for helpful discussions during this study and Mike Hasagawa and Dilap Shah for providing osmotin and MsDef1. This work was supported by a grant from the National Research Initiative of the USDA Cooperative State Research, Education, and Extension Service (2007-35319-102681) and the 111 Project from the Ministry of Education of China (B07049).
References
- Baidyaroy D., Brosch G., Ahn J.H., Graessle S., Wegener S., Tonukari N.J., Caballero O., Loidl P., Walton J.D. (2001). A gene related to yeast HOS2 histone deacetylase affects extracellular depolymerase expression and virulence in a plant pathogenic fungus. Plant Cell 13: 1609–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhambra G.K., Wang Z.Y., Soanes D.M., Wakley G.E., Talbot N.J. (2006). Peroxisomal carnitine acetyl transferase is required for elaboration of penetration hyphae during plant infection by Magnaporthe grisea. Mol. Microbiol. 61: 46–60 [DOI] [PubMed] [Google Scholar]
- Bourett T.M., Sweigard J.A., Czymmek K.J., Carroll A., Howard R.J. (2002). Reef coral fluorescent proteins for visualizing fungal pathogens. Fungal Genet. Biol. 37: 211–220 [DOI] [PubMed] [Google Scholar]
- Briggs S.D., Bryk M., Strahl B.D., Cheung W.L., Davie J.K., Dent S.Y.R., Winston F., Allis C.D. (2001). Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15: 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch G., Loidl P., Graessle S. (2008). Histone modifications and chromatin dynamics: A focus on filamentous fungi. FEMS Microbiol. Rev. 32: 409–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno K.S., Tenjo F., Li L., Hamer J.E., Xu J.R. (2004). Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 3: 1525–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerna D., Wilson D.K. (2005). The structure of sif2p, a WD repeat protein functioning in the SET3 corepressor complex. J. Mol. Biol. 351: 923–935 [DOI] [PubMed] [Google Scholar]
- Chao C.C.T., Ellingboe A.H. (1991). Selection for mating competence in Magnaporthe grisea pathogenic to rice. Can. J. Bot. 69: 2130–2134 [Google Scholar]
- Chi M.H., Park S.Y., Kim S., Lee Y.H. (2009). A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog. 5: e1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clergeot P.H., Gourgues M., Cots J., Laurans F., Latorse M.P., Pepin R., Tharreau D., Notteghem J.L., Lebrun M.H. (2001). PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea. Proc. Natl. Acad. Sci. USA 98: 6963–6968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coca M.A., Damsz B., Yun D.J., Hasegawa P.M., Bressan R.A., Narasimhan M.L. (2000). Heterotrimeric G-proteins of a filamentous fungus regulate cell wall composition and susceptibility to a plant PR-5 protein. Plant J. 22: 61–69 [DOI] [PubMed] [Google Scholar]
- Cohen T.J., Mallory M.J., Strich R., Yao T.P. (2008). Hos2p/Set3p deacetylase complex signals secretory stress through the Mpk1p cell integrity pathway. Eukaryot. Cell 7: 1191–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean R.A., et al. (2005). The genome sequence of the rice blast fungus Magnaporthe grisea. Nature 434: 980–986 [DOI] [PubMed] [Google Scholar]
- Ding S.L., Mehrabi R., Koten C., Kang Z.S., Wei Y.D., Seong K.Y., Kistler H.C., Xu J.R. (2009). Transducin beta-like gene FTL1 is essential for pathogenesis in Fusarium graminearum. Eukaryot. Cell 8: 867–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbole D.J. (2007). Magnaporthe as a model for understanding host-pathogen interactions. Annu. Rev. Phytopathol. 45: 437–456 [DOI] [PubMed] [Google Scholar]
- Fang E.G.C., Dean R.A. (2000). Site-directed mutagenesis of the MAGB gene affects growth and development in Magnaporthe grisea. Mol. Plant Microbe Interact. 13: 1214–1227 [DOI] [PubMed] [Google Scholar]
- Ficner R. (2009). Novel structural insights into class I and II histone deacetylases. Curr. Top. Med. Chem. 9: 235–240 [DOI] [PubMed] [Google Scholar]
- Flaherty J.E., Dunkle L.D. (2005). Identification and expression analysis of regulatory genes induced during conidiation in Exserohilum turcicum. Fungal Genet. Biol. 42: 471–481 [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Barak O., Lazar M.A. (2001). The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol. 21: 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J., Goh J., Yoo S., Chi M.H., Choi J., Rho H.S., Park J., Han S.S., Kim B.R., Park S.Y., Kim S., Lee Y.H. (2008). A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Mol. Plant Microbe Interact. 21: 525–534 [DOI] [PubMed] [Google Scholar]
- Kankanala P., Czymmek K., Valent B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19: 706–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Park S.Y., Kim K.S., Rho H.S., Chi M.H., Choi J., Park J., Kong S., Park J., Goh J., Lee Y.H. (2009). Homeobox transcription factors are required for conidiation and appressorium development in the rice blast fungus Magnaporthe oryzae. PLoS Genet. 5: e1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Ding S.L., Sharon A., Orbach M., Xu J.R. (2007). Mir1 is highly upregulated and localized to nuclei during infectious hyphal growth in the rice blast fungus. Mol. Plant Microbe Interact. 20: 448–458 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Mehrabi R., Ding S., Xu J.R. (2008). MADS-box transcription factor Mig1 is required for infectious growth in Magnaporthe grisea. Eukaryot. Cell 7: 791–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell T.K., Dean R.A. (1995). The cAMP-dependent protein kinase catalytic subunit is required for appressorium formation and pathogenesis by the rice blast pathogen Magnaporthe grisea. Plant Cell 7: 1869–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera G., Giraldo M.C., Khang C.H., Coughlan S., Valent B. (2009). Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell 21: 1273–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z.M., Kenny A.E., Curcio M.J. (2006). Hos2 and Set3 promote integration of Ty1 retrotransposons at tRNA genes in Saccharomyces cerevisiae. Genetics 172: 2157–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G., Bruno K.S., Staiger C.J., Talbot N.J., Xu J.R. (2004). Independent genetic mechanisms mediate turgor generation and penetration peg formation during plant infection in the rice blast fungus. Mol. Microbiol. 53: 1695–1707 [DOI] [PubMed] [Google Scholar]
- Park G., Xue C., Zhao X., Kim Y., Orbach M., Xu J.R. (2006). Multiple upstream signals converge on an adaptor protein Mst50 to activate the PMK1 pathway in Magnaporthe grisea. Plant Cell 18: 2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel W., Schaft D., Roguev A., Shevchenko A., Tekotte H., Wilm M., Rigaut G., Seraphin B., Aasland R., Stewart A.F. (2001). The Saccharomyces cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15: 2991–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy V., Zhao X.H., Snyder A.K., Xu J.R., Shah D.M. (2007). Two mitogen-activated protein kinase signalling cascades mediate basal resistance to antifungal plant defensins in Fusarium graminearum. Cell. Microbiol. 9: 1491–1506 [DOI] [PubMed] [Google Scholar]
- Rispail N., et al. (2009). Comparative genomics of MAP kinase and calcium–calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet. Biol. 46: 287–298 [DOI] [PubMed] [Google Scholar]
- Sesma A., Osbourn A.E. (2004). The rice leaf blast pathogen undergoes developmental processes typical of root-infecting fungi. Nature 431: 582–586 [DOI] [PubMed] [Google Scholar]
- Sweigard J.A., Carroll A.M., Farrall L., Chumley F.G., Valent B. (1998). Magnaporthe grisea pathogenicity genes obtained through insertional mutagenesis. Mol. Plant Microbe Interact. 11: 404–412 [DOI] [PubMed] [Google Scholar]
- Tabb D.L., Eng J.K., Yates J.R. (2001). Protein identification by SEQUEST. Proteome Research: Mass Spectometry, James P., (Berlin: Springer-Verlag; ). 125–142 [Google Scholar]
- Talbot N.J., Ebbole D.J., Hamer J.E. (1993). Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5: 1575–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W.A., Wollscheid B., O'Brien R., Eng J., Li X., Bodenmiller B., Watts J., Hood L., Aebersold R. (2005). Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and mass spectrometry. Nat. Methods 2: 591–598 [DOI] [PubMed] [Google Scholar]
- Thines E., Weber R.W.S., Talbot N.J. (2000). MAP kinase and protein kinase A-dependent mobilizationof triacylglycerol and glycogen during appressorium tugor generation by Magnaporthe grisea. Plant Cell 12: 1703–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trojer P., Brandtner E.M., Brosch G., Loidl P., Galehr J., Linzmaier R., Haas H., Mair K., Tribus M., Graessle S. (2003). Histone deacetylases in fungi: novel members, new facts. Nucleic Acids Res. 31: 3971–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker S.L., Talbot N.J. (2001). Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu. Rev. Phytopathol. 39: 385–419 [DOI] [PubMed] [Google Scholar]
- Tucker S.L., Thornton C.R., Tasker K., Jacob C., Giles G., Egan M., Talbot N.J. (2004). A fungal metallothionein is required for pathogenicity of Magnaporthe grisea. Plant Cell 16: 1575–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax J.A., Woodbury E.L., Quang P.N., Paraz M., Blethrow J.D., Shah K., Shokat K.M., Morgan D.O. (2003). Targets of the cyclin-dependent kinase Cdk1. Nature 425: 859–864 [DOI] [PubMed] [Google Scholar]
- Urban M., Bhargava T., Hamer J.E. (1999). An ATP-driven efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J. 18: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergne E., Ballini E., Marques S., Mammar B.S., Droc G., Gaillard S., Bourot S., DeRose R., Tharreau D., Notteghem J.L., Lebrun M.H., Morel J.B. (2007). Early and specific gene expression triggered by rice resistance gene Pi33 in response to infection by ACE1 avirulent blast fungus. New Phytol. 174: 159–171 [DOI] [PubMed] [Google Scholar]
- Viaud M.C., Balhadere P.V., Talbot N.J. (2002). A Magnaporthe grisea cyclophilin acts as a virulence determinant during plant infection. Plant Cell 14: 917–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba F., Collemare J., Landraud P., Lambou K., Brozek V., Cirer B., Morin D., Bruel C., Beffa R., Lebrun M.H. (2008). Improved gene targeting in Magnaporthe grisea by inactivation of MgKU80 required for non-homologous end joining. Fungal Genet. Biol. 45: 68–75 [DOI] [PubMed] [Google Scholar]
- Wilson R.A., Jenkinson J.M., Gibson R.P., Littlechild J.A., Wang Z.Y., Talbot N.J. (2007). Tps1 regulates the pentose phosphate pathway, nitrogen metabolism and fungal virulence. EMBO J. 26: 3673–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R.A., Talbot N.J. (2009). Under pressure: Investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7: 185–195 [DOI] [PubMed] [Google Scholar]
- Xu J.R., Hamer J.E. (1996). MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10: 2696–2706 [DOI] [PubMed] [Google Scholar]
- Xu J.R., Staiger C.J., Hamer J.E. (1998). Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. USA 95: 12713–12718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.R., Zhao X., Dean R.A. (2007). From genes to genomes; A new paradigm for studying fungal pathogenesis in Magnaporthe oryzae. Adv. Genet. 57: 175–218 [DOI] [PubMed] [Google Scholar]
- Yoon H.G., Chan D.W., Huang Z.Q., Li J.W., Fondell J.D., Qin J., Wong J.M. (2003). Purification and functional characterization of the human N-CoR complex: The roles of HDAC3, TBL1 and TBLR1. EMBO J. 22: 1336–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-M., Chang Q., Zeng L., Gu J., Brown S., Basch R.S. (2006). TBLR1 regulates the expression of nuclear hormone receptor co-repressors. BMC Cell Biol. 7: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Kim Y., Park G., Xu J.R. (2005). A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe grisea. Plant Cell 17: 1317–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Mehrabi R., Xu J.-R. (2007). Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryot. Cell 6: 1701–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.H., Xu J.R. (2007). A highly conserved MAPK-docking site in Mst7 is essential for Pmk1 activation in Magnaporthe grisea. Mol. Microbiol. 63: 881–894 [DOI] [PubMed] [Google Scholar]
- Zhao X.H., Xue C., Kim Y., Xu J.R. (2004). A ligation-PCR approach for generating gene replacement constructs in Magnaporthe grisea. Fungal Genet. Newsl. 51: 17–18 [Google Scholar]
- Zhou F., Galan J., Geahlen R.L., Tao W.A. (2007). A novel quantitative proteomics strategy to study phosphorylation-dependent peptide-protein interactions. J. Proteome Res. 6: 133–140 [DOI] [PubMed] [Google Scholar]