This study shows that Arabidopsis CSAat1A, which encodes a Cockayne Syndrome A-like protein, and its homolog, CSAat1B, form a heterotetramer in response to DNA damage and thereby play a key role in the plant’s response to UV-B irradiation.
Abstract
In plants, as in animals, DNA is constantly subject to chemical modification. UV-B irradiation is a major genotoxic agent and has significant effects on plant growth and development. Through forward genetic screening, we identified a UV-B–sensitive mutant (csaat1a-3) in Arabidopsis thaliana, in which expression of CSAat1A, encoding a Cockayne Syndrome A-like protein, is reduced due to insertion of a T-DNA in the promoter region. Arabidopsis lacking CSAat1A or its homolog CSAat1B is more sensitive to UV-B and the genotoxic drug methyl methanesulfonate and exhibits reduced transcription-coupled repair activity. Yeast two-hybrid analysis indicated that both CSAat1A and B interact with DDB1A (UV-Damage DNA Binding Protein1). Coimmunoprecipitation assays demonstrated that CSAat1A and B associate with the CULLIN4 (CUL4)-DDB1A complex in Arabidopsis. A split-yellow fluorescent protein assay showed that this interaction occurs in the nucleus, consistent with the idea that the CUL4-DDB1A-CSA complex functions as a nuclear E3 ubiquitin ligase. CSAat1A and B formed heterotetramers in Arabidopsis. Taken together, our data suggest that the plant CUL4-DDB1ACSAat1A and B complex represents a unique mechanism to promote ubiquitination of substrates in response to DNA damage.
INTRODUCTION
Due to their sessile nature, plants have developed unique and efficient biochemical mechanisms to cope with many environmental stresses. While UV-C radiation (200 to 280 nm) is completely absorbed by atmospheric gases, a significant amount of UV-B (280 to 320 nm) reaches the earth’s surface. High doses of UV-B irradiation reduce plant growth and development by inducing structural damage to DNA and membranes and disrupting processes including photosynthesis, secondary metabolism, responses to stress, and photomorphogenesis (Frohnmeyer and Staiger, 2003). To prevent damage from UV irradiation, plants have developed various mechanisms, including increased accumulation of UV-absorbing pigments (flavonoids and hydroxycinnamic acid derivatives) (Li et al., 1993; Landry et al., 1995; Jin et al., 2000; Bieza and Lois, 2001; Kliebenstein et al., 2002; Oravecz et al., 2006; Zhao et al., 2007; Bashandy et al., 2009), activation of reactive oxygen species (Mackerness et al., 2001; Gao et al., 2008), and generation of DNA damage repair systems (Gallego et al., 2000; Liu et al., 2000, 2003; Ulm et al., 2001; Sakamoto et al., 2003; Liang et al., 2006; Vonarx et al., 2006; Wang and Liu, 2006; Hidema et al., 2007).
UV irradiation induces two major classes of DNA damage products: cyclobutane-pyrimidine dimers (CPDs) and 6-4 photoproducts (6-4 PPs). To survive and avoid changes in genome stability, plants recover from these lesions through photoreactivation and light-independent repair pathways. In Arabidopsis thaliana, UV Resistance 2 (UVR2), which encodes a photolyase, is specific for CPD damage repair (Ahmad et al., 1997; Jiang et al., 1997), while UVR3, another type of photolyase protein, functions in 6-4 PP damage repair (Nakajima et al., 1998). In addition to their function during light-dependent damage, DNA damage repair pathways also play important roles in excision repair, mutagenic repair or dimer bypass, recombinational repair, and the regulation of cell cycle checkpoints (Sinha and Häder, 2002). Among the DNA damage repair pathways, the nucleotide excision repair (NER) pathway in mammalian cells is involved in both global genome repair (GGR) and transcription-coupled repair (TCR). Classical GGR and TCR factors include Damage-specific DNA Binding Protein 2 (DDB2) and Cockayne Syndrome A (CSA) proteins, which are the substrate receptors for the CUL4-DDB1 E3 ligase (O’Connell and Harper, 2007). CUL4-DDB1DDB2 regulates the ubiquitination of the xeroderma pigmentosa complementation group C protein (Sugasawa et al., 2005), while CUL4-DDB1CSA regulates the ubiquitination of CSB (Groisman et al., 2006) in the DNA damage repair process.
A CUL4-DDB1 complex has recently been identified in plants and functions in many biological processes (Bernhardt et al., 2006; Lee et al., 2008; Molinier et al., 2008; Wang et al., 2008; Zhang et al., 2008). It has been shown that an Arabidopsis mutant lacking DDB2 has enhanced sensitivity to UV irradiation, methyl methanesulfonate (MMS), and H2O2 stress (Koga et al., 2006) and that overexpression of Arabidopsis DDB1A increases plant UV tolerance (Al Khateeb and Schroeder, 2009). Moreover, DDB2 associates with the CUL4-DDB1A complex to form an E3 ligase that modulates GGR-type DNA damage repair upon UV stress (Molinier et al., 2008). However, it has not been determined if a CUL4-DDB1CSA-like complex exists in the plant or how it functions.
Using a forward genetic screen, we isolated a CSA-like gene (CSAat1A) from Arabidopsis. CSAat1A associates with the CUL4-DDB1A complex in the nucleus and functions in UV damage repair. The Arabidopsis genome encodes two CSA-like genes that form heterotetramers, suggesting that a CUL4-DDB1ACSA E3 ligase is present in plants and plays a critical role in the response of the plant to UV-B irradiation.
RESULTS
Genetic Screening for Arabidopsis Mutants That Are Defective in Their Response to UV-B
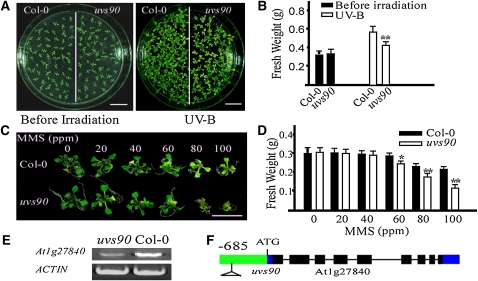
To identify novel components that regulate the response of Arabidopsis to UV-B, we screened a T-DNA activation-tagged pool by monitoring seedling growth rate after UV-B irradiation. Several mutants defective in their response to UV-B were recovered. One of them, uvs90, was more sensitive to UV-B stress than the wild type. When 12-d-old uvs90 mutant and wild-type seedlings were exposed to UV-B (130 μmol m−2 s−1) for 30 min and then incubated in a growth chamber for 5 d, the cotyledons of uvs90 plants were bleached and plant growth was reduced relative to that of the wild type (Figures 1A and 1B). Consistent with this observation, when wild-type and uvs90 plants were treated with MMS, a genotoxic chemical that produces heat-labile DNA damage, the reduction in growth of the uvs90 mutant was more severe than that of the wild type (Figures 1C and 1D). Using thermal asymmetric interlaced-PCR, insertion of the T-DNA fragment, which leads to the decreased transcription of UVS90 (Figure 1E), was detected in the promoter of At1g27840, 685 bp upstream of the translation start codon (Figure 1F; see Supplemental Figure 1 online).
Figure 1.
The uvs90 Mutant Is Sensitive to UV-B and MMS.
(A) Twelve-day-old wild-type (Col-0) and uvs90 seedlings were treated with UV-B (130 μmol m−2 s−1) for 30 min. After treatment, plants were transferred to a growth chamber for 5 d and then photographed. Bars = 1 cm.
(B) The fresh weight of 20 seedlings from each group shown in (A) was measured. All data represent means ± se of at least five replicate experiments (Student’s t test, **P < 0.01).
(C) Six-day-old wild-type and uvs90 seedlings were treated with the indicated concentrations of the genotoxic drug MMS. Photographs were taken 10 d after treatment. Bar = 1 cm.
(D) The fresh weight of 10 seedlings from each group shown in (C) was measured. All data represent means ± se of at least five replicate experiments (Student’s t test, *P < 0.05 and **P<0.01).
(E) Expression of At1g27840 in wild-type and uvs90 mutant seedlings. RT-PCR products after 30 cycles using gene-specific primers. ACTIN was included as a loading control. Three replicate experiments were performed.
(F) The T-DNA insertion site of the uvs90 mutant in At1g27840. Exons, filled black rectangles; introns, solid lines; untranslated regions, filled blue rectangles; promoter, filled green rectangle.
UVS90 Is a WD40-Repeat Protein and Shares Significant Sequence Similarity to the Human CSA Protein
The UVS90 gene contains nine exons and eight introns and encodes a 451–amino acid protein with a predicted molecular mass of 49.75 kD and an isoelectric point of 8.8. The UVS90 protein contains four WD40 repeats and shares 54% sequence similarity with human CSA (see Supplemental Figure 2 online). Hereafter, we refer to UVS90 as CSAat1A and the uvs90 mutant as csaat1a-3. The Arabidopsis genome encodes another protein (At1g19750, CSAat1B), which is 92% identical to CSAat1A (see Supplemental Figure 2 online).
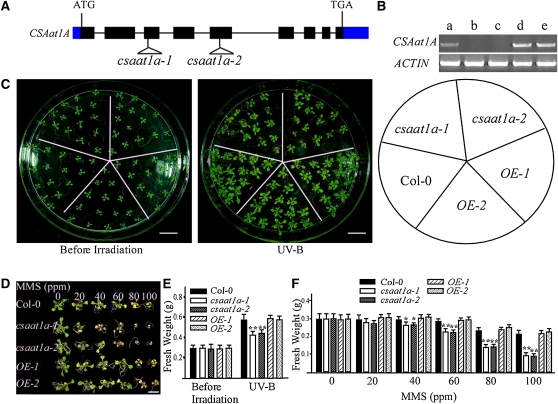
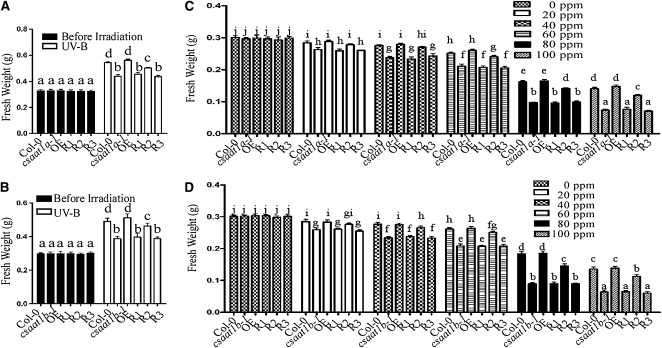
In mammalian cells, CSA is known to play a critical role in DNA damage repair (reviewed in Svejstrup, 2002). To determine whether the UV-B sensitive phenotype of the csaat1a-3 mutant is due to the decreased expression of CSAat1A, two Arabidopsis mutant lines (SALK_028416/csaat1a-1 and SALK_151258/csaat1a-2) with T-DNA insertions in the CSAat1A gene were obtained from the ABRC (Alonso et al., 2003). The T-DNA insertion sites in these mutants were determined by PCR using CSAat1A-specific primers, and the T-DNA left border primers were found in the third or fifth exons of the CSAat1A gene, respectively (Figure 2A). The absence of a full-length CSAat1A transcript in the csaat1a-1 and csaat1a-2 mutants was confirmed by RT-PCR analysis (Figure 2B). When 12-d-old mutant and wild-type seedlings were exposed to UV-B (130 μmol m−2 s−1) for 30 min and then incubated in a growth chamber for 5 d, the mutants were more sensitive to UV-B than the wild type, with more bleached cotyledons and a slower growth rate (Figures 2C); this defect could be complemented by expression of the CSAat1A cDNA in the mutants (Figure 2C; OE-1 and OE-2). To determine if DNA repair is involved in csaat1a UV-B sensitivity, wild-type and csaat1a plants were treated with MMS at 0, 20, 40, 60, 80, and 100 ppm. The growth of the csaat1a mutants was dramatically reduced relative to that of the wild type; this defect was also complemented by expression of the CSAat1A cDNA in the mutants (Figures 2D and 2F; OE-1 and OE-2). Taken together, these results suggest that the csaat1a UV-B–sensitive phenotype may be due to a difference in DNA repair ability in the mutant relative to in the wild type.
Figure 2.
csaat1a Knockout Mutants Are Sensitive to UV-B and MMS.
(A) Gene structure of CSAat1A with T-DNA insertion sites found in the csaat1a mutants. Exons, filled black rectangles; introns, solid lines; untranslated regions, filled blue rectangles.
(B) Expression of CSAat1A in wild-type (Col-0) and csaat1a mutant seedlings, and mutant seedlings overexpressing CSAat1A (OE); RT-PCR products after 30 cycles with gene-specific primers and ACTIN as a loading control. Three replicate experiments were performed. a, Col-0; b, csaat1a-1; c, csaat1a-2; d, 35Spro-CSAat1A-Flag in csaat1a-1 (OE-1); e, 35Spro-CSAat1A-Flag in csaat1a-2 (OE-2).
(C) Twelve-day-old wild-type, csaat1a mutant, and CSAat1A OE seedlings were treated with UV-B (130 μmol m−2 s−1) for 30 min. After treatment, plants were transferred to a growth chamber for 5 d and then photographed. Bars = 1 cm.
(D) Six-day-old wild-type, csaat1a mutant, and CSAat1A OE seedlings were treated with the indicated concentrations of MMS. Photographs were taken 10 d after treatment. Bar = 1 cm.
(E) The fresh weight of 20 seedlings from each group shown in (C) was measured. All data represent means ± se of at least five replicate experiments (Student’s t test, **P < 0.01).
(F) The fresh weight of 10 seedlings from each group shown in (D) was measured. All data represent means ± se of at least five replicate experiments (Student’s t test, *P < 0.05 and **P<0.01).
The Arabidopsis csaat1b Mutant Is Also Sensitive to UV-B and MMS
The amino acid sequence of CSAat1A is 92% identical to that of CSAat1B (see Supplemental Figure 2 online). However, single csaat1a mutants are sensitive to UV stress, suggesting that the functions of these two proteins do not fully overlap. To determine if CSAat1B also plays a role in DNA damage repair, two T-DNA insertion lines, SALK_152568/csaat1b-1 and SALK_144623/csaat1b-2, in CSAat1B were obtained from the ABRC, and the T-DNA insertion sites and the absence of full-length transcripts were determined (see Supplemental Figures 3A and 3B online). When mutant and wild-type seedlings were treated with UV-B and MMS, sensitive phenotypes were also observed in the csaat1b mutants (see Supplemental Figures 3C to 3F online). Expression of CSAat1B cDNA in the csaat1b mutants under the control of the 35S promoter (OE-1 and OE-2) complemented the mutant phenotypes (see Supplemental Figures 3C to 3F online), indicating that CSAat1B is also involved in the response of Arabidopsis to UV-B irradiation.
Tissue-Specific Expression and Subcellular Localization of CSAat1A and CSAat1B
Our data demonstrate that CSAat1A and CSAat1B function in the response of the plant to UV-B irradiation. To determine the tissue-specific expression of these two genes, promoter regions of CSAat1A (1.55 kb upstream of the start codon) and CSAat1B (1.5 kb upstream of the start codon) were fused to the β-glucuronidase (GUS) reporter gene. The resulting constructs were transferred into wild-type Arabidopsis (Columbia-0 [Col-0]). For each construct, 10 independent F2 transgenic lines were analyzed for GUS expression. While the intensity of GUS staining in each of the independent lines was variable, the tissue-specific localization was the same. As shown in Supplemental Figures 4A to 4L online, GUS driven by either the CSAat1A or CSAat1B promoter was expressed throughout young seedlings and in leaves, stems, flowers, and siliques.
To determine the subcellular localization of CSAat1A and CSAat1B, green florescent protein (GFP) was fused to the N terminus of each protein, and expression was driven by the cauliflower mosaic virus 35S (35S) promoter. When the fusion constructs were transferred to Col-0 plants, GFP-CSAat1A and GFP-CSAat1B signals were observed in the nucleus (see Supplemental Figures 4M and 4N online).
CSAat1A and B Form a Complex with CUL4-DDB1A in the Nucleus
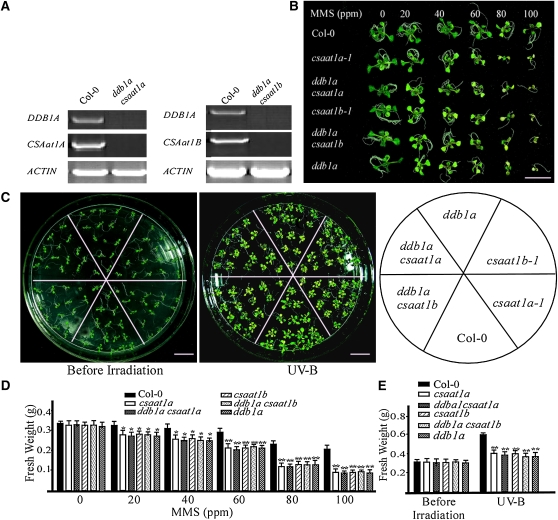
In mammalian cells, CSA associates with CUL4-DDB1 to form an E3 ubiquitin ligase that promotes the ubiquitination of CSB during the regulation of DNA damage repair through TCR (O’Connell and Harper, 2007). In Arabidopsis, CUL4-DDB1A-DDB2 was recently shown to function in GGR during UV stress (Molinier et al., 2008). To determine if the CUL4-DDB1A complex and CSAat1A and B interact genetically, we generated ddb1a csaat1a and ddb1a csaat1b double mutants (Figure 3A). Six-day-old wild-type, single, and double mutant seedlings were treated with different concentrations of MMS (Figures 3B and 3D), and 12-d-old plants were treated with UV-B (Figures 3C and 3E). For each treatment, growth reductions of the single and double mutants were not significantly different from each other, but were more severe than that of the wild type. These data suggest that CSAat1A and B may function in the same pathway as DDB1A.
Figure 3.
CSAat1A and B Genetically Interact with DDB1A.
(A) Expression of DDB1A, CSAat1A, and B in wild-type plants and the ddb1a csaat1a and ddb1a csaat1b double mutants. RT-PCR products after 30 cycles with gene-specific primers and ACTIN as a loading control. Three replicate experiments were performed.
(B) Six-day-old wild-type, csaat1a, ddb1a csaat1a, csaat1b, ddb1a csaat1b, and ddb1a seedlings were treated with the indicated concentrations of MMS. Photographs were taken 10 d after treatment. Bar = 1 cm.
(C) Twelve-day-old wild-type (Col-0), csaat1a, ddb1a csaat1a, csaat1b, ddb1a csaat1b, and ddb1a seedlings were treated with UV-B (130 μmol m−2 s−1) for 30 min. After treatment, plants were transferred to a growth chamber for 5 d and then photographed. Bars = 1 cm.
(D) The fresh weight of 10 seedlings from each group shown in (B) was measured. All data represent means ± se of at least five replicate experiments (Student’s t test, *P < 0.05 and **P < 0.01).
(E) The fresh weight of 20 seedlings from each group shown in (C) was measured. All data represent means ± se of at least five replicate experiments (Student’s t test, **P < 0.01).
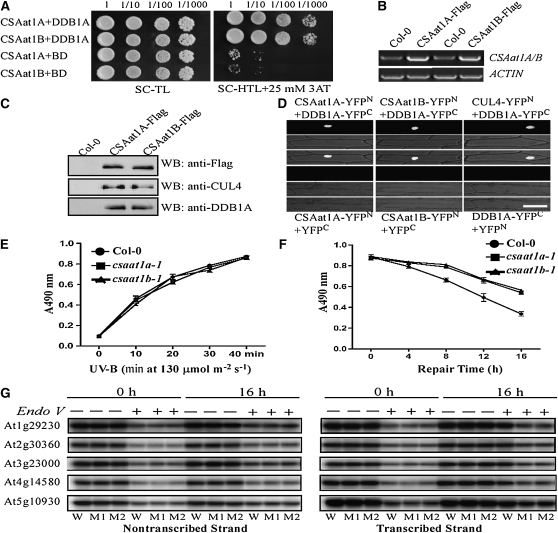
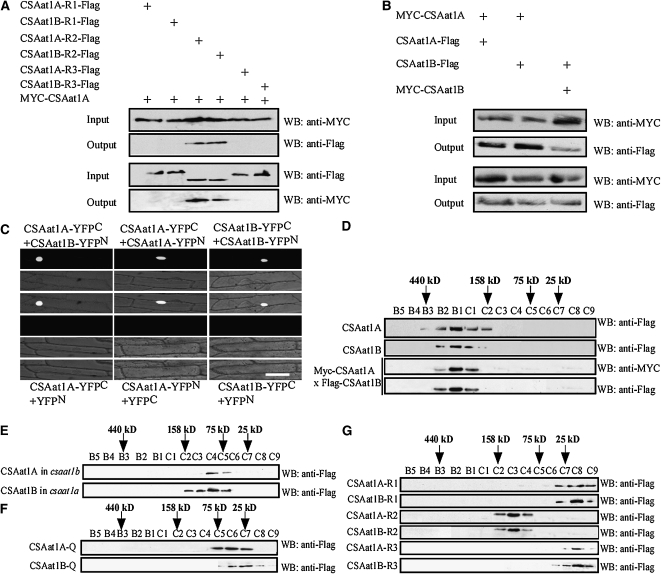
We then determined if CSAat1A and B interact with DDB1A, as CSA protein does in mammalian cells. Arabidopsis DDB1A was cotransformed with either CSAat1A or B into yeast, and yeast two-hybrid (Y2H) analysis showed that both interact with DDB1A (Figure 4A). To determine if CSAat1A and B can form a complex with CUL4 and DDB1A in planta, a Flag-tag in a cluster of three tandem repeats was fused to the C terminus of either CSAat1A or B under the control of the 35S promoter, and the constructs were transformed into their corresponding mutants. Transgenic lines that rescued the mutant phenotypes (Figure 2C; see Supplemental Figure 3C online) and had relatively low expression levels of the CSAat1A or B transgene were used for coimmunoprecipitation assays. Proteins were extracted from the transgenic plants, and CSAat1A- or B-Flag was immunoprecipitated with anti-Flag–conjugated agarose. Pull-down products were analyzed on immunoblots with anti-CUL4 and anti-DDB1A antibodies. The results demonstrate that both CSAat1A and B pulled down CUL4 and DDB1A, indicating that both form complexes with CUL4 and DDB1A (Figure 4C).
Figure 4.
CSAat1A and B Associate with the CUL4-DDB1A Complex in the Nucleus.
(A) Interaction between CSAat1A or B and DDB1A in yeast. pGADT7-CSAat1A or B plasmids were cotransformed with pGBKT7-DDB1A into yeast strain AH109. Growth of the transformed yeast was assayed on media minus Trp and Leu (left panel) or minus Trp, Leu, and His with 25 mM 3-amino-1,2,4-triazole (3AT; right panel). Columns in each panel represent serial decimal dilutions.
(B) The expression levels of CSAat1A or B in the transgenic plants analyzed in (C). RT-PCR products after 30 cycles with gene-specific primers and ACTIN as a loading control. Three replicate experiments were performed.
(C) Interaction between CSAat1A or B and DDB1A and CUL4 in transgenic plants. Protein was extracted from 15-d-old wild-type or 35Spro-CSAat1A- or B-Flag transgenic plants (each in their corresponding knockout mutant background). CSAat1A or B was immunoprecipitated with anti-Flag–conjugated agarose, and the pull-down products were detected by immunoblot analysis using anti-Flag, anti-DDB1A, or anti-CUL4 antibodies. Bar = 100 μm.
(D) Split YFP analysis. Plasmid combinations CSAat1A-YFPN + DDB1A-YFPC, CSAat1B-YFPN + DDB1A-YFPC, CUL4-YFPN + DDB1A-YFPC, CSAat1A-YFPN + YFPC, CSAat1B-YFPN + YFPC, or DDB1A-YFPC + YFPN were transiently expressed in onion epidermal cells. Top panels, confocal YFP images; middle panels, bright-field images; bottom panels, combined bright-field and YFP images. CSAat1A and B and CUL4 were fused to the YFP N terminus (YFPN) and DDB1A was fused to the YFP C terminus (YFPC). All constructs were under the control of the 35S promoter. WB, immunoblot.
(E) and (F) CPD accumulation. Twelve-day-old seedlings grown on MS medium were irradiated with five different doses of UV-B light (0, 10, 20, 30, and 40 min, 130 μmol m−2 s−1). The plants were immediately harvested under green light to avoid CPD repair by photolyases. For DNA repair assays, 12-d-old seedlings grown on MS medium were irradiated with UV-B light (130 μmol m−2 s−1) for 40 min. The plants were immediately transferred to continuous white light and harvested under green light after different repair times (0, 4, 8, 12, and 16 h). The DNA was extracted using CTAB buffer, and CPD levels were determined with ELISA assays using an anti-TDM-2 antibody. The absorbance at 490 nm (A490 nm) was measured with a microplate photometer. Wild type (circles), csaat1a-1 (squares), and csaat1b-1 (triangles). Data represent means ± se of three independent experiments.
(G) Analysis of strand-specific DNA repair. Wild-type (W), csaat1a-1 (M1), and csaat1b-1 (M2) seedlings were irradiated with UV-B, and samples were harvested at 0 or 16 h after treatment. Ten milligrams of DNA from each sample was digested with BamHІ and SpeI (for At1g29230 probes); EcoRI and SalI (for At2g30360 probes); EcoRI and XhoI (for At3g23000 probes); PstI and XhoI (for At4g14580 probes); and BamHI and XbaI (for At5g10930 probes) and then treated with (+) or without (−) T4 endonuclease V. DNA samples were analyzed by DNA gel blot hybridization using strand-specific probes.
If CUL4-DDB1ACSAat1A and B functions in DNA damage repair in Arabidopsis, the complex should be located in the nucleus. To determine if this is the case, we cloned the CSAat1A, CUL4, or CSAat1B cDNAs into the N terminus of the split-yellow fluorescent protein (YFP) vector pUC-SPYNE (YFPN) and cloned DDB1A into the C terminus of the pUC-SPYCE vector (YFPC) (Walter et al., 2004). Different combinations of CUL4/CSAat1A and B/pUC-SPYNE and DDB1A/pUC-SPYCE were cotransfected into onion (Allium cepa) epidermal cells, which were analyzed by confocal microscopy to detect YFP bimolecular florescence complementation. The YFP signal was detected in the nucleus in CSAat1A-YFPN/DDB1A-YFPC, CSAat1B-YFPN/DDB1A-YFPC, and CUL4-YFPN/DDB1A-YFPC cotransfection assays (Figure 4D). We did not observe YFP signal with any construct individually or with combinations of any gene and empty pUC-YFPN/C vector. These results indicate that the CUL4-DDB1ACSAat1A and B complex is localized to the nucleus.
Genomic DNA is one of the major molecules affected by UV-B irradiation. UV-B irradiation results in phototransformation and causes CPDs and 6-4 PPs to accumulate, which turns on DNA damage repair processes. To compare DNA damage in the mutants and wild-type plants, wild-type, csaat1a, and csaat1b mutant seedlings were exposed to increasing doses of UV-B radiation for 40 min, transferred to a growth chamber, and analyzed for DNA damage repair after 0, 4, 8, 12, and 16 h. Genomic DNA was extracted from the plants and subjected to an ELISA assay to measure CPD induction (Figures 4E and 4F). The wild type and the mutants accumulated similar amounts of CPDs immediately after the plants were exposed to increasing doses of UV-B; however, the wild type accumulated less CPDs than the mutants after DNA repair. These results indicate that the wild type and mutant had similar levels of UV-B–based DNA damage and that the DNA damage repair machinery is impaired in the csaat1a and csaat1b mutants. In mammalian cells, CUL4-DDB1CSA E3 ligase functions in DNA damage repair through TCR. To determine if CSAat1A and B are also involved in TCR, genomic DNA was extracted from these plants either immediately after treatment or after a 16-h incubation to allow DNA repair. The DNA was digested with BamHI and SpeI, EcoRI and SalI, EcoRI and XhoI, PstI and XhoI, or BamHI and XbaI to release 4.1-, 4.9-, 5.9-, 5.75-, or 2.5-kb fragments, respectively, containing the intronless At1g29230, At2g30360, At3g23000, At4g14580, or At5g10930 genes, and then treated with T4 endonuclease V to remove the damaged DNA. The DNAs were subsequently analyzed by DNA gel blot analysis and probed with either the transcribed or nontranscribed strands. As shown in Figure 4G, after the 16-h incubation, UV-B–induced DNA damage for the transcribed strands was repaired more efficiently in the wild type than in either of the two mutants, while DNA damage repair for the nontranscribed strand was similar for the wild type and the mutants. These results indicate that CSAat1A and B function in TCR.
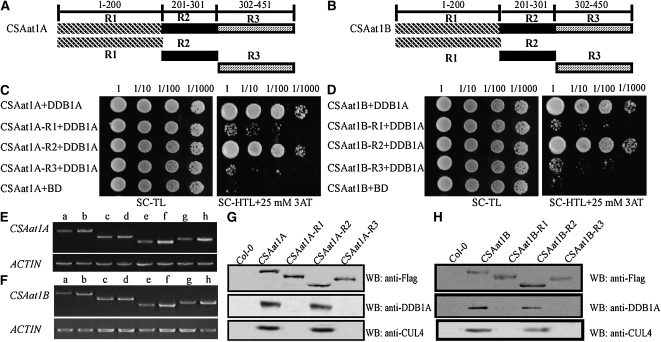
The WDxR Motif in the WD40 Domain Is Essential for CSAat1A and B Interaction with CUL4-DDB1A
Previous reports demonstrated that the WDxR motif in the WD40 domain is required for DDB1-CUL4 associated factor (DCAF) proteins to physically bind to DDB1A in Arabidopsis (Lee et al., 2008; Zhang et al., 2008). To identify the specific domain that interacts with DDB1A, CSAat1A and B were divided into three fragments: amino acid 1-200, which contains one WD40 domain (R1); amino acid 201-301, which contains the second WD40 domain that harbors a WDxR motif (R2); and amino acid 302 to the C terminus, which contains the last two WD40 domains (R3) (Figures 5A and 5B). Full-length CSAat1A and B and the three peptides corresponding to each protein were cotransformed with DDB1A into yeast for Y2H analysis. DDB1A interaction strength was similar with full-length CSAat1A and B and the R2 peptides (Figures 5C and 5D). To determine whether R2 interacts with DDB1A in planta, the 3×Flag-tag was fused to the R1, R2, or R3 peptides for each gene under the control of the 35S promoter. The six resulting plasmids were transformed into their corresponding knockout mutants (Figures 5E and 5F). Proteins were extracted from the transgenic plants and Flag-tagged proteins were immunoprecipitated with anti-Flag–conjugated agarose. The pull-down products were then analyzed on immunoblots with anti-CUL4 or anti-DDB1A antibodies. Similar amounts of CUL4 and DDB1A were pulled down by CSAat1A and B and CSAat1A- and B-R2 but not by the R1 and R3 peptides (Figures 5G and 5H), suggesting that the R2 region is responsible for the interaction of CSAat1A and B with the CUL4-DDB1A complex.
Figure 5.
The Second WD40 Domain in CSAat1A and B Interacts with DDB1A.
(A) and (B) Schematic representations of regions of the CSA1at1A and B proteins.
(C) and (D) Different regions of CSAat1A and B interact with DDB1A in yeast. Whole CSAat1A and B proteins as well as specific regions of the proteins (R1, R2, and R3) were cloned into pGADT7 and cotransformed with DDB1A into yeast. Growth of the transformed yeast was assayed on media minus Trp and Leu (left panels) or minus Trp, Leu, and His with 25 mM 3-amino-1,2,4-triazole (right panels). Columns in each panel represent serial decimal dilutions.
(E) and (F) Expression levels of CSAat1A and B and different fragments of the genes in their corresponding knockout mutants. a, c, e, and g, wild type; b, 35Spro-CSAat1A- or B-Flag; d, 35Spro-CSAat1A- or B-R1-Flag; f, 35Spro-CSAat1A- or B-R2-Flag; h, 35Spro-CSAat1A- or B-R3-Flag. RT-PCR products after 30 cycles with gene-specific primers and ACTIN as a loading control. Three replicate experiments were performed.
(G) and (H) Flag-tagged R1, R2, and R3 regions of CSAat1A and B interact with DDB1A and CUL4 in vivo. CSAat1A- and B-Flag proteins were immunoprecipitated with anti-Flag-agarose. The pull-down products were analyzed via immunoblots with anti-Flag, anti-DDB1A, or anti-CUL4 antibodies. WB, immunoblot.
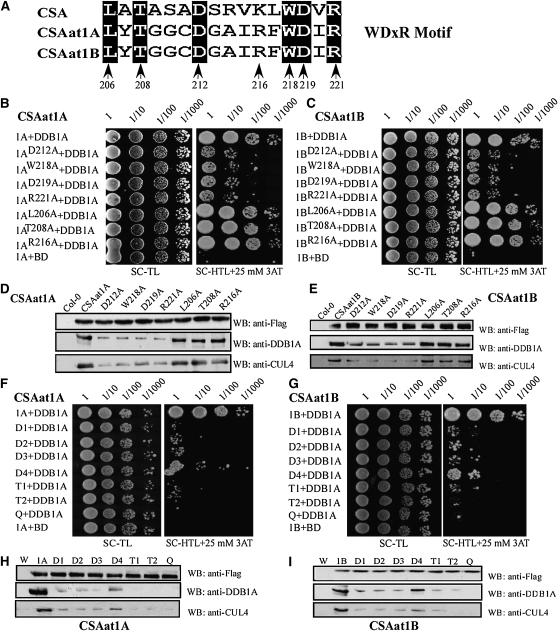
Both CSAat1A and B contain a WDxR motif in the WD40 domain of the R2 region (see Supplemental Figure 2 online). To determine if this WDxR motif is critical for CSAat1A and B interaction with DDB1A, six conserved amino acids (Leu-206, Thr-208, Asp-212, Trp-218, Asp-219, and Arg-221) in the WDxR motif were converted to Ala, and Arg-216 was changed to Ala as a control (Figure 6A). Interactions between the mutant proteins and their corresponding wild-type proteins were analyzed in Y2H assays or fused to 3×Flag-tags under control of the 35S promoter and expressed in their corresponding mutant backgrounds (see Supplemental Figure 5 online) for coimmunoprecipitation assays.
Figure 6.
The WDxR Motif Is Essential for CSAat1A and B Interaction with DDB1A.
(A) Positions of single mutations generated in the WDxR motif. The numbers indicate the amino acid residue in CSAat1A and B. All indicated sites were mutated to Ala (A).
(B) and (C) Effect of single amino acid mutations in the WDxR motif on the interaction between CSAat1A or B and DDB1A in yeast. CSA1at1A or B, CSAat1AD212A or BD212A, CSAat1AW218A or BW218A, CSAat1AD219A or BD219A, CSAat1AR221A or BR221A, CSAat1AL206A or BL206A, CSAat1AT208A, or BT208A and CSAat1AR216A or BR216A in pGADT7 were cotransformed with pGBKT7-DDB1A into yeast. Growth of the transformed yeast was assayed on media minus Trp and Leu (left panels) or minus Trp, Leu, and His with 25 mM 3-amino-1,2,4-triazole (right panels). Columns in each panel represent serial decimal dilutions. BD, binding domain.
(D) and (E) Effect of single amino acid mutations in the WDxR motif on the interaction between CSAat1A or B and DDB1A-CUL4 in planta. The CSAat1A- or B-Flag proteins were immunoprecipitated with anti-Flag agarose. The pull-down products were analyzed via immunoblots with anti-Flag, anti-DDB1A, or anti-CUL4 antibodies. WB, immunoblot.
(F) and (G) Effect of multiple amino acid mutations in the WDxR motif on the interaction between CSAat1A or B and DDB1A in yeast. CSA1at1A or CSA1at1B (B); D1, CSAat1AD212AW218A or BD212AW218A; D2, CSAat1AD212AD219A or BD212AD219A; D3, CSAat1AD212AR221A or BD212AR221A; D4, CSAat1AL206AD212A or BL206AD212A; T1, CSAat1AD212AW218AD219A or BD212AW218AD219A; T2, CSAat1AW218AD219AR221A or BW218AD219AR221A; and Q, CSAat1A D212AW218AD219AR221A or B D212AW218AD219AR221A in pGADT7 were cotransformed with pGBKT7-DDB1A into yeast. Growth of the transformed yeast was assayed on media minus Trp and Leu (left panels) or minus Trp, Leu, and His with 25 mM 3-amino-1, 2,4-triazole (right panels). Columns in each panel represent serial decimal dilutions. BD, vector pGBKT7.
(H) and (I) Effect of multiple amino acid mutations in the WDxR motif on the interaction between CSAat1A or B and DDB1A-CUL4 in planta. The CSAat1A- or B-Flag proteins were immunoprecipitated with anti-Flag agarose. The pull-down products were analyzed via immunoblot analysis with anti-Flag, anti-DDB1A, or anti-CUL4 antibodies. W, wild type; 1A and 1B, 35Spro-CSAat1A- or B-Flag, respectively; D1, 35S-pro-CSAat1AD212AW218A- or BD212AW218A-Flag; D2, 35Spro-CSAat1AD212AD219A- or BD212AD219A-Flag; D3, 35Spro-CSAat1AD212AR221A- or BD212AR221A-Flag; D4, 35Spro-CSAat1AD212AL206A- or BD212AL206A-Flag; T1, 35Spro-CSAat1AD212AW218AD219A- or BD212AW218AD219A-Flag; T2, 35Spro-CSAat1AW218AD219AR221A- or BW218AD219AR221A-Flag; Q, 35Spro-CSAat1AD212AW218AD219AR221A- or BD212AW218AD219AR221A-Flag. WB, immunoblot.
In yeast, the interaction between DDB1A and CSAat1A and B was reduced by mutations at positions D212, W218, D219, or R221 when compared with interactions with the wild-type proteins (Figures 6B and 6C). However, mutations in Leu-206, Thr-208, or Arg-216 did not alter the interaction (Figures 6B and 6C). To further determine if these mutations affect the interaction between CSAat1A and B and the CUL4-DDB1A complex, the seven CSAat1A mutant proteins used above were expressed individually in either csaat1a-1 or csaatlb-1 plants. Transgenic lines with similar CSAat1A and B expression levels were used for further studies (see Supplemental Figure 5 online). As was observed in Y2H assays, mutations at positions Asp-212, Trp-218, Asp-219, or Arg-221 reduced the interaction between CSAat1A and B and the CUL4-DDB1A complex, but mutations at Leu-206, Thr-208, or Arg-216 had no effect compared with wild-type CSAat1A and B (Figures 6D and 6E). These results indicate that the WDxR motif in CSAat1A and B is also important for the formation of the CUL4-DDB1ACSAat1A and B complex in Arabidopsis.
Single mutations at positions Asp-212, Trp-218, Asp-219, or Arg-221 in CSAat1A and B reduced their ability to interact with DDB1A (Figures 6B to 6E). To determine if the WDxR motif is essential for the interaction between CSAat1A and B and the CUL4-DDB1A complex, we generated four CSAat1A and B mutant genes each containing two mutations (D212AW218A, D212AD219A, D212AR221A, or L206AD212A); two mutant genes each containing three mutations (D212AW218AD219A or W218AD219AR221A); and one mutant gene combining four mutations (D212AW218AD219AR221A) for analysis in yeast and in planta. In yeast, when compared with the single mutation constructs, the double mutations (D212W218, D212D219, and D212R221) further reduced the interaction between CSAat1A and B and DDB1A. However, the L206D212 mutant had less effect than the other three. The two triple mutants (D212W218D219 or W218D219R221) were still able to interact with DDB1A, but the interaction was weaker than that of the three double mutants. Mutations in all four residues abolished any interaction (Figures 6F and 6G). Our results demonstrate that D212, W218, D219, and R221 are important for forming the CUL4-DDB1ACSAat1A and B complex. Similar results were observed in planta (Figures 6H and 6I); the interaction between CSAat1A and B and the CUL4-DDB1A complex decreased more significantly with increasing mutations and was abolished with quadruple mutations.
The WDxR Motif Is Critical for CSAat1A and B Function in Response to UV-B Irradiation
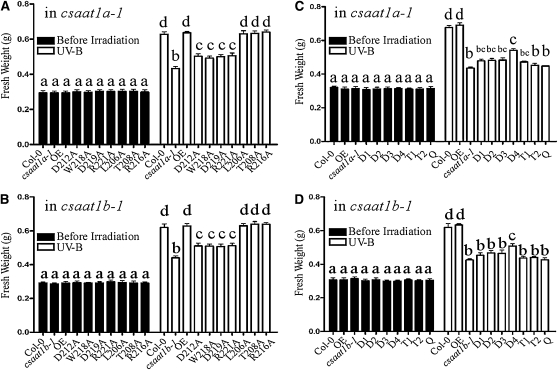
To determine if interaction between CSAat1A and B and CUL4-DDB1A is required for UV-B and MMS tolerance in Arabidopsis, the transgenic lines used for coimmunoprecipitation assays (Figures 5G and 5H) were treated with UV-B or MMS. When we determined if the expression of the CSAat1A- or B-R1, -R2, or -R3 peptides rescued the csaat1a and b UV-B and MMS phenotypes, surprisingly, we found that CSAat1A- and B-R2 but not -R1 and -R3 could partially suppress the corresponding mutant phenotypes (Figure 7; see Supplemental Figure 6 online). Based on the model that WD40 domains provide an interaction platform for the substrates, our data suggest that the presence of only one WD40 domain with a WDxR motif is sufficient for CSAat1A and B interaction with DDB1A but not for UV-B and MMS responses and that, while the WDxR motif is sufficient for CSAat1A and B to form the CUL4-DDB1ACSAat1A and B complex, the other three WD40 domains may be required for interaction with CSAat1A and B substrates. When the CSAat1A and B genes with single mutations in the WDxR motif were expressed in their corresponding mutants, the interaction between CSAat1A and B and the CUL4-DDB1A complex was reduced and the UV-B– or MMS-sensitive phenotypes of the mutants were only partially rescued (Figures 8A and 8B; see Supplemental Figures 7A, 7B, 8A, 8B, 9A, and 9B online). CSAat1A and B with mutations L206A, T208A, or R216A, which did not affect the interaction between CSAat1A or B and CUL4-DDB1A complex, complemented the mutant phenotypes to the level seen for wild-type CSAat1A and B (Figures 8A and 8B; see Supplemental Figures 7A, 7B, 8A, 8B, 9A, and 9B online). Transgenic plants expressing CSAat1A and B containing additional mutations in the WDxR motif (Asp-212, Trp-218, Asp-219, or Arg-221) showed reductions in the rescue of the UV-B– or MMS-sensitive phenotypes that correlated with increasing mutation numbers (Figures 8C and 8D; see Supplemental Figures 7C, 7D, 8C, 8D, 9C, and 9D online).
Figure 7.
Fresh Weight of Transgenic Plants Expressing Different CSAat1A or B Regions after UV-B and MMS Treatment.
(A) and (B) Col-0, 12-d-old wild type; csaat1a or b; OE, 35Spro-CSAat1A- or B- Flag; R1, 35Spro-CSAat1A - or B-R1-Flag; R2, 35Spro-CSAat1A- or B-R2-Flag; R3, 35Spro-CSAat1A- or B-R3-Flag seedlings in the csaat1a or b background were treated with UV-B (130 μmol m−2 s−1) for 30 min. After treatment, plants were transferred to a growth chamber for 5 d, and the fresh weight of 20 seedlings per group was measured. Significant differences (P ≤ 0.05, Student’s t test) are indicated by different lowercase letters.
(C) and (D) Col-0, 6-d-old wild type; csaat1a or b; OE, 35Spro-CSAat1A- or B- Flag; R1, 35Spro-CSAat1A- or B-R1-Flag; R2, 35Spro-CSAat1A- or B-R2- Flag; R3, 35Spro-CSAat1A- or B-R3-Flag seedlings in the csaat1a or b background were treated with the indicated concentrations of MMS. The fresh weight of 10 seedlings in each group was measured. All data represent means ± se of at least five replicate experiments. Significant differences (P ≤ 0.05, Student’s t test) are indicated by different lowercase letters.
Figure 8.
Fresh Weight of Transgenic Plants Expressing Different Point Mutations in the WDxR Motif of CSAat1A after UV-B Treatment.
(A) and (B) Col-0, wild type; csaat1a or b; OE, 35Spro-CSAat1A- or B-Flag; D212A, 35Spro-CSAat1AD212A- or BD212A-Flag; W218A, 35Spro-CSAat1AW218A- or BW218A-Flag; D219A, 35Spro-CSAat1AD219A- or BD219A-Flag; R221A, 35Spro-CSAat1AR221A- or BR221A-Flag; L206A, 35Spro-CSAat1AL206A- or BL206A-Flag; T208A, 35Spro-CSAat1AT208A- or BT208A-Flag; and R216A, 35Spro-CSAat1AR216- or BR216-Flag transgenic plants in the csaat1a or b background were treated as described in Figure 7. All data represent means ± se of at least five replicate experiments. Significant differences (P ≤ 0.05, Student’s t test) are indicated by different lowercase letters.
(C) and (D) Col-0, wild type; OE, 35Spro-CSAat1A- or B-Flag; csaat1a or b; D1, 35Spro-CSAat1AD212AW218A- or BD212AW218A-Flag; D2, 35Spro-CSAat1AD212AD219A- or BD212AD219A-Flag; D3, 35Spro-CSAat1AD212AR221A- or BD212AR221A-Flag; D4, 35Spro-CSAat1AL206AD212A- or BL206AD212A-Flag; T1, 35Spro-CSAat1AD212AW218AD219A- or BD212AW218AD219A-Flag; T2, 35Spro-CSAat1AW218AD219AR221A- or BW218AD219AR221A-Flag; Q, 35Spro-CSAat1AD212AW218AD219AR221A- or BD212AW218AD219AR221A-Flag transgenic plants in the csaat1a or b background were treated with UV-B as described in Figure 7. All data represent means ± se of at least five replicate experiments. Significant differences (P ≤ 0.05, Student’s t test) are indicated by different lowercase letters.
CSAat1A and B Form a Complex in Arabidopsis
The csaat1a, csaat1b, and double ddb1a csaat1a and b mutants had similar UV-B– and MMS-sensitive phenotypes, suggesting that CSAat1A and B function in the same pathway with DDB1A and that their functions in response to UV-B irradiation may not fully overlap. Because CSAat1A and B are both found in the nucleus, share significant sequence identity, and their transcripts are also present in similar tissues, one possible explanation for the csaat1a and b mutant phenotypes is that CSAat1A and B form a complex. Because the R2 peptides partially rescue their mutant phenotypes and contain a WDxR motif, it is possible that the WDxR motif also plays a role in the formation of a CSAat1A and B complex. To test this, the 35Spro-Myc-CSAat1A plasmid was cotransformed with the 35Spro-CSAat1B-R1, -R2, or -R3-3×Flag plasmids into Arabidopsis leaf protoplasts. The CSAat1A protein was immunoprecipitated with anti-Myc or anti-Flag conjugated agarose. The pull-down products were then analyzed on immunoblots with anti-Flag or anti-Myc antibodies. As expected, CSAat1B-R2 but not -R1 or -R3 was pulled down by CSAat1A (Figure 9A). Because CSAat1A and B share 92% amino acid sequence similarity, they may also be capable of forming homodimers in planta. To test this, combinations of epitope-tagged (Myc or Flag) CSAat1A and B or CSAat1A or B fragments were cotransfected into wild-type leaf protoplasts. Proteins were extracted, immunoprecipitated, and analyzed on immunoblots. The results demonstrate that CSAat1A and B interacted with each other and also could self-interact through their R2 peptides (Figures 9A and 9B). Using a split-YFP assay in onion cells, the interactions between CSAat1A and B and the self-interactions for CSAat1A and B were shown to take place in the nucleus (Figure 9C). These results suggest that CSAat1A and B are capable of forming complexes in the plant.
Figure 9.
CSA1at1A and B Form Heterotetramers in Planta.
(A) The R2 region of CSA1at1A and B is required for their interaction in vivo. The 35Spro-Myc-CSAat1A plasmid was cotransformed with 35Spro-CSAat1A- or B-R1-Flag, 35Spro-CSAat1A- or B-R2-Flag, or 35Spro-CSAat1A- or B-R3-Flag into wild-type protoplasts. Total protein extracts were analyzed using protein gel blots with monoclonal anti-Myc or anti-Flag antibodies to detect the presence of Myc-CSAat1A and CSAat1A- or B-R1, -R2, or R3-Flag. Immunoprecipitation was performed using anti-Myc or anti-Flag agarose conjugate, and pull-down products were analyzed with anti-Flag or anti-Myc antibodies. Input, crude protein extract. Output, products pulled down by the anti-Myc or anti-Flag agarose conjugate. WB, immunoblot.
(B) Interaction between CSAat1A and CSAat1B in vivo. The 35Spro-Myc-CSAat1A or B plasmid was cotransformed into wild-type protoplasts with 35Spro-CSAat1A-Flag or 35Spro-CSAat1B-Flag. Total protein extracts were analyzed using protein gel blots with monoclonal anti-Myc and anti-Flag antibodies to detect the presence of Myc-CSAat1A or B and CSAat1A- or B-Flag. Immunoprecipitation was performed using anti-Myc or anti-Flag agarose conjugate, and pull-down products were analyzed with anti-Myc or anti-Flag antibodies. WB, immunoblot.
(C) Split YFP analysis. Plasmid combinations CSAat1B-YFPN + CSAat1A-YFPC, CSAat1B-YFPN + CSAat1B-YFPC, CSAat1A-YFPN + CSAat1A-YFPC, CSAat1A-YFPC + YFPN, CSAat1A-YFPN + YFPC, or CSAat1B-YFPC + YFPN were transiently expressed in onion epidermal cells. All genes were expressed under the control of the 35S promoter. Top panels, confocal YFP images; middle panels, bright-field images; bottom panels, combined bright-field and YFP images. Bar = 100 μm.
(D) to (G) Gel filtration analysis.
(D) Transgenic plants were used to detect the formation of heterotetramers: CSAat1A or B; 35Spro-CSAat1A- or B-Flag in the csaat1a or b backgrounds; Myc-1A× Flag-1B, a Myc-CSAat1A transgenic plant was crossed into a 35Spro-CSAat1B-Flag transgenic plant (F1 seedlings were analyzed).
(E) CSAat1A in csaat1b or CSAat1B in csaat1a, the 35Spro-CSAat1A-Flag or 35Spro-CSAat1B-Flag plasmid was transformed into csaat1b or csaat1a.
(F) CSAat1A- or B-Q, 35Spro-CSAat1AD212AW218AD219AR221A- or BD212AW218AD219AR221A-Flag constructs were transformed into the csaat1a or b mutant.
(G) CSAat1A- or B-R1, -R2, -R3, 35Spro-CSAat1A- or B-R1, -R2, and -R3-Flag in their corresponding mutant backgrounds. Soluble protein was extracted from seedlings and loaded onto a Superdex-200 column. The elution fractions (0.5 mL each, labeled B5 to C9) were collected and concentrated to 50 μL. Protein from 10 μL of each sample was separated by SDS-PAGE (12% gel), followed by immunoblot analysis with anti-Flag or anti-Myc antibodies. WB, immunoblot.
To assess the oligomerization state of CSAat1A and B in planta, soluble proteins were extracted from the transgenic plants harboring Myc or Flag CSAat1A or B and loaded onto a Superdex-200 gel filtration column. The major CSAat1A and B proteins eluted in a peak corresponding to a species with a molecular mass of 200 kD in these three transgenic lines, in agreement with the tetramer size of the CSAat1A and B proteins (Figure 9D). Taken together, our data suggest that the CSAat1A and B proteins likely form heterotetramers rather than homotetramers in vivo. To further confirm this, 35Spro-CSAat1A-3×Flag was transformed into the csaat1b-1 mutant and 35Spro-CSAat1B-3×Flag was transformed into the csaatla-1 mutant, and the proteins were analyzed by gel filtration chromatography. The major CSAat1A- and B-Flag–containing fraction eluted at 100 kD (Figure 9E). These data show that, in the absence of either CSAat1A or CSAat1B, the remaining CSA protein likely forms a dimer.
The WDxR motif is not only essential for the interaction with DDB1A but is also required for CSAat1A and B self-interaction. If this is the case, mutations in the motif should lead to disassociation of the CSAat1A and B dimers. To test this, proteins were extracted from transgenic plants harboring 35Spro-CSAat1AD212AW218AD219AR221A- or BD212AW218AD219AR221A-3×Flag. The major CSAat1A- and B-Flag–containing gel filtration fraction eluted at 50 kD (Figure 9F), in agreement with the predicted size of the monomer proteins. This result is consistent with the Y2H analysis that demonstrated that mutations in these four amino acids abolished CSAat1A interaction with CSAat1B and also with itself (see Supplemental Figure 10 online). In the CSAat1A or B protein, only the WD40 domain in the R2 region contains the WDxR motif. The R2 peptides interact with DDB1A and partially rescue their corresponding mutant phenotypes. When proteins were extracted from the 35Spro-CSAat1A- or B-R1-3×Flag, -R2, or -R3 transgenic plants, the major CSAat1A- or B-R2-Flag–containing gel filtration fraction eluted at 120 kD. However, both the CSAat1-A or B-R1-Flag and -R3 containing fractions eluted at 15 kD (Figure 9G). These results indicate that CSAat1A and B form heterotetramers in the plant.
DISCUSSION
Cullin-based E3 ligases are a major subfamily of ubiquitin ligases and function in many biological processes. CUL4A and CUL4B, two paralogs in mammalian cells, coexpress in many cell types, share high sequence similarity, form separate E3 ligase complexes with the adapter proteins DDB1 and DDB2, and have redundant and distinct functions in promoting efficient NER (O’Connell and Harper, 2007; Guerrero-Santoro et al., 2008; Liu et al., 2009). CUL4A-DDB1 also forms an E3 ligase complex with CSA. This complex interacts with CSB and functions in the TCR pathway (Fousteri et al., 2006). DCAF proteins interact directly with DDB1 through the WDxR motif in the WD40 domain in mammalian cells. The Arabidopsis genome encodes 119 putative DCAF proteins with 165 WDxR motifs (Lee et al., 2008; Zhang et al., 2008). However, only few of these proteins have been shown functionally to interact with CUL4-DDB1A (Bernhardt et al., 2006; Lee et al., 2008; Zhang et al., 2008; Molinier et al., 2008). In this study, we identified two Arabidopsis DCAF proteins (CSAat1A and B) that form heterotetramers, associate with the CUL4-DDB1A complex in planta, and function in response to UV stress through regulation of TCR. Consistent with this notion, CSAat1A and B localized and interacted with DDB1A in the nucleus. Moreover, knockdown of a putative Arabidopsis CSB-like gene (a SWI2/SNF2 chromatin remodeling gene) by double-stranded RNA interference led to transgenic plants that were more sensitive to UV treatment than the wild type (Shaked et al., 2006). Taken together, these data indicate that the CUL4-DDB1CSACSB pathway exists in plants and functions in UV-induced DNA damage repair.
As with the other DCAF proteins identified, CSAat1A and B interact with DDB1A through the WDxR motif in their WD40 domain. Mutations in either of the two conserved amino acids in this motif (DxxxxxWDxR) reduced the interaction with the CUL4-DDB1A complex and the ability of these proteins to function in the response of the plant to UV irradiation. The R2 region containing the second WD40 domain with a WDxR motif had an interaction activity with CUL4-DDB1A equal to the intact proteins but only partially rescued the mutant phenotypes (see Supplemental Figure 6 online), suggesting that formation of a complex with CUL4-DDB1A is necessary but not sufficient for CSAat1A and B function in response to UV stress. CSA interacts with CSB in mammalian cells and is required for recruitment of XPA binding protein 2 (XAB2), a nucleosome binding protein (HMGN1), and Transcription factor S-II (TFIIS) to the DNA lesion, while CSB recruits p300 and NER proteins to the UV-stalled RNA polymerase II (RNAPIIo) (Fousteri et al., 2006). Our data suggest that the R2 region of CSAat1A and B is not only required for association with the CUL4-DDB1A complex but may be critical for interaction with Arabidopsis CSB that, in turn, recruits the NER proteins to damaged regions of DNA.
CUL4A and CUL4B partially overlap in their function, but they form individual E3 ligases with DDB1-DDB2 in mammalian cells (Guerrero-Santoro et al., 2008). CSAat1A and B share significant sequence identity and have overlapping subcellular localization and expression patterns (see Supplemental Figures 2 and 4 online), suggesting that they functionally overlap. However, the absence of either of the genes in Arabidopsis results in a mutant that is more sensitive to UV stress than the wild type and overexpression of either in the mutant background of the other does not rescue the UV-sensitive phenotype (Figure 2; see Supplemental Figure 3 online), indicating that they have unique roles in the UV response.
Our data show that CSAat1A and B elute in a 200-kD complex, suggesting that they may predominantly form a heterotetramer in planta and that formation of this heterotetramer is critical for their full functioning in DNA damage repair during UV stress (Figure 9; see Supplemental Figures 6A, 6B, and 7 online). Our data indicate that CSAat1A and B also have self-interaction activity (Figures 9A and 9B). With the loss of either gene in Arabidopsis, the remaining protein can form a homodimer (Figure 9E), which eliminates or reduces the efficiency of functional CUL4-DDB1ACSAat1A or B complex in response to UV stress. It is not known if CSA forms a homodimer in mammalian cells; results from our study may provide insights into how this protein functions in other organisms. We do not know if the csaat1a csaat1b double mutant has a more severe UV-sensitive phenotype than the corresponding single mutants as we were unable to generate the double mutant because both genes are located in the same region of chromosome 1.
The WDxR motif is required and sufficient for the formation of the CSAat1A and B heterotetramer. Mutations in conserved amino acids abolished their interaction, and the R2 domain could form heterotetramers with the other proteins (Figures 9F and 9G). Interestingly, the WDxR motif is also required for interaction with DDB1A-CUL4 and may also interact with unknown targets, such as CSB. Because the R2 peptide interacts with CUL4-DDB1A but only partially rescues the mutant phenotype (see Supplemental Figure 6 online), the R1 and R3 domains must play a role in the interaction with downstream targets. How the interacting surface linked by the four WDxR motifs in the heterotetramer can assemble with both the DDB1A and CSB proteins remains to be determined. However, four CSAat1A and B proteins could provide a more efficient platform to recruit the UV damage repair machinery to the DNA lesion region and for degradation of CSB for post TCR recovery.
Taken together, the results from our study demonstrate that CSAat1A and B predominantly form a heterotetramer in planta and that the formation of this heterotetramer is critical for their full functioning in DNA damage repair. In addition, our study provides insight into how CSA functions in the plant and demonstrates that the plant has a unique and complex CUL4-DDB1ACSA E3 ligase system that functions in the plant’s response to UV stress.
METHODS
Plant Growth and Mutant Screening
All Arabidopsis thaliana wild-type plants used in this study were of the Col-0 ecotype. Arabidopsis seeds were surface sterilized by washing in a 20% sodium hypochlorite solution for 10 min, rinsed five times with sterile water, spread on Murashige and Skoog (MS) medium with 0.3% agar (Sigma-Aldrich), and grown in a growth chamber (Philips TLD 30W/54 YZ30RR25) at 23°C.
Twelve-day-old seedlings were used to screen UV-B–sensitive mutants from a T-DNA insertion pool (pSKI015) in the Col-0 background. Seedlings were treated with UV-B light (UVP CL-1000M 34-0042-01, 302 nm, 130 μmol m−2 s−1) for 30 min and subsequently incubated for 5 d in a growth chamber. At the end of this incubation, candidate mutants were identified and photographed. Assays were repeated three times to confirm the phenotypes of the candidate mutants. The uvs90 mutant was isolated, and the T-DNA insertion localized to the promoter of At1g27840 using thermal asymmetric interlaced-PCR. Three T-DNA left border specific primers (LB1, LB2, and LB3) and one degenerate (AD) primer were used (Liu et al., 1995). The T-DNA insertion site was confirmed using gene-specific primers (uvs90 T-DNAF, uvs90 T-DNAR). All primers used above are listed in Supplemental Table 1 online.
MMS Treatment
Six-day-old seedlings grown on MS medium with 0.3% agar were transferred to Petri dishes containing liquid MS medium supplemented with different concentrations (0, 20, 40, 60, 80, and 100 ppm) of MMS. The plants were incubated in a growth chamber. Photographs were taken, and fresh weight was measured 10 d after transfer to the growth chamber.
Homozygous Mutant Identification
All loss-of-function mutants of CSAat1A and B genes, including csaat1a-1 (salk_024816), csaat1a-2 (Salk_151258), csaat1b-1 (Salk_152568), and csaat1b-2 (salk_144623), were obtained from the ABRC. The homozygous mutants were identified by PCR-based genotyping using gene-specific primers (see Supplemental Table 1 online) and the T-DNA left border primers (LBa1 and LBb1).
The csaat1a ddb1a and csaat1b ddb1a double mutants were obtained by crossing csaat1a-1 and csaat1b-1 to ddb1a (SALK_041255), respectively. From F2 plants, csaat1a and csaat1b homozygotes were identified using the primers described above. The ddb1a homozygotes were identified using DDB1A gene-specific primers (DDB1A T-DNAF and DDB1A T-DNAR) and LBa1 and LBb1. The primers used to identify homozygous mutants are listed in Supplemental Table 1 online.
Subcellular Localization
CSAat1A and B cDNA coding regions were amplified by RT-PCR using the following gene-specific primers: CSAat1A cDNAF and CSAat1A cDNAR; CSAat1B cDNAF and CSAat1B cDNAR (see Supplemental Table 1 online). Products were cloned into the BamHI and KpnI sites of the binary vector pCAMBIA1205-GFP downstream of GFP and confirmed by sequencing. The resulting plasmids were transformed into wild-type plants using Agrobacterium tumefaciens strain GV3101. T2 transgenic plants were used for subcellular localization of CSAat1A and B. Images of 7-d-old T2 transgenic seedlings were taken with a Zeiss LSM510 Meta confocal microscope with GFP excitation at 488 nm.
GUS Staining
The CSAat1A and B promoters (1554 and 1500 bp upstream of the translation start sites, respectively) were amplified from wild-type DNA and cloned into the pCAMBIA1381 vector. The CSAat1A promoter was amplified with the CSAat1A PF and CSAat1A PR primers (see Supplemental Table 1 online) and was cloned into the PstI and EcoRI sites of pCAMBIA1381. The CSAat1B promoter was amplified with the following primers, CSAat1B PF and CSAat1B PR (see Supplemental Table 1 online), and was cloned into the EcoRI and SalI sites. The constructs were introduced into A. tumefaciens strain GV3101 and transformed into wild-type Arabidopsis plants. GUS staining of the T2 transgenic lines was performed as described (Zhao et al., 2007).
Tissue from pCSAat1A:GUS and pCSAat1B:GUS transgenic plants was immersed in reaction buffer containing 100 mM sodium phosphate, pH 7.0, 0.1% Triton X-100, 3 mM X-Gluc, and 8 mM β-mercaptoethanol. Samples were incubated in the dark for 12 h at 37°C, rinsed with sterile water, and then treated with 100% ethanol at 37°C to extract chlorophyll.
Generation of Transgenic Plants
Full-length coding sequences for CSAat1A and B, CSAat1AL206A and BL206A, CSAat1AT208A and BT208A, CSAat1AD212A and BD212A, CSAat1AR216A and BR216A, CSAat1AW218A and BW218A, CSAat1AD219A and BD219A, CSAat1AR221A and BR221A, CSAat1AD212AL206A and BD212AL206A, CSAat1AD212AW218A and BD212AW218A, CSAat1AD212AD219A and BD212AD219A, CSAat1AD212AR221A and BD212AR221A, CSAat1AD212AW218AD219A and BD212AW218AD219A, CSAat1AD212AW218AR221A and BD212AW218AR221A, and CSAat1AD212AW218AD219AR221A and BD212AW218AD219AR221A, and for fragments (R1, R2, and R3) were cloned into the pCAMBIA1307-Flag binary vector upstream of the Flag-tag coding sequence. All primers and plasmid constructs used in these studies are listed in Supplemental Table 2 online. All fragments were cloned into CSAat1A-related plasmids using SacI and BamHI sites and into CSAat1B-related plasmids using SacI and SalI sites.
The resulting constructs were sequenced and introduced into A. tumefaciens strain GV3101 and transformed into Col-0 or csaat1a-1 or csaat1b-1; T3 homozygous lines were used for the analysis.
Y2H Assays
Full-length coding sequences for CSAat1A and B, CSAat1AL206A and BL206A, CSAat1AT208A and BT208A, CSAat1AD212A and BD212A, CSAat1AR216A and BR216A, CSAat1AW218A and BW218A, CSAat1AD219A and BD219A, CSAat1AR221A and BR221A, CSAat1AD212AL206A and BD212AL206A, CSAat1AD212AW218A and BD212AW218A, CSAat1AD212AD219A and BD212AD219A, CSAat1AD212AR221A and BD212AR221A, CSAat1AD212AW218AD219A and BD212AW218AD219A, CSAat1AD212AW218AR221A and BD212AW218AR221A, and CSAat1A D212AW218AD219AR221A and BD212AW218AD219AR221A were amplified using the Flag-tag mutation plasmids as templates and were individually cloned into the pGADT7 vector using the following primers: CSAat1A ADF, CSAat1A ADR; and CSAat1B ADF, CSAat1B ADR (see Supplemental Table 1 online). DDB1A was cloned into pGBKT7 using the primer pairs DDB1A BDF and DDB1A BDR (see Supplemental Table 1 online). Fragments (R1, R2, and R3) were cloned into the pGADT7 vector; primers used for these constructs are listed in Supplemental Table 3 online.
Yeast strain AH109 expressing the CSAat1A/B plasmids (prey) were transformed with pGBKT7-DDB1A (bait). Transformed yeast cells were selected on synthetic complete medium lacking Trp and Leu. Empty prey or bait vector was transformed with CSAat1A and B or DDB1A as negative controls. Interaction was determined on synthetic complete medium lacking Trp, Leu, and His and supplemented with 25 mM 3-amino-1,2,4-triazole (Sigma-Aldrich). All bait and prey proteins were tested for self-activation in the same conditions; none were found to activate the two reporter genes His3 or LacZ.
RT-PCR Analysis
Total RNA from 15-d-old seedlings grown on MS medium was extracted using RNAVzol reagent (Vigorous Biotechnology) and treated with RNase-free DNase I (TAKARA) to remove DNA. Ten micrograms of RNA was used for reverse transcription with M-MLV reverse transcriptase (Promega) according to the manufacturer’s instructions. The cDNAs were then used for PCR amplification.
For testing csaat1a and b mutants and different Flag-tag mutation-containing transgenic plants, the following primers were used: CSAat1A RTF, CSAat1A RTR; and CSAat1B RTF, CSAat1B RTR (see Supplemental Table 1 online).
Primers used for to test for the presence of different fragments of CSAat1A and B (R1, R2, and R3) are listed in Supplemental Table 3 online.
ACTIN (used as an internal control) was amplified using the ACTINF and ACTINR primers (see Supplemental Table 1 online).
Coimmunoprecipitation and Immunoblot Analysis
The coding sequences of CSAat1A and B, CSA1at1A- and B-R1, CSA1at1A- and B-R2, and CSAat1A- and B-R3 were translationally fused downstream of the Myc or upstream of Flag tags and cloned into the pCAMBIA1307 (Lin et al., 2009) vector. All plasmids were purified by CsCl gradient centrifugation, and the coimmunoprecipitation experiments were performed as previously described (Lin et al., 2009).
Transgenic Arabidopsis seedlings were homogenized in extraction buffer consisting of 10 mM Tris-HCl, pH 7.6, 150 mM NaCl, 2 mM EDTA, 0.5% (v/v) Nonidet P-40, and 2× protease inhibitor (Roche). Extracts were centrifuged at 13,000 rpm for 15 min at 4°C; the resulting supernatant was analyzed. Anti-Flag agarose conjugate (Sigma-Aldrich) (20 μL) was incubated with the extract supernatant for 2 h at 4°C. After washing five times in 1 mL of extraction buffer, the immunoprecipitation products were detected via immunoblot analysis. The blots were probed with primary anti-Flag (Sigma-Aldrich), anti-Myc (Sigma-Aldrich), anti-DDB1A (Zhang et al., 2008), or anti-CUL4 (Chen et al., 2006) antibodies, and chemiluminescence signals were detected by autoradiography.
Split YFP
To detect the interaction between CSAat1A and B with DDB1A and CUL4 and between CSAat1A and CSAat1B, full-length coding sequences of CSAat1A and B and DDB1A were individually cloned in frame into pSPYNE-35S/pUC-SYNE or pSPYCE-35S/pUC-SYCE (Walter et al., 2004) to generate pUC-SYNE-CSAat1A, pUC-SYNE-CSAat1B, pUC-SYNE-CUL4, pUC-SYCE-DDB1A, pUC-SYCE-CSAat1A, and pUC-SYCE-CSAat1B. The primers used to construct the plasmids are listed in Supplemental Table 3 online.
For transient expression in onion epidermal cells, live onion epidermal cells were bombarded with 10 μg plasmid DNA using the Biolistic PDS-1000/He Gene Gun System (Bio-Rad). Bombarded epidermal cells were incubated for 12 h at 23°C under continuous white light and then YFP fluorescence was imaged with a Zeiss LSM510 Meta confocal microscope with excitation at 513 nm.
Gel Filtration
Arabidopsis seedlings were homogenized in buffer containing 10 mM Tris-HCl, pH 7.6, 500 mM NaCl, 2 mM EDTA, 0.5% (v/v) Nonidet P-40, and 2× protease inhibitor (Roche). The homogenate was microcentrifuged at 150,000g at 4°C for 60 min, and the supernatant was filtered through a 0.45-μm filter (Millpore) before loading onto a Superdex-200 gel filtration column (GE). The column was equilibrated with homogenization buffer and the proteins were eluted in the same buffer at a flow rate of 0.2 mL/min. All manipulations were performed at 4°C. Fractions of 0.5 mL were collected, each fraction was concentrated, and proteins were separated using SDS-PAGE, followed by immunoblot analysis. The molecular mass standards used for gel filtration size estimation were ferritin (440 kD), aldolase (158 kD), conalbumin (75 kD), and carbonic anhydrase (25 kD).
ELISA Assays
For DNA damage assays, 12-d-old seedlings grown on MS medium were irradiated with five different doses of UV-B light (0, 10, 20, 30, and 40 min, 130 μmol m−2 s−1). The plants were immediately harvested under green light to avoid CPD repair by photolyases. For DNA repair assays, 12-d-old seedlings grown on MS medium were irradiated with UV-B light (130 μmol m−2 s−1) for 40 min. The plants were immediately transferred to continuous white light and harvested under green light after different repair times (0, 4, 8, 12, and 16 h). The DNA was extracted in CTAB buffer with 0.1 mol/L Tris-HCl, pH 8.0, 20 mmol/L EDTA, pH 8.0, 1.4 mol/L NaCl, 2% cetyl trimethyl ammonium bromide (w/v; Amresco), and 1% polyvinyl pyrrolidone (w/v; Amresco). CPDs were determined by ELISA using an anti-TDM-2 antibody (Tanaka et al., 2002) according to the manufacturer’s instructions. The ELISA assay procedure was performed as previously described (Zhao et al., 2007).
DNA Gel Blot and DNA Repair Analysis
Twelve-day-old Arabidopsis seedlings were treated with UV-B light (130 μmol m−2 s−1) for 30 min and subsequently incubated in a growth chamber for 0 h or 16 h. Plants were harvested and immediately frozen in liquid nitrogen and genomic DNA was extracted using CTAB buffer. The pyrimidine dimer content was examined by digesting the DNA with T4 endonuclease V as previously described (Mellon et al., 1987). For DNA gel blot analysis, 10 mg of genomic DNA was digested with different combinations of restriction endonucleases, including BamHІ and SpeI (for At1g29230 probes); EcoRI and SalI (for At2g30360 probes); EcoRI and XhoI (for At3g23000 probes); PstI and XhoI (for At4g14580 probes); and BamHI and XbaI (for At5g10930 probes); and fractionated on an agarose gel (1% [w/v]). The separated DNA was transferred to Hybond N+ membranes (Amersham). The nontranscribed probes were radiolabeled with 32P using the following gene-specific primers: At1g29230N, At2g30360N, At3G23000N, At4g14580N, and At5g10930N (see Supplemental Table 1 online). DNA gel blot hybridization was performed at 65°C for 16 h in hybridization solution (200 mM sodium phosphate buffer, pH 7.2, 1 mM EDTA, pH 8.0, 50% formamide, 10% BSA, and 7% SDS), followed by washing at 65°C sequentially in 2× SSC and 0.5% SDS, 1×SSC and 0.5% SDS, for 15 min each. The membranes were exposed to a phosphor screen (Amersham) for 24 h after which signals were captured with a Typhoon 9410 phosphor imager (Amersham). The nontranscribed probes were washed at 90°C in 0.5× SSC, 1 mM EDTA, pH 8.0, and 0.1% SDS for 30 min. The transcribed probes were also radiolabeled with 32P using the following gene-specific primers: At1g29230T, At2g30360T, At3G23000T, At4g14580T, and At5g10930T (see Supplemental Table 1 online). The hybridization was performed as described above for the nontranscribed probes.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CSAat1A (At1g27840, NM_102549), CSAat1B (At1g19750, NM_101831), DDB1A (At4g05420, NM_116781), and CUL4 (At5g46210, NM_123990). The accession numbers of the T-DNA insertion mutants are SALK_024816 (csaat1a-1), SALK_151258 (csaat1a-2), SALK_152568 (csaat1b-1), and SALK_144623 (csaat1b-2).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. uvs90 Gene Cloning.
Supplemental Figure 2. Sequence Alignment of CSA, CSAat1A, and CSAat1B.
Supplemental Figure 3. csaat1b Knockout Mutants Are Sensitive to UV-B and MMS.
Supplemental Figure 4. Tissue-Specific Expression and Subcellular Localization of CSAat1A and B.
Supplemental Figure 5. Expression of CSAat1A and B Point Mutations in Transgenic Arabidopsis Plants.
Supplemental Figure 6. Expression of the R2 Region of CSAat1A or B in csaat1a or b Mutants Partially Rescues Their UV-B– and MMS-Sensitive Phenotypes.
Supplemental Figure 7. Mutations in the WDxR Motif of CSAat1A and B Impair UV-B Tolerance in Arabidopsis.
Supplemental Figure 8. Mutations in the WDxR Motif Impair MMS Tolerance in Arabidopsis.
Supplemental Figure 9. Fresh Weight of Transgenic Plants Expressing Different Point Mutations in the WDxR Motif of CSAat1A or B after MMS Treatment.
Supplemental Figure 10. CSAat1AD212AW218AD219AR221A Does Not Interact with CSAat1A or CSAat1B.
Supplemental Table 1. Primers Used for RT-PCR and Homozygous Mutant Identification.
Supplemental Table 2. Primers Used for Plasmid Constructions.
Supplemental Table 3. Primers Used for Yeast Two-Hybrid Assay.
Supplementary Material
Acknowledgments
We thank Feng Shao for critical reading of the manuscript and stimulating discussions, Shidi Li for excellent technical assistance, and the ABRC (Ohio State University) for the T-DNA insertion lines. This work was supported by National Basic Research Program of China Grant 2006CB100100, National High Technology Research and Development Program of China 863 Grant 2003AA210100 to Y.G., and by U.S. Department of Energy/Energy Biosciences Grant DE-FG02-04ER15616 to K.S.S.
References
- Ahmad M., Jarillo J.A., Klimczak L.J., Landry L.G., Peng T., Last R.L., Cashmore A.R. (1997). An enzyme similar to animal type II photolyases mediates photoreactivation in Arabidopsis. Plant Cell 9: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Khateeb W.M., Schroeder D.F. (2009). Overexpression of Arabidopsis damaged DNA binding protein 1A (DDB1A) enhances UV tolerance. Plant Mol. Biol. 70: 371–383 [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bashandy T., Taconnat L., Renou J.P., Meyer Y., Reichheld J.P. (2009). Accumulation of flavonoids in an ntra ntrb mutant leads to tolerance to UV-C. Mol. Plant 2: 249–258 [DOI] [PubMed] [Google Scholar]
- Bernhardt A., Lechner E., Hano P., Schade V., Dieterle M., Anders M., Dubin M.J., Benvenuto G., Bowler C., Genschik P., Hellmann H. (2006). CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J. 47: 591–603 [DOI] [PubMed] [Google Scholar]
- Bieza K., Lois R. (2001). An Arabidopsis mutant tolerant to lethal ultraviolet-B levels shows constitutively elevated accumulation of flavonoids and other phenolics. Plant Physiol. 126: 1105–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Shen Y., Tang X., Yu L., Wang J., Guo L., Zhang Y., Zhang H., Feng S., Strickland E., Zheng N., Deng X.W. (2006). Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri M., Vermeulen W., van Zeeland A.A., Mullenders L.H. (2006). Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell 23: 471–482 [DOI] [PubMed] [Google Scholar]
- Frohnmeyer H., Staiger D. (2003). Ultraviolet-B radiation-mediated responses in plants. Balancing damage and protection. Plant Physiol. 133: 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego F., Fleck O., Li A., Wyrzykowska J., Tinland B. (2000). AtRAD1, a plant homologue of human and yeast nucleotide excision repair endonucleases, is involved in dark repair of UV damages and recombination. Plant J. 21: 507–518 [DOI] [PubMed] [Google Scholar]
- Gao C., Zhang L., Wen F., Xing D. (2008). Sorting out the role of reactive oxygen species during plant programmed cell death induced by ultraviolet-C overexposure. Plant Signal. Behav. 3: 197–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R., Kuraoka I., Chevallier O., Gaye N., Magnaldo T., Tanaka K., Kisselev A.F., Harel-Bellan A., Nakatani Y. (2006). CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 20: 1429–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Santoro J., Kapetanaki M.G., Hsieh C.L., Gorbachinsky I., Levine A.S., Rapic-Otrin V. (2008). The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res. 68: 5014–5022 [DOI] [PubMed] [Google Scholar]
- Hidema J., Taguchi T., Ono T., Teranishi M., Yamamoto K., Kumagai T. (2007). Increase in CPD photolyase activity functions effectively to prevent growth inhibition caused by UVB radiation. Plant J. 50: 70–79 [DOI] [PubMed] [Google Scholar]
- Jiang C.Z., Yee J., Mitchell D.L., Britt A.B. (1997). Photorepair mutants of Arabidopsis. Proc. Natl. Acad. Sci. USA 94: 7441–7445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., Tonelli C., Weisshaar B., Martin C. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19: 6150–6161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D.J., Lim J.E., Landry L.G., Last R.L. (2002). Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 130: 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga A., Ishibashi T., Kimura S., Uchiyama Y., Sakaguchi K. (2006). Characterization of T-DNA insertion mutants and RNAi silenced plants of Arabidopsis thaliana UV-damaged DNA binding protein 2 (AtUV-DDB2). Plant Mol. Biol. 61: 227–240 [DOI] [PubMed] [Google Scholar]
- Landry L.G., Chapple C.C., Last R.L. (1995). Arabidopsis mutants lacking phenolic sunscreens exhibit enhanced ultraviolet-B injury and oxidative damage. Plant Physiol. 109: 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Terzaghi W., Gusmaroli G., Charron J.B., Yoon H.J., Chen H., He Y.J., Xiong Y., Deng X.W. (2008). Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20: 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Ou-Lee T.M., Raba R., Amundson R.G., Last R.L. (1993). Arabidopsis flavonoid mutants are hypersensitive to UV-B irradiation. Plant Cell 5: 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Flury S., Kalck V., Hohn B., Molinier J. (2006). CENTRIN2 interacts with the Arabidopsis homolog of the human XPC protein (AtRAD4) and contributes to efficient synthesis-dependent repair of bulky DNA lesions. Plant Mol. Biol. 61: 345–356 [DOI] [PubMed] [Google Scholar]
- Lin H., Yang Y., Quan R., Mendoza I., Wu Y., Du W., Zhao S., Schumaker K., Pardo J., Guo Y. (2009). Phosphorylation of SOS3-LIKE CALCIUM BINDING PROTEIN8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 21: 1607–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Lee S., Zhang J., Peters S.B., Hannah J., Zhang Y., Yin Y., Koff A., Ma L., Zhou P. (2009). CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol. Cell 34: 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.G., Mitsukawa N., Oosumi T., Whittier R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Liu Z., Hong S.W., Escobar M., Vierling E., Mitchell D.L., Mount D.W., Hall J.D. (2003). Arabidopsis UVH6, a homolog of human XPD and yeast RAD3 DNA repair genes, functions in DNA repair and is essential for plant growth. Plant Physiol. 132: 1405–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Hossain G.S., Islas-Osuna M.A., Mitchell D.L., Mount D.W. (2000). Repair of UV damage in plants by nucleotide excision repair: Arabidopsis UVH1 DNA repair gene is a homolog of Saccharomyces cerevisiae Rad1. Plant J. 21: 519–528 [DOI] [PubMed] [Google Scholar]
- Mackerness A.H.S., John C.F., Jordan B., Thomas B. (2001). Early signaling components in ultraviolet-B responses: Distinct roles for different reactive oxygen species and nitric oxide. FEBS Lett. 489: 237–242 [DOI] [PubMed] [Google Scholar]
- Mellon I., Spivak G., Hanawalt P.C. (1987). Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51: 241–249 [DOI] [PubMed] [Google Scholar]
- Molinier J., Lechner E., Dumbliauskas E., Genschik P. (2008). Regulation and role of Arabidopsis CUL4-DDB1A-DDB2 in maintaining genome integrity upon UV stress. PLoS Genet. 4: e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima S., Sugiyama M., Iwai S., Hitomi K., Otoshi E., Kim S.T., Jiang C.Z., Todo T., Britt A.B., Yamamoto K. (1998). Cloning and characterization of a gene (UVR3) required for photorepair of 6-4 photoproducts in Arabidopsis thaliana. Nucleic Acids Res. 26: 638–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell B.C., Harper J.W. (2007). Ubiquitin proteasome system (UPS): What can chromatin do for you? Curr. Opin. Cell Biol. 19: 206–214 [DOI] [PubMed] [Google Scholar]
- Oravecz A., Baumann A., Mate Z., Brzezinska A., Molinier J., Oakeley E.J., Adam E., Schafer E., Nagy F., Ulm R. (2006). CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto A., Lan V.T., Hase Y., Shikazono N., Matsunaga T., Tanaka A. (2003). Disruption of the AtREV3 gene causes hypersensitivity to ultraviolet B light and gamma-rays in Arabidopsis: Implication of the presence of a translesion synthesis mechanism in plants. Plant Cell 15: 2042–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H., Avivi-Ragolsky N., Levy A.A. (2006). Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics 173: 985–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R.P., Häder D.P. (2002). UV-induced DNA damage and repair: A review. Photochem. Photobiol. Sci. 1: 225–236 [DOI] [PubMed] [Google Scholar]
- Sugasawa K., Okuda Y., Saijo M., Nishi R., Matsuda N., Chu G., Mori T., Iwai S., Tanaka K., Hanaoka F. (2005). UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121: 387–400 [DOI] [PubMed] [Google Scholar]
- Svejstrup J.Q. (2002). Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell Biol. 3: 21–29 [DOI] [PubMed] [Google Scholar]
- Tanaka A., Sakamoto A., Ishigaki Y., Nikaido O., Sun G., Hase Y., Shikazono N., Tano S., Watanabe H. (2002). An ultraviolet-B-resistant mutant with enhanced DNA repair in Arabidopsis. Plant Physiol. 129: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm R., Ichimura K., Mizoguchi T., Peck S.C., Zhu T., Wang X., Shinozaki K., Paszkowski J. (2001). Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 21: 6483–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonarx E.J., Tabone E.K., Osmond M.J., Anderson H.J., Kunz B.A. (2006). Arabidopsis homologue of human transcription factor IIH/ nucleotide excision repair factor p44 can function in transcription and DNA repair and interacts with AtXPD. Plant J. 46: 512–521 [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schutze K., Batistic O., Weckermann K., Nake C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang C., Liu Z. (2006). Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 8: 350–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Liu J., Feng Y., Niu X., Giovannoni J., Liu Y. (2008). Altered plastid levels and potential for improved fruit nutrient content by downregulation of the tomato DDB1-interacting protein CUL4. Plant J. 55: 89–103 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng S., Chen F., Chen H., Wang J., McCall C., Xiong Y., Deng X.W. (2008). Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell 20: 1437–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhang W., Zhao Y., Gong X., Guo L., Zhu G., Wang X., Gong Z., Schumaker K.S., Guo Y. (2007). SAD2, an importin-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 19: 3805–3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.