Pc-G proteins are key regulators of plant cell fate and development. This study examines the Pc-G interacting protein BLISTER, which has Pc-G related and unrelated functions and might link Pc-G proteins to specific developmental processes.
Abstract
Polycomb-group (Pc-G) proteins are important regulators of many developmental processes in plants and animals and repress gene expression by imparting histone H3 lysine 27 trimethylation (H3K27me3). Here, we present the identification of the novel, plant-specific Arabidopsis thaliana protein BLISTER (BLI), which interacts with the Pc-G histone methyltransferase CURLY LEAF (CLF). We map the interaction of BLI with CLF to a predicted coiled-coil domain in BLI that shares similarity with STRUCTURAL MAINTENANCE OF CHROMOSOMES proteins. BLI colocalizes with CLF in the nucleus, shows an overlapping expression pattern with CLF throughout plant development that is strongest in dividing cells, and represses a subset of Pc-G target genes. Loss of BLI results in a pleiotropic developmental mutant phenotype, indicating that BLI prevents premature differentiation. Furthermore, bli mutants exhibit severe epidermal defects, including loss of cell adhesion, outgrowth of cells, and increased cotyledon cell size. As these phenotypes have not been observed in Pc-G mutants, we propose that BLI has functions related to Pc-G proteins but can also act independently in Arabidopsis development.
INTRODUCTION
In plants and animals, Polycomb-group (Pc-G) proteins and the antagonistically acting Trithorax-group proteins are essential regulators of developmental processes and the control of cell fate. These proteins are epigenetic regulators (i.e., they confer mitotically stable gene expression patterns that are usually reset during meiosis) (Schuettengruber et al., 2007; Schatlowski et al., 2008).
In animals, various Pc-G complexes have been purified, and their biochemical function has been partially resolved. Drosophila melanogaster Polycomb repressive complex 2 (PRC2) consists of the four core members ENHANCER OF ZESTE [E(z)], SUPPRESSOR OF ZESTE 12 [Su(z)12], EXTRA SEX COMBS (ESC), and P55 (reviewed in Schuettengruber and Cavalli, 2009) and possesses histone methyltransferase activity toward Lys-9 and -27 of histone H3 (Cao et al., 2002; Czermin et al., 2002; Kuzmichev et al., 2002; Muller et al., 2002). The 1-MD PRC1 complex is recruited to methylated Lys-27 and/or Lys-9 via its chromodomain protein POLYCOMB and thought to confer long-term silencing by nucleosome compaction, Histone H2A monoubiquitination, and inhibition of SWI/SNF-mediated chromatin remodeling (Saurin et al., 2001; de Napoles et al., 2004; Francis et al., 2004; Wang et al., 2004a). A third complex, PhoRC, has been isolated from Drosophila and contains the sequence-specific DNA binding factor PLEIOHOMEOTIC (Klymenko et al., 2006). Many interactions of PRCs with additional proteins have been identified in animals that in some cases modify the enzymatic activity (reviewed in Schuettengruber and Cavalli, 2009). In addition, a recent study also showed a role for the PRC2 member EMBRYONIC ECTODERM DEVELOPMENT (the mammalian homolog of ESC) in binding of trimethylated Histone H3 Lysine 27 (H3K27me3), possibly allowing the propagation of the epigenetic mark through mitosis without a requirement for PRC1 (Margueron et al., 2009). Although DNA elements (the so-called Polycomb response elements) that are required for recruitment of Pc-G proteins and Polycomb silencing have been identified both in Drosophila and recently also in mammals, it is still unclear how recruitment is achieved at the molecular level (Simon et al., 1993; Sing et al., 2009). In plants, no such DNA elements have been identified so far. However, all members of the PRC2 and their function in H3K27 methylation are conserved in plants and have been initially identified in forward genetic screens studying diverse developmental processes. Three different PRC2-like complexes have been proposed, based on the common phenotypes observed in the loss-of-function alleles of plant PRC2 members: the FERTILIZATION INDEPENDENT SEED (FIS) proteins are involved in the suppression of seed development without fertilization, the EMBRYONIC FLOWER (EMF) complex prevents precocious flowering, and the VERNALIZATION (VRN) complex is involved in the vernalization response (the acquisition of the competence to flower after winter) (Chanvivattana et al., 2004; reviewed in Schatlowski et al., 2008). Only the homolog of ESC, FERTILIZATION INDEPENDENT ENDOSPERM (FIE), is present as a single-copy gene in the Arabidopsis thaliana genome and participates in all three PRC2 complexes (Ohad et al., 1999; Kinoshita et al., 2001; Wood et al., 2006). The Drosophila PRC2 members E(z) and Su(z)12 are represented by small three-member gene families in Arabidopsis [CURLY LEAF (CLF), SWINGER (SWN), and MEDEA for E(z) and FIS2, VRN2, and EMF2 for Su(z)12]. Although they were mostly identified as specific regulators of certain developmental processes, they share redundant functions (Chanvivattana et al., 2004; Schubert et al., 2006; Wang et al., 2006). A biochemical complex purification has so far only been successful for the VRN complex, which confirmed previous genetic and biochemical interactions (Wood et al., 2006; De Lucia et al., 2008). This complex also contains members of the plant-specific VERNALIZATION-LIKE family and may be similar to Drosophila Polycomb-like-PRC2, which is required for high levels of H3K27me3 (Nekrasov et al., 2007; De Lucia et al., 2008).
Although PRC2 and its function are largely conserved in plants, much less is known how the PRC2 mark H3K27me3 results in stable gene silencing. Most of the PRC1 proteins are not conserved in plants, and only the Arabidopsis homologs RING1a and RING1b of the Drosophila PRC1 protein RING1 have been linked to Pc-G silencing (Xu and Shen, 2008). However, in diverse genetic screens that revealed PRC2 members, additional plant-specific proteins with PRC1-related functions have been identified. These include the chromodomain protein terminal FLOWER2/LIKE HETEROCHROMATIN PROTEIN1 (TFL2/LHP1), EMBRYONIC FLOWER1, and VERNALIZATION1 (Larsson et al., 1998; Aubert et al., 2001; Gaudin et al., 2001; Levy et al., 2002; Calonje et al., 2008). Although the initial genetic screens indicated a relatively specific developmental function for PRC2 members, whole-genome analyses of H3K27me3 presence revealed several thousand target genes (Turck et al., 2007; Zhang et al., 2007; Oh et al., 2008). Many of these are involved in other developmental processes than flowering time control, seed and floral development, and numerous target genes control abiotic and biotic responses. Still many factors in Pc-G regulation await discovery, as it is unclear how specificity for certain target genes is achieved. In addition, how Pc-G proteins are recruited to their target genes is completely unknown, especially in plants. It also can be envisaged that specificity factors link Pc-G proteins to control certain developmental programs like flowering induction and flower development, and these factors will likely be plant specific.
Here, we present the isolation of the plant-specific CURLY LEAF (CLF) interacting protein BLISTER (BLI). BLI interacts via its highly conserved coiled-coil domain with the CXC domain of CLF. BLI is required for normal seed, leaf, and flower development and controls cotyledon and leaf patterning by inhibiting premature differentiation. Flower-specific Pc-G target genes are elevated in a bli mutant, but transcript levels of the floral repressor FLOWERING LOCUS C (FLC) are reduced. As bli mutants display phenotypes that have not been reported for Pc-G mutants, but expression of several Pc-G target genes is controlled by BLI, we propose that BLI conducts Pc-G–related and independent functions and may link the Pc-G machinery to specific developmental processes.
RESULTS
BLI Interacts with CLF
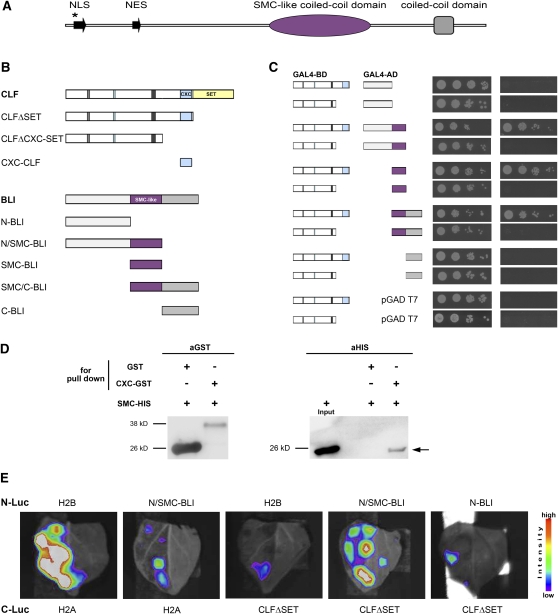
To identify Arabidopsis proteins that interact with plant Pc-G proteins, we performed a yeast two-hybrid screen with a CLF clone lacking the SET (the histone methyltransferase) domain as bait (i.e., fused to the GAL4-DNA binding domain [GAL4-BD]). We focused on putative interaction partners that were found at least twice in the screen or were known to be involved in chromatin regulation. The validity of the screen was confirmed by the identification of the known PRC2 members EMF2 and VRN2 (Chanvivattana et al., 2004). In addition, we identified clones corresponding to three closely related homologs of NUCLEOSOME ASSEMBLY PROTEIN1, to Golgin-candidate 5, to the Chromatin assembly factor 1 subunit FASCIATA1 (FAS1), and to two unknown proteins (see Supplemental Table 1 online). We focused on the genes for which no mutant phenotype had been previously described and analyzed T-DNA insertion lines for all genes (see Supplemental Table 1 online). Only an insertion in at3g23980, which we named BLI, caused a developmental phenotype (see below); therefore, BLI was chosen for further analyses. The BLI protein is predicted to contain 714 amino acids and is unique in the Arabidopsis genome. However, its C-terminal half has high sequence similarity to proteins from diverse plant species, including the moss Physcomitrella patens, the monocot rice (Oryza sativa), and the dicot grapevine (Vitis vinifera), but has no apparent homologs in animals (see Supplemental Figure 1 online). The region containing amino acids 354 to 527 shares similarity with a part of bacterial STRUCTURAL MAINTENANCE OF CHROMOSOMES proteins, which are involved in chromosome segregation, and is predicted to contain a coiled-coil domain (Rose et al., 2004). In addition, nuclear localization and nuclear export signals and a cyclin binding motif are predicted in the BLI N-terminal protein sequence (Figure 1).
Figure 1.
Protein Structure of BLI and Interaction with CLF.
(A) Protein structure of BLI. NLS, nuclear localization signal; NES, nuclear export signal; SMC, region of similarity to STRUCTURAL MAINTENANCE OF CHROMOSOMES. The asterisk marks the position of a putative cyclin binding motif (RRKL).
(B) Schemes display full-length protein structures of CLF and BLI as well as truncated versions used for interaction assays in (C) to (E).
(C) Truncated versions of CLF and BLI were used for interaction studies using the GAL4 based yeast two-hybrid system. Growth on nonselective media was unaffected (left), but growth of serial dilutions on selective media (-LWHA, right) was only detected when CLF fusions including the CXC domain were cotransformed with BLI truncations containing the SMC-like domain. L, Leu; W, Trp; H, His; A, adenine.
(D) In vitro pull-down assay: GST and CXC-GST protein samples were incubated with SMC-His6 extracts. CXC-GST fusion protein (CLF) but not GST coprecipitates with SMC-His6-fusion protein (BLI) (indicated by the arrow).
(E) Transient split-luciferase assay in N. benthamiana: Agrobacteria containing inducible i35Spro fusion proteins of the N-terminal half of luciferase and BLI (N/SMC or N) or H2B were coinfiltrated with inducible i35Spro fusion proteins of the C-terminal half of luciferase and CLFΔSET or H2A. Expression was induced with β-estradiol for 3 h, and luciferase substrate was infiltrated in the left half of leaves prior monitoring.
To verify the interaction and map the relevant domains of BLI and CLF, we tested several truncations of CLF (Chanvivattana et al., 2004) as bait and BLI as prey (i.e., as fusions to the GAL4-BD and GAL4-activation domain [GAL4-AD], respectively) (Figure 1). Yeast carrying CLF truncations fused to GAL4-BD and the GAL4-AD vectors without insert did not grow on selective media (i.e., lacking Trp, Leu, His, and adenine). However, when the CLF truncations were combined with partial clones of BLI fused to GAL4-AD, growth on selective media was observed with CLF versions carrying the highly conserved CXC (Pre-SET) domain. For BLI GAL4-AD fusions, growth on selective media and therefore an interaction with CLF was only obtained with the BLI truncations harboring the highly conserved SMC-like coiled-coil domain (Figure 1C). To find additional evidence for the interaction of BLI and CLF, we performed in vitro pull-down analyses. The SMC domain of BLI was fused to a His6-tag and the CXC domain to glutathione S-transferase (GST-CXC), and both fusions were expressed in Escherichia coli. Subsequently, cultures were mixed and GST fusion proteins and associated proteins were purified. Whereas no copurification of the SMC domain and GST were observed, the SMC domain specifically copurified with GST-CXC (Figure 1D). Thus, CLF and BLI interact in yeast and in vitro via their highly conserved CXC and coiled-coil SMC-like domains, respectively.
To confirm the interaction in planta, we made use of the split-luciferase complementation assay (Fujikawa and Kato, 2007). We transferred the N- and C-terminal parts of Renilla Luciferase (N-Luc or C-Luc, respectively) in a β-estradiol vector system to limit overexpression artifacts and unspecific interactions (Zuo et al., 2000; Curtis and Grossniklaus, 2003) and constructed various fusions. After Agrobacterium tumefaciens–mediated transient transformation of Nicotiana benthamiana leaves, β-estradiol was added to induce expression followed by infiltration of the luciferase substrate ViviRen. Luminescence and, therefore, interaction was detected for H2B and H2A, which was previously shown (Fujikawa and Kato, 2007), confirming the functionality of the system to detect interactions. Similarly, luminescence was detected in a combination of BLI and CLF both lacking their C-terminal parts. The interaction was specific since strongly reduced luminescence was revealed when the BLI fusion was combined with a H2A fusion or when the CLF fusion was coexpressed with a H2B fusion. In addition, a BLI fusion lacking both the C-terminal and the SMC domain did not show interaction with CLF (Figure 1E). Thus, also in planta BLI and CLF show a specific interaction that requires the SMC domain.
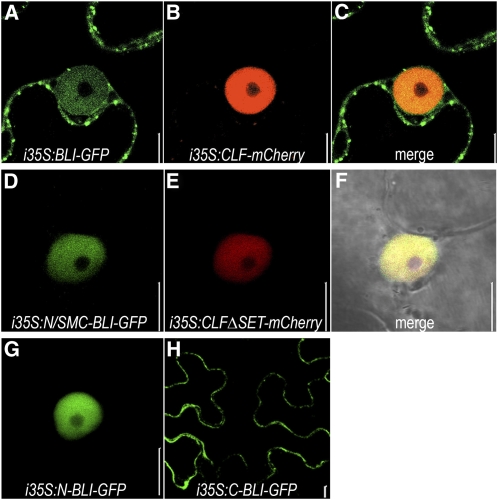
To verify that the truncations all localized to overlapping compartments, they were fused to green fluorescent protein (GFP) or mCherry and transiently expressed using the β-estradiol–inducible system (Bleckmann et al., 2010). All fusions showed nuclear localization, suggesting that an interaction is not impeded by different subcellular distribution (Figure 2). Interestingly, the full-length BLI-GFP fusion also localized to cytoplasmatic vesicles. Only weak cytoplasmic localization and no vesicles were observed in fusion proteins lacking the C-terminal part. In addition, a fusion containing just the C-terminal region preferentially localized to the cytoplasm but did not reveal vesicle formation (Figure 2). Thus, BLI may shuttle between the nucleus and the cytoplasm, which indicates functions for BLI both in- and outside the nucleus. Our data indicate that cytoplasmic localization and vesicle formation depends on the C-terminal region of BLI but may not require the predicted nuclear export signal.
Figure 2.
Subcellular Localization of Full Length and Truncated Versions of BLI and CLF.
(A) to (C) Transient expression assays in N. benthamiana using β-estradiol–inducible i35Spro:BLI-GFP (A) and i35Spro:CLF-mCherry fusions (B). (C) is an overlay of (A) and (B).
(D) to (F) i35Spro fusion proteins containing only the N/SMC domain of BLI (D) and CLFΔSET (E) colocalize in the nucleus (F) (see Figure 1B).
(G) and (H) Subcellular localization of i35SproGFP fusion proteins containing the N terminus (G) or the C terminus (H) of BLI, respectively (see Figure 1B).
Bars = 10 μm.
Expression of BLI in Arabidopsis Development
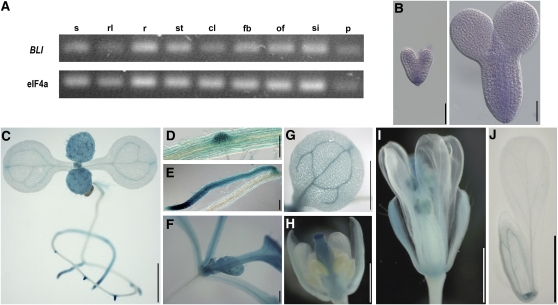
According to publically available microarray data sets from a variety of different Arabidopsis tissues, BLI is ubiquitously expressed with a peak in mature pollen (Schmid et al., 2005). We analyzed BLI expression by RT-PCR in different tissues and did not observe any obvious differences in expression level, even in mature pollen (Figure 3A). To determine the spatial expression pattern of BLI in more detail, we cloned a genomic fragment of BLI, including the promoter region as translational fusion to the uidA-Gene (BLIpro:BLI-GUS). In 2-week old seedlings, expression was observed in the shoot apical meristem, in true leaves, and in the vasculature of cotyledons (Figure 3). In the root system, emerging lateral roots and the tip of the primary root showed strong β-glucuronidase (GUS) expression that declined in the differentiation zone of the root. In flowers, GUS expression was detected in sepals and in carpels (Figure 3). Using whole-mount in situ hybridization on isolated embryos, BLI expression was determined to be predominantly in the basal part and the vasculature of heart shape and torpedo stage embryos (Figure 3B). Thus, both in embryo and vegetative development, BLI expression was strongest in dividing tissue and in the vasculature.
Figure 3.
BLI Expression Pattern.
(A) Detection of BLI transcript by RT-PCR in all tissues analyzed. s, seedlings; rl, rosette leaves; r, root; st, stem; cl, cauline leaves; fb, flower bud; of, open flower; si, silique; p, mature pollen. eIF4a was used as control.
(B) Whole-mount mRNA in situ hybridization analysis of embryos in heart stage (left) and torpedo stage (right).
(C) to (J) BLIpro:BLI-GUS fusion protein in bli-1 mutant background: expression in 14-d-old seedlings ([C] to [G]), emerging side roots ([D] and [E]), and close-up on region containing the shoot apical meristem (F) and cotyledon (G). Flowers ([H] and [I]) and sepal and petal (J) from 25-d-old plant. Bar = 50 μm in (B), 1000 μm in (C) and (G) to (J), and 200 μm in (D) to (F). Note that staining of anthers (I) cannot be distinguished between BLIpro:BLI-GUS and LAT52pro:GUS, which is present on the T-DNA inserted in BLI.
Identification of bli Mutants
The BLI gene is predicted to consist of 13 exons (see The Arabidopsis Information Resource), supported by a full-length cDNA from the RIKEN collection (Seki et al., 2002). We analyzed several T-DNA insertion lines mapping to the transcribed region of BLI to study the role of BLI in plant development. Only insertions in introns or the 3′ untranslated region are available. In bli-1 (N805222), which carries an insertion in intron 6, the transcript 5′ of the insertion is severely reduced (<10%). Although we detected a truncated transcript 3′ of the insertion, full-length BLI cDNA was not detectable (see Supplemental Figure 2 online; Figure 4). Importantly, the insertion in bli-1 disrupts the highly conserved SMC-like domain, which is required for the CLF interaction, suggesting that this allele is likely to be null or severe loss of function. In bli-2 (N505565, intron 4), we only observed a slight reduction (up to 50%) compared with the wild-type transcript level. Unlike bli-1 plants, the phenotype of bli-2 mutants did not differ from the wild type under our growth conditions; thus, we conclude that bli-2 is not a null allele (Figure 4B). The other available T-DNA insertions at BLI had no detectable effect on BLI transcript levels (see Supplemental Figure 2 online).
Figure 4.
Analysis of bli Mutant Alleles.
(A) Gene structure of BLI. Purple boxes indicate exons, and arrowheads mark the position of T-DNA insertions of bli-1 (N805222) and bli-2 (N505565).
(B) Detection of cDNA corresponding to the full-length coding sequence of BLI by RT-PCR. eIF4a was taken as control.
(C) Four-week-old wild-type plant (left) compared with bli-1 (right) and bli-1 complemented by BLIpro:BLI–GUS (middle).
(D) Three-week-old plants: wild type, left; bli-1, right. Arrows point at leaves taken for close-up in (E).
(E) Size difference of true leaf (wild type, left; bli-1, right). Bars = 1 cm.
(F) and (G) Wild-type inflorescence (F) and bli-1 inflorescence (G) containing flowers with strongly bent and narrow sepals. Bars = 500 μM.
(H) Size and seed set of wild-type (left) and bli-1 opened siliques (middle and right); arrowheads point at seeds with altered shape, and arrow points at one example of an unfertilized ovule. Bar = 500 μm.
(I) and (J) Twenty-day-old plants: the wild type (I) and variable bli-1 mutant seedling phenotype (J). Bars = 1 cm.
(K) and (L) Delayed germination of bli-1 seeds (L) compared with Col-0 (K) 4 d after imbibition.
(M) to (O) Comparison of seed shapes of the wild type (M), bli-1 (N), and bli-1 BLIpro:BLI-GUS (O). Bars = 200 μM.
Plants in (C) and (F) to (L) were grown under 16-h-light/8-h-dark cycles at 20°C, and those in (D) and (E) were grown under 8-h-light/16-h-dark cycles at 20°C.
[See online article for color version of this figure.]
Loss of BLI1 Impairs Arabidopsis Development
The bli-1 mutant displayed a pleiotropic phenotype with seed, leaf, and flower development affected and it generally grew slower than the wild type (Figure 4). Only 12% bli-1 mutants were recovered in the progeny of bli-1/BLI plants (in contrast with the expected 25%) (n = 208 for phenotypic bli-1 mutants, and n = 1512 for phenotypic wild-type seedlings; P ≤ 0.001). We also observed reduced transmission through the male germline in reciprocal crosses of bli-1/BLI and BLI/BLI (see Supplemental Table 2 online). bli-1 mutant seeds were shrivelled and showed delayed germination (after 2 d imbibitions, only 21% of bli-1 seeds [n = 405] had germinated compared with 92% of the wild type [n = 302]) (Figure 4).
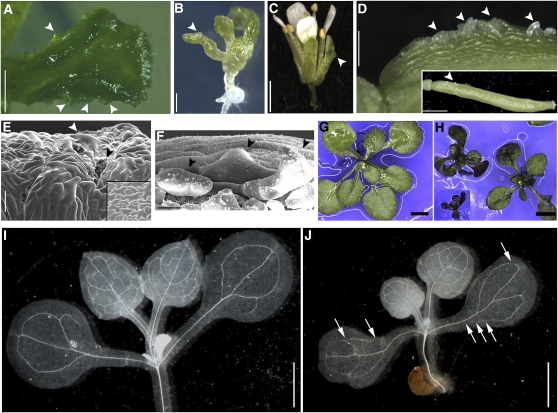
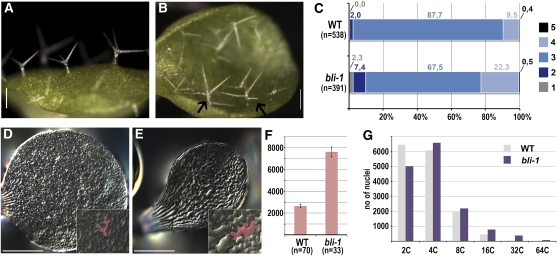
After germination, the phenotypic severity of bli-1 mutants varied strongly. Typically, mutants were smaller than the wild type, and nearly infertile, producing very few seed (Figure 4). As only one bli mutant allele could be identified that showed a strong reduction of BLI transcript and a developmental phenotype, it was important to complement the mutant phenotype. Therefore, the BLIpro:BLI-GUS construct was introduced into bli-1 mutants and was observed to fully complement the mutant phenotype (Figure 4), confirming that a reduction in at3g23980 transcript is responsible for the bli-1 phenotype. Cotyledon, leaf, and flower defects of bli-1 were analyzed in more detail by light and electron microscopy. Flowers appeared not properly closed in the bli-1 mutant due to smaller sepals, which revealed the underlying petals (Figure 4). Compared with the wild type, the cotyledons and leaves of bli-1 were narrow and small and frequently exhibited outgrowths of cells from the epidermis, resembling blisters (Figures 4 to 6). In general, the epidermis of both leaves and sepals appeared abnormal with irregular cell size and shape and loss of cell adhesion or cell death (Figure 5). To find further evidence for defects of the epidermis and the cuticle, we stained seedlings with the hydrophobic dye toluidine blue (TB). TB is repelled by an intact epidermis; thus, it only stains organs of mutants with defective epidermis (Tanaka et al., 2004). Indeed, whereas wild-type seedlings showed no purple TB staining, bli-1 mutants displayed strong staining of leaves and cotyledons (Figures 5G and 5H). As BLI was strongly expressed in the vasculature of cotyledons (Figure 3), we analyzed the vascular pattern of bli-1 cotyledons and leaves by chloral hydrate clearing. Wild-type cotyledons show a simple, interconnected vascular system. In bli-1, however, we frequently observed nonconnected vascular strands (Figures 5I and 5J). We also realized that the size of cotyledon epidermal cells was greatly increased (Figures 6D to 6F). As the cotyledons of bli-1 mutants were smaller than those of the wild type, the number of at least the epidermal cells is strongly reduced in bli-1. In many cases, cell size is positively correlated with the level of nuclear ploidy resulting from successive rounds of endoreduplication (reviewed in Sugimoto-Shirasu and Roberts, 2003). To analyze whether similar correlations are observed in bli-1 mutants, we measured ploidy in wild-type and bli-1 seedlings and found a marked increase in cells with higher ploidy levels (especially of nuclei with a DNA content of 32C and 64C) (Figure 6G). In addition, trichome branching number is often positively associated with higher ploidy levels (reviewed in Ishida et al., 2008). Although overall trichome numbers are decreased in bli-1 mutants, there was also a strong reduction in the proportion of three-branched trichomes (the most common form in the wild type) in the first two leaves. A larger fraction of bli-1 mutant trichomes was four-branched, although the number of trichomes with less than three branches was also increased (Figures 6A to 6C).
Figure 5.
Absence of BLI Causes Severe Cotyledon and Leaf Defects.
(A) to (D) Epidermal defects in bli-1. Outgrowing cells (arrowheads) on a true leaf (A), on a cotyledon of a seedling (B) (bar = 250 μm in [A] and 500 μm in [B]), on a sepal ([C]; bar = 1 mm), and silique ([D]; bar = 200 μm). Inset displays the complete silique (bar = 1 mm).
(E) and (F) Scanning electron micrographs of bli-1 and Col-0 (inset) cotyledon adaxial surface (E) and close-up on misshapen cells on a silique of bli-1 (F). White arrowhead in (E) marks enlarged cell, and black arrow depicts a hole in the epidermis. Arrowheads in (F) mark outgrowing cells. Bar = 100 μm in (E) and 30 μM in (F).
(G) and (H) TB staining of Col-0 (G) and bli-1 seedlings (H). Bars = 0.2 cm.
(I) and (J) Clearing of seedlings reveals vasculature defects of bli-1 cotyledons: Col-0 (I) and bli-1 (J). Arrows point at discontinuous veins. Bars = 1 mm.
Figure 6.
Increased Number of Trichome Branches and Cell Size Correlate with Higher Levels of Endoreduplication.
(A) and (B) Trichome branching: wild-type leaf (A) and bli-1 leaf (B). Arrows point at four-branched trichomes. Bars = 200 μm.
(C) Number of trichomes and branches per trichome were counted for 20 wild-type and mutant plants in leaves 1 and 2; n represents total number of trichomes; numbers display percentages of trichomes with different branch numbers.
(D) and (E) Size of cotyledon cells: Cotyledons of 5-d-old seedlings were printed with agarose: wild-type cotyledon (D) and bli-1 cotyledon (E). Insets are close-ups of central areas in (D) and (E). Bars = 500 μm.
(F) Cell sizes of four wild-type and bli-1 cotyledons were measured inside a defined, central square. Numbers are average cell area in μm2. Error bars are ±se.
(G) Ploidy analysis of seedlings 10 d after germination. Ploidy levels of bli-1 are displayed as total number of nuclei.
[See online article for color version of this figure.]
Thus, BLI is required to prevent premature differentiation of at least the cotyledon epidermis and vascular system and promotes cell division and growth of the organs and tissues in which BLI is expressed, including cotyledons, leaves, and sepals.
Genetic Interactions of bli-1 with tfl2/lhp1 and clf
Pc-G proteins act in at least two complexes: PRC2 acts as the histone methyltransferase complex and contains CLF, whereas PRC1 is involved in binding of the methylated histones and likely includes TFL2/LHP1 in Arabidopsis. We sought to analyze the genetic interaction of bli-1 with mutants deficient in PRC1 or PRC2 by introducing the bli-1 mutant into tfl2-2 or clf-28 backgrounds, respectively. clf and tfl2/lhp1 mutants produce upward curled leaves and are early flowering and smaller than the wild type. tfl2-2, but not clf-28, mutants frequently form terminal flowers (Goodrich et al., 1997; Larsson et al., 1998; Gaudin et al., 2001; Doyle and Amasino, 2009). Under long-day conditions, bli-1 clf-28 and bli-1 tfl2-2 had narrow and small cotyledons like those of bli-1 mutants but were very early flowering and had upward curled leaves similar to clf or tfl2 mutants. However, both double mutants were minute, forming a very small rosette and showed reduced plant height (Figure 7). As leaf curling of clf mutants is largely suppressed under short-day conditions, we analyzed bli-1 clf-28 mutants also under these conditions. Similar to long-day-grown plants, the double mutant displayed strong leaf curling and was minute, whereas none of the single mutants showed leaf curling or was strongly reduced in size compared with the wild type. Thus, the very reduced sizes of the double mutants compared with either single mutant strongly suggest a synergistic genetic interaction between bli-1 and tfl2 or clf, respectively, similar to the double mutant phenotype of clf and tfl2 (Barrero et al., 2007). However, some aspects of the bli-1 tfl2 and bli-1 clf double mutants appear to be additive, suggesting that BLI and CLF or TFL2 act both in common and in separate pathways to control Arabidopsis development.
Figure 7.
Genetic Interaction of bli-1 with clf and tfl2 Mutants.
(A) and (B) Phenotype of 25-d-old Col-0, tfl2-2, bli-1 tfl2-2, and bli-1 mutants (A) and Col-0, bli-1, bli-1 clf-28, and clf-28 mutants (B). Plants were grown at 16-h-light/8-h-dark cycles at 20°C.
(C) Phenotype of Col-0, bli-1, bli-1 clf-28, and clf-28 mutants grown under short-day conditions (10-h-light/14-h-dark cycles at 20°C).
[See online article for color version of this figure.]
BLI Regulates Expression of a Subset of Pc-G Target Genes
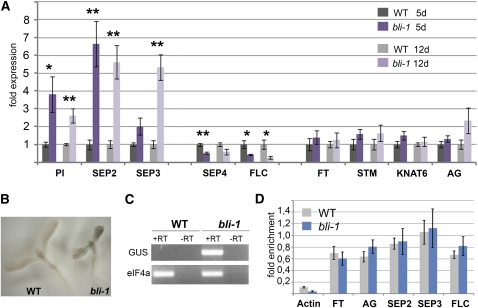
Pc-G proteins prevent precocious expression of many floral homeotic genes, including the MADS box transcription factor genes AGAMOUS (AG), SEPALLATA3 (SEP3), PISTILLATA (PI), and APETALA3 (Goodrich et al., 1997; Barrero et al., 2007). In addition, homeobox genes like SHOOT MERISTEMLESS (STM) and KNOTTED-LIKE FROM ARABIDOPSIS THALIANA2 (KNAT2), whose expression is confined to the shoot apical meristem in the wild type, are misexpressed in leaves of several Pc-G mutants (Katz et al., 2004). Also, the floral repressor FLC and the key floral integrator FLOWERING LOCUS T (FT) are severely misregulated in clf mutants (Jiang et al., 2008). All these genes are covered by the Pc-G mark H3K27me3 in the wild type (Schubert et al., 2006; Turck et al., 2007; Zhang et al., 2007). At least for AG, STM, FLC, and FT, H3K27me3 levels were shown to depend on CLF or combined action of CLF/SWN (Schubert et al., 2006; Jiang et al., 2008). To reveal whether BLI is involved in the regulation of Pc-G target genes, we analyzed expression of several Pc-G target genes in bli-1 mutants. As germination is delayed in bli-1 mutants (Figure 4), bli-1 seeds were imbibed 2 d before the wild type, and plant material was harvested for RNA extraction at the same developmental stage (5 and 12 d after germination). We observed moderate but reproducible misexpression of PI, SEP2, and SEP3, whereas no significant changes were observed for AG, KNAT6, STM, SEP4, and FT (Figure 8A). Interestingly, we found a marked downregulation of FLC in the bli-1 mutant. Whereas PI and SEP2 showed similar misexpression values in 5-d- and 12-d-old seedlings, the effect on SEP3 was much more pronounced in 12-d-old seedlings. In contrast with the 5-d-old seedlings, which only harbored cotyledons, the 12-d-old seedlings had visible leaves, so the differences in expression level might be a result of tissue-specific differences in gene misexpression.
Figure 8.
Expression and H3K27me3 Levels of Pc-G Target Genes in bli-1.
(A) Quantitative RT-PCR analysis of expression levels of Pc-G target genes. RNA was extracted from seedlings 5 and 12 d after germination. Values were normalized to the reference gene At4g34270 (Czechowski et al., 2005) and are displayed as fold difference to the wild type. Bars display the mean of three biological replicates, and error bars indicate ±se. Significance of increased or decreased mRNA levels in bli-1 mutants compared with the wild type was determined by two-tailed Student's t test: **P < 0.01 and *P < 0.05.
(B) and (C) Ectopic expression of LAT52pro:GUS in bli-1: GUS staining of 10-d-old seedlings (B) and RT-PCR analysis (C). +/− RT, +/− reverse transcriptase in cDNA synthesis. RNA was extracted from 10-d-old seedlings.
(D) Chromatin immunoprecipitation assay with antibodies against H3K27me3 on 10-d-old wild-type and bli-1 seedlings. Chromatin immunoprecipitation samples were analyzed by quantitative PCR using oligonucleotides matching intronic regions of the genes tested. Results were normalized to a H3K27me3-ChIP quantitative PCR on the FUS3 locus as reference. Actin was used as a negative control showing no enrichment. Experiment was performed twice with similar results; error bars indicate ±se of technical replicates.
As bli-1 mutants show reduced transmission through the male germline (see Supplemental Table 2 online), we speculated that BLI might regulate expression of pollen-specific genes. We took advantage of the fact that the T-DNA inserted in bli-1 is derived from the binary vector pCSA110, which carries a LAT52pro:GUS reporter gene (Sessions et al., 2002). The LAT52 gene was initially isolated in tomato (Solanum lycopersicum), and this reporter confers pollen-specific expression in tomato and Arabidopsis (Twell et al., 1990). Indeed, GUS expression could be detected in emerging leaves and cotyledons in bli-1 mutants (Figure 8B), suggesting that BLI represses the LAT52 promoter during vegetative growth.
The observed misexpression was not due to inappropriate staining conditions as no GUS staining was detected in wild-type plants or plants hemizygous for the insertion in BLI. In addition, RT-PCR analyses confirmed strong misexpression of the GUS gene (Figure 8C). Thus, besides the floral specific genes PI, SEP2, and SEP3, BLI also represses the pollen-specific reporter LAT52pro:GUS in leaves. The latter may indicate a general role for BLI in restricting the expression of pollen-specific genes, which will need to be studied in more detail. In addition, BLI does not only act as a repressor of Pc-G targets but in the case of FLC is required for activation.
As misexpression of the Pc-G target genes AG and STM is correlated with a loss of H3K27me3 in clf or clf swn mutants, respectively (Schubert et al., 2006), we studied the H3K27me3 profile of AG, FLC, FT, SEP2, and SEP3 by chromatin immunoprecipitation (Figure 8D). To account for differences in precipitation efficiency, we normalized to FUSCA3 (FUS3) H3K27me3 levels, as this gene is a H3K27me3 target (Makarevich et al., 2006), and we were unable to detect its expression in bli-1 and wild-type seedlings consistent with the reported exclusive expression of FUS3 during seed development (Gazzarrini et al., 2004). We observed similar levels of H3K27me3 in bli-1 and the wild type for all genes analyzed; thus, BLI may not be required for H3K27me3 of these genes in the conditions tested (Figure 8D). However, as the misexpression of the Pc-G target genes is relatively mild in bli-1 mutants and possibly is restricted to a limited number of cells, we cannot exclude that H3K27me3 levels might be different in the cells where the H3K27me3 targets are misexpressed.
DISCUSSION
Conservation of BLI in the Plant Kingdom and Function of the CLF–BLI Interaction Domains
In this study, we analyzed the BLI protein and its role in Arabidopsis growth and development as an interacting partner of the Pc-G protein CLF. BLI homologs are found in the sequenced genomes of higher plants and even in that of the moss P. patens (see Supplemental Figure 1 online). Recently, the Physcomitrella CLF and FIE homologs were analyzed and shown to be important for development and stem cell maintenance (Mosquna et al., 2009; Okano et al., 2009). Like the Pc-G proteins, BLI is widely conserved in plant species, consistent with BLI being important for Pc-G protein function in plants. The C-terminal domain of BLI, containing a coiled-coil domain with moderate similarity to prokaryotic SMC-like proteins, is most highly conserved. SMC proteins are involved in various processes of chromatin biology, including chromosome condensation and DNA repair (Schubert, 2009). However, BLI lacks several other domains that are found in SMC proteins. In addition, the TITAN proteins have been identified as the homologs of SMC proteins in Arabidopsis (Liu et al., 2002). We revealed the SMC-related coiled-coil domain of BLI as the region that mediates the interaction with the CXC domain of CLF by yeast two-hybrid and in vitro pull-down analyses (Figure 1). Also in planta, lack of the SMC domain prevents interaction with CLF (Figure 1). In Drosophila, mutations in the highly conserved CXC domain of the CLF homolog E(z) result in loss-of-function phenotypes and global reduction in H3K27me3, which is correlated with reduced H3K27 methyltransferase activity in vitro (Cao et al., 2002; Wang et al., 2004b; Ketel et al., 2005). In addition, the domain is required for an interaction with the sequence-specific DNA binding factor PLEIOHOMEOTIC (Wang et al., 2004b). In vitro binding studies also revealed that the CXC domains of E(z) and CLF bind to single-stranded DNA and therefore might be involved in the recruitment or stabilization of PRC2 (Krajewski et al., 2005).
Although many studies show an important role of the CXC domain, its molecular function is still unclear, and it is an attractive possibility that BLI interacts with the CXC domain to modulate histone methyltransferase activity or recruitment to Pc-G target genes.
Role of BLI in Arabidopsis Development
To study the role of BLI in Arabidopsis development, we identified a bli allele that showed a severe reduction in BLI full-length transcript and a developmental phenotype (bli-1) (Figure 4). Although we cannot definitively show that bli-1 is a null allele, all CLF-related functions of BLI are likely depleted by the insertion as the CLF interaction domain is disrupted. bli-1 mutants revealed a pleiotropic mutant phenotype, which includes defects in leaf, flower, and embryo development (Figures 4 to 6). Consistent with its role in various organs and tissues, BLI is expressed throughout plant development in a subset of tissues (Figure 3). Particularly pronounced defects of the bli-1 mutant are observed in cotyledon, leaf, and sepal development where we found strong BLI expression (Figure 5). The epidermal layer showed the most prominent defects: outgrowing cells indicating a loss of cellular identity. Similar phenotypes are observed in clf swn Pc-G double mutants, which develop ectopic meristems on cotyledons and eventually show complete loss of cell identity (Chanvivattana et al., 2004). Further analyses will be needed to reveal the fate and identity of the outgrowing cells in bli-1 mutants. bli-1 mutants also display other severe defects in organ patterning, cellular identity, and differentiation: vascular strands are frequently unconnected and trichome numbers and patterning are altered in bli-1 (Figures 5 and 6). Vascular development is controlled by many different pathways, including an important role of the phytohormone auxin (reviewed in Sieburth and Deyholos, 2006), but also cyclophilin71 mutants, which are possibly linked to Pc-G silencing display similar alterations (Li et al., 2007). As cell size at least in the cotyledon epidermis is also strongly increased, cell numbers are decreased and a higher level of endoreduplication is observed in bli-1 mutants (Figure 6), this reveals an important role for BLI to prevent premature cellular differentiation and promote cell division. Other chromatin regulators like the fas mutants that carry mutations in chromatin assembly factor 1 (CAF-1) subunits show similar defects as bli-1 mutants (Exner et al., 2006; Ramirez-Parra and Gutierrez, 2007). CAF-1 is required for replication-dependent histone assembly and transmission of epigenetic silencing through mitosis and shares the MSI1 protein with PRC2 (Kaya et al., 2001; Ono et al., 2006; Schonrock et al., 2006). Interestingly, FAS1 shows a similar expression pattern as BLI (Ramirez-Parra and Gutierrez, 2007) and was also identified in the yeast two, hybrid screen with CLF (see Supplemental Table 1 online). However, whereas fas mutants show misregulation of cell cycle–regulating genes, no differences were detected in bli-1 mutants in preliminary whole-genome transcriptome analyses (N. Schatlowski, unpublished data). However, BLI may act posttranslationally on cyclins as it contains a putative cyclin binding motif (Figure 1). So far, plant Pc-G proteins have not been linked to cell cycle control, whereas there is extensive evidence from Drosophila and vertebrates for a regulation of the cell cycle by Pc-G proteins, both by transcriptional regulation of cyclin genes and interaction with proteins involved in cell cycle control (reviewed in Martinez and Cavalli, 2006). Thus, BLI might be an important link between the plant Pc-G machinery and the control of cell cycle progression to faithfully transmit epigenetic information through mitosis, possibly with relation to the CAF-1 complex.
Role of BLI in Pc-G Silencing
BLI and CLF interact in yeast, in vitro, and in planta, share nuclear localization and an overlapping expression pattern (Figures 2 and 3; Goodrich et al., 1997). In addition, bli-1 mutants exhibit misexpression of the Polycomb-target genes PI, SEP2, and SEP3, which are also misexpressed in various Pc-G mutants, including clf and tfl2/lhp1 mutants (Barrero et al., 2007). Although several other well-studied Pc-G target genes showed no misexpression in bli-1, it is important to note that most Arabidopsis Pc-G mutants show only misexpression of relatively few, specific target genes. This is particularly the case for mutants that have been linked to PRC1 like tfl2/lhp1 and ring1a/b mutants (Nakahigashi et al., 2005; Xu and Shen, 2008). Misexpression of Pc-G target genes in tfl2/lhp1 and ring1a/b mutants is not correlated with a decrease in H3K27me3, which we also observe in bli-1 mutants (Figures 8 and 9) (Turck et al., 2007; Xu and Shen, 2008). The isolation of BLI-containing complexes will be fundamental in revealing the association of BLI to PRC2- or PRC1-like complexes. Similar to tfl2 clf double mutants (Barrero et al., 2007), bli clf and bli tfl2 double mutants are minute (Figure 7), but bli mutants show additional phenotypes that are not observed in Pc-G mutants. In addition to repression of Pc-G target genes, BLI promotes expression of the Pc-G target gene FLC, the key floral repressor. Interestingly, suppression of FLC by CLF is reported for the dominant clf-59 allele that carries a point mutation in the CXC domain (Doyle and Amasino, 2009), in contrast with clf loss-of-function mutants that have elevated FLC expression (Jiang et al., 2008). A disruption of the interaction with BLI by the residue that is mutated in clf-59 may therefore be responsible for the gain-of-function phenotype observed in clf-59.
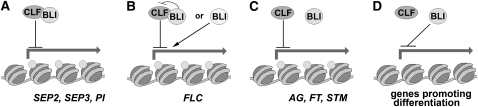
Figure 9.
Model for Pc-G–Related and Independent Functions of BLI.
(A) to (C) Action of BLI and CLF on Pc-G target genes.
(A) CLF and BLI interact to cooperatively repress SEP2, SEP3, and PI. As H3K27me3 levels of SEP2 and SEP3 are not changed in bli-1 mutants, BLI may only transiently interact with CLF and function downstream of CLF and H3K27me3.
(B) Antagonistic action of CLF and BLI on FLC expression. BLI may bind to CLF to inhibit CLF or independently of CLF promote FLC expression.
(C) CLF acts independently of BLI to repress AG, FT, and STM.
(D) Based on the bli-1 single and clf bli-1 double mutant phenotype, BLI acts independently of CLF to inhibit genes that promote differentiation. Gray circles indicate H3K27me3.
We propose that BLI specifically interacts with CLF and possibly modulates its activity to control the coordinated and timely expression of SEP2, SEP3, PI, and FLC that act antagonistically on floral commitment (Figure 9). Further analyses will be required to reveal whether BLI is generally associated with a plant PRC2 or whether the interaction occurs only transiently. BLI likely also has additional functions besides Pc-G silencing as several aspects of the bli-1 mutant phenotype have not been described in Pc-G mutants yet and BLI localizes to cytoplasmic vesicles besides its presence in the nucleus.
METHODS
Biological Material
All BLI T-DNA insertion lines were identified using the SIGNAL database (http://signal.salk.edu/cgi-bin/tdnaexpress) and provided by the Nottingham Arabidopsis Stock Centre: bli-1 (N805222) and bli-2 (N505565) (Sessions et al., 2002; Alonso et al., 2003) (see Supplemental Figure 2 online for the analyses of additional T-DNA insertions in BLI). Homozygous mutants were isolated by PCR-based genotyping (for oligonucleotide sequences, see Supplemental Table 3 online), and the site of insertion was confirmed by sequencing. For analysis of genetic interaction with clf and tfl2/lhp1, crosses were performed using clf-28 (Doyle and Amasino, 2009) and tfl2-2 (Larsson et al., 1998). All genotypes used in this study are in the Columbia-0 (Col-0) background.
All seeds were sterilized (70% ethanol for 5 min, 96% ethanol + 0.05% Triton X-100 for 5 min), sown on GM media (half-strength Murashige and Skoog medium and 0.5% sucrose), and stratified for 2 d at 4°C and transferred to soil after 10 to 12 d. Plants were grown at either long-day conditions (16-h-light/8-h-dark cycles at 20°C), short-day conditions (10-h-light/14-h-dark cycles at 20°C) or continuous light conditions (24 h light at 20°C). bli-1 seeds were imbibed 2 d earlier than the wild type when directly compared.
Transformation of Arabidopsis thaliana was performed after the floral dip method using the Agrobacterium tumefaciens strain GV3101 pMP90 (Clough and Bent, 1998).
Complementation of the bli-1 Phenotype
To generate the BLIpro:BLI-GUS construct, the genomic locus of BLI, including 1.7 kb upstream of the transcription start site, was amplified using oligonucleotides with attached attB sites (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTGAACTGGCA ATTCAGAATCGGG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTG GAGAAGCTTGCTTGTCCTTCTTTTC-3′), recombined into pDONR201 (Invitrogen), and subsequently cloned into pMDC163 (Curtis and Grossniklaus, 2003) as translational fusion with the uidA gene, following the manufacturer’s instructions (Invitrogen; Gateway cloning manual). Using the floral dip method, bli-1/+ plants were transformed with A. tumefaciens carrying the T-DNA vectors. Plants positive for the transgene were selected on GM medium containing hygromycin (15 μg/mL) and genotyped for bli-1−/−.
Yeast Two-Hybrid Interaction Studies
For the yeast two-hybrid screen, a truncated version of CLF lacking the SET domain was used as fusion to GAL4-BD (CMGB2) as previously described (Chanvivattana et al., 2004). A library of cDNA fragments that were generated from flower RNA and introduced in pGAD10 (Clontech) (generously provided by José Luis Riechmann) was transformed into yeast AH109 carrying CMGB2. Four million transformants were screened, and 75 clones that showed growth on selective media (−His [H], −Trp [W], −Leu [L]) were identified. Plasmids were isolated and the inserts sequenced. All yeast techniques and interaction studies were performed as described in the yeast protocols handbook of the Matchmaker system (Clontech).
For interaction studies of CLF and BLI, GAL4-BD vectors containing CLF cDNA fragments were used as described (Chanvivattana et al., 2004). To generate BLI-GAL4-AD vectors, a full-length cDNA was ordered (Riken RAFL-07-13-G11) and used as template to generate GAL4-AD clones with truncations of BLI. The complete coding sequence was PCR amplified and recombined into pDONR201 using Gateway technology (Invitrogen). Truncations of BLI were generated by PCR (for oligonucleotides, see Supplemental Table 3 online), inserted into pCR8/GW-TOPO (Invitrogen), and recombined into the vector pGADT7-DEST (Horak et al., 2008) to generate GAL4-AD vectors. BD and AD clones were cotransformed into yeast strain AH109, and transformants were selected on medium lacking Trp and Leu. Interaction was studied by plating serial dilutions of yeasts on quadruple dropout medium (−L, −W, −H, −adenine).
In Vitro Pull-Down Assay
The CXC domain of CLF was amplified from a GAL4-BD vector containing a CLF cDNA fragment (Chanvivattana et al., 2004) (for oligonucleotide sequences, see Supplemental Table 3 online). The PCR fragment was inserted into pCR8/GW-TOPO (Invitrogen) and recombined, according to the manufacturer’s instructions, into a modified pGEX4T3 (GE Healthcare) where the complete Gateway cassette was inserted into the XhoI site. For the SMC-His6-fusion gene, the pCR8/GW-TOPO entry clone (see above) was used for recombination into pDEST17 (Invitrogen). Escherichia coli BL21.DE3 cells were transformed with plasmids, grown at 20°C to OD600 0.4, and induced with 0.5 mM isopropyl-β-thiogalactopyranosid for 6 to 8 h. In vitro pull-down assays were performed using the MagneGST protein purification kit (Promega). For SMC-His6 protein, the lysed cell extracts were centrifuged for 10 min at 4°C, and the supernatant was incubated with the GST proteins bound to magnetic particles overnight at 4°C. Washing and elution was performed according to the manufacturer’s instructions. For immunoblot analyses, 6× SDS loading buffer (0125 M Tris-HCl, pH 6.8, 2% SDS, 20% [v/v] glycerol, and 0.2% [w/v] bromophenol blue) was added to input and eluate samples, separated by 15% SDS-PAGE, and transferred onto polyvinylidenfluorid membrane. For immunodetection, anti-GST antibody (Promega) or anti-His antibody (Roche) were used as primary antibodies and detected with horseradish peroxidase–conjugated secondary antibodies (anti-rabbit, Sigma-Aldrich; anti-mouse, Upstate).
Transient Colocalization Assay
Modified versions of pMDC7 carrying the GFP (pABindGFP) or mCherry (pABindCherry) coding sequence (Bleckmann et al., 2010) were used to insert the complete coding sequence of BLI and CLF as well as various truncations of BLI or CLF via Gateway-mediated cloning.
Vectors were transformed in A. tumefaciens GV3101 pMP90. For transient expression assays, abaxial sides of leaves of 4-week-old Nicotiana benthamiana plants were infiltrated essentially as described by Bleckmann et al. (2010). To avoid gene silencing, an A. tumefaciens culture containing a T-DNA with the gene for the silencing suppressor p19 was mixed in a 1:1:1 ratio. About 48 to 72 h after A. tumefaciens infiltration, expression was induced by brushing 20 μM β-estradiol and 0.1% Tween onto infiltrated leaves. Fluorescence was monitored in leaf epidermis cells 2 to 6 h after induction using a Zeiss LSM 510 Meta confocal microscope with ×40 1.3 numerical aperture Zeiss oil immersion objective.
Split Renilla Luciferase Complementation Assay
The destination vectors pYS40 (RLuc-N) and pYS39 (RLuc-C) were constructed using vectors pDuExP and pDuExB (Fujikawa and Kato, 2007) as templates to PCR amplify the N-terminal (amino acids 1 to 229) and C-terminal (amino acids 230 to 311) region of Renilla reniformis luciferase (RLuc), introducing a 5′ PacI and a 3′ SpeI restriction site. Both templates contain at their C terminus the flexible linker sequence 2xGGGGS, and the C-terminal RLuc also contains a tetracysteine-tag (CCPGCC) 5′ of the stop codon.
The amplicons were introduced by PacI/SpeI sites 3′ of the Gateway cassette of the β-estradiol–inducible plant transformation vector pMDC7 (Curtis and Grossniklaus, 2003). Entry clones in pCR8/GW-TOPO carrying the gene or gene fragment of interest without stop codon were recombined by LR reaction with pYS40 and pYS39 to result in expression vectors containing C-terminal fusions to N-RLuc and C-RLuc, respectively.
Transient expression of the split RLuc constructs in N. benthamiana and β-estradiol induction were performed as described by Bleckmann et al. (2010).
Luminescence detection was performed infiltrating 10 μM ViviRen live-cell substrate (Promega) dissolved in deionized water into N. benthamiana leaves that had been induced with β-estradiol for 3 h. After a period of 10 min in darkness, luminescence was measured with a NightOwl system (Berthold) for 30 min with 8 × 8 pixel binning.
Gene Expression Analyses
Detection of GUS activity was performed as described previously (Colon-Carmona et al., 1999).
For whole-mount in situ hybridization in embryos, BLI probes against the C-terminal part of BLI were generated using oligonucleotides 5′-TTGGAAGATAAGGCTCTCAG-3′ and 5′-CCAAGCTTCTAATACGACTCACTATAGGGAGATTAGAGAAGCTTG CTTG-3′. Labeling with digoxygenin was performed according to the manufacturer’s instructions (Roche). Tissue preparation and RNA in situ hybridization were performed as described (Hejatko et al., 2006; Stahl et al., 2009).
Five- and 10-d-old seedlings were used for total RNA extraction (RNeasy plant mini kit; Qiagen). RNA was resuspended in 30 μL RNase-free water, treated with DNase (Fermentas), transcribed into cDNA using SuperScriptII following the manufacturer’s instructions (Invitrogen), and subjected to real-time PCR. Real-time PCR was performed in biological and technical triplicates using oligonucleotides spanning exon-exon borders (for oligonucleotides, see Supplemental Table 3 online) with Mesa Blue Sybr Mix (Eurogentec) in a Chromo4 real-time PCR machine (Bio-Rad). Expression levels were normalized to the reference gene At4g34270 (Czechowski et al., 2005).
Phenotypic Analysis
For scanning electron microscopy, plant material was treated as described previously (Kwiatkowska, 2004).
TB staining was performed as described by Tanaka et al. (2004).
To compare cell sizes, cotyledons of 5-d-old seedlings were printed with 1.5% agarose, negatives were examined using a Zeiss Axioskop Mot Plus and photographed, and cell areas were measured with ImageJ.
For ploidy analysis, 10-d-old seedlings were chopped in nuclei isolation buffer (15 mM Tris, 2 mM Na2EDTA, 0.5 mM Spermin, 80 mM KCl, 20 mM NaCl, 15 mM β-mercaptoethanol, and 0.1% Triton X-100, pH 7.5) and stained with propidium iodide (0005 mg/mL), and DNA content was measured using a FACS Aria (BD Bioscience). For examination of vascular defects, tissue was fixed for 12 h in ethanol:acetic acid (6:1), washed twice 30 min in 100% ethanol and 30 min with 70% ethanol, and cleared with chloral hydrate:glycerol:water (8 g:1 mL:2 mL) for several hours.
Photographs were taken with an AxioCam ICC1 camera (Zeiss) mounted onto a Zeiss Stemi 2000C. Collages of digital photographs were assembled using Adobe Photoshop.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation analysis was performed with seedlings 10 d after germination as described previously (Schubert et al., 2006) using antibodies against H3K27me3 (Millipore; 07-449). Input and immunoprecipitation DNA was diluted 1:10, and 1 μL was used for real-time PCR. Oligonucleotides were designed to match intronic regions (for oligonucleotides, see Supplemental Table 3 online). Values for immunoprecipitation were referenced to input values. To account for differences in immunoprecipitation efficiencies, %IP values were normalized to the FUS3 locus, which carries H3K27me3 and is not expressed in wild-type and bli-1 seedlings.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers at3g23980 (BLI), at2g23380 (CLF), and at5g17690 (TFL2/LHP1). Additional accession numbers are found in Figure 1 and Supplemental Table 1 online.
Author Contributions
N.S., J.G., and D.S. designed the research, Y.S. supplied vectors and established the split-luciferase complementation assay, M.L.H. cloned vectors for in vitro pull-down analyses and the split-luciferase complementation assay, and N.S. and D.S. performed the experiments and wrote the article.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Conservation of BLI in Plants.
Supplemental Figure 2. Analyses of T-DNA Insertions in BLI.
Supplemental Table 1. Clones Identified in the Yeast Two-Hybrid Screen with CLF-SET and T-DNA Insertion Lines Analyzed.
Supplemental Table 2. Transmission of the bli-1 Mutant Allele.
Supplemental Table 3. Oligonucleotides Used in This Study.
Supplementary Material
Acknowledgments
Work in the Schubert lab is supported by the Deutsche Forschungsgemeinschaft through Grant SFB590. We thank Nora Lorberg for excellent technical assistance, Ben Hunter for initial characterization of the bli-1 allele, José Luis Riechmann for providing the yeast two-hybrid cDNA library, Naohiro Kato for providing vectors for the split-luciferase complementation system, Andrea Bleckmann for the modified pMDC7 vectors and help with confocal microscopy, Madlen Rast for help with whole mount in situ hybridization, Klaus Harter for pGADT7-Dest vector, Klaus Meyer for help with the fluorescence-activated cell sorting analyses, and the Nottingham Arabidopsis Stock Centre for seeds and the ABRC for DNA stocks. We thank Rüdiger Simon and members of the Schubert lab for critical reading of the manuscript.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aubert D., Chen L., Moon Y.H., Martin D., Castle L.A., Yang C.H., Sung Z.R. (2001). EMF1, a novel protein involved in the control of shoot architecture and flowering in Arabidopsis. Plant Cell 13: 1865–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero J.M., Gonzalez-Bayon R., del Pozo J.C., Ponce M.R., Micol J.L. (2007). INCURVATA2 encodes the catalytic subunit of DNA Polymerase alpha and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana. Plant Cell 19: 2822–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckmann A., Weidtkamp-Peters S., Seidel C.A., Simon R. (2010). Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152: 166–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonje M., Sanchez R., Chen L., Sung Z.R. (2008). EMBRYONIC FLOWER1 participates in polycomb group-mediated AG gene silencing in Arabidopsis. Plant Cell 20: 277–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. (2002). Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y., Bishopp A., Schubert D., Stock C., Moon Y.H., Sung Z.R., Goodrich J. (2004). Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colon-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T., Stitt M., Altmann T., Udvardi M.K., Scheible W.R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V. (2002). Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- De Lucia F., Crevillen P., Jones A.M., Greb T., Dean C. (2008). A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc. Natl. Acad. Sci. USA 105: 16831–16836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M., Mermoud J.E., Wakao R., Tang Y.A., Endoh M., Appanah R., Nesterova T.B., Silva J., Otte A.P., Vidal M., Koseki H., Brockdorff N. (2004). Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev. Cell 7: 663–676 [DOI] [PubMed] [Google Scholar]
- Doyle M.R., Amasino R.M. (2009). A single amino acid change in the enhancer of zeste ortholog CURLY LEAF results in vernalization-independent, rapid flowering in Arabidopsis. Plant Physiol. 151: 1688–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exner V., Taranto P., Schonrock N., Gruissem W., Hennig L. (2006). Chromatin assembly factor CAF-1 is required for cellular differentiation during plant development. Development 133: 4163–4172 [DOI] [PubMed] [Google Scholar]
- Francis N.J., Kingston R.E., Woodcock C.L. (2004). Chromatin compaction by a polycomb group protein complex. Science 306: 1574–1577 [DOI] [PubMed] [Google Scholar]
- Fujikawa Y., Kato N. (2007). Split luciferase complementation assay to study protein-protein interactions in Arabidopsis protoplasts. Plant J. 52: 185–195 [DOI] [PubMed] [Google Scholar]
- Gaudin V., Libault M., Pouteau S., Juul T., Zhao G., Lefebvre D., Grandjean O. (2001). Mutations in LIKE HETEROCHROMATIN PROTEIN 1 affect flowering time and plant architecture in Arabidopsis. Development 128: 4847–4858 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S., Tsuchiya Y., Lumba S., Okamoto M., McCourt P. (2004). The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 7: 373–385 [DOI] [PubMed] [Google Scholar]
- Goodrich J., Puangsomlee P., Martin M., Long D., Meyerowitz E.M., Coupland G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51 [DOI] [PubMed] [Google Scholar]
- Hejatko J., Blilou I., Brewer P.B., Friml J., Scheres B., Benkova E. (2006). In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat. Protoc. 1: 1939–1946 [DOI] [PubMed] [Google Scholar]
- Horak J., Grefen C., Berendzen K.W., Hahn A., Stierhof Y.D., Stadelhofer B., Stahl M., Koncz C., Harter K. (2008). The Arabidopsis thaliana response regulator ARR22 is a putative AHP phospho-histidine phosphatase expressed in the chalaza of developing seeds. BMC Plant Biol. 8: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Kurata T., Okada K., Wada T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Jiang D., Wang Y., Wang Y., He Y. (2008). Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz A., Oliva M., Mosquna A., Hakim O., Ohad N. (2004). FIE and CURLY LEAF polycomb proteins interact in the regulation of homeobox gene expression during sporophyte development. Plant J. 37: 707–719 [DOI] [PubMed] [Google Scholar]
- Kaya H., Shibahara K.I., Taoka K.I., Iwabuchi M., Stillman B., Araki T. (2001). FASCIATA genes for chromatin assembly factor-1 in arabidopsis maintain the cellular organization of apical meristems. Cell 104: 131–142 [DOI] [PubMed] [Google Scholar]
- Ketel C.S., Andersen E.F., Vargas M.L., Suh J., Strome S., Simon J.A. (2005). Subunit contributions to histone methyltransferase activities of fly and worm polycomb group complexes. Mol. Cell. Biol. 25: 6857–6868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T., Harada J.J., Goldberg R.B., Fischer R.L. (2001). Polycomb repression of flowering during early plant development. Proc. Natl. Acad. Sci. USA 98: 14156–14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T., Papp B., Fischle W., Kocher T., Schelder M., Fritsch C., Wild B., Wilm M., Muller J. (2006). A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev. 20: 1110–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski W.A., Nakamura T., Mazo A., Canaani E. (2005). A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol. Cell. Biol. 25: 1891–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D. (2002). Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowska D. (2004). Surface growth at the reproductive shoot apex of Arabidopsis thaliana pin-formed 1 and wild type. J. Exp. Bot. 55: 1021–1032 [DOI] [PubMed] [Google Scholar]
- Larsson A.S., Landberg K., Meeks-Wagner D.R. (1998). The TERMINAL FLOWER2 (TFL2) gene controls the reproductive transition and meristem identity in Arabidopsis thaliana. Genetics 149: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy Y.Y., Mesnage S., Mylne J.S., Gendall A.R., Dean C. (2002). Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297: 243–246 [DOI] [PubMed] [Google Scholar]
- Li H., He Z., Lu G., Lee S.C., Alonso J., Ecker J.R., Luan S. (2007). A WD40 domain cyclophilin interacts with histone H3 and functions in gene repression and organogenesis in Arabidopsis. Plant Cell 19: 2403–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.M., McElver J., Tzafrir I., Joosen R., Wittich P., Patton D., Van Lammeren A.A., Meinke D. (2002). Condensin and cohesin knockouts in Arabidopsis exhibit a titan seed phenotype. Plant J. 29: 405–415 [DOI] [PubMed] [Google Scholar]
- Makarevich G., Leroy O., Akinci U., Schubert D., Clarenz O., Goodrich J., Grossniklaus U., Kohler C. (2006). Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 7: 947–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., et al. (2009). Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A.M., Cavalli G. (2006). The role of polycomb group proteins in cell cycle regulation during development. Cell Cycle 5: 1189–1197 [DOI] [PubMed] [Google Scholar]
- Mosquna A., Katz A., Decker E.L., Rensing S.A., Reski R., Ohad N. (2009). Regulation of stem cell maintenance by the Polycomb protein FIE has been conserved during land plant evolution. Development 136: 2433–2444 [DOI] [PubMed] [Google Scholar]
- Muller J., Hart C.M., Francis N.J., Vargas M.L., Sengupta A., Wild B., Miller E.L., O'Connor M.B., Kingston R.E., Simon J.A. (2002). Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Nakahigashi K., Jasencakova Z., Schubert I., Goto K. (2005). The Arabidopsis heterochromatin protein1 homolog (TERMINAL FLOWER2) silences genes within the euchromatic region but not genes positioned in heterochromatin. Plant Cell Physiol. 46: 1747–1756 [DOI] [PubMed] [Google Scholar]
- Nekrasov M., Klymenko T., Fraterman S., Papp B., Oktaba K., Kocher T., Cohen A., Stunnenberg H.G., Wilm M., Muller J. (2007). Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 26: 4078–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S., Park S., van Nocker S. (2008). Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genet. 4: e1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N., Yadegari R., Margossian L., Hannon M., Michaeli D., Harada J.J., Goldberg R.B., Fischer R.L. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano Y., Aono N., Hiwatashi Y., Murata T., Nishiyama T., Ishikawa T., Kubo M., Hasebe M. (2009). A polycomb repressive complex 2 gene regulates apogamy and gives evolutionary insights into early land plant evolution. Proc. Natl. Acad. Sci. USA 106: 16321–16326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Kaya H., Takeda S., Abe M., Ogawa Y., Kato M., Kakutani T., Mittelsten S.O., Araki T., Shibahara K. (2006). Chromatin assembly factor 1 ensures the stable maintenance of silent chromatin states in Arabidopsis. Genes Cells 11: 153–162 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E., Gutierrez C. (2007). E2F regulates FASCIATA1, a chromatin assembly gene whose loss switches on the endocycle and activates gene expression by changing the epigenetic status. Plant Physiol. 144: 105–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose A., Manikantan S., Schraegle S.J., Maloy M.A., Stahlberg E.A., Meier I. (2004). Genome-wide identification of Arabidopsis coiled-coil proteins and establishment of the ARABI-COIL database. Plant Physiol. 134: 927–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A.J., Shao Z., Erdjument-Bromage H., Tempst P., Kingston R.E. (2001). A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412: 655–660 [DOI] [PubMed] [Google Scholar]
- Schatlowski N., Creasey K., Goodrich J., Schubert D. (2008). Keeping plants in shape: Polycomb-group genes and histone methylation. Semin. Cell Dev. Biol. 19: 547–553 [DOI] [PubMed] [Google Scholar]
- Schmid M., Davison T.S., Henz S.R., Pape U.J., Demar M., Vingron M., Scholkopf B., Weigel D., Lohmann J.U. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schonrock N., Exner V., Probst A., Gruissem W., Hennig L. (2006). Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. J. Biol. Chem. 281: 9560–9568 [DOI] [PubMed] [Google Scholar]
- Schubert D., Primavesi L., Bishopp A., Roberts G., Doonan J., Jenuwein T., Goodrich J. (2006). Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J. 25: 4638–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert V. (2009). SMC proteins and their multiple functions in higher plants. Cytogenet. Genome Res. 124: 202–214 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Cavalli G. (2009). Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136: 3531–3542 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. (2007). Genome regulation by polycomb and trithorax proteins. Cell 128: 735–745 [DOI] [PubMed] [Google Scholar]
- Seki M., et al. (2002). Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Sessions A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth L.E., Deyholos M.K. (2006). Vascular development: The long and winding road. Curr. Opin. Plant Biol. 9: 48–54 [DOI] [PubMed] [Google Scholar]
- Simon J., Chiang A., Bender W., Shimell M.J., O'Connor M. (1993). Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev. Biol. 158: 131–144 [DOI] [PubMed] [Google Scholar]
- Sing A., Pannell D., Karaiskakis A., Sturgeon K., Djabali M., Ellis J., Lipshitz H.D., Cordes S.P. (2009). A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell 138: 885–897 [DOI] [PubMed] [Google Scholar]
- Stahl Y., Wink R.H., Ingram G.C., Simon R. (2009). A signaling module controlling the stem cell niche in Arabidopsis root meristems. Curr. Biol. 19: 909–914 [DOI] [PubMed] [Google Scholar]
- Sugimoto-Shirasu K., Roberts K. (2003). “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant Biol. 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Tanaka T., Tanaka H., Machida C., Watanabe M., Machida Y. (2004). A new method for rapid visualization of defects in leaf cuticle reveals five intrinsic patterns of surface defects in Arabidopsis. Plant J. 37: 139–146 [DOI] [PubMed] [Google Scholar]
- Turck F., Roudier F., Farrona S., Martin-Magniette M.L., Guillaume E., Buisine N., Gagnot S., Martienssen R.A., Coupland G., Colot V. (2007). Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D., Yamaguchi J., McCormick S. (1990). Pollen-specific gene expression in transgenic plants: Coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109: 705–713 [DOI] [PubMed] [Google Scholar]
- Wang D., Tyson M.D., Jackson S.S., Yadegari R. (2006). Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 13244–13249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y. (2004a). Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878 [DOI] [PubMed] [Google Scholar]
- Wang L., Brown J.L., Cao R., Zhang Y., Kassis J.A., Jones R.S. (2004b). Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 14: 637–646 [DOI] [PubMed] [Google Scholar]
- Wood C.C., Robertson M., Tanner G., Peacock W.J., Dennis E.S., Helliwell C.A. (2006). The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc. Natl. Acad. Sci. USA 103: 14631–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Shen W.H. (2008). Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr. Biol. 18: 1966–1971 [DOI] [PubMed] [Google Scholar]
- Zhang X., Clarenz O., Cokus S., Bernatavichute Y.V., Pellegrini M., Goodrich J., Jacobsen S.E. (2007). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Chua N.H. (2000). Technical advance: An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265–273 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.